Open Access Article
Open Access ArticleLabel-free quantification of passive membrane permeability of cyclic peptides across lipid bilayers: penetration speed of cyclosporin A across lipid bilayers†
Takahiro
Ono
 a,
Kazuhito V.
Tabata
a,
Kazuhito V.
Tabata
 b,
Yuki
Goto
b,
Yuki
Goto
 c,
Yutaro
Saito
c,
Yutaro
Saito
 a,
Hiroaki
Suga
a,
Hiroaki
Suga
 c,
Hiroyuki
Noji
c,
Hiroyuki
Noji
 bd,
Jumpei
Morimoto
bd,
Jumpei
Morimoto
 *a and
Shinsuke
Sando
*a and
Shinsuke
Sando
 *ad
*ad
aDepartment of Chemistry & Biotechnology, Graduate School of Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8656, Japan. E-mail: jmorimoto@chembio.t.u-tokyo.ac.jp; ssando@chembio.t.u-tokyo.ac.jp
bDepartment of Applied Chemistry, Graduate School of Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8656, Japan
cDepartment of Chemistry, Graduate School of Science, The University of Tokyo, Bunkyo-ku, Tokyo 113-0033, Japan
dDepartment of Bioengineering, Graduate School of Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8656, Japan
First published on 6th December 2022
Abstract
Cyclic peptides that passively penetrate cell membranes are under active investigation in drug discovery research. PAMPA (Parallel Artificial Membrane Permeability Assay) and Caco-2 assay are mainly used for permeability measurements in these studies. However, permeability rates across the artificial membrane and the cell monolayer used for these assays are intrinsically different from the ones across pure lipid bilayers. There are also membrane permeability assays for peptides using reconstructed lipid bilayers, but they require labeling for detection, and the absolute membrane permeability of the natural peptides themselves could not be determined. Here, we constructed a lipid bilayer permeability assay and realized the first label-free measurements of the lipid bilayer permeability of cyclic peptides. Quantitative permeability values across lipid bilayers were determined for model cyclic hexapeptides and an important natural product, cyclosporin A (CsA). The obtained quantitative permeability values will provide new and advanced knowledge about the passive permeability of cyclic peptides.
Introduction
The membrane permeability of cyclic peptides has been actively investigated.1–3 Although cyclic peptides are poorly membrane permeable in general, some cyclic peptides from natural sources, such as cyclosporin A (CsA), exhibit high membrane permeability. These cyclic peptides with high passive membrane permeability are of significance in medicinal chemistry because they can target intracellular protein–protein interactions that are difficult to be targeted by other drug modalities.The passive membrane permeability of cyclic peptides is ideally evaluated using lipid bilayers because they need to cross the lipid bilayer membrane to penetrate into intracellular space passively. However, the permeability of cyclic peptides has been investigated mostly based on the two permeability assays, which use membranes that are not lipid bilayers. One of the assays is PAMPA (Parallel Artificial Membrane Permeability Assay),4 in which artificial membranes approximately 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times thicker than cell membranes are used (Fig. 1, top). The other type of assay is the cell monolayer assay, such as the Caco-2 assay.5 This assay evaluates permeability across cell monolayers, which is different from the pure passive membrane permeability across lipid bilayers because of many factors like degradation, efflux, active transport, and passage through intercellular space involved in the permeability across cell monolayers.
000 times thicker than cell membranes are used (Fig. 1, top). The other type of assay is the cell monolayer assay, such as the Caco-2 assay.5 This assay evaluates permeability across cell monolayers, which is different from the pure passive membrane permeability across lipid bilayers because of many factors like degradation, efflux, active transport, and passage through intercellular space involved in the permeability across cell monolayers.
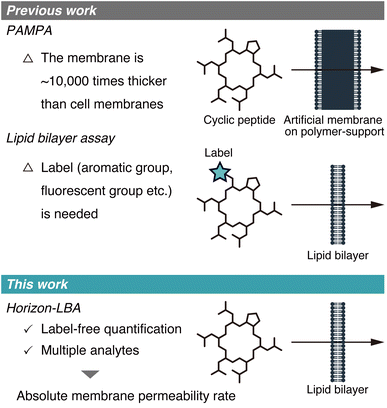 | ||
| Fig. 1 Schematic illustration of the membrane permeability assays developed in previous studies (top) and this study (bottom). | ||
To obtain the quantitative passive permeability rates of cyclic peptides across cell membranes, assays that can directly measure permeability across lipid bilayers are required.
