Open Access Article
Open Access ArticleRiboflavin secreted by Shewanella sp. FDL-2 facilitates its reduction of Se(IV) and Te(IV) by promoting electron transfer†
Manman Cheng ab,
Haikun Zhangc,
Yan Li*b and
Wenhao Chen*a
ab,
Haikun Zhangc,
Yan Li*b and
Wenhao Chen*a
aSolid-State Fermentation Resource Utilization Key Laboratory of Sichuan Province, Yibin University, Yibin City, Sichuan Province 644000, China. E-mail: 84460462@qq.com
bCollege of Life Sciences, Yantai University, Yantai 264000, China. E-mail: liyan0709@hotmail.com
cYantai Institute of Costal Zone Research, Chinese Academy of Sciences, Yantai 264000, China
First published on 24th November 2023
Abstract
The biological reduction of selenite (Se(IV)) or tellurite (Te(IV)) to Se0 or Te0 has received increasing attention, as related studies have favored the development of Se/Te pollution control methods. In the presence of the electron donor, the microbes acquired energy and transferred electrons to Se(IV) or Te(IV) to achieve their detoxication. However, the microbial electron transfer pathways involved in this process are still not fully understood. In this study, we reported that marine Shewanella sp. FDL-2 (FDL-2) was capable of reducing Se(IV) and Te(IV) through a novel riboflavin-involved pathway. The results showed that FDL-2 can effectively reduce 10 mM Se(IV) and 5 mM Te(IV) to Se0 and Te0, which was further confirmed by XPS and XRD analyses. RT-qPCR results indicate the upregulation of genes coding flavin-related proteins, and the production of flavin-related substances by strain FDL-2 during Se(IV)/Te(IV) bioreduction was proven by fluorescence chromatography analysis. In addition, the presence of riboflavin enhanced the electron transfer efficiency, indicating its promoting effect on the bioreduction of Se(IV)/Te(IV). Overall, our results highlight a riboflavin-involved electron transfer pathway during Se(IV)/Te(IV) bioreduction and thus deepen our understanding of the corresponding mechanism.
1. Introduction
Selenium (Se) and tellurium (Te), which are homologous elements belonging to chalcogen metalloids and have an intermediate nature between metallic and nonmetallic elements, have found many uses in optical devices, glasses, metal alloys and semiconductors.1–5 Their widespread use has resulted in elevated concentrations in the environment, including water, soil and sediment, causing potential environmental risks to humans, fish, and plants.6,7 Selenium and tellurium have a very diverse range of valence states, including 0, II, IV and VI, with Se(IV) and Te(IV) as their most dangerous forms.2,8 Thus, one of the promising strategies is to reduce their environmental toxicity by transforming Se(IV) and Te(IV) to less toxic and immobile Se0 and Te0.Microorganisms are important driving forces for Se and Te cycles in the natural environment. To date, many microorganisms are capable of reducing Se(IV)/Te(IV) to Se0/Te0 in the presence of electron donors. For example, members belonging to the genera Pseudomonas, Pseudoalteromonas, Stenotrophomonas, Acinetobacter, Shewanella, Bacillus, Sulfurospirillum barnesii, Thauera, Raoultella, Escherichia and Ochrobactrum have been reported to form in-/or extracellular Se0/Te0 solids with different physicochemical properties.6,8–12 There are a variety of mechanisms for Se(IV) and Te(IV) reduction in various microorganisms. In summary, the following five reductive pathways have been reported: (i) thiol group-involved Painter-type reaction; (ii) thioredoxin reductase-mediated reduction; (iii) siderophore-mediated reduction; (iv) sulfide-mediated reduction; and (v) dissimilatory reduction.13 However, intriguingly, it seems that there are other pathways besides these identified ones, since cutting off the above reduction pathways did not stop the subsequent Se(IV) or Te(IV) reduction by those bacterial strains. Therefore, some unknown Se(IV)/Te(IV) reduction pathways remain to be revealed to better understand the corresponding mechanism of Se(IV)/Te(IV) bioreduction.
In our preliminary work, we screened a large number of Se(IV)/Te(IV)-reducing bacteria from marine environments, of which the genus Shewanella is the most efficient since they have diverse electron transport pathways. In this study, a highly effective Se(IV)/Te(IV)-reducing bacterium, Shewanella sp. FDL-2 (FDL-2), was employed to investigate the effect of riboflavin secreted by FDL-2 cells on Se(IV)/Te(IV) bioreduction. The relevant findings will enrich our understanding of the mechanism of biological reduction of Se(IV) and Te(IV).
2. Materials and methods
2.1. Culture conditions
Na2SeO3 and K2TeO3 were purchased from Macklin Biochemical Co., Ltd. (Shanghai, China). Shewanella sp. FDL-2 was isolated from marine sediment in Dalian, Liaoning Province, China (39.23° N, 122.70° E) and stored in the China Center for Type Culture Collection (CCTCC No. 2020910). 2216E medium (g L−1): peptone 5, yeast extract 1, ferric citrate 0.1, NaCl 19.45, MgCl2 5.98, Na2SO4 3.24, CaCl2 1.8, KCl 0.55, Na2CO3 0.16, KBr 0.08, SrCl2 0.034, H3BO3 0.022, Na2SiO3 0.004, NaF 0.0024, NaNO3 0.0016, Na2HPO4 0.008. Mineral salt medium (MSM, g L−1): NH4Cl 0.5, KCl 0.5, MgCl2·7H2O 0.85, lactate 2, HEPES 5.67, NaCl 20 (pH 7.2 ± 0.2).2.2. Optimization of growth conditions for strain FDL-2
Different environmental factors, including dissolved oxygen concentrations (adjusted by 100–500 mL of medium volumes), carbon sources (C1–C6 compounds, including sodium formate, sodium acetate, sodium pyruvate, sodium lactate, glucose), salt concentrations (0–5%, m/v) and pH values (3–10), were applied to investigate their effects on the growth of strain FDL-2. Samples were taken at intervals to record the growth of cells.2.3. Se(IV)/Te(IV) reduction by strain FDL-2
Strain FDL-2 was cultured in 2216E liquid medium in a rotary incubation shaker (150 rpm and 30 °C were applied, taking into account the dissolved oxygen and enzyme activity, respectively) overnight to an OD600 value of ∼2 (late stationary phase, to ensure that we could obtain enough FDL-2 cells). FDL-2 cells were centrifuged at 8000 rpm for 5 min, washed twice, and resuspended in 135 mL serum bottles containing 100 mL MSM. The obtained optimal conditions for FDL-2 growth were applied for the subsequent experiment. Then, 1, 5, and 10 mM Se(IV) and 1, 2, and 5 mM Te(IV) were added to the reaction systems, followed by 15 min of nitrogen exposure. A sterile needle and a syringe were used for periodic sampling.The obtained samples were first centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and 4 °C for 5 min; then, the liquid supernatants were used to analyze Se/Te species and riboflavin; the pellets were freeze-dried overnight and used for X-ray photoelectron spectroscopy (XPS, ESCALAB 250Xi, England), X-ray power diffraction (XRD, BRUKER D8 ADVANCE, Germany) and Fourier transform infrared spectroscopy (FTIR, Jasco FT/IR-4100, Japan) analyses. Fluorescence excitation emission matrix (FEEM) analysis of the riboflavin in liquid supernatants was performed with a fluorescence spectrophotometer (Hitachi F4000, Japan). The operating parameters were as follows: scan rate, 240 nm min−1; excitation/emission slit band width, 2.5 nm; scanning field, emission/excitation spectra from 300/220 nm to 550/400 nm; contour figures, interval of 0.1.
