Open Access Article
Open Access ArticleEvaluation of the biological responses of silver nanoparticles synthesized using Pelargonium x hortorum extract
Christian Andrea Lopez-Ayusoab,
Rene Garcia-Contrerasb,
Ravichandran Manisekaran b,
Mario Figueroa
b,
Mario Figueroa c,
Ma. Concepción Arenas-Arrocenab,
Genoveva Hernandez-Padrond,
Amaury Pozos-Guilléne and
Laura Susana Acosta-Torres
c,
Ma. Concepción Arenas-Arrocenab,
Genoveva Hernandez-Padrond,
Amaury Pozos-Guilléne and
Laura Susana Acosta-Torres *b
*b
aPrograma de Doctorado en Ciencias Odontológicas, Universidad Nacional Autónoma de México (UNAM), Mexico
bInterdisciplinary Research Laboratory (LII), Nanostructures and Biomaterials Area, Escuela Nacional de Estudios Superiores (ENES) Unidad León, Universidad Nacional Autónoma de México, Predio el Saucillo y el Potrero, Comunidad de los Tepetates, 37684, León, Mexico. E-mail: lacosta@enes.unam.mx
cFacultad de Química, UNAM, Ciudad de México, 04510, Mexico
dCentro de Física Aplicada y Tecnología Avanzada (CFATA), Departamento de Nanotecnología, Universidad Nacional Autónoma de México, Campus Juriquilla, Juriquilla 76230, Mexico
eBasic Science Laboratory, Faculty of Stomatology, San Luis Potosí University, Av. Dr. Manuel Nava #2, Zona Universitaria, 78290, San Luis Potosí, SLP, Mexico
First published on 11th October 2023
Abstract
Silver nanoparticles (AgNPs) are one of the widely studied nanomaterials for diverse biomedical applications, in particular, as antimicrobial agents to kill bacteria, fungi, and viruses. In this report, AgNPs were synthesized using a geranium (Pelargonium x hortorum) leaves extract and tested for their antimicrobial and cytotoxic activity and reactive oxygen species (ROS) production. Using green biosynthesis, the leaves extract was employed as a reducing and stabilizing agent. Synthesis parameters like reaction time and precursor (silver nitrate AgNO3) volume final were modified, and the products were tested against Streptococcus mutans. For the first time, the metabolomic analysis of extract, we have identified more than 50 metabolites. The UV-Vis analysis showed a peak ranging from 410–430 nm, and TEM confirmed their nearly spherical morphology for all NPs. The antimicrobial activity of the NPs revealed a minimum inhibitory concentration (MIC) of 10 μg mL−1. Concerning cytotoxicity, a dose-time-dependent effect was observed with a 50% cellular cytotoxicity concentration (CC50) of 4.51 μg mL−1 at 24 h. Interestingly, the cell nuclei were visualized using fluorescence microscopy, and no significant changes were observed. These results suggest that synthesized spherical AgNPs are promising potential candidates for medical applications.
1. Introduction
Nanotechnology is an interdisciplinary science that combines various scientific knowledge of materials, physics, chemistry, and biology, among other areas.1 It is a novel field to study diverse nanomaterials, which can be classified by their dimensions, such as nanoparticles (NPs), nanowires, nanorods, and 2D nanostructures with sizes of less than 100 nm.2 NPs have innovative applications in different fields, such as the food industry, medicine, cosmetics, agriculture, and the environment.3,4 In the biomedical area, NPs have a wide range of applications due to their antimicrobial, antibiofilm, anti-inflammatory, and anticancer properties.5–7Because of the above-mentioned outstanding properties, they are applied in ventricular catheters,8 wound dressings, cavity filler,9 neurosurgical shunts, hand gels, and creams,10 to mention a few.
From the medical point of view, the development of resistance mechanisms against antibiotics by pathogenic microorganisms has been a cause for concern.11 These resistance mechanisms have motivated researchers to design new antimicrobials.12 Metal NPs are an effective way to control many pathogenic and antibiotic-resistant microorganisms. In particular, silver nanoparticles (AgNPs) have presented significant advantages in using them as antimicrobial agents to treat bacteria, fungi, and viruses.13
To obtain AgNPs, there are various methods, such as chemical, physical, and biological; however, it has been described that the first two can present difficulty in purification and the presence of toxic chemical species that can trigger adverse physical effects.14 Biological synthesis, especially those mediated by plants, has aroused great interest due to its simplicity, non-toxic, economical, safe, and ecological features.14–16 The plant extracts allow the generation of NPs with narrow distribution by controlling the biosynthesis method.17–19 In recent times, several plants, such as Aloe vera,20 Berberis vulgaris,21 Salvia spinosa,22 Diospyros lotus,23 Calliandra haematocephala,15 Pelargonium hortorum,24 Pelargonium graveolens,25 Geranium maculatum26 between others, have been processed for green synthesis of AgNPs, to reduce the use of chemicals and toxic solvents:3 for this reason, plant extracts are used in the generation of various materials in nanomedicine.