Recently, a few studies about measurements of membrane permeability of peptides using lipid bilayers were reported (Fig. 1, middle). A. Hennig group, W. M. Nau group, J. Hochman group, and P. S. Dittrich group used liposomes to measure the permeability of peptides.6–9 N. Rodriguez group, B. M. Paegel group, and A. R. Thiam group utilized DIB (droplet interface bilayer) for permeability evaluation of peptides.10–12 However, the methods used in these studies require labels, such as aromatic groups, fluorescent groups, and azide/alkyne groups, in the analyte structures for detection, which hampers measuring the permeability of non-modified cyclic peptides. As a result, to date, the absolute passive permeability rates of critically important natural peptides such as CsA have never been quantitatively determined, presenting a major challenge in peptide drug discovery research.
Here, we report the first label-free quantification of the permeability of cyclic peptides, including CsA, across lipid bilayers using a newly established permeability assay for peptides (Fig. 1, bottom).
Results and discussion
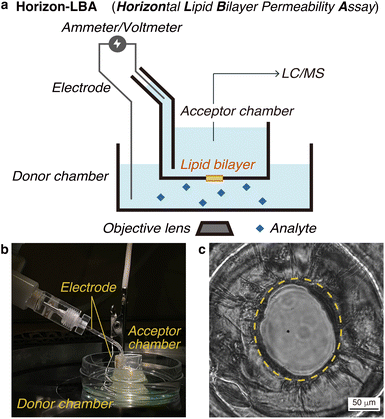
First, we constructed a new lipid bilayer permeability assay, “Horizon-LBA (horizontal lipid bilayer permeability assay),” based on a method to build a black lipid membrane (BLM) in a pore on the bottom of a chamber developed by Ide and coworkers (Fig. 2a and b).13–15 The apparatus consists of two chambers on a microscope. We used the lower chamber as a donor chamber and the upper chamber as an acceptor chamber in the permeability assay. After adding the compounds to the donor chamber and incubation, the compounds that crossed the lipid bilayers and reached the acceptor chamber are quantified by LC/MS to determine the lipid bilayer permeability. The LC/MS-based quantification method enables the measurement of lipid bilayer permeability of label-free cyclic peptides. As a setup for the permeability assay, the horizontal lipid bilayer setup has an advantage over other BLM setups in that the condition of the lipid bilayer can be monitored based on electrochemical information and microscopic observations. They are useful for confirming the area and integrity of the fragile lipid bilayer during permeability experiments (Fig. 2c).We measured the thickness of the lipid bilayers in Horizon-LBA and the artificial membranes in PAMPA for comparison. The thickness of membranes was calculated from electrostatic capacities by regarding the donor/acceptor chambers that sandwich the membrane as a condenser (Table S1†). The thickness of the lipid bilayers in Horizon-LBA was 8 ± 1 nm. A lipid bilayer was suggested to be formed in our system since this value was similar to the thickness of a lipid bilayer of DOPC reported by B. A. Lewis and D. M. Engelman (3.8 ± 0.1 nm).16 On the other hand, the thickness of the artificial membrane of PAMPA was determined to be 43 ± 7 μm. It is about 103–104 times thicker than a lipid bilayer. This result experimentally confirmed the difference in membrane thickness between PAMPA and Horizon-LBA. According to the result, considering membrane thickness, Horizon-LBA is more representative of biological systems than PAMPA.
We conducted permeability measurements of small molecules to confirm that our new system can quantify the permeability of analytes. Permeability measurements were performed using a mixture of 10 analytes as the donor solution. After incubating the system for 20 minutes at room temperature, we recovered the solution from the acceptor chamber and quantified the analytes in the solution by LC/MS. We calculated permeability coefficients, Papp, for the analytes detected from the acceptor chamber (Table S2†). Caffeine was used to verify our system. R. Strutt and coworkers measured the lipid bilayer permeability using their permeability assay based on DIB of DOPC and a UV detector.17 In this assay, a lipid bilayer is formed at the droplet interface and the permeability of an analyte is determined based on the UV absorbance of the analyte. In their report, the permeability coefficient of caffeine was determined to be 2.81 ± 0.0920 × 10−4 cm s−1. In our system, the permeability coefficient of caffeine was 3.0 ± 0.6 × 10−4 cm s−1. The similarity of the permeability coefficients of these assays suggests that our system, Horizon-LBA, properly works as a lipid bilayer permeability assay.