000 rpm and 4 °C for 5 min; then, the liquid supernatants were used to analyze Se/Te species and riboflavin; the pellets were freeze-dried overnight and used for X-ray photoelectron spectroscopy (XPS, ESCALAB 250Xi, England), X-ray power diffraction (XRD, BRUKER D8 ADVANCE, Germany) and Fourier transform infrared spectroscopy (FTIR, Jasco FT/IR-4100, Japan) analyses. Fluorescence excitation emission matrix (FEEM) analysis of the riboflavin in liquid supernatants was performed with a fluorescence spectrophotometer (Hitachi F4000, Japan). The operating parameters were as follows: scan rate, 240 nm min−1; excitation/emission slit band width, 2.5 nm; scanning field, emission/excitation spectra from 300/220 nm to 550/400 nm; contour figures, interval of 0.1.
2.4. 16S rRNA and genome sequencing
DNA extraction for strain FDL-2 was performed by using a bacterial DNA Extraction Kit (TransGen, China). The quality of DNA extracts was analyzed by a NanoDrop 2000 (Thermo, US). The 16S rRNA gene primers are 27F and 1492R (5′-AGRGTTTGATYMTGGCTCAG-3′, 5′-GGYTACCTTGTTACGACTT-3′).14 Sequencing was performed at Qingke Biotechnology Co., Ltd. (Qingdao, China). Phylogenetic analysis was performed by MEGA software (version 11.0) based on the neighbor-joining method. Genome sequencing was performed by BGI Genomics Co., Ltd. through Illumina sequencing platforms. Whole-genome annotation was performed using prokka,15 and the whole-genome circle map was created using R software at https://www.chiplot.online/circos.html.2.5. RT-qPCR analysis
FDL-2 cells in the 1 mM Se(IV)/Te(IV)-supplemented and control (without Se(IV) or Te(IV)) systems were prepared for RT-qPCR assays. RNA extraction and cDNA synthesis were carried out using a TransZol Up Kit and cDNA Synthesis SuperMix (TransGen, China), respectively. A real-time PCR system (Light Cycle 480 II, Roche, Switzerland) was employed to run the protocol with reaction systems prepared using Perfect Start Green qPCR SuperMix. The primers of functional and 16S rRNA (control) genes were synthesized at Rui-biotechnology Co. Ltd. (Qingdao, China). The related results were analyzed by the 2−ΔΔCt method.2.6. Effects of riboflavin on Se(IV)/Te(IV) bioreduction
Accelerating experiments and cyclic voltammetry (CV) methods were applied to investigate the role of riboflavin in Se(IV)/Te(IV) bioreduction. For accelerating experiments, 0, 0.2, 0.4, and 0.6 mM riboflavin were added to the 1 mM Se(IV)/Te(IV) reduction systems as described in Section 2.3. Se(IV) and Te(IV) concentrations were periodically recorded for further analysis. After 6 h of inoculation, the supernatants in the riboflavin-supplemented system and riboflavin-free system were sampled by centrifugation (8000 rpm, 5 min) and filtered through a 0.2 μm membrane (Millipore) for CV analysis. The CV analysis was performed by an electrochemical workstation (CHI660E, Chenhua, China) with a scan rate of 5 mV s−1 and a voltage of −1.2 to 0.6 V (working electrode: carbon felt, reference electrode: Ag/AgCl).2.7. Analytical methods
The concentrations of Se and Te were measured using a spectrophotometric method. The pretreatment procedure of the samples to be tested was as follows: a 1 mL sample was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 8 min. The supernatant was filtered using a 0.45 μm membrane (Millipore) and stored at 4 °C for Se(IV) or Te(IV) measurement. For Se0 and Te0 measurements, the pellets were washed with 1 M NaCl three times to remove Se(IV)/Te(IV) and then treated with sonication (30 s sonication, 30 s pause, 30 min) in an ice bath; after that, the pellet was centrifuged (6000 rpm, 8 min), the supernatant was discarded, and the pellet was washed twice with 10 mL of NaCl (1 M), resuspended in 1 mL of Na2S (1 M, red solution for Se0 measurement) or 1 mL of KOH (1 M, black solution for Te0 measurement), and finally determined at 500 nm and 340 nm.9 Se(IV) and Te(IV) concentrations were measured at 500 nm and 340 nm following the methods described by Arindam et al. (2022)16 and Mal et al. (2017),17 respectively. In addition, the electron transport system activity (ETSA) was detected according to a previously reported method.18
000 rpm for 8 min. The supernatant was filtered using a 0.45 μm membrane (Millipore) and stored at 4 °C for Se(IV) or Te(IV) measurement. For Se0 and Te0 measurements, the pellets were washed with 1 M NaCl three times to remove Se(IV)/Te(IV) and then treated with sonication (30 s sonication, 30 s pause, 30 min) in an ice bath; after that, the pellet was centrifuged (6000 rpm, 8 min), the supernatant was discarded, and the pellet was washed twice with 10 mL of NaCl (1 M), resuspended in 1 mL of Na2S (1 M, red solution for Se0 measurement) or 1 mL of KOH (1 M, black solution for Te0 measurement), and finally determined at 500 nm and 340 nm.9 Se(IV) and Te(IV) concentrations were measured at 500 nm and 340 nm following the methods described by Arindam et al. (2022)16 and Mal et al. (2017),17 respectively. In addition, the electron transport system activity (ETSA) was detected according to a previously reported method.18