The geranium Pelargonium x hortorum is widely used in traditional medicine for its properties as antimicrobial, anti-inflammatory, cell regeneration, and antioxidant.27 The antimicrobial effect is one of the reasons why it is used for the synthesis of AgNPs. This effect is related to high concentrations of oxygenated metabolites in this plant's aqueous extract which have also been associated with the ability to damage the membrane integrity of bacteria and fungi.27,28
With Pelargonium extracts, the formation of AgNPs of spherical,24 triangular, and hexagonal morphology with sizes between 16–40 nm has been reported.27,28 However, some characteristics related to oxidation and cytotoxicity continue to cause controversy. In this sense, research continues to be carried out to evaluate that the AgNPs obtained by biosynthesis can increase the antimicrobial potential and antibiofilm activity and, in turn, maintain the biological effects in eukaryotic cells.17
Therefore, this research aims to synthesize AgNPs using Pelargonium x hortorum leaves extract and to evaluate its various biological response through antimicrobial profile in Streptococcus mutans (S. mutans), cytotoxicity, oxidative stress, and genotoxic effects in Human gingival fibroblast (HGF). The reduction of the Ag metal and the stabilization of resulting NPs was done by the leaves extract without adding other chemical agents. In addition, untargeted metabolomic analysis using liquid chromatography-tandem mass spectrometry (LC-MS/MS) data of the leaves extract revealed the presence of several classes of compounds, including flavonoids, lignans, phenyl propanoinds, among others. Considering this investigation, the methodology is entirely green, which exhibits enhanced antibacterial and less cytotoxic behaviors.
2. Chemicals and experimental methods
All the chemical reagents and assay kits were purchased from Sigma-Aldrich® St Louis, Missouri, USA and used without modification unless otherwise mentioned.2.1 Preparation of Pelargonium hortorum leaves extract
The plants of Pelargonium x hortorum L.H. Bailey (Geraniaceae) were cultivated in Atlixco, Puebla (18°54′N 98°27′O) and fresh leaves were collected at the beginning of March, in the spring season, in Leon, Mexico (21°02′41.6′′N 101°40′13.2′′ W). A specimen was taxonomically identified as Pelargonium x hortorum by Dr. Sol Cristians Niizawa (voucher specimen No. 182314; Faculty of Science Herbarium – UNAM, FCME). To obtain the leaves extract (∼12%), approximately 12.15 g of fresh leaves were weighed, rinsed, and added to 100 mL of deionized water (DI water). The solution was maintained in a boiling condition for 15 min at a constant temperature of 90–92 °C. Then, by decantation, the solution was filtered by using Whatman filter paper 4–12 μm to obtain an aqueous extract without any leaves debris.2.2 Metabolomic analysis of Pelargonium x hortorum leaves extract
Leaves extract was analyzed using an Acquity ultra-performance liquid chromatographic (UPLC) system (Waters Corp., Milford, MA, USA) equipped with a photodiode array detector (PDA, λ 190–700 nm) and coupled to a Q-Exactive Plus (Thermo Fisher Scientific, Waltham, MA, USA) high-resolution mass spectrometer. The UPLC column used was an Acquity BEH C18 (Waters 50 mm × 2.1 mm I.D., 1.7 μm, 130 Å) at 40 °C, with a gradient system from 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 85 CH3CN–0.1% aqueous formic acid to 100% of CH3CN in 8 min, then held for 1.5 min with CH3CN, and returned to the starting conditions. The flow rate was 0.3 mL min−1, and the injection volume of 3.0 μL. High-resolution tandem MS data (HRMS-MS/MS) data were obtained using an electrospray source (ESI) in positive and negative modes at a full scan range (m/z 150–2000), followed by a top 5 ions MS/MS fragmentation analysis during the full run, with the following settings: capillary voltage, 5 V; capillary temperature, 300 °C; tube lens offset, 35 V; spray voltage, 3.80 kV; sheath and auxiliary gas flow, 30 arbitrary units.29,30
85 CH3CN–0.1% aqueous formic acid to 100% of CH3CN in 8 min, then held for 1.5 min with CH3CN, and returned to the starting conditions. The flow rate was 0.3 mL min−1, and the injection volume of 3.0 μL. High-resolution tandem MS data (HRMS-MS/MS) data were obtained using an electrospray source (ESI) in positive and negative modes at a full scan range (m/z 150–2000), followed by a top 5 ions MS/MS fragmentation analysis during the full run, with the following settings: capillary voltage, 5 V; capillary temperature, 300 °C; tube lens offset, 35 V; spray voltage, 3.80 kV; sheath and auxiliary gas flow, 30 arbitrary units.29,30
The HRMS-MS/MS raw data was converted to .mzXML using MSConvert (ProteoWizard) and preprocessed using MZmine 3 software version 3.3.0 (ref. 31) according to the following workflow: mass detection at MS1 and MS2, ADAP chromatogram building29,32 chromatogram resolving, deisotoping, join aligner, feature list rows filtering, gap-filling and duplicate feature filtering. Features that had less than threefold difference to blank were removed. The group intensity threshold, min highest intensity and minimum absolute height parameters were adjusted based on the chosen noise level for MS1. In addition, other parameters adjusted were minimum group size of scans, m/z tolerance, min ratio of peak, peak duration ranges, isotopes peaks grouper, m/z tolerance for peak alignment, weight for m/z, and peak-list rows filter. The resulting MS1 feature list was exported to Excel (.csv), and the MS2 feature data was exported as a .mgf file for feature analysis in the GNPS platform.