Comparison of the permeabilities of the small molecules measured by Horizon-LBA with the reported permeabilities by PAMPA and Caco-2 assay (Table S2†) demonstrate that the permeability rates across lipid bilayers differ from those measured by PAMPA and Caco-2 assay. This indicates the need for a permeability assay system using lipid bilayers, such as Horizon-LBA, to measure pure passive membrane permeability across lipid bilayers.
Using the established permeability assay, we conducted label-free quantification of the lipid bilayer permeability of cyclic peptides.
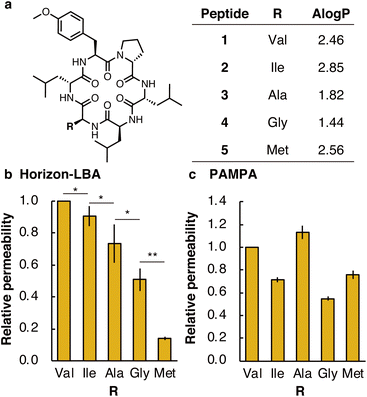
First, we measured the membrane permeability of a model cyclic hexapeptide (1) (Fig. 3a and Table S3†).18 The permeability coefficient of 1 across lipid bilayers was determined to be 3 ± 1 × 10−4 cm s−1. The permeability coefficient of 1 was also measured by PAMPA and determined to be 0.165 ± 0.009 × 10−4 cm s−1. The significant difference in permeability coefficients between Horizon-LBA and PAMPA may be derived from the difference in the thickness and composition of the artificial membrane and lipid bilayers.
We next compared the permeability of 1 and its derivatives in which the Val residue of 1 is replaced with a side chain of different lipophilicity. Previous studies by PAMPA have shown a correlation between membrane permeability and calculated lipophilicity.19 We synthesized cyclic peptide 1 and its derivatives in which Val in 1 was changed to Ile (2), Ala (3), Gly (4), or Met (5) (Fig. 3a). The synthesized five peptides were mixed and analyzed by Horizon-LBA and PAMPA. In Horizon-LBA, since the standard deviations of permeability coefficients between multiple experiments were relatively large, we normalized the permeability of each compound with the Papp of 1 in each experiment as 1.0 (Fig. 3b and Table S3†). The relative permeabilities were 0.90 ± 0.06 (2, R = Ile), 0.7 ± 0.1 (3, R = Ala), 0.51 ± 0.07 (4, R = Gly), and 0.14 ± 0.01 (5, R = Met). There was a statistically significant difference between the relative permeability values of each cyclic peptide (p < 0.05). The permeability coefficients of the cyclic hexapeptides were also measured by PAMPA and the values were compared with those obtained by Horizon-LBA. The rank order of the permeability values between each cyclic peptide in PAMPA was different from Horizon-LBA (Fig. 3c and Table S3†). In PAMPA, the order of permeability was R = Ala > Val > Met > Ile > Gly, which is different from the order in Horizon-LBA (R = Val > Ile > Ala > Gly > Met). In particular, the fact that 5 (R = Met), which had notably lower permeability in Horizon-LBA than the other four peptides, has the same level of permeability in PAMPA as 2 (R = Ile) is a remarkable difference. This difference may be attributed to the different physicochemical properties of artificial and lipid bilayer membranes. The artificial membrane is ∼104 times thicker than lipid bilayers, as discussed earlier. Besides, the artificial membrane of PAMPA consists of ∼1% phospholipid and ∼99% of decane, which is intrinsically different from lipid bilayer membranes. Therefore, the peptide is expected to interact with an artificial membrane differently with the lipid bilayer.
Our results so far showed that the absolute permeability coefficients of a cyclic peptide as well as relative permeability among cyclic peptides are significantly different between lipid bilayer-based assays and PAMPA. This demonstrates the importance of determining the permeability coefficients using lipid bilayers to understand the actual permeation speed across lipid bilayers.