3. Results and discussion
3.1. Description of strain FDL-2
The characteristics of strain FDL-2 are shown in Fig. 1. Strain FDL-2 is a Gram-negative strain that can grow rapidly in solid 2216E media to form a round colony in orange-yellow. The phylogenetic analysis showed that strain FDL-2 used in this study belonged to the genus Shewanella in the family Shewanellaceae. As previously reported,8,9 the genus Shewanella is one of the most important dissimilating metal-reducing bacteria and has also been reported to have the ability to reduce Se(IV) and Te(IV), including Shewanella oneidensis MR-1, Shewanella sp. CNZ-1, Shewanella sp. FDA-1 and Shewanella sp. O23S. This may be due to the diverse electron transfer pathways of the bacteria belonging to the genus Shewanella.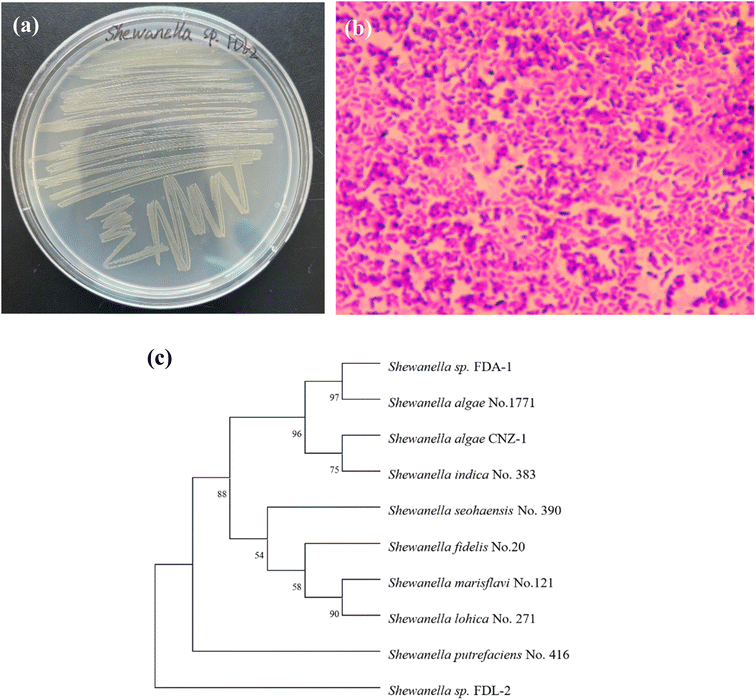 | ||
| Fig. 1 Characterization and identification of strain FDL-2: colony morphology (a); microscopy image (b); phylogenetic tree (c). | ||
3.2. Effects of environmental factors on FDL-2 cell growth
The effects of environmental factors on the growth of FDL-2 cells were tested at 30 °C (Fig. S1†), which is the most suitable temperature for bacterial enzyme activity. Although strain FDL-2 is a facultative anaerobic bacterium, the higher the concentration of dissolved oxygen in the medium is, the faster the growth rate. The OD600 value of FDL-2 cells reached ∼1.1 after 8 h in the system with 100 mL culture medium, while the OD600 value was only ∼0.3 in the system with 500 mL of medium added. Strain FDL-2 was able to use all five tested carbon sources, with the highest growth efficiency in the lactate-supplemented system, followed by the pyruvate- and acetate-supplemented systems. Sodium formate and glucose did not work well for the growth of strain FDL-2. In addition, FDL-2 cells were able to adapt to a very broad pH range, from 5 to 10, with pH 7–8 growing best and pH 3 without bacterial growth. Similarly, strain FDL-2 can grow in a salinity range of 0–5%, growing best in the presence of 3% NaCl and worst without any salinity. Accordingly, the final MSM medium for FDL-2 cultivation was designed as (g L−1): sodium lactate 2, NaCl 30, NH4Cl 0.5, KCl 0.5, MgCl2·7H2O 0.85, HEPS 5.67 and pH ∼7.2 ± 0.2.3.3. Se(IV)/Te(IV) bioreduction by strain FDL-2
Se(IV) and Te(IV) reduction by strain FDL-2 was conducted under the optimized conditions. As shown in Fig. 2a, approximately 94%, 82% and 85% Se(IV) were removed from the liquid phase of the 1, 2 and 10 mM Se(IV)-added systems, respectively, within 48 h. At the same time, the corresponding Se0 concentrations were detected in different reaction systems, indicating the reduction of Se(IV) to Se0 by strain FDL-2. For Te(IV) reduction systems, over 97% Te(IV) (in all Te(IV)-containing systems) was removed from the liquid phase with the detection of Te0 after 48 h of reaction (Fig. 2b), which also confirmed the reduction of Te(IV) to Te0 by strain FDL-2. It is worth noting that for both Se(IV) and Te(IV) bioreduction, Se(IV) removal by FDL-2 cells could be completed in the first six hours, but the entire reduction process lasted 12 to 48 hours later (Fig. 2). One possible explanation is that the FDL-2 cells first adsorbed Se(IV) or Te(IV) rapidly, and then the subsequent reduction reaction continued for a longer time because of the involvement of related enzymes. The adsorption and reduction processes were dominated by some polysaccharides and reductases in strain FDL-2, respectively. Trace amounts of Se and Te may play beneficial roles in various metabolic activities and oxidative stress mitigation, while high concentrations are toxic to microorganisms. Since Te(IV) is much more biotoxic than Se(IV),8 this results in the same number of FDL-2 cells that can only reduce 5 mM of Te(IV) but 10 mM of Se(IV).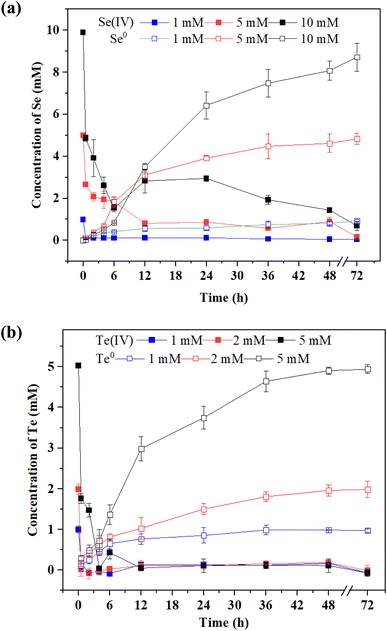 | ||
| Fig. 2 Reduction of (a) Se(IV) and (b) Te(IV) by strain FDL-2 at different initial concentrations (OD600 = 0.3). | ||
XPS analysis showed that Se0 and Te0 were formed in the Se(IV)- and Te(VI)-supplemented systems, respectively (Fig. 3a and b). It is worth noting that the reduction degrees of Se(IV) and Te(IV) are different under the same experimental conditions. The Se(IV)-reductive products contain mainly Se0, while the Te(IV)-reduction products contain both Te(IV) and Te0. This may be because the biological toxicity of Te(IV) is greater than that of Se(IV), resulting in the incomplete reduction of Te(IV). The production of Se0 and Te0 was further confirmed by XRD analysis. The appearance of characteristic absorption peaks at (100), (101), (100), and (101) was consistent with the standard patterns of Se0 (JCPDS 06-0362) and Te0 (JCPDS 36-1452) (Fig. 3c and d).
