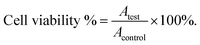Open Access Article
Open Access ArticleEnhancing the inhibition of cell proliferation and induction of apoptosis in H22 hepatoma cells through biotransformation of notoginsenoside R1 by Lactiplantibacillus plantarum S165 into 20(S/R)-notoginsenoside R2
Penghui Wanga,
Yansong Gaob,
Ge Yangb,
Yujuan Zhaob,
Zijian Zhaob,
Ge Gaoa,
Lei Zhao*a and
Shengyu Li *b
*b
aSchool of Pharmaceutical Sciences, Changchun University of Chinese Medicine, Changchun 130117, P. R. China
bInstitute of Agro-Food Technology, Jilin Academy of Agricultural Sciences, Changchun 130033, P. R. China. E-mail: lisy720@126.com; Fax: +86 431 87063075; Tel: +86 431 87063289
First published on 11th October 2023
Abstract
Notoginsenoside R2 is a crucial active saponin in Panax notoginseng (Burk.) F. H. Chen, but its natural content is relatively low. In this study, we investigated the biotransformation of notoginsenoside R1 to 20(S/R)-notoginsenoside R2 using Lactiplantibacillus plantarum S165, compared the inhibitory effects on cancer cell proliferation and conducted a mechanistic study. Notoginsenoside R1 was transformed using Lactiplantibacillus plantarum S165 at 37 °C for 21 days. The fermentation products were identified using a combination of HPLC, UPLC-MS/MS, and 13C-NMR methods. The inhibition effects of 20(S/R)-notoginsenoside R2 on H22 hepatoma cells were assessed by CCK-8 and TUNEL assays, and the underlying mechanism was investigated by Western blotting. Lactiplantibacillus plantarum S165 could effectively transform notoginsenoside R1 to 20(S/R)-notoginsenoside R2 with a conversion yield of 82.85%. Our results showed that 20(S/R)-notoginsenoside R2 inhibited H22 hepatoma cells proliferation and promoted apoptosis. The apoptosis of H22 hepatoma cells was promoted by 20(S/R)-notoginsenoside R2 through the blockade of the PI3K/AKT/mTOR signaling pathway. The biotransformation method used in this study resulted in the production of 20(S)-notoginsenoside R2 and 20(R)-notoginsenoside R2 from notoginsenoside R1, and the anti-tumor activity of the transformed substance markedly improved.
1. Introduction
Hepatocellular carcinoma (HCC) is a primary liver cancer type with a high mortality rate, accounting for about 90% of all cases.1 It is one of the deadliest cancers and poses a significant global health burden. Statistics suggest that HCC is the third leading cause of cancer-related deaths worldwide, only behind gastric cancer and lung cancer.2 Despite advances in treatment options such as surgery, chemotherapy, and radiation therapy, the overall survival rate of HCC patients remains low.3 Although significant progress has been made in the development of new anti-tumor drugs, the lack of tissue-specificity and the occurrence of drug-related side effects remain major challenges in the field of cancer treatment. These issues highlight the need for the discovery of low-toxicity, natural anti-tumor drugs as a potential solution to this problem.4Panax notoginseng (Burk.) F. H. Chen is a traditional medicinal plant belonging to the Panax genus of the Araliaceae family. It is known as Sanqi or Tianqi in China and has been used for several hundred years to treat cardiovascular diseases, inflammation, and trauma.5 In recent years, extensive chemical and pharmacological research on Sanqi has revealed that its main components are triterpenoid saponins, including gypenosides, ginsenosides, and notoginsenosides.6 Among the isolated saponins, major constituents include ginsenosides Rb1, Rg1, Re, and notoginsenoside R1.7 However, most studies have focused on the pharmacological activities of minor saponins, as their activities are considered more valuable than those of major saponins.8 Minor saponins have attracted widespread attention due to their stronger physiological activities in vivo and good physical or chemical properties in crossing cell membranes.9 For instance, minor saponins like ginsenosides F1, F2, Rg3, compound K, and notoginsenoside R2 are produced by the hydrolysis of glycosides from major saponins, namely Rg1, Re, Rb1, and notoginsenoside R1. Therefore, some studies aimed to transform major saponins into minor saponins with higher activities. For instance, Wang et al. prepared a potential agent for anti-herpes simplex virus infection namely notoginsenoside ST-4 by enzymatically transforming notoginsenoside R7 using a novel new recombinant glycoside hydrolase from Herpetosiphon aurantiacus.10
In order to obtain minor saponins, in recent years, many researchers have focused on transforming major saponins by removing the glucose residues using traditional methods, such as through acid, base, and thermal conversion treatments. However, these methods have several drawbacks, including intense reactions, high costs, long processing times, low specificity, and environmental concerns.11 Consequently, the microbial transformation approach has gained attention owing its high specificity, great yield, and eco-friendly nature.12 Previous study have explored the transformation of saponins using various microorganisms, such as Cordyceps sinensis, Burkholderia sp., Lentilactobacillus buchneri, Aspergillus sp. and Bacillus sp.13–17 Lactobacillus, as a food-grade probiotic, possesses several significant advantages.18 Its remarkable resilience in acidic and bile salt environments makes it a strong contender for diverse applications.19 Additionally, Lactobacillus is easily obtainable and cultivatable, which enhances its suitability as a safer and more convenient choice for biotransformation processes.20 Based on the aforementioned reasons, in this study, Lactiplantibacillus plantarum S165 was obtained by screening strains in fermented foods. Subsequently, this strain was used to biotransform notoginsenoside R1 to obtain notoginsenoside R2. Also as the PPT-type saponins, notoginsenoside R2 is rarer than notoginsenoside R1, although only one glucose is missing (Fig. 1), and notoginsenoside R2 is divided into 20(S)-notoginsenoside R2 and 20(R)-notoginsenoside R2 due to the chiral carbon of position 20. It has been suggested that notoginsenoside R2 can alleviate Alzheimer's disease.21,22 Differently, we investigated the anti-tumor effect and underlying apoptosis mechanism of notoginsenoside R2. These impressive results demonstrated the high specificity of Lactiplantibacillus plantarum S165 for the hydrolysis of glucose at position C-20, making it a promising candidate for notoginsenoside R2 production in both the pharmaceutical and food industries. This finding has important implications for the development of safe, effective, and environmentally friendly natural products for cancer therapy and other applications.