Finally, we measured the permeability of an important natural product, CsA, across lipid bilayers (Fig. 4). CsA is known to have immunosuppressive effects and is used as a drug.20 CsA penetrates into intracellular space and interacts with intracellular proteins, cyclophilin and calcineurin. Previous studies using PAMPA and Caco-2 assays indicate that CsA passively permeates cell membranes. However, due to the lack of experimental methods, the permeability across pure lipid bilayers has not been directly observed, and the absolute passive permeation rate across lipid bilayers are not known. Using Horizon-LBA, we measured the permeability of CsA. The permeability coefficient (Papp) was determined to be 3 ± 1 × 10−6 cm s−1 (Fig. 4b). Although CsA permeates the lipid bilayers, the rate is approximately 100 times slower than that of small molecules, e.g., caffeine (3.0 × 10−4 cm s−1). Interestingly, the permeability coefficient of CsA across lipid bilayers is close to the reported Papp values in cell monolayer assays (e.g., apical to basolateral: Papp = 1.51 × 10−6 cm s−1, basolateral to apical: Papp = 4.51 × 10−6 cm s−1 in Caco-2 assay by J. Seo group, Papp = 1.4 × 10−6 cm s−1 in MDCK-LE by R. S. Lokey group).19,21 This similarity suggests that passive permeation is the primary pathway for CsA penetration into cells. This has been indirectly declared by Caco-2 assay,22 but our result is the first direct evidence of passive permeation of CsA across lipid bilayers.
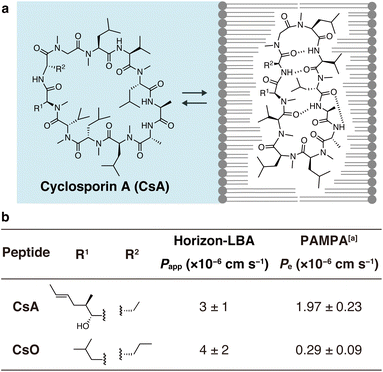 | ||
| Fig. 4 Permeability of cyclosporin A (CsA) and cyclosporin O (CsO) measured in Horizon-LBA. (a) The chemical structure of CsA. (b) Side chain structures and the permeability coefficients of CsA and CsO across the lipid bilayers. The incubation was conducted for 4 h at room temperature. The experiment was conducted in triplicate. Data are presented as means ± standard deviation. aThe values are from ref. 21. | ||
We also evaluated the permeability of CsO, a derivative of CsA. CsA has been suggested to change its conformations in the process of passive membrane permeation (Fig. 4a). The chameleonic change between open and closed conformations was hypothesized to be important for effective membrane permeation. One of the experimental supports for the hypothesis is the fact that CsO, which rigidly forms a closed conformational state, shows a much lower permeability coefficient on PAMPA.21 We revisited this using the lipid bilayer permeability assay. As a result, the permeability coefficient of the CsO was similar to CsA (Fig. 4b). This result of Horizon-LBA is not consistent with the previously reported observation based on PAMPA, which suggested that CsO is less permeable than CsA. Although further investigations are needed to clarify the reason for the difference in the trend of results in the artificial membrane and the lipid bilayer, the result of Horizon-LBA may suggest that CsA and CsO penetrate cell membranes passively at a similar rate.
Conclusions
In conclusion, we here reported the first label-free quantitative measurements of cyclic peptide permeability across lipid bilayers. The LC/MS-based quantification method, in combination with horizontal lipid bilayer fabrication technology, enabled the measurement of the permeability of natural CsA itself, which is not modified with any chromophores or fluorescent groups. This is the first direct evidence that CsA passively crosses cell membranes, and the quantitative value of the permeation speed was determined at last. The coefficient of variation (CV) in Horizon-LBA was often larger than 30%, but similar large CVs are often observed in other planar lipid bilayer permeability assays.12,23 The large CV may reflect the variation of the lipid bilayer condition in each experiment.Unlike PAMPA, Horizon-LBA uses reconstructed lipid bilayers. Permeability assays using lipid bilayers are useful to understand the actual speed of passive permeation of compounds across cell membranes. PAMPA membrane is about 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times thicker than lipid bilayers. Therefore, analytes must pass through the long path of organic solvent (normally dodecane) in PAMPA, and the main obstacle for the analytes may be this organic solvent layer. PAMPA may be useful to compare the passive permeability of compounds but does not provide information about the actual permeation speed of the compounds across lipid bilayers. In cell monolayer assays, analytes must pass through a lipid bilayer twice, i.e., the apical and basolateral cell membranes, and also pass through the cytosol. Besides, the permeability coefficients in the cell monolayer assays are affected by various factors such as degradation, efflux, active transport, and passage through intercellular space. The permeation speed determined by Horizon-LBA will provide insights into how peptides passively permeate the cell membrane. The insights would eventually be useful for predicting the passive membrane permeability of a given peptide and designing highly permeable peptides in the future.