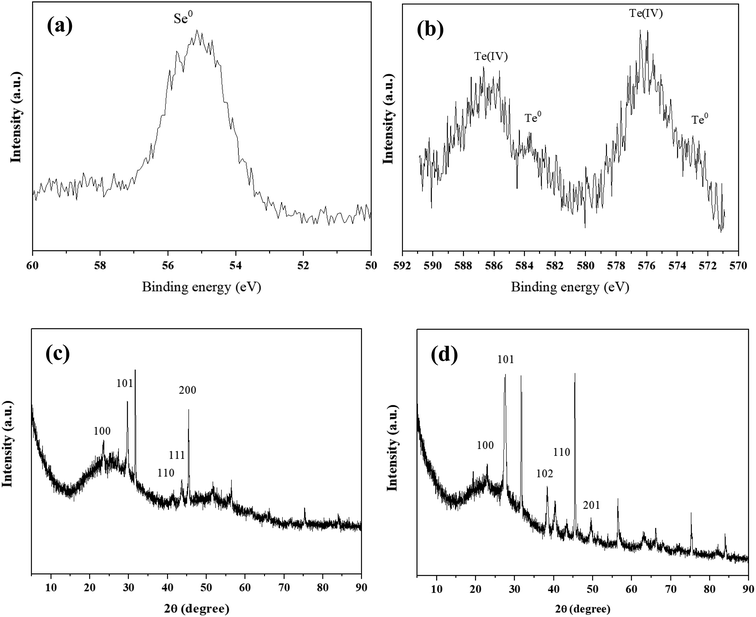 | ||
| Fig. 3 XPS patterns (a and b) and XRD patterns (c and d) of precipitates obtained from 1 mM Se(IV)- and Te(IV)-treated systems at 6 h. | ||
3.4. Upregulation of riboflavin-related genes
The genome sequencing of FDL-2 was obtained with a length of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 860
860![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 565 bp and a GC content of 52.98%. A total of 4395 protein-coding genes and 99 RNA genes, including 91 tRNAs, 7 rRNAs and 1 tmRNA, were predicted (BioProject accession number: PRJNA1021365, Fig. 4a). Of these, we did not find important genes involved in the classical electron transfer pathway, such as cymA, mtrABC, omcA, etc., similar to the model strain of Shewanella oneidensis MR-1. However, strain FDL-2 can still efficiently reduce Se(IV) and Te(IV). Accordingly, we speculate that strain FDL-2 may have other Se(IV)/Te(IV) reduction pathways that are different from those of strain MR-1.
565 bp and a GC content of 52.98%. A total of 4395 protein-coding genes and 99 RNA genes, including 91 tRNAs, 7 rRNAs and 1 tmRNA, were predicted (BioProject accession number: PRJNA1021365, Fig. 4a). Of these, we did not find important genes involved in the classical electron transfer pathway, such as cymA, mtrABC, omcA, etc., similar to the model strain of Shewanella oneidensis MR-1. However, strain FDL-2 can still efficiently reduce Se(IV) and Te(IV). Accordingly, we speculate that strain FDL-2 may have other Se(IV)/Te(IV) reduction pathways that are different from those of strain MR-1.