A molecular network was created following the GNPS (https://gnps.ucsd.edu),33 workflow, where edges were filtered to have a cosine score above 0.7 and more than 6 matched peaks. Further, edges between two nodes were kept in the network if and only if each node appeared in each other's respective top 10 most similar nodes. The maximum size of a molecular family was set to 100, and the lowest-scoring edges were removed from molecular families until the molecular family size was below this threshold. The spectra in the network were then searched against GNPS spectral libraries.33–35 The library spectra were filtered in the same manner as the input data. All matches kept between network spectra and library spectra were required to have a score above 0.7 and at least 6 matched peaks. Visualization of the molecular networks was completed using Cytoscape 3.10.36 Formula predictions and mass accuracy were calculated using Formula Calculator v.1.2.3. Finally, the annotation of compounds was at confidence levels 2 and 3 according to the metabolomics standards initiative and exact mass accuracy <5 ppm.37
2.3 Biosynthesis of AgNPs
Green synthesis was done using the leaves extract by mixing with silver nitrate (AgNO3) precursor of 25 mM concentration. 25 mM precursor solution was prepared with 0.1061 g of AgNO3 (purity ≥ 99%) dissolved in 25 mL of DI water. Ethylene-glycol (EG) was used as a solvent, and leaves extract as a reducing and stabilizing agent. The procedure has been detailed as follows; a three-neck round bottom flask (250 mL) was connected to a condenser and filled with 20 mL of EG. The solution was boiled to a temperature of 161 °C and then mixed with 10 mL of extract reducing the temperature to 110 °C and keeping it constant until the end of the synthesis. Five min later, the AgNO3 solution was added in varying volumes and times.38Thus, 7 different experimental groups resulted, represented in Table 1. The obtained AgNPs were washed using centrifugation (4600 rpm) and sonication for 10 min, with the combination of organic solvents, groups A, B, & C (ethanol 100%, twice), D, E, F & G (acetone 100%, twice and ethanol 100% once) and finally DI water. The final product was dispersed in DI water and kept in at 4 °C for various experiments.
| Compounds | Observed ion (m/z)a | Adduct | Molecular formula [adduct] | Exact mass (calculated) | Mass accuracy (ppm) |
|---|---|---|---|---|---|
| a Values taken from GNPS analysis. | |||||
| Galactaric acid | 209.030 | [M − H]− | C6H9O8 | 209.0303 | −1.4 |
| 5-Megastigmene-3,9-diol | 213.185 | [M + H]+ | C13H25O2 | 213.1849 | +0.4 |
| Bisabolene-1,4-endoperoxide | 219.174 | [M − H2O + H]+ | C15H23O | 219.1743 | −1.6 |
| Pantothenic acid | 220.119 | [M + H]+ | C9H18NO5 | 220.1179 | +4.8 |
| Palmitoleic acid | 255.232 | [M + H]+ | C16H31O2 | 255.2318 | +0.6 |
| Phloretin | 275.091 | [M + H]+ | C15H15O5 | 275.0914 | −1.5 |
| Malic acid | 289.018 | [2M + Na − 2H]− | C8H10O10Na | 289.0177 | +1.0 |
| Quercetin | 301.035 | [M − H]− | C15H9O7 | 301.0354 | −1.2 |
| 2-Phenylethyl β-D-glucopyranoside | 302.160 | [M + NH4]+ | C14H24NO6 | 302.1598 | +0.6 |
| Coumaroyl hexoside | 325.092 | [M − H]− | C15H17O8 | 325.0929 | −2.7 |
| Coumaroyl quinic acid | 337.093 | [M − H]− | C16H17O8 | 337.0929 | +0.3 |
| 2-Coumaroylquinic acid | 339.108 | [M + H]+ | C16H19O8 | 339.1074 | +1.6 |
| Coumaric acid 4-O-glucoside | 344.134 | [M + NH4]+ | C15H22NO8 | 344.1340 | 0.0 |
| Chlorogenic acid | 353.088 | [M − H]− | C16H17O9 | 353.0878 | +0.6 |
| Coumaroyl + C6H9O8 | 355.067 | [M − H]− | C15H15O10 | 355.0671 | −0.2 |
| 3-O-Feruloylquinic acid | 367.103 | [M − H]− | C17H19O9 | 367.1034 | −1.2 |
| NP-006680_3-(Benzoyloxy)-2-hydroxypropyl β-D-glucopyranosiduronic acid | 371.098 | [M − H]− | C16H19O10 | 371.0984 | −1.0 |