000 times thicker than lipid bilayers. Therefore, analytes must pass through the long path of organic solvent (normally dodecane) in PAMPA, and the main obstacle for the analytes may be this organic solvent layer. PAMPA may be useful to compare the passive permeability of compounds but does not provide information about the actual permeation speed of the compounds across lipid bilayers. In cell monolayer assays, analytes must pass through a lipid bilayer twice, i.e., the apical and basolateral cell membranes, and also pass through the cytosol. Besides, the permeability coefficients in the cell monolayer assays are affected by various factors such as degradation, efflux, active transport, and passage through intercellular space. The permeation speed determined by Horizon-LBA will provide insights into how peptides passively permeate the cell membrane. The insights would eventually be useful for predicting the passive membrane permeability of a given peptide and designing highly permeable peptides in the future.
Previously, lipid composition has been reported to affect the lipid bilayer permeability of small molecules.7,24 Therefore, permeability measurements with various lipid compositions in Horizon-LBA will greatly contribute to the elucidation of the interaction of peptides with various membranes and their permeability.
Currently, our setup cannot measure permeability in parallel like PAMPA. However, the system that can form parallel BLMs has been previously reported, and it will realize a parallel lipid bilayer permeability assay and high-throughput measurements of a larger set of compounds.25 The quantitative values of lipid bilayer permeability obtained by Horizon-LBA are not only important for studying the pharmacokinetics of cyclic peptides for drug discovery but also improve our fundamental knowledge of peptide membrane permeability.
Data availability
The authors declare that the data supporting the findings of this study are available within the article and its ESI.†Author contributions
T. Ono: investigation, methodology, formal analysis, writing – original draft. K. V. Tabata: methodology, resources, supervision. Y. Goto: methodology, resources. Y. Saito: investigation. H. Suga: resources, supervision. H. Noji: resources, supervision. J. Morimoto: project administration, resources, supervision, validation, writing – original draft. S. Sando: conceptualization, funding acquisition, project administration, resources, supervision, writing – original draft. All the authors contributed to review and editing of the manuscript.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
T. O. was supported by a Program for Leading Graduate Schools (MERIT). The authors acknowledge the One-stop Sharing Facility Center for Future Drug Discoveries at the University of Tokyo for the use of micrOTOF II. This work was supported by CREST, Japan Science and Technology Agency, Grant Numbers JPMJCR21N5 (S. S.), JPMJCR18S6 (K. V. T.), PRESTO, Japan Science and Technology Agency, Grant Number JPMJPR21AF (J. M.), and JSPS Grant Numbers JP20H05618 (H. S.), JP17H06355 (K. V. T.) and JP20H02866 (Y. G.).Notes and references
- A. Zorzi, K. Deyle and C. Heinis, Curr. Opin. Chem. Biol., 2017, 38, 24–29 CrossRef CAS PubMed.
- P. G. Dougherty, A. Sahni and D. Pei, Chem. Rev., 2019, 119, 10241–10287 CrossRef CAS PubMed.
- A. A. Vinogradov, Y. Yin and H. Suga, J. Am. Chem. Soc., 2019, 141, 4167–4181 CrossRef CAS PubMed.
- M. Kansy, F. Senner and K. Gubernator, J. Med. Chem., 1998, 41, 1007–1010 CrossRef CAS PubMed.
- P. Artursson and J. Karlsson, Biochem. Biophys. Res. Commun., 1991, 175, 880–885 CrossRef CAS PubMed.
- A. Barba-Bon, Y.-C. Pan, F. Biedermann, D.-S. Guo, W. M. Nau and A. Hennig, J. Am. Chem. Soc., 2019, 141, 20137–20145 CrossRef CAS PubMed.
- F. Biedermann, G. Ghale, A. Hennig and W. M. Nau, Commun. Biol., 2020, 3, 1–10 CrossRef PubMed.