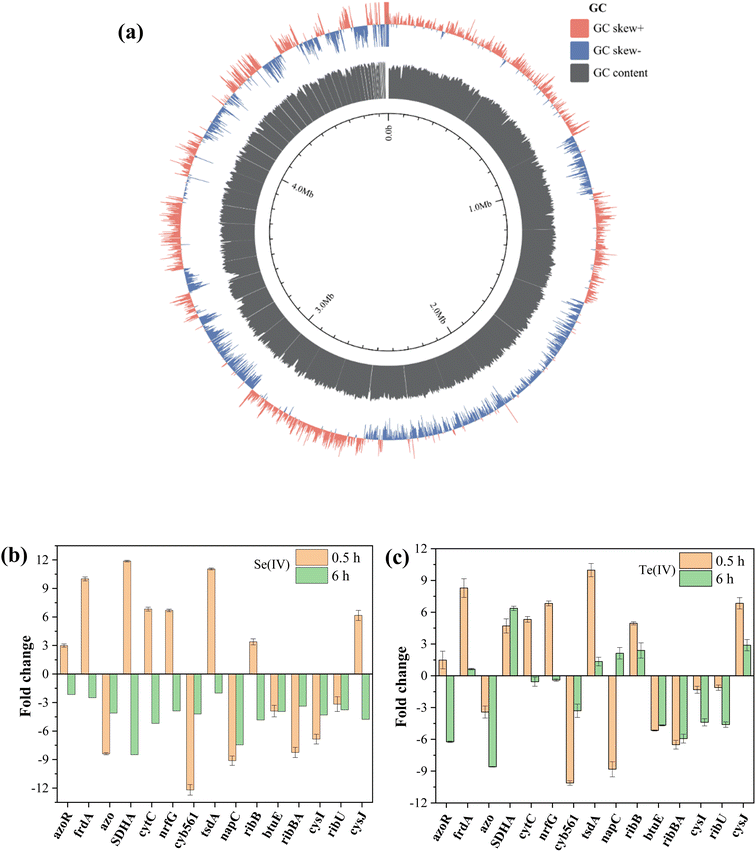 | ||
| Fig. 4 Genome analysis of strain FDL-2 (a) and RT-qPCR analyses of samples obtained from 1 mM (b) Se(IV)- and (c) Te(IV)-treated systems at 0.5/6 h. | ||
Based on the annotation results of the genome of strain FDL-2, cytochrome and reductase coding genes associated with electron transport have received major attention. Combined with previous studies, we finally selected 15 potential functional genes that may be involved in Se(IV)/Te(IV) bioreduction for RT-qPCR analysis. The primers of functional genes are listed in Table 1, and the primers of all selected genes for RT-qPCR assays are shown in Table S1.†
| Gene name | Annotated gene name | KO ID/locus tag | Protein name |
|---|---|---|---|
| FDL-2_00165 | azoR | SBAL678_RS25030 | FMN-dependent NADH-azoreductase |
| FDL-2_02437 | frdA | K00244 | Fumarate reductase flavoprotein subunit |
| FDL-2_01864 | azo | A0A379YYD5 | FMN-dependent NADPH-azoreductase |
| FDL-2_01688 | SDHA | K00234 | Succinate dehydrogenase flavoprotein subunit |
| FDL-2_04257 | cytC | K03532 | Cytochrome c5 |
| FDL-2_02760 | nrfG | GMX02_RS01375 | Format-dependent nitrite reductase complex |
| FDL-2_01463 | cyb561 | K08360 | Cytochrome b561 |
| FDL-2_00533 | tsdA | At1D132_RS11615 | Thiosulfate dehydrogenase |
| FDL-2_01215 | napC | K02569 | Nitrate reductase, cytochrome c-type protein |
| FDL-2_01396 | ribB | K02858 | Riboflavin biosynthesis protein |
| FDL-2_01436 | btuE | b1710 | Thioredoxin/glutathione peroxidase |
| FDL-2_01397 | ribA | K14652 | Riboflavin synthase |
| FDL-2_04281 | ribU | K23675 | Riboflavin transporter |
| FDL-2_01902 | cysI | K00381 | Sulfite reductase [NADPH] hemoprotein bate-component |
| FDL-2_01903 | cysJ | K00380 | Sulfite reductase [NADPH] flavoprotein α-component |
The selected genes showed varying degrees of upregulated expression in the presence of Se(IV) or Te(IV) after 0.5 and 6 h of inoculation (Fig. 4b and c). The upregulated expression of functional genes was mainly concentrated in the early stage of reaction (0.5 h), and many genes began to downregulate expression after 6 hours of induction. These results are consistent with the reaction process (Fig. 2) since the reaction is basically complete approximately 6 h in the 1 mM Se(IV)- or Te(IV)-added systems. In the Te(IV)-supplemented system, the upregulated expression of functional genes of strain FDL-2 lagged slightly behind that in the Se(IV)- supplemented system, which may be attributed to the toxicity of Te(IV) being greater than that of Se(IV). Similar to previous reports,9 some reductases and cytochromes, including frdA (10.0-/8.3-fold), SDHA (11.9-/4.7-fold), cytC (6.8-/5.3-fold) nrfG (6.7-/6.8-fold), tsdA (11.1-/10.0-fold) etc. showed significantly upregulated expression during both Se(IV) and Te(IV) exposure (fold change in Se(IV)-added system/fold change in Te(IV)-added system). The associated proteins encoded by these genes are all involved in the conventional dissimilation reduction pathway. Previous studies have shown that nitrate reductase is involved in the reduction of many metals, including Se(IV), V(V) and Sb(V).19 For strain FDL-2, the upregulated expression of the narG gene encoding a nitrite reductase was observed, indicating that it may have played a similar role to the napC gene.
Of particular concern, two different types of genes were also significantly upregulated, namely ribB (3.4-/5.0-fold, riboflavin biosynthesis protein) and cysJ (6.2-/6.8-fold, sulfite reductase [NADPH] flavoprotein α-component). The two upregulated genes are related to riboflavin, one encoding riboflavin synthase, which can catalyze the dismutation of two molecules of 6,7-dimethyl-8-ribityllumazine to produce riboflavin,20 and the other is sulfite reductases that use riboflavin as an accessory factor. These results highlight the role of riboflavin in the biological reduction of Se(IV) and Te(IV) by strain FDL-2.