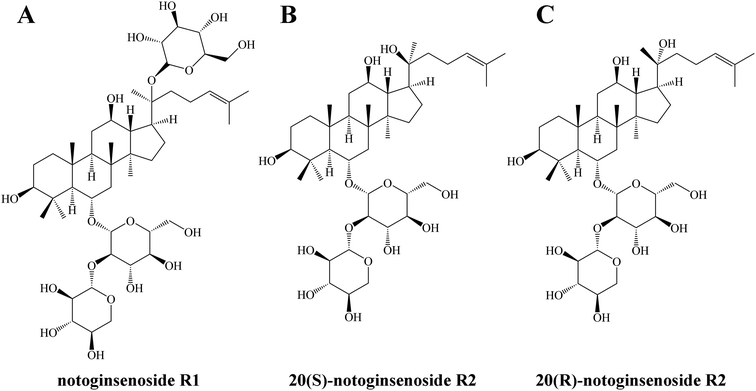 | ||
| Fig. 1 Structure of notoginsenoside R1 (A), 20(S)-notoginsenoside R2 (B) and 20(R)-notoginsenoside R2 (C). | ||
2. Methods and materials
2.1 Material and reagents
Standard notoginsenoside R1, 20(S)-notoginsenoside R2 and 20(R)-notoginsenoside R2 were supplied by Chengdu Alfa Biotechnology Co., Ltd. (Chengdu, China). Trypsin–EDTA was purchased from Thermo Fisher Scientific (China) Co., Ltd. Fetal Bovine Serum (FBS) was obtained from Cyagen Biosciences (Guangzhou) Inc.2.2 Strain and culture conditions
In this study, the strain S165 was isolated from sauerkraut, which was preliminarily identified as Lactiplantibacillus plantarum. The strain has been stored in China Center for Type Culture Collection (CCTCC no. M2022783). Before use, single colonies were subcultured in MRS broth at 37 °C for 15 h, the culture in the exponential phase was then inoculated into MRS broth and incubated at 37 °C for 20 h.232.3 Preparation of 20(S/R)-notoginsenoside R2
A 2000 mL silk mouth reagent flask containing 1000 mL fermentation medium consisting of 30 g glucose and 10 g yeast extract was incubated at 121 °C for 15 min. After cooling to room temperature, 1 g notoginsenoside R1 was added to the fermentation medium before filtration with a 0.22 μm filter (Millex®-GP, Merck Millipore Ltd. Tullagreen, Cork, Ireland). The prepared Lactiplantibacillus plantarum S165 strain was inoculated into the above fermentation medium to obtain an initial number of viable bacteria of 1.0 × 107 CFU mL−1. The inoculated notoginsenoside R1 solution was then incubated at 37 °C for 21 d.24After 21 d of fermentation, the culture was freeze-dried, completely dissolved in distilled water, and subjected to absorption on a D101 macroporous resin column for 10 h after completion of loading at a rate of 0.4 BV h−1. Gradient elution with ethanol at various concentrations (0, 30%, 50%, 70%, and 90%) was used for column elution. The resin chromatography column was loaded; the flow rate was 0.4 BV h−1 and the elution volume of each gradient was 5 BV.25 The eluted fractions of 50% ethanol rich in our target notoginsenosides were vacuum concentrated and lyophilized to obtain 828.53 mg mixture powders of 20(S)-notoginsenoside R2 and 20(R)-notoginsenoside R2, and named it 20(S/R)-notoginsenoside R2.
2.4 Detection of OD600, pH, viable bacteria and β-glucosidase activity
Solution OD600 was tested at 600 nm with the SpectraMax ABS Plus microplate reader (Molecular Devices, Shanghai, China), pH was detected by pH meter and viable bacteria by plate counting every 4 h. To determine β-glucosidase activity according to reference by Li et al., pNP-β-D-glucopyranoside (pNPG) was used as a substrate to detect the activity of β-glucosidase.26 Detection was performed within 48 h of the beginning of biotransformation and at the moment before the end of biotransformation.2.5 High-performance liquid chromatography (HPLC) analysis
HPLC was performed using an Agilent 1260 Infinity system (Agilent Corporation, USA). A reverse phase column (Pursuit 5 C18, 4.6 mm × 250 mm, 5 μm) was used at 30 °C. HPLC-grade water (A) and acetonitrile (B) were used as the mobile phase. The analysis was performed with the mobile phase at a flow rate of 1 mL min−1 with the following conditions: 0–40 min 18–21% (B); 40–42 min 21–26% (B); 42–46 min 26–32% (B); 46–66 min 32–34% (B); 66–71 min 34–38% (B); 71–77.70 min 38.0–49.1% (B); 77.70–82 min 49.1% (B); 82–83 min 49.1–50.6% (B); 83–88 min 50.6–59.6% (B); 88–89.80 min 59.6–65.0% (B); 89.80–97 min 65% (B); 97–102 min 65–75% (B); 102–110 min 75–85% (B); 110–115 min 85% (B); 115–125 min 85–18% (B), and 125–130 min 18.0% (B). The injection volume was 20 μL, the column temperature was maintained at 30 °C, and UV absorption was measured at 203 nm.27 The transformation of notoginsenosides was monitored by comparing their chromatograms to those of standard notoginsenosides.2.6 Ultra-performance liquid chromatography (UPLC)-mass spectrometry (MS)/MS analysis