- T. J. Desai, B. Habulihaz, J. R. Cannon, A. Chandramohan, H. Y. K. Kaan, A. Sadruddin, T. Y. Yuen, C. Johannes, D. Thean, C. J. Brown, D. P. Lane, A. W. Partridge, R. Evers, T. K. Sawyer and J. Hochman, Pharm. Res., 2021, 38, 843–850 CrossRef CAS PubMed.
- C.-C. Lin, M. Bachmann, S. Bachler, K. Venkatesan and P. S. Dittrich, ACS Appl. Mater. Interfaces, 2018, 10, 41909–41916 CrossRef CAS PubMed.
- P. Gehan, S. Kulifaj, P. Soule, J. B. Bodin, M. Amoura, A. Walrant, S. Sagan, A. R. Thiam, K. Ngo, V. Vivier, S. Cribier and N. Rodriguez, Biochim. Biophys. Acta, Biomembr., 2020, 1862, 183415 CrossRef CAS PubMed.
- J. Hu, W. G. Cochrane, A. X. Jones, D. G. Blackmond and B. M. Paegel, Nat. Chem., 2021, 13, 786–791 CrossRef CAS PubMed.
- V. Faugeras, O. Duclos, D. Bazile and A. R. Thiam, Langmuir, 2022, 38, 5682–5691 CrossRef CAS PubMed.
- T. Ide and T. Yanagida, Biochem. Biophys. Res. Commun., 1999, 265, 595–599 CrossRef CAS PubMed.
- K. V. Tabata, K. Sato, T. Ide, T. Nishizaka, A. Nakano and H. Noji, EMBO J., 2009, 28, 3279–3289 CrossRef CAS PubMed.
- K. Sato, R. Sasaki, R. Matsuda, M. Nakagawa, T. Ekimoto, T. Yamane, M. Ikeguchi, K. V. Tabata, H. Noji and K. Kinbara, J. Am. Chem. Soc., 2022, 144, 11802–11809 CrossRef CAS PubMed.
- B. A. Lewis and D. M. Engelman, J. Mol. Biol., 1983, 166, 211–217 CrossRef CAS PubMed.
- R. Strutt, F. Sheffield, N. E. Barlow, A. J. Flemming, J. D. Harling, R. V. Law, N. J. Brooks, L. M. C. Barter and O. Ces, Lab Chip, 2022, 22, 972–985 RSC.
- W. M. Hewitt, S. S. F. Leung, C. R. Pye, A. R. Ponkey, M. Bednarek, M. P. Jacobson and R. S. Lokey, J. Am. Chem. Soc., 2015, 137, 715–721 CrossRef CAS PubMed.
- M. R. Naylor, A. M. Ly, M. J. Handford, D. P. Ramos, C. R. Pye, A. Furukawa, V. G. Klein, R. P. Noland, Q. Edmondson, A. C. Turmon, W. M. Hewitt, J. Schwochert, C. E. Townsend, C. N. Kelly, M. J. Blanco and R. S. Lokey, J. Med. Chem., 2018, 61, 11169–11182 CrossRef CAS PubMed.
- K. M. Corbett, L. Ford, D. B. Warren, C. W. Pouton and D. K. Chalmers, J. Med. Chem., 2021, 64, 13131–13151 CrossRef CAS PubMed.
- D. Lee, S. Lee, J. Choi, Y.-K. Song, M. J. Kim, D.-S. Shin, M. A. Bae, Y.-C. Kim, C.-J. Park, K.-R. Lee, J.-H. Choi and J. Seo, J. Med. Chem., 2021, 64, 8272–8286 CrossRef CAS PubMed.
- P. F. Augustijns, T. P. Bradshaw, L.-S. L. Gan, R. W. Hendren and D. R. Thakker, Biochem. Biophys. Res. Commun., 1993, 197, 360–365 CrossRef CAS PubMed.
- Z. Aminipour, M. Khorshid, H. Keshvari, S. Bonakdar, P. Wagner and B. Van der Bruggen, Eur. J. Pharm. Biopharm., 2020, 146, 133–142 CrossRef CAS PubMed.
- S. Purushothaman, J. Cama and U. F. Keyser, Soft Matter, 2016, 12, 2135–2144 RSC.
- B. Le Pioufle, H. Suzuki, K. V. Tabata, H. Noji and S. Takeuchi, Anal. Chem., 2008, 80, 328–332 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details and supplementary figures & tables (PDF). See DOI: https://doi.org/10.1039/d2sc05785a |
| This journal is © The Royal Society of Chemistry 2023 |