| NP-016624_3,5,5-Trimethyl-4-[3-(β-D-glucopyranosyloxy)butyl]-2-cyclohexene-1-one | 373.222 | [M + H]+ | C19H33O7 | 373.2221 | −0.2 |
| NP-016360_3,5,5-Trimethyl-4-[3-[(β-D-glucopyranosyl)oxy]butyl]-3-cyclohexen-1-ol | 375.238 | [M + H]+ | C19H35O7 | 375.2377 | +0.7 |
| NP-001173_[5-Hydroxy-2-(3-hydroxybutyl)-3,3-dimethylcyclohexyl]methyl β-D-glucopyranoside | 393.249 | [M + H]+ | C19H37O8 | 393.2483 | +1.8 |
| Kaempferol 3-α-L-arabinopyranoside | 417.083 | [M − H]− | C20H17O10 | 417.0827 | +0.7 |
| Kaempferol 3-O-β-D-xyloside | 419.097 | [M + H]+ | C20H19O10 | 419.0973 | −0.7 |
| Icariside F2 | 420.187 | [M + NH4]+ | C18H30NO10 | 420.1864 | +1.4 |
| Isovitexin | 433.113 | [M + H]+ | C21H21O10 | 433.1129 | +0.2 |
| Avicularin | 435.092 | [M + H]+ | C20H19O11 | 435.0922 | −0.4 |
| Phloretin 2′-O-glucoside | 435.130 | [M − H]− | C21H23O10 | 435.1298 | +0.8 |
| (2R,3R,4S,5S,6R)-2-Octoxy-6-[[(2S,3R,4S,5R)-3,4,5-trihydroxyoxan-2-yl]oxymethyl]oxane-3,4,5-triol | 442.264 | [M + NH4]+ | C19H40NO10 | 442.2647 | −1.5 |
| Luteolin 4′-O-glucoside | 447.093 | [M − H]− | C21H19O11 | 447.0933 | −0.6 |
| Myricetin-3-O-xyloside | 449.073 | [M − H]− | C20H17O12 | 449.0726 | +1.0 |
| Astragalin | 449.108 | [M + H]+ | C21H21O11 | 449.1078 | +0.4 |
| Isoquercetin | 463.088 | [M − H]− | C21H19O12 | 463.0882 | −0.4 |
| Kaempferol-4′-glucoside | 471.090 | [M + Na]+ | C21H20O11Na | 471.0898 | +0.5 |
| NP-007396_6-O-[(2E)-3-(4-Hydroxyphenyl)-2-propenoyl]-1-O-(3,4,5-trihydroxybenzoyl)hexopyranose | 477.104 | [M − H]− | C22H21O12 | 477.1038 | +0.3 |
| Myricetin 3-O-β-D-galactopyranoside | 479.083 | [M − H]− | C21H19O13 | 479.0831 | −0.2 |
| 1,6-Digalloyl-β-D-glucopyranose | 502.119 | [M + NH4]+ | C20H24NO14 | 502.1191 | −0.3 |
| Quercetin-3-O-glucose-6′′-acetate | 507.114 | [M + H]+ | C23H23O13 | 507.1133 | +1.3 |
| NP-017433_(3,4,5-Trihydroxy-6-((4-(2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl)butan-2-yl)oxy)tetrahydro-2H-pyran-2-yl)methyl 3,4,5-trihydroxybenzoate | 525.234 | [M + H]+ | C26H37O11 | 525.2330 | +1.8 |
| Luteolin 7-(6′′-malonylglucoside) | 535.109 | [M + H]+ | C24H23O14 | 535.1082 | +1.4 |
| Procyanidin B1 | 577.136 | [M − H]− | C30H25O12 | 577.1352 | +1.5 |
| Procyanidin B2 | 579.151 | [M + H]+ | C30H27O12 | 579.1497 | +2.2 |
| Tiliroside | 593.130 | [M − H]− | C30H25O13 | 593.1301 | −0.1 |
| 6′′-O-(3-Hydroxy-3-methylglutaroyl)astragalin | 593.151 | [M + H]+ | C27H29O15 | 593.1501 | +1.5 |
| Nicotiflorin | 593.151 | [M − H]− | C27H29O15 | 593.1512 | −0.3 |
| Rutin | 611.163 | [M + H]+ | C27H31O16 | 611.1607 | +3.8 |
| Cyanidin-3,5-di-O-glucoside | 611.164 | [M]+ | C27H31O16+ | 611.1607 | +5.5 |
| 2′′-O-Galloylhyperin | 617.116 | [M + H]+ | C28H25O16 | 617.1137 | +3.7 |
| 1,3,6-Tri-O-galloylglucose | 635.089 | [M − H]− | C27H23O18 | 635.0890 | 0.0 |
| 5-Hydroxy-3-[(2S,3R,4R,5S)-3-hydroxy-5-(hydroxymethyl)-4-[(2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxyoxolan-2-yl]oxy-2-(4-hydroxyphenyl)-7-[(2S,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methyloxan-2-yl]oxychromen-4-one | 727.210 | [M + H]+ | C32H39O19 | 727.2080 | +2.7 |
| Kaempferol 3-O-(2,6-di-O-α-L-rhamnopyranosyl)-β-D-galactopyranoside | 739.210 | [M − H]− | C33H39O19 | 739.2091 | +1.2 |
| Quercetin 3-(2R-apiosylrutinoside) | 741.189 | [M − H]− | C32H37O20 | 741.1884 | +0.9 |
| Mauritianin | 741.226 | [M + H]+ | C33H41O19 | 741.2236 | +3.2 |
| Manghaslin | 757.221 | [M + H]+ | C33H41O20 | 757.2186 | +3.2 |
| 1,2,3,6-Tetragalloylglucose | 787.100 | [M − H]− | C34H27O22 | 787.0999 | +0.1 |
| Hyperoside | 927.184 | [2M − H]− | C42H39O24 | 927.1837 | +0.3 |
| Pentagalloylglucose | 939.111 | [M − H]− | C41H31O26 | 939.1109 | +0.1 |
2.4 Characterization techniques