UPLC was conducted on Dionex UltiMate 3000 HPLC platform (Thermo Fisher Scientific, USA) and chromatographic separations were through Sigma HPLC Column C18 (50 mm × 3.0 mm × 2.7 μm). Mobile phase A was 0.1% formic acid water and B was acetonitrile. The column oven was kept at 30 °C. The injection volume was 5 μL and the flow rate was 0.2 mL min−1 for each run. The elution gradient is presented in Section 2.5.Thermo Scientific™ Q Exactive™ (Thermo Fisher Scientific, USA) was used for MSE acquisition. Conditions for MS were as follows: ESI source, negative ion mode; sheath gas velocity, 4 × 106 Pa; auxiliary gas velocity, 1 × 106 Pa; auxiliary gas temperature, 300 °C; dry gas temperature, 350 °C; scan mode, full scan-ddMS2 (m/z 150–2000) and resolution, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 FWHM.28
000 FWHM.28
2.7 Nuclear magnetic resonance (NMR) analysis
The 13C-NMR experiment was performed using the Bruker 500 MHz NMR Spectrometer (Avance III 500MR) at 125 MHz with 13C-NMR with tetramethylsilane (TMS) as internal standard. Approximately 10–20 mg samples were dissolved in pyridine-d5 to record the NMR spectra using the lowest field signals of pyridine-d5 (13C, δ 149.9) as an internal reference.292.8 Cell culture and assessing the viability
H22 hepatoma (H22) cells were purchased from Jiangsu Keygen Biotech Crop., Ltd. (Jiangsu, China). Cells were cultured in RPMI 1640 medium (Jiangsu Keygen Biotech Corp., Ltd.) supplemented with heat-inactivated FBS (10%) and penicillin/streptomycin (1%). The cells were incubated in an incubator (Shanghai Yiheng Technology Instrument Co., Ltd.) at 37 °C with 5% CO2 and 95% air.Cell viability was determined by CCK-8 assay (U.S. Everbright Inc.). H22 cells were seeded in 96-well plates at a density of 1 × 104 cells per well and incubated in a CO2 incubator (5% CO2 and 95% air, 37 °C) for 24 h until they reached 80–90% confluence. Further, cells were treated with various concentrations of notoginsenoside R1 and 20(S/R)-notoginsenoside R2 (0, 20, 40, 60, 80, and 100 μg mL−1) as indicated for 24 h. Finally, the cells were incubated with 10% (v/v) of CCK-8 solution for 3 h and the absorbance at 450 nm was recorded with the SpectraMax ABS Plus microplate reader (Molecular Devices, Shanghai, China).30
Logistic regression analysis was used to create cell viability rate curves.
2.9 Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay
TUNEL assay was preformed to evaluate the apoptosis state of H22 cells. Briefly, the treated cells were fixed with 4% paraformaldehyde after washing thrice with PBS and then incubated with a TUNEL reaction mixture for 2 h in 37 °C. After washing thrice with PBS, the nuclei of cells were stained with DAPI (Wuhan Servicebio Technology CO., LTD). Three randomly chosen fields were assessed for each group.312.10 Western blotting
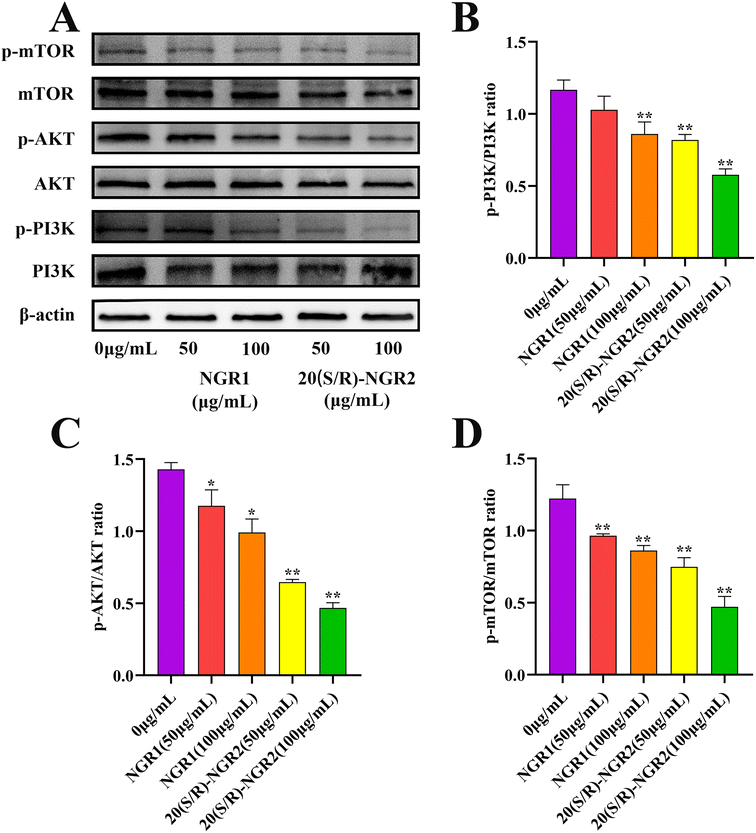
H22 cells were seeded in 6-well plates at a density of 2 × 105 cells per well and treated with the various concentrations of notoginsenoside R1 and 20(S/R)-notoginsenoside R2 (0, 50, and 100 μg mL−1) as indicated. After 24 h incubation, whole cell extracts were prepared with RIPA buffer containing 1 mM PMSF. Proteins in the cell extracts were electrophoresed and transferred onto a PVDF membrane. The PVDF membrane was blocked with 3% BSA in Tris-buffered saline with 0.05% Tween-20 (TBST) for 1 h at room temperature and incubated with antibodies overnight at 4 °C, including rabbit anti-β-actin (Affinity #AF7018), rabbit anti-PI3K (Abcam ab40776), rabbit anti-p-PI3K (Abcam ab278545), rabbit anti-AKT (GeneTex N3C2), rabbit anti-p-AKT (GTX 128414), rabbit anti-mTOR (Bioss, bsm-54471R), and rabbit anti-p-mTOR (Bioss, bs-3495R). After washing with TBST, the membrane was incubated with a horseradish peroxidase-conjugated secondary antibody for 1 h at 37 °C.32 The intensity of the protein bands was then detected using an Image Quant LAS 4000 system (Fuji Film, Tokyo, Japan) and β-actin was used as a loading control for normalization.2.11 Statistical analysis