3.5. The role of riboflavin secreted by FDL-2 cells
FEEM analyses revealed that humic acid substances were secreted by strain FDL-2 in both Se(IV)- and Te(IV)-supplemented systems, which can be confirmed by different fluorophores at Ex/Em = 320–400/400–550 nm (Fig. 5). Previous studies have reported that the characteristic peaks in these regions represent the presence of fulvic acid-like, polyaromatic type humic acid-like and polycarboxylate type humic acid-like matter.21 Humic acid substances have been reported as a mediator for metal reduction and electron transfer due to their quinyl groups and amino groups.22,23 Accordingly, we speculated that riboflavin secreted by strain FDL-2 was involved in Se(IV) and Te(IV) bioreduction based on genomic and RT-qPCR analyses. In addition, we further verified that the presence of riboflavin can promote electron transfer (Fig. 5c). The electron transfer efficiency was increased by a factor of 4.01 in the presence of 0.1 mM riboflavin. These findings further support our hypothesis.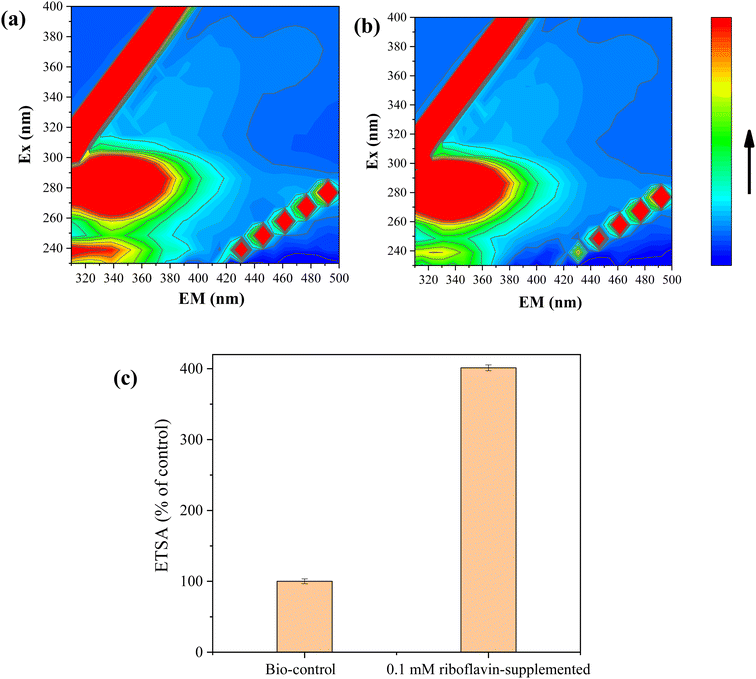 | ||
| Fig. 5 FEEM analyses of liquid supernatant obtained in 1 mM (a) Se(IV)- and (b) Te(IV)-supplemented reaction systems; (c) effect of riboflavin on the electron transfer efficiency (OD600 = 0.3). | ||
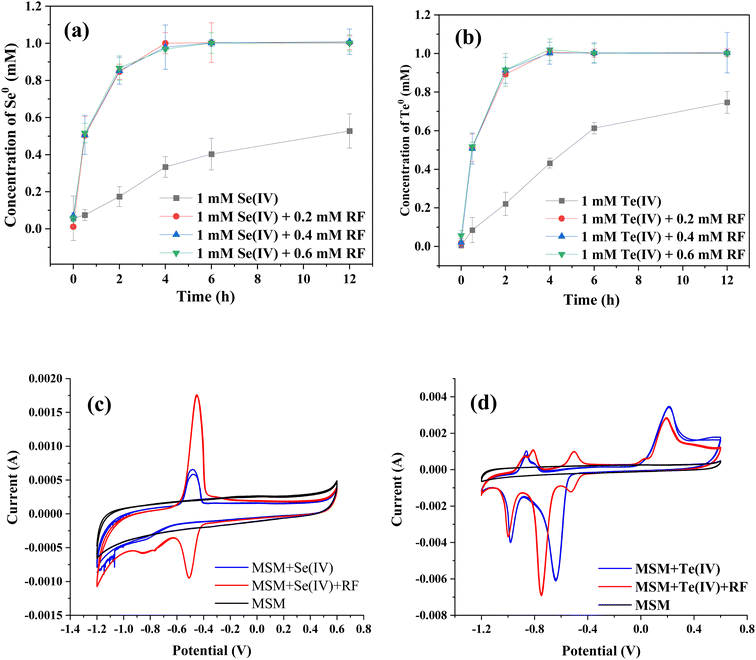
Fig. 6a and b show the effects of riboflavin on Se(IV) and Te(IV) reduction (calculated as the amounts of generated Se0 and Te0). In the presence of riboflavin, 1 mM Se(IV) and Te(IV) could be completely reduced within 4 h, while 12 h was used for the control system. It should be noted that the catalytic effect of 0.2 mM riboflavin is similar to that of 0.6 mM riboflavin, which may be due to the limited electrons produced by bacteria in a certain period of time. The color change of the whole reaction process is shown in Fig. S2,† which also shows that the Se(IV) and Te(IV) reduction efficiencies of the riboflavin-added systems were faster than that of the control system. This finding is similar to those of previous studies in that riboflavin was able to mediate or support extracellular electron transfer for microorganisms.24–26
The CV results showed that strain FDL-2 did secrete riboflavin since the characteristic reductive peak of riboflavin was observed (Fig. 6c). Moreover, the supernatant obtained from the Se(IV)-added system (without exogenous riboflavin) showed stronger reducibility than oxidizability since the riboflavin secreted by strain FDL-2 was reduced to riboflavin-H2 in the absence of oxygen. We have confirmed the accelerated effect of riboflavin on Se(IV) and Te(IV) reduction (Fig. 6a and b), so it is more certain, combined with the CV results, that the accelerated electron transfer enhanced the transfer of electrons from the donor to Se(IV) and Te(IV) via riboflavin. Interestingly, multiple pairs of redox peaks were observed for the supernatant obtained from the Te(IV)-supplemented system (Fig. 6d), which may depend on the electrochemical properties of Te itself and require further investigation.
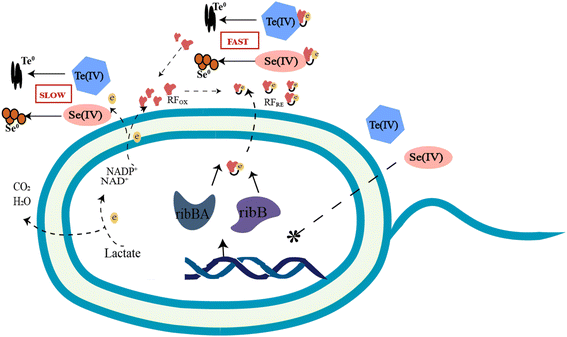
To date, increasing terrestrial and marine bacterial resources have been identified to be capable of reducing Se(IV) and Te(IV) to Se0 and Te0.8–13 Some of the more studied bacterial genera include Shewanella, Pseudomonas, Bacillus, Pseudoalteromonas, Acinetobacter, etc. They differ not only in their ability to reduce Se(IV) and Te(IV) but also in their reduction mechanisms. Three possible Se(IV)/Te(IV)-induced response mechanisms for bacteria have been previously reported: (i) assimilation reduction; (ii) dissimilatory reduction; and (iii) detoxification.5 Since 2020, we have carried out several studies8,9 on the reduction of Se(IV) and Te(IV) and revealed several key genes and proteins that may be involved in the reduction of Se(IV)/Te(IV) at the transcriptomic and proteomic levels, respectively. Based on the results of this study, we proposed a riboflavin-involved electron transfer pathway from electron donors to Se(IV) and Te(IV). Riboflavin and its analogs, which have been widely reported for their potential antiviral, anticancer and antibacterial effects, are involved in many biological activities.27–29 In addition to the previous studies, this study provides the first solid evidence to show that riboflavin plays a key role in the bioreduction of Se(IV)/Te(IV). As shown in Fig. 7, riboflavin secreted by strain FDL-2 served as a redox mediator, which can shuttle electrons from reductases, including nitrate reductase, nitrite reductase, fumarate reductase and some other terminal reductases, to Se(IV) or Te(IV).