The optical property of biosynthesized AgNPs was monitored in a UV-visible spectrophotometer (Multiskan Go, Thermo Fisher Scientific™, Finland) at the wavelength range of 300–600 nm at a resolution of 1 nm. The morphology was investigated using transmission electron microscopy (TEM, JEOL, JEM-1010 Tokyo, Japan) at an accelerating voltage of 80 kV. The 30 μL of AgNPs sample was dispersed in 1 mL of isopropanol, and later 10 μL was drop-casted on the carbon-coated copper grids, followed by solvent evaporation at room temperature. The size distribution of the resulting AgNPs was estimated based on the obtained TEM micrograph using Image J software (NIH, Maryland, USA). The Pelargonium x hortorum extract and functional group of the AgNPs were analyzed by using a Fourier transform infrared spectroscopy (FTIR, PerkinElmer™ Frontier Spectrometer – ATR mode, Massachusetts, USA) with a resolution of 4 cm−1 in the region from 4000 to 400 cm−1. The zeta potential analysis was carried out by using folded capillary cell cuvettes with the Zetasizer Nano ZS90 Size Analyzer (Malvern Panalytical, Malvern, UK).2.5 Antimicrobial studies
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) to obtain a final 1 × 104 CFU mL−1 concentration. From the disk diffusion method results, we chose group F (which will be used for further experiments) based on the ZOI and other characterization techniques. Different concentrations of AgNPs (0, 2.5, 5, 10, 20 and 40 μg mL−1) were added to a 96-well plate. The positive and negative controls were 2% chlorhexidine and sterile Mueller Hinton broth. The plate was incubated for 24 h in a shaking incubator. Finally, the absorbance at 600 nm was determined using a Multiskan GO™ reader. The determination of the MIC (NCCLS) was based on turbidity, determined as the minimum concentration at which no viable cells were observed (absence of turbidity) compared to the negative control. The optical density was normalized and converted to percentages considering the negative control value as 100% of bacterial growth.
1000) to obtain a final 1 × 104 CFU mL−1 concentration. From the disk diffusion method results, we chose group F (which will be used for further experiments) based on the ZOI and other characterization techniques. Different concentrations of AgNPs (0, 2.5, 5, 10, 20 and 40 μg mL−1) were added to a 96-well plate. The positive and negative controls were 2% chlorhexidine and sterile Mueller Hinton broth. The plate was incubated for 24 h in a shaking incubator. Finally, the absorbance at 600 nm was determined using a Multiskan GO™ reader. The determination of the MIC (NCCLS) was based on turbidity, determined as the minimum concentration at which no viable cells were observed (absence of turbidity) compared to the negative control. The optical density was normalized and converted to percentages considering the negative control value as 100% of bacterial growth.2.6 Biological response studies
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 IU mL−1 penicillin G & 10
000 IU mL−1 penicillin G & 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mg mL−1 streptomycin, Sigma-Aldrich, Mexico), and 1% glutamine (Glutamax, Gibco® Thermo Fisher Scientific™, Massachusetts, USA). The Petri dishes were incubated at 37 °C with 5% CO2 and 95% relative humidity (Thermo Fisher Scientific™) for 3 weeks until a cell confluence of 80% was obtained. The MEM medium was replaced every 3 days, and the cell subcultures were performed by washing twice with 1× phosphate buffer solution (PBS, 2 mL) and 1 mL of trypsin; 0.05% EDTA-2Na was added and maintained in the incubator. Then, on the four-cell division, the HGFs were grown on electrocharged slides until a cell confluence of 90% was obtained, and later were sent to the Oral and Maxillofacial Pathology area Pathology, ENES, Leon campus, where immunohistochemical staining was performed in accordance with Kokko's technique40 and a set of samples was used to optimize vimentin antibody (BioSB, Santa Barbara, California, USA) and collagen detection by Masson's trichromic staining (Hycel, Zapopan, Jalisco, Mexico). Finally, the cells were observed in an optical microscope Leica DM750 (Leica Microsystems, Wetzlar, Germany) at 40×. The HGF were inoculated at a 1