All experiments were performed in triplicates. Data were analyzed by SPSS 16 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism 8 software (GraphPad, La Jolla, CA, USA). One-way analysis of variance (ANOVA) and Tukey's post hoc test were employed to compare the results and their significance, respectively. Finally, the results were expressed as means ± standard deviation. A P-value less than 0.05 was considered statistically significant.3. Results
3.1 Identification of products bioconversion
The transformation products of notoginsenoside R1 by Lactiplantibacillus plantarum S165 were identified by HPLC, UPLC-MS/MS, and 13C-NMR analysis. Through HPLC analysis and comparison with established standards, it was observed that the notoginsenoside R1 content in the solution gradually decreased over time, while the content of 20(S)-notoginsenoside R2 and its epimer increased (Fig. 2A and B). Ultimately, the biotransformation product attained an impressive purity of 99.99%, accompanied by a fermentation yield of 82.85% (Fig. 2C). For comprehensive profiling of bioconversion products from notoginsenoside R1, an analytical method based on UPLC-MS/MS was conducted. As shown in Table 1 and Fig. 3, notoginsenoside R1 (tR = 9.02, m/z 931.52), 20(S)-notoginsenoside R2 (tR = 45.26, m/z 769.47), 20(R)-notoginsenoside R2 (tR = 46.83, m/z 769.47) were identified. Although the retention times of 20(S)-notoginsenoside R2 and 20(R)-notoginsenoside R2 were different, the molecular ion peaks are same. The same results were founded in the 13C-NMR experiment (Table 2), and other carbon signals were almost completely overlapping, except for the significant difference in the chemical shifts of C17, C21, and C22. Compared to 20(R)-notoginsenoside R2, the chemical shift of C17 and C21 of 20(S)-notoginsenoside R2 moved to the low field, while the chemical shift of C22 moved to the high field. Accordingly, the biotransformation pathway of notoginsenoside R1 was determined as “notoginsenoside R1 → 20(S)-notoginsenoside R2 and 20(R)-notoginsenoside R2” (Fig. 4), this is a hydrolysis process about the glucose of C-20 position.| No. | Product | tR (min) | Measured mass (m/z) | Theoretical mass | Mass error (ppm) |
|---|---|---|---|---|---|
| 1 | Notoginsenoside R1 | 9.02 | 931.52 | 931.52 | 0.00 |
| 2 | 20(S)-Notoginsenoside R2 | 45.26 | 769.47 | 769.47 | 0.00 |
| 3 | 20(R)-Notoginsenoside R2 | 46.83 | 769.47 | 769.47 | 0.00 |
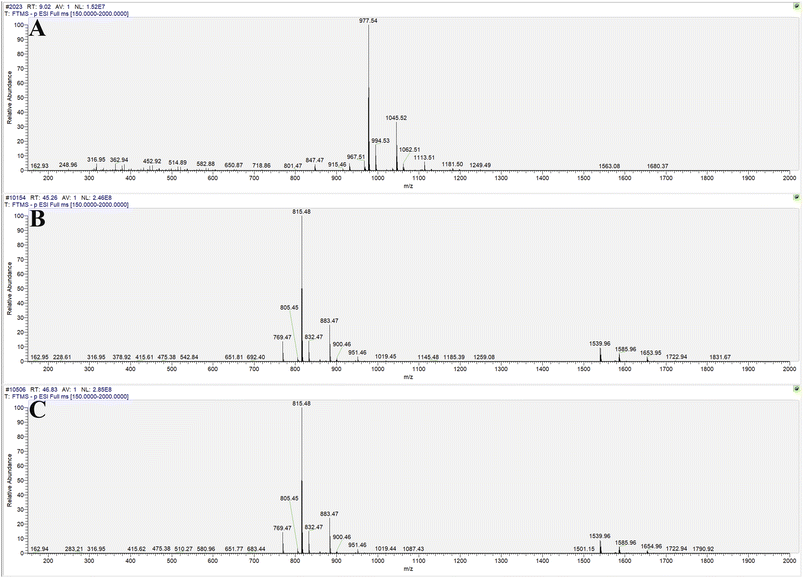 | ||
| Fig. 3 Molecular ion peaks of UPLC-MS/MS: notoginsenoside R1 (A), 20(S)-notoginsenoside R2 (B) and 20(R)-notoginsenoside R2 (C). | ||
| Position | 20(S)-Notoginsenoside R2 | 20(R)-Notoginsenoside R2 | |
|---|---|---|---|
| 1 | 39.07 | 39.07 | |
| 2 | 29.99 | 29.99 | |
| 3 | 76.22 | 76.22 | |
| 4 | 40.54 | 40.54 | |
| 5 | 60.50 | 60.50 | |
| 6 | 78.69 | 78.69 | |
| 7 | 43.87 | 43.87 | |
| 8 | 39.04 | 39.04 | |
| 9 | 49.52 | 49.52 | |
| 10 | 38.85 | 38.85 | |
| 11 | 31.90 | 31.90 | |
| 12 | 70.55 | 70.55 | |
| 13 | 48.12 | 48.12 | |
| 14 | 51.16 | 51.16 | |
| 15 | 30.55 | 30.55 | |
| 16 | 25.11 | 25.11 | |
| 17 | 53.73 | 54.70 | |
| 18 | 16.16 | 16.16 | |
| 19 | 15.64 | 15.64 | |
| 20 | 73.19 | 73.19 | |
| 21 | 26.04 | 26.11 | |
| 22 | 35.60 | 34.87 | |
| 23 | 21.88 | 21.88 | |
| 24 | 124.56 | 124.79 | |
| 25 | 130.60 | 130.60 | |
| 26 | 24.50 | 24.50 | |
| 27 | 16.30 | 16.30 | |
| 28 | 31.70 | 31.70 | |
| 29 | 16.45 | 16.45 | |
| 30 | 15.13 | 15.13 | |
| Glc | 1′ | 102.47 | 102.47 |
| 2′ | 79.32 | 79.32 | |
| 3′ | 77.92 | 77.92 | |
| 4′ | 70.69 | 70.69 | |
| 5′ | 78.29 | 78.29 | |
| 6′ | 61.53 | 61.53 | |
| Xyl | 1′ | 102.49 | 102.49 |
| 2′ | 74.18 | 74.18 | |
| 3′ | 76.88 | 76.88 | |
| 4′ | 73.04 | 73.04 | |
| 5′ | 65.53 | 65.53 |
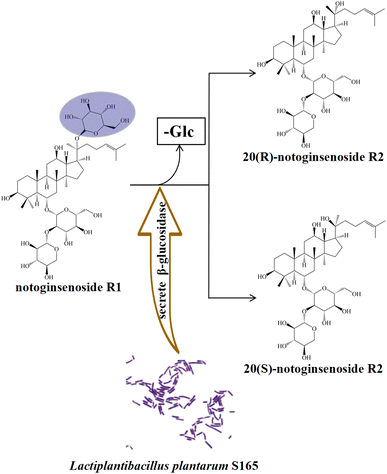 | ||
| Fig. 4 Biotransformation pathways for notoginsenoside R1. “-Glc” indicates the loss of glucosyl group. | ||
3.2 Growth curves and the β-glucosidase activity
As biotransformation progressed, the strain in the culture medium grew rapidly within 24 h and produced large amounts of β-glucosidase and acid. After 24 h, the growth rate decreased and gradually died. The maximum number of viable bacteria was 1.80 × 109 CFU mL−1, pH was stable at 4.00, OD600 was about 1.50, and β-glucosidase activity was stable at 105.00 U mL−1 (Fig. 5).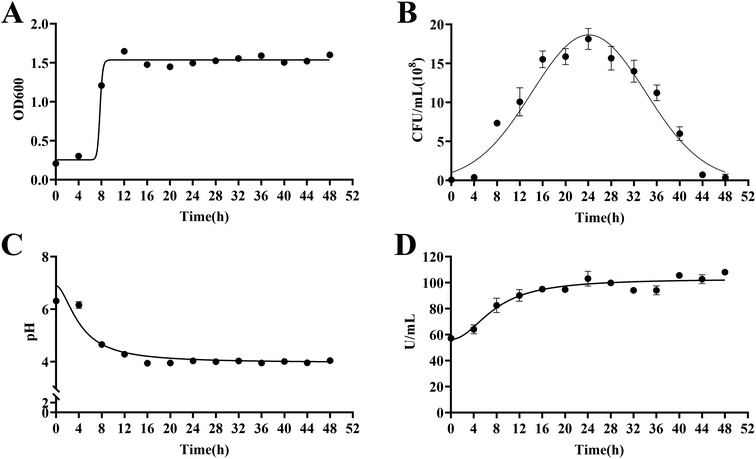 | ||