During this process, the presence of riboflavin enhanced the electron transfer efficiency and thus facilitated the reduction of Se(IV) and Te(IV). Thus, this mechanism can be classified as type II as “mediated dissimilation reduction”. The secretion of FMN and riboflavin by Shewanella strains has been previously reported, and such substances can facilitate electron transport out of the cell.30–32 However, there is no direct evidence that these substances are involved in the biological reduction of Se(IV) or Te(IV). For example, Shewanella denitrificans can secrete flavins, but it cannot reduce metal oxides and electrodes.33 Notably, strain FDL-2 does not have a well-known “MtrABC + cytochrome C” electron transport pathway as Shewanella oneidensis MR-1. Therefore, the riboflavin-involved electron transport pathway may compensate for this deficiency. These findings enrich our understanding of the biological reduction mechanism of Se(IV) and Te(IV).
4. Conclusions
The biological reduction mechanism of Se(IV)/Te(IV) remains not fully discovered. We now report a riboflavin-involved pathway for Se(IV)/Te(IV) bioreduction. During this process, the riboflavin can shuttle electrons from electron donors to Se(IV) and Te(IV), which enhance the electron transfer efficiency and thus facilitate the reduction of Se(IV) and Te(IV). Overall, these findings deepen the understanding of the biological reduction of Se(IV) and Te(IV) and provide guidance for the development of Se(IV) and Te(IV) pollution remediation technologies.Author contributions
Manman Cheng: conceptualization, data curation, investigation, writing – original draft, writing – review & editing. Haikun Zhang: investigation, writing – review & editing. Yan Li: formal analysis, methodology, writing – review & editing. Wenhao Chen: writing – review & editing.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
The authors want to thank the support from the National Natural Science Foundation of China (42077305) and the Solid-state Fermentation Resource Utilization Key Laboratory of Sichuan Province (No. 2018GTY001).References
- S. Huang, Y. Wang, C. Tang, H. Jia and L. Wu, Speeding up selenite bioremediation using the highly selenite-tolerant strain Providencia rettgeri HF16-A novel mechanism of selenite reduction based on proteomic analysis, J. Hazard. Mater., 2021, 406, 124690 CrossRef CAS PubMed.
- D. H. Kim, M. G. Kim, S. Jiang, J. H. Lee and H. G. Hur, Promoted reduction of tellurite and formation of extracellular tellurium nanorods by concerted reaction between iron and Shewanella oneidensis MR-1, Environ. Sci. Technol., 2013, 47(15), 8709–8715 CrossRef CAS PubMed.
- S. Wang, Tellurium, its resourcefulness and recovery, Jom, 2011, 63(8), 90–93 CrossRef CAS.
- T. Wang, L. Yang, B. Zhang and J. Liu, Extracellular biosynthesis and transformation of selenium nanoparticles and application in H2O2 biosensor, Colloids Surf., B, 2010, 80(1), 94–102 CrossRef CAS PubMed.
- L. Manna, E. C. Scher and A. P. Alivisatos, Synthesis of soluble and processable rod-, arrow-, teardrop-, and tetrapod-shaped CdSe nanocrystals, J. Am. Chem. Soc., 2000, 122(51), 12700–12706 CrossRef CAS.
- Y. Dong, H. Zhang, L. Hawthorn, H. E. Ganther and C. Ip, Delineation of the molecular basis for selenium-induced growth arrest in human prostate cancer cells by oligonucleotide array, Cancer Res., 2003, 63(1), 52–59 CAS.
- R. J. Turner, J. H. Weiner and D. E. Taylor, Selenium metabolism in Escherichia coli, BioMetals, 1998, 11(3), 223–227 CrossRef CAS PubMed.
- M. Cheng, L. Liang, Y. Sun, H. K. Zhang and X. K. Hu, Reduction of selenite and tellurite by a highly metal-tolerant marine bacterium, Int. Microbiol., 2023 DOI:10.1007/s10123-023-00382-w.
- M. Cheng, Y. Sun, X. Sui and H. Zhang, Characterization of the differentiated reduction of selenite and tellurite by a halotolerant bacterium: process and mechanism, J. Water Process. Eng., 2022, 47, 102809 CrossRef.
- J. Kessi and K. W. Hanselmann, Similarities between the abiotic reduction of selenite with glutathione and the dissimilatory reaction mediated by Rhodospirillum rubrum and Escherichia coli, J. Biol. Chem., 2004, 279(49), 50662–50669 CrossRef CAS PubMed.
- T. Kagami, A. Fudemoto, N. Fujimoto, E. Notaguchi, M. Kanzaki, M. Kuroda, S. Soda, M. Yamashita and M. Ike, Isolation and characterization of bacteria capable of reducing tellurium oxyanions to insoluble elemental tellurium for tellurium recovery from wastewater, Waste Biomass Valorization, 2012, 3(4), 409–418 CrossRef CAS.
- W. D. Bonificio and D. R. Clarke, Bacterial recovery and recycling of tellurium from tellurium-containing compounds by P. seudoalteromonas sp. EPR 3, J. Appl. Microbiol., 2014, 117(5), 1293–1304 CrossRef CAS PubMed.
- S. L. Wadgaonkar, J. Mal, Y. V. Nancharaiah, N. O. Maheshwari, G. Esposito and P. N. L. Lens, Formation of Se(0), Te(0), and Se(0)-Te(0) nanostructures during simultaneous bioreduction of selenite and tellurite in a UASB reactor, Appl. Microbiol. Biotechnol., 2018, 102(6), 2899–2911 CrossRef CAS PubMed.