000 mg mL−1 streptomycin, Sigma-Aldrich, Mexico), and 1% glutamine (Glutamax, Gibco® Thermo Fisher Scientific™, Massachusetts, USA). The Petri dishes were incubated at 37 °C with 5% CO2 and 95% relative humidity (Thermo Fisher Scientific™) for 3 weeks until a cell confluence of 80% was obtained. The MEM medium was replaced every 3 days, and the cell subcultures were performed by washing twice with 1× phosphate buffer solution (PBS, 2 mL) and 1 mL of trypsin; 0.05% EDTA-2Na was added and maintained in the incubator. Then, on the four-cell division, the HGFs were grown on electrocharged slides until a cell confluence of 90% was obtained, and later were sent to the Oral and Maxillofacial Pathology area Pathology, ENES, Leon campus, where immunohistochemical staining was performed in accordance with Kokko's technique40 and a set of samples was used to optimize vimentin antibody (BioSB, Santa Barbara, California, USA) and collagen detection by Masson's trichromic staining (Hycel, Zapopan, Jalisco, Mexico). Finally, the cells were observed in an optical microscope Leica DM750 (Leica Microsystems, Wetzlar, Germany) at 40×. The HGF were inoculated at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio in 96-microwell plates and incubated for 24 h in a fresh culture medium to allow the attachment for cell-based studies.
3 ratio in 96-microwell plates and incubated for 24 h in a fresh culture medium to allow the attachment for cell-based studies.2.7 Statistical analysis
The obtained data from all the experiments are expressed as a mean and standard deviation of at least triplicate independent experiments. The data analysis and distribution were performed using ANOVA and Tukey post hoc test. p < 0.05 were statistically significant.3. Results and discussion
3.1 Chemical compositions of Pelargonium x hortorum leaves extract
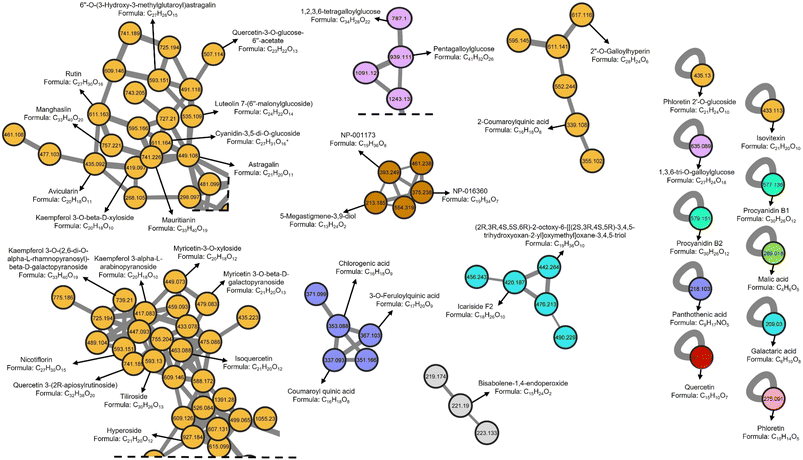
In the literature, there are a few chemical studies on the chemical composition of the genus Pelargonium, and even less on Pelargonium x hortorum. Thus, it has been reported the presence of the alkaloids elaeocarpidine and its 20-H isomer epieiaeocarpidine by Dragendorff's reaction;41,42 tanins, flavonoids, sesquiterpenes, phenolic acids, cinnamic acids, coumarins and monoterpenes by GC-MS;43 and polyphenols and flavonoids measured using the Folin-Ciocalteu and ammonium chloride methods, and terpenoids by GC-MS44 analysis in the leaves of the plant; anacardin acids in the trichome exudate of the plant;45 gallic acid and catechin in the roots and flowers;46 and anthocyanins pelargonidin glucoside derivative, peonidin glucoside derivative, cyanidin-3,5-diglucoside, petunidin-3-O-(6-acetyl)-5-O-diglucoside, malvidin-3-O-(6-O-acetyl)-5-O-diglucoside, peonidin-3, 5-O-diglucoside, and malvidin-3,5-O-diglucoside determined by UPLC-MS in flowers.47In this work, untargeted metabolomic analysis of the leaves extract of Pelargonium x hortorum was performed using UPLC-HRESIMS-MS/MS data and the Global Natural Products Social molecular networking (GNPS) platform (Fig. 1). The metabolite features present in the extract were grouped into 1101 nodes arranged in 57 clusters with >3 nodes per cluster, 56 with two nodes, and 499 singletons. Chemical ontology analysis revealed the presence of 18 classes of compounds (Fig. 1). Detailed analysis of the clusters allowed automatic annotation of 56 metabolites indicated in Table 1 and Fig. 2. These results represent the first report of a comprehensive chemical analysis of leaves extract of Pelargonium x hortorum by HRESIMS-MS/MS data.