| Fig. 5 Growth curves and the β-glucosidase activity of Lactiplantibacillus plantarum S165. OD600 (A), viable bacteria (B), pH value (C) and β-glucosidase activity (D). | ||
3.3 20(S/R)-Notoginsenoside R2 reduced the viability of H22 cells
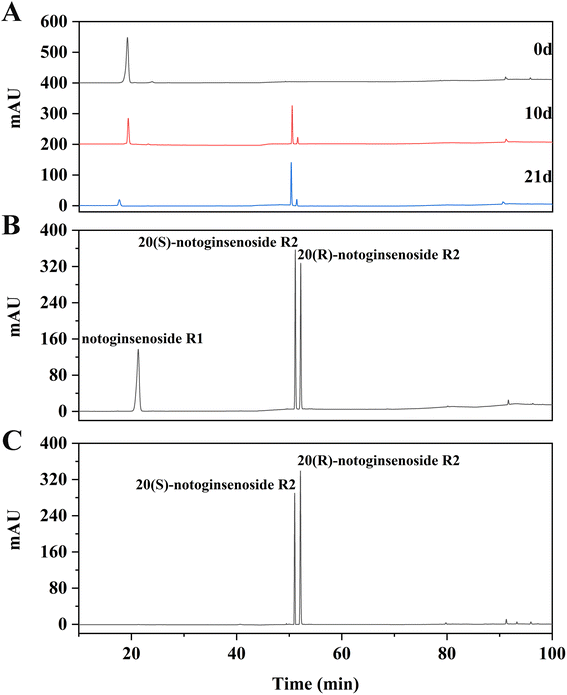
As shown in the results of the CCK-8 assay in Fig. 6, after 24 h of 20(S/R)-notoginsenoside R2 treatment, the proliferation of H22 cells was suppressed and the inhibition rate was dose-dependent within the concentration ranges tested. Through the comparison, 20(S/R)-notoginsenoside R2 was founded to be significantly better than the control group. The IC50 values of notoginsenoside R1 and 20(S/R)-notoginsenoside R2 are 121.50 μg mL−1 and 65.91 μg mL−1 respectively. Among these, when the concentration of 20(S/R)-notoginsenoside R2 reached 100 μg mL−1, a significant decrease in cell growth and viability was observed, which was 19.52 ± 3.09% (P < 0.01).3.4 20(S/R)-Notoginsenoside R2 led to apoptosis
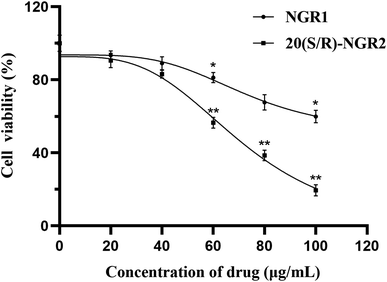
As presented in Fig. 7, TUNEL staining analysis showed that cell apoptosis was dramatically induced by 20(S/R)-notoginsenoside R2 compared to control. There was increased apoptosis of H22 cells induced by 20(S/R)-notoginsenoside R2; specifically, 50 μg mL−1 and 100 μg mL−1 of notoginsenoside R1 led to a 1.94 ± 0.31% and 10.85 ± 1.66% (P < 0.05) apoptosis rate at 24 h, whereas 50 and 100 μg mL−1 of 20(S/R)-notoginsenoside R2 led to apoptosis rates of 25.03 ± 1.31% (P < 0.01) and 60.10 ± 1.48% (P < 0.01), respectively.3.5 20(S/R)-Notoginsenoside R2 promoted the apoptosis of H22 cells by modulating PI3K/AKT/mTOR pathway
In order to investigate the signaling pathway activated by 20(S/R)-notoginsenoside R2 in H22 cells, the expression of PI3K/AKT/mTOR pathway related proteins was detected (Fig. 8). The expression of phosphorylated PI3K, AKT, and mTOR proteins was significantly reduced in the 20(S/R)-notoginsenoside R2 treated group compared to the control group (P < 0.01), indicating a significant inhibition of the PI3K signaling pathway. The results showed that treatment with 20(S/R)-notoginsenoside R2 significantly increased apoptosis in H22 cells.4. Discussion
Notoginsenoside R1 is a bioactive compound that was isolated from Sanqi and has been extensively studied for its various therapeutic properties, including anti-inflammatory and anti-oxidative effects.33 However, its clinical applications are limited owing to low bioavailability and poor solubility. Therefore, several studies have explored the transformation of notoginsenoside into its metabolites. For instance, Li Qi et al. showed the transformation of notoginsenoside R1 into ginsenoside Rg1 by Dictyoglomus thermophilum β-xylosidase Xln-DT and demonstrated an enhanced in vivo anti-fatigue activity.34 K. C. Shin et al. showed the conversion of notoginsenoside R1 into ginsenoside Rg1 with β-xylosidase of Thermoanaerobacterium thermosaccharolyticum.35 Notoginsenoside R1 has three glycosidic linkages at C-6 and C-20 and can be hydrolyzed by glycosidase through different reaction pathways. Nevertheless, only C-20 glycosidic linkages was specifically hydrolyzed in the biotransformation process in the present study. During the biotransformation process of the study, Lactiplantibacillus plantarum S165 grew rapidly and produced large amounts of acid and β-glucosidase within 24 h. Soon afterwards, Lactiplantibacillus plantarum S165 began to die after 24 h, but the pH value of fermentation broth and the activity of β-glucosidase remained stable, ensuring the continuous biotransformation of notoginsenoside R1. Finally, notoginsenoside R1 was transformed into 20(S/R)-notoginsenoside R2 through glycosidases secreted by Lactiplantibacillus plantarum S165 and the resulting transformation product was structurally characterized by HPLC, UPLC-MS/MS, and 13C-NMR spectroscopy analyses. Furthermore, CCK-8 and TUNEL experiments showed that 20(S/R)-notoginsenoside R2 had a significant inhibitory effect on cancer cell proliferation