- G. W. Tyson, C. Jarrod, H. Philip, E. A. Eric, J. R. Rachna, M. R. Paul, V. S. Victor, M. R. Edward, S. R. Daniel and F. B. Jillian, Community structure and metabolism through reconstruction of microbial genomes from the environment, Nature, 2004, 428, 37–43 CrossRef CAS PubMed.
- T. Seemann, Prokka: rapid prokaryotic genome annotation, Bioinformatics, 2014, 30(14), 2068–2069 CrossRef CAS PubMed.
- S. Arindam, K. Manoj, C. Rayanee, S. Sudeshna and P. Kannan, Simultaneous removal of selenite and heavy metals from wastewater and their recovery as nanoparticles using an inverse fluidized bed bioreactor, J. Cleaner Prod., 2022, 376, 134248 CrossRef.
- J. Mal, V. N. Yarlagadda, M. Neeraj, D. van H. Eric and N. L. L. Piet, Continuous removal and recovery of tellurium in an upflow anaerobic granular sludge bed reactor, J. Hazard. Mater., 2017, 327, 79–88 CrossRef CAS PubMed.
- Y. He, J. Guo, Y. Song, Z. Chen, C. Lu, Y. Han and R. Zhao, Acceleration mechanism of bioavailable Fe (III) on Te (IV) bioreduction of Shewanella oneidensis MR-1: promotion of electron generation, electron transfer and energy level, J. Hazard. Mater., 2021, 403, 123728 CrossRef CAS PubMed.
- H. Zhang, Y. Sun, M. Cheng, X. Sui, Y. Huang and X. Hu, How iron-bearing minerals affect the biological reduction of Sb (V): a newly discovered function of nitrate reductase, Sci. Total Environ., 2023, 167001 CrossRef CAS PubMed.
- R.-R. Kim, B. Illarionov, M. Joshi, M. Cushman, C. Y. Lee, W. Eisenreich, M. Fischer and A. Bacher, Mechanistic insights on riboflavin synthase inspired by selective binding of the 6,7-Dimethyl-8-ribityllumazine exomethylene anion, J. Am. Chem. Soc., 2010, 132(9), 2983–2990 CrossRef CAS PubMed.
- W. Chen, P. Westerhoff, J. A. Leenheer and K. Booksh, Fluorescence excitation-emission matrix regional integration to quantify spectra for dissolved organic matter, Environ. Sci. Technol., 2003, 37, 5701–5710 CrossRef CAS PubMed.
- D. R. Lovley, J. L. Fraga, E. L. Blunt-Harris, L. A. Hayes, E. J. P. Phillips and J. D. Coates, Humic substances as a mediator for microbially catalyzed metal reduction, Acta Hydrochim. Hydrobiol., 1998, 26, 152–157 CrossRef CAS.
- E. E. Roden, A. Kappler, I. Bauer, J. Jiang, A. Paul, R. Stoesser, K. Knishi and H. Xu, Extracellular electron transfer through microbial reduction of solid-phase humic substances, Nat. Geosci., 2010, 3, 417–421 CrossRef CAS.
- S. J. Fuller, D. G. McMillan, M. B. Renz, M. Schmidt, I. T. Burke and D. I. Stewart, Extracellular electron transport-mediated Fe(III) reduction by a community of alkaliphilic bacteria that use flavins as electron shuttles, Appl. Environ. Microbiol., 2014, 80, 128–137 CrossRef CAS PubMed.
- E. Marsili, D. B. Baron, I. D. Shikhare, D. Coursolle, J. A. Gralnick and D. R. Bond, Shewanella secretes flavins that mediate extracellular electron transfer, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 3968–3973 CrossRef CAS PubMed.
- D. M. Gurumurthy, R. N. Bharagava, A. Kumar, B. Singh, M. Ashfaq, G. D. Saratale and S. I. Mulla, EPS bound flavins driven mediated electron transfer in thermophilic Geobacillus sp, Microbiol. Res., 2019, 229, 126324 CrossRef CAS PubMed.
- X. Li and Y. Song, Structure and function of SARS-CoV and SARS-CoV-2 main proteases and their inhibition: a comprehensive review, Eur. J. Med. Chem., 2023, 115772 CrossRef CAS PubMed.
- J. Qin, X. Li, C. Hunt, W. Wang, H. Wang and R. Zhang, Natural products targeting the p53-MDM2 pathway and mutant p53: recent advances and implications in cancer medicine, Genes Dis., 2018, 5(3), 204–219 CrossRef CAS PubMed.
- X. Li and S. Ma, Advances in the discovery of novel antimicrobials targeting the assembly of bacterial cell division protein FtsZ, Eur. J. Med. Chem., 2015, 95, 1–15 CrossRef PubMed.
- A. S. Beliaev, D. A. Saffarini, J. L. McLaughlin and D. Hunnicutt, MtrC, an outer membrane decahaem c cytochrome required for metal reduction in Shewanella putrefaciens MR-1, Mol. Microbiol., 2001, 39, 722–730 CrossRef CAS PubMed.
- O. Bretschger, A. Obraztsova, C. A. Sturm, I. S. Chang, Y. A. Gorby, S. B. Reed, D. E. Culley, C. L. Reardon, S. Barua, M. F. Romine, J. Zhou, A. S. Beliaev, R. Bouhenni, D. Saffarini, F. Mansfeld, B. Kim, J. K. Fredrickson and K. H. Nealson, Current production and metal oxide reduction by Shewanella oneidensis MR-1 wild type and mutants, Appl. Environ. Microbiol., 2008, 73, 7003–7012 CrossRef PubMed.
- L. Shi, C. S. Thomas, M. Z. John and K. F. James, Respiration of metal (hydr)oxides by Shewanella and Geobacter: a key role for multihaem c-type cytochromes, Mol. Microbiol., 2007, 65, 12–20 CrossRef CAS PubMed.
- E. D. Brutinel and A. G. Jeffrey, Shuttling happens: soluble flavin mediators of extracellular electron transfer in Shewanella, Appl. Microbiol. Biotechnol., 2012, 93, 41–48 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra07093j |
| This journal is © The Royal Society of Chemistry 2023 |