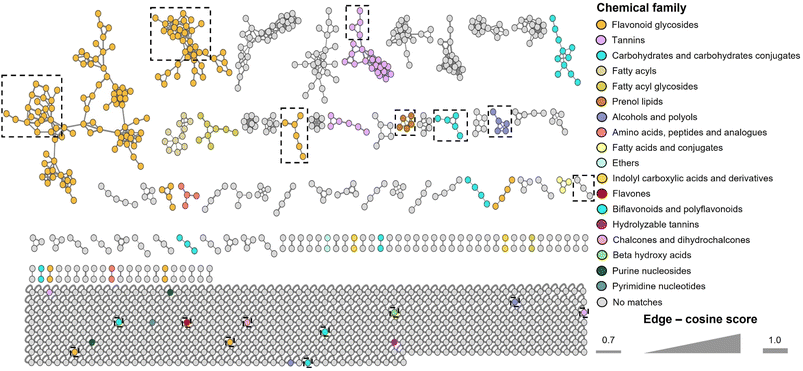 | ||
| Fig. 1 Classic molecular networking of Pelargonium x hortorum leaves extract including chemical classification. | ||
3.2 Biosynthesis and characterization of AgNPs
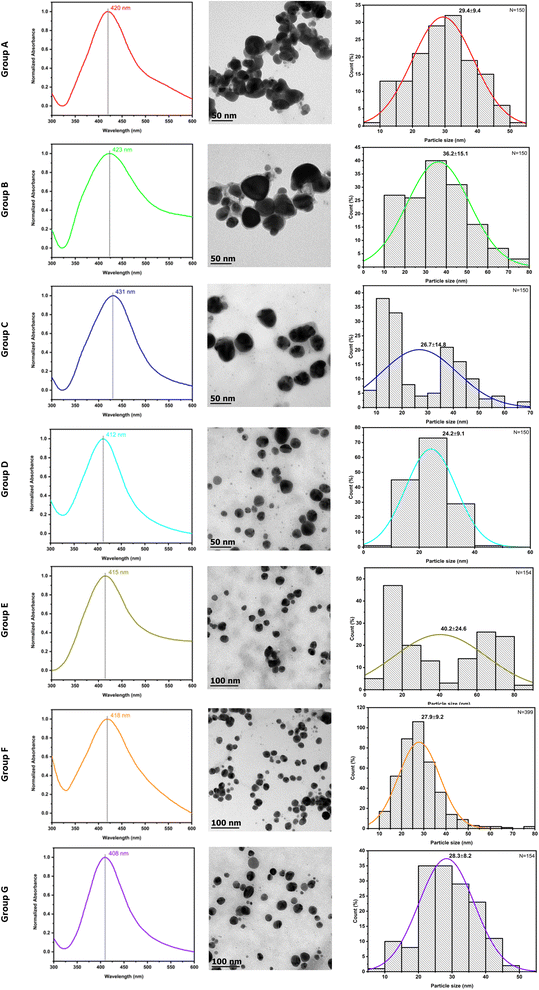
The visual color change of the reaction solution from light green to amber and the appearance of surface plasmon resonance (SPR) absorption between 400–450 nm from the UV-vis spectrum demonstrates the typical formation of AgNPs.48,49 The results obtained through the proposed synthesis methodology show an evident color change when the concentration of AgNO3 exceeds 400 μg mL−1, which intensifies to an amber tone and gets darker concerning the precursor volume. The absorption bands depicted between 407 and 431 nm corroborates with the previous report that confirms the reduction of Ag+ to Ag0.50–52 According to Mie's theory, spherical-shaped NPs usually show a single SPR band; thus, Fig. 3 column shows similar characteristics.53 Mittal et al. reported that the biosynthesis of AgNPs using Pelargonium graveolens belonging to the Geraniaceae family resulted in a size of 16 and 40 nm, which coincides with our results where AgNPs are in the range of 24 to 40 nm.16It has been reported that the various plant extract favors the formation of AgNPs synthesis due to the presence of macromolecules and phytochemicals, such as alkaloids, polyphenols, flavonoids, phenolics, amino acids, carbohydrates, enzymes, and vitamins, which function as reducing and stabilizing agent.54 In the case of the Pelargonium x hortorum aqueous extract, flavonoids, phenolic acid, and cinnamic acid act mainly to reduce the metal ions and monoterpenes & sesquiterpenes as a stabilizing agent.28 In addition, the presence of tannins, phenolic acid, cinnamic acid, coumarin, and sesquiterpenes act as biological reagents for the rapid biosynthesis of metal or metallic oxide NPs.27
In this study, the seven groups of AgNPs with different precursor concentrations and synthesis times were employed to obtain an optimal condition, and their respective SPR peaks, along with the average particle size, have been represented in Table 2. The resulting SPR band of group A: 420 nm, group B: 423 nm, group C 431 nm, group D: 412 nm, group E: 407 nm, group F: 415 ± 3.5 nm (n = 3), and group G: 408 nm as shown in Fig. 3 column a, from the TEM analysis (Fig. 3 column b) all the obtained groups showed spherical morphology with the average particle size of group A: 29.4 ± 9.4 nm, group B: 36.2 ± 10.7 nm, group C: 26.7 ± 14.8 nm, group D: 24.2 ± 9.1 nm, group E: 40.2 ± 24.6 nm, group F: 27.9 ± 9.2 nm and group G: 28.3 ± 8.2 nm (Fig. 3 column c) which is calculated using ImageJ software using the TEM micrograph. It is observed that the NPs size is determined based on the precursor volume and reaction time. Also, the combination of organic solvents in AgNPs washing shown the elimination of a thin layer of organic coating from the plant extract (Fig. 3 column b, groups D–G). But with a particular concentration (groups F and G), the NPs size is highly homogeneous without aggregation, which is determined to be an optimal parameter.