and exhibited a stronger inhibitory effect than notoginsenoside R1. The activity of the PI3K/AKT/mTOR pathway may underlie the mechanism. These results provide novel insights into the potential therapeutic applications of 20(S/R)-notoginsenoside R2 in cancer treatment.Notoginsenoside R2 is an active saponin molecule extracted from the root of Sanqi, and has two types of structures, namely the R-type and S-type.36 Our finding showed the conversion of notoginsenoside R1 into 20(S/R)-notoginsenoside R2 by Lactiplantibacillus plantarum S165, which is in agreement with previous results.37 The presence of both S and R configurations is prevalent in this species. Liu et al. demonstrated that the S-type exhibits superior activity compared to the R-type, with an elevated cardioprotective effect against DOX-induced cell injury.13 There is a possibility that the activity of our notoginsenoside R2 may follow a similar pattern but due to similar polarity, unfortunately, our study could not differentiate between the two epimers. Thus, our study was unable to provide evidence for the configuration that exhibited better activity.
The presence of sugar moieties is crucial in regulating the activity of saponins, with an increasing number of sugar moieties leading to a better reduction in the cytotoxic effect of the compound.38 Observations made by Kai Quan et al. suggest an inverse relationship between the number of sugar moieties and the anticancer effects of saponins.39 Our study also verifies that the removal of the 20-C sugar moiety enhances the toxicity of notoginsenosides toward H22 cells and increases their apoptosis. Hydrophobicity plays a crucial role in the absorption of compounds and the removal of sugar moieties can increase the hydrophobic nature of these compounds, thus enhancing their permeability through cell membranes. This property can explain why the anticancer activities of these compounds tend to increase as the number of sugar moieties in the molecule decreases.38 This phenomenon also may be attributed to steric hindrance resulting from the attachment of sugar molecules. Our study suggests that a reduction in glycosylation decreases the steric hindrance of molecules, which in turn increases their anti H22 hepatoma cells activity. Furthermore, through molecular modeling and docking studies, Chen et al. confirmed that the variance in Na+/K+-ATPase inhibitory potency among ginsenosides is due to the steric hindrance caused by the sugar attachment at the C-6 and C-20 positions of the steroid-like structure.40 The activity of saponins is also affected by the positioning of sugar groups. In our experiment, the structure of 20(S/R)-notoginsenoside R2 lacks a sugar group at C-20 but has an S-configuration of the hydroxyl moiety and a carbon chain with a dimethyl allyl group. Yang et al. confirmed that this structure exerted the most significant impact on cell viability by arresting the DNA cycle and inducing apoptosis.41
The PI3K/AKT/mTOR pathway plays a critical role in regulating numerous cellular processes, including mechanisms underlying survival and apoptosis by controlling cell signal transduction.42 In our study, we observed that 20(S/R)-notoginsenoside R2 could significantly inhibit the proliferation of H22 cells. Furthermore, this inhibitory effect may be due to the activity of the PI3K pathway. Tao et al. indicated that cell viability, proliferation, and tube formation of primary human umbilical vein endothelial cells were inhibited by notoginsenoside R2 through the blockade of the Rap1GAP/PI3K/Akt signaling pathway.43 This also supports our hypothesis that 20(S/R)-notoginsenoside R2 effectively inhibits the PI3K pathway and decreases the proliferation of H22 cells in vitro. The Western blotting results also showed that 20(S/R)-notoginsenoside R2 effectively inhibited the phosphorylation of PI3K pathway proteins, leading to reduced p-mTOR content, inhibition of cellular proliferation, and induction of apoptosis in H22 hepatoma cells. This suggests that 20(S/R)-notoginsenoside R2 has potential therapeutic effects for inhibiting tumor cells.
In summary, glycosidases secreted by Lactiplantibacillus plantarum S165 was used to efficiently transform notoginsenoside R1 for 20(S/R)-notoginsenoside R2 production. The results of our study indicate that 20(S/R)-notoginsenoside R2 exhibits higher activity than R1 and effectively inhibits the proliferation of H22 cells. Mechanistically, the PI3K/AKT/mTOR pathway may underlie the inhibitory effect on cell proliferation. Our experiment demonstrates that the preparation method of 20(S/R)-notoginsenoside R2 is simple and feasible, further providing a new avenue for the structural modification of existing compounds through biotransformation.
5. Conclusions
In this study, the substances transformed from notoginsenoside R1 by Lactiplantibacillus plantarum S165 were identified by HPLC, UPLC-MS/MS, and 13C-NMR analyses as 20(S/R)-notoginsenoside R2. It is noteworthy that the glycosidases secreted by Lactiplantibacillus plantarum S165 specifically hydrolyzed the β-(1→6)-glucosidic linkage at C-20, leaving the other linkages unaffected. This provides a simple and environmentally friendly method for obtaining notoginsenoside R2 and its epimer, which showed inhibition of H22 cells proliferation. In conclusion, these findings suggest that microbial transformation of saponins is a promising approach for generating desired compounds.Author contributions
Penghui Wang: conceptualization, methodology, software, writing-reviewing and editing. Yansong Gao: validation, supervision. Ge Yang: investigation, data curation. Yujuan Zhao: resources, investigation. Zijian Zhao: validation, methodology. Ge Gao: software. Lei Zhao: supervision, visualization. Shengyu Li: project administration, funding acquisition. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare that there is no potential conflict of interest.Acknowledgements
This work was financially supported by Basic Scientific Research Projects of Jilin Academy of Agricultural Sciences (KYJF2021ZR108).References
- R. Donne and A. Lujambio, Hepatology, 2023, 77, 1773–1796 CrossRef PubMed.
- Y. H. Liu, H. Y. Qin, Y. Y. Zhong, S. Li, H. J. Wang, H. Wang, L. L. Chen, X. Tang, Y. L. Li, Z. Y. Qian, H. Y. Li, L. Zhang and T. Chen, BMC Cancer, 2021, 21, 37 CrossRef CAS PubMed.
- H. Zhu, Y. Shan, K. Ge, J. Lu, W. Kong and C. Jia, Cell. Oncol., 2020, 43, 1203–1214 CrossRef CAS PubMed.
- Y. Sun, Y. Yang, S. Liu, S. Yang, C. Chen, M. Lin, Q. Zeng, J. Long, J. Yao, F. Yi, L. Meng, Q. Ai and N. Chen, Cells, 2022, 11, 2529 CrossRef CAS PubMed.
- J. Y. Xue, Y. Y. Wu, Y. L. Han, X. Y. Song, M. Y. Zhang, J. Cheng, B. Lin, M. Y. Xia and Y. X. Zhang, J. Ethnopharmacol., 2023, 312, 116457 CrossRef CAS PubMed.
- D. H. Kim, J. Ginseng Res., 2012, 36, 1–15 CrossRef CAS PubMed.
- K. Sun, C. S. Wang, J. Guo, Y. Horie, S. P. Fang, F. Wang, Y. Y. Liu, L. Y. Liu, J. Y. Yang, J. Y. Fan and J. Y. Han, Life Sci., 2007, 81, 509–518 CrossRef CAS PubMed.
- Z. Chen, Z. Zhang, J. Liu, H. Qi, J. Li, J. Chen, Q. Huang, Q. Liu, J. Mi and X. Li, Front. Cell. Infect. Microbiol., 2022, 12, 853981 CrossRef CAS PubMed.
- M. Murugesan, R. Mathiyalagan, V. Boopathi, B. M. Kong, S. K. Choi, C. S. Lee, D. C. Yang, S. C. Kang and T. Thambi, Nanomaterials, 2022, 12, 3427 CrossRef CAS PubMed.
- R. F. Wang, M. M. Zheng, Y. D. Cao, H. Li, C. X. Li, J. H. Xu and Z. T. Wang, Appl. Microbiol. Biotechnol., 2015, 99, 3433–3442 CrossRef CAS PubMed.
- B. Nan, Y. L. Liu, Y. You, W. C. Li, J. J. Fan, Y. S. Wang, C. H. Piao, D. L. Hu, G. J. Lu and Y. H. Wang, Food Funct., 2018, 9, 6020–6028 RSC.
- Y. Han, B. Sun, B. Jiang, X. Hu, M. I. Spranger, Y. Zhang and Y. Zhao, J. Appl. Microbiol., 2010, 109, 792–798 CrossRef CAS PubMed.
- J. Liu, Y. Xin, Z. Qiu, Q. Zhang, T. He, Y. Qiu and W. Wang, RSC Adv., 2022, 12, 12938–12946 RSC.