| Group | Extract addition time (min) | Precursor volume total (μL) | Precursor volume (μL) & time (min) | Reaction time | SPR band | Particle size (nm) |
|---|---|---|---|---|---|---|
| A | 5 | 800 | 20/1.30 | 1 h | 420 | 29.4 |
| B | 1600 | 20/2 | 2 h 38 min | 423 | 36.2 | |
| C | 1000 | 1 h 38 min | 431 | 26.7 | ||
| D | 15 | 1200 | 12 min | 412 | 24.2 | |
| E | 1600 | 200/2 | 16 min | 407 | 40.2 | |
| F | 1200 | 30 min | 415 | 27.9 | ||
| G | 1200 | 1 h | 408 | 28.3 |
FTIR analysis was carried out for the AgNPs to understand or determine the various functional compounds that aid in reducing and stabilizing organic leaves content. Fig. 4A shows several characteristic peaks of extract (753, 1026, 1188, 1315, 1443, 1596, 1706, 2838, 2915 and 3204 cm−1) and NPs (702, 1060, 1374, 1605, 2847, and 3187 cm−1). Each peak corresponds to diverse functional groups of the metabolites like alcohols, aldehydes, phenolics, hydroxyls, and carboxylic acid compounds.55 The peaks at various positions belong to several bioactive compounds in the leaves extract, like tannins, flavonoids, acids (phenolic and cinnamic), monoterpenes, sesquiterpenes, and other aromatic compounds.56 The bands at 753 cm−1 represent the aromatic rings, 1026 and 1188 correspond to alcohol groups, 1315 and 1706 cm−1 are characteristics of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group, and 1443 cm−1 relates to C–H scissors vibration, 1596 cm−1 denoted C@C double bond vibration, 2838 and 2915 cm−1 assigned to the C–H stretching, and finally broad absorbance at 3204 cm−1 of OH group. The absorption bands seen in the AgNPs spectra are slightly shifted from the signature peaks of the extract, confirming that the presence of the organic residual of leaves extract is solely responsible for the reduction and stabilization agent.57
O group, and 1443 cm−1 relates to C–H scissors vibration, 1596 cm−1 denoted C@C double bond vibration, 2838 and 2915 cm−1 assigned to the C–H stretching, and finally broad absorbance at 3204 cm−1 of OH group. The absorption bands seen in the AgNPs spectra are slightly shifted from the signature peaks of the extract, confirming that the presence of the organic residual of leaves extract is solely responsible for the reduction and stabilization agent.57
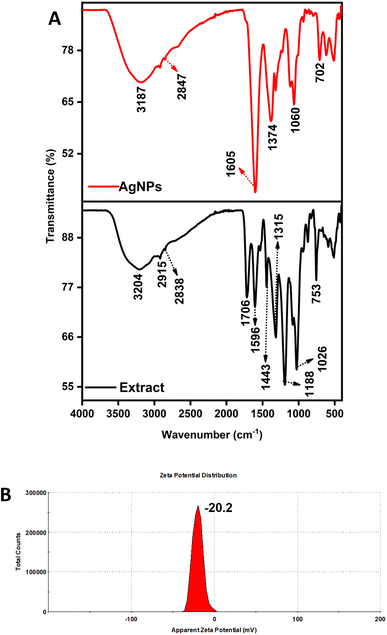 | ||
| Fig. 4 (A) FTIR analysis of the extract and as-synthesized AgNPs, (B) zeta potential analysis of AgNPs (group F). | ||
Based on the above characteristics, group F AgNPs, an optimized condition, further analysis to determine the surface charge by zeta potential was carried out, and the value was found to be −20.2 mV (Fig. 4B), which is moderately stable. This proves that the NPs have negligible aggregation due to electrostatic repulsion by the capping of pelargonium functional groups and are pretty stable with extended shelf-life. This data also corroborates with the other investigation of plant extract-mediated AgNPs.58–60
3.3 Antimicrobial activity of biosynthesized AgNPs