- Y. Fu, Z. H. Yin and C. Y. Yin, J. Appl. Microbiol., 2017, 122, 1579–1585 CrossRef CAS PubMed.
- G. Renchinkhand, U. Magsar, H. C. Bae, S. H. Choi and M. S. Nam, Foods, 2022, 11, 529 CrossRef CAS PubMed.
- K. H. Chang, M. N. Jo, K. T. Kim and H. D. Paik, J. Ginseng Res., 2014, 38, 47–51 CrossRef PubMed.
- C. Zhang, Y. Xu, M. Gu, Z. Liu, J. Zhang, Q. Zeng and D. Zhu, Antonie van Leeuwenhoek, 2021, 114, 437–444 CrossRef CAS PubMed.
- J. M. Landete, Crit. Rev. Biotechnol., 2017, 37, 296–308 CrossRef CAS PubMed.
- Z. Zhang, X. Tao, N. P. Shah and H. Wei, J. Dairy Sci., 2016, 99, 2666–2674 CrossRef CAS PubMed.
- Y. Chen, Y. Wang, A. Zhu, L. Zhang, X. Zhang, J. Zhang and C. Zhang, Front. Microbiol., 2022, 13, 1022200 CrossRef PubMed.
- X. B. Meng, G. B. Sun, M. Wang, J. Sun, M. Qin and X. B. Sun, Evid.-Based Complementary Altern. Med., 2013, 2013, 971712 Search PubMed.
- Y. Hu, L. Wu, L. Jiang, N. Liang, X. Zhu, Q. He, H. Qin and W. Chen, Hum. Exp. Toxicol., 2021, 40, S347–s358 CrossRef CAS PubMed.
- L. Zhao, Y. Shen, Y. Wang, L. Wang, L. Zhang, Z. Zhao and S. Li, Sci. Rep., 2022, 12, 15490 CrossRef CAS PubMed.
- Y. W. Lin, Y. C. Mou, C. C. Su and B. H. Chiang, J. Agric. Food Chem., 2010, 58, 8528–8534 CrossRef CAS PubMed.
- F. Liu, N. Ma, F. B. Xia, P. Li, C. He, Z. Wu and J. B. Wan, J. Ginseng Res., 2019, 43, 105–115 CrossRef PubMed.
- Y. F. Li, Y. Z. Liang, X. M. Cui, L. J. Shao, D. J. Lou and X. Y. Yang, Molecules, 2022, 27, 6615 CrossRef CAS PubMed.
- Z. Wang, X. Sun, Y. Zhao, L. Ga, Q. Li, Q. Li, X. Wang and C. Yang, Front. Pharmacol., 2022, 13, 949757 CrossRef CAS.
- Z. R. Yang, Z. H. Wang, J. F. Tang, Y. Yan, S. J. Yue, W. W. Feng, Z. Y. Shi, X. T. Meng, C. Peng, C. Y. Wang, D. L. Meng and D. Yan, Front. Pharmacol., 2018, 9, 633 CrossRef.
- D. Yoon, W. C. Shin, S. M. Oh, B. R. Choi and D. Young Lee, Food Res. Int., 2022, 159, 111610 CrossRef CAS PubMed.
- Y. Li, B. Xu, X. Ren, L. Wang, Y. Xu, Y. Zhao, C. Yang, C. Yuan, H. Li, X. Tong, Y. Wang and J. Du, Cell. Mol. Biol. Lett., 2022, 27, 81 CrossRef CAS.
- M. Sun, Y. Chen, X. Liu and Y. Cui, Bioengineered, 2021, 12, 8006–8019 CrossRef CAS PubMed.
- L. Zhao, Y. Jiang, Y. X. Ni, T. Z. Zhang, C. C. Duan, C. Huang, Y. J. Zhao, L. Gao and S. Y. Li, J. Funct. Foods, 2017, 35, 97–104 CrossRef CAS.
- R. Shao, W. Li, R. Chen, K. Li, Y. Cao, G. Chen and L. Jiang, Front. Pharmacol., 2023, 14, 1101240 CrossRef CAS PubMed.
- Q. Li, L. Wang, X. Fang and L. Zhao, J. Microbiol. Biotechnol., 2022, 32, 447–457 CrossRef CAS PubMed.
- K. C. Shin, M. J. Seo and D. K. Oh, Biotechnol. Lett., 2014, 36, 2275–2281 CrossRef CAS PubMed.
- S. Sun, C. Z. Wang, R. Tong, X. L. Li, A. Fishbein, Q. Wang, T. C. He, W. Du and C. S. Yuan, Food Chem., 2010, 118, 307–314 CrossRef CAS.
- P. Zhong, Y. Xiu, K. Zhou, H. Zhao, N. Wang, F. Zheng and S. Yu, 3 Biotech, 2022, 12, 289 CrossRef PubMed.
- L. Jia and Y. Zhao, Curr. Med. Chem., 2009, 16, 2475–2484 CrossRef CAS PubMed.
- K. Quan, Q. Liu, J. Y. Wan, Y. J. Zhao, R. Z. Guo, R. N. Alolga, P. Li and L. W. Qi, Sci. Rep., 2015, 5, 8598 CrossRef CAS PubMed.
- R. J. Chen, T. Y. Chung, F. Y. Li, N. H. Lin and J. T. Tzen, Acta Pharmacol. Sin., 2009, 30, 61–69 CrossRef PubMed.
- H. Yang, G. Yoo, H. S. Kim, J. Y. Kim, S. O. Kim, Y. H. Yoo and S. H. Sung, J. Agric. Food Chem., 2012, 60, 11759–11764 CrossRef CAS PubMed.
- L. Wang, S. Y. Li, Y. Jiang, Z. J. Zhao, Y. J. Shen, J. J. Zhang and L. Zhao, J. Funct. Foods, 2021, 85, 9 Search PubMed.
- P. Tao, J. Lin, B. Zhang and S. Wang, Ann. Transl. Med., 2021, 9, 1743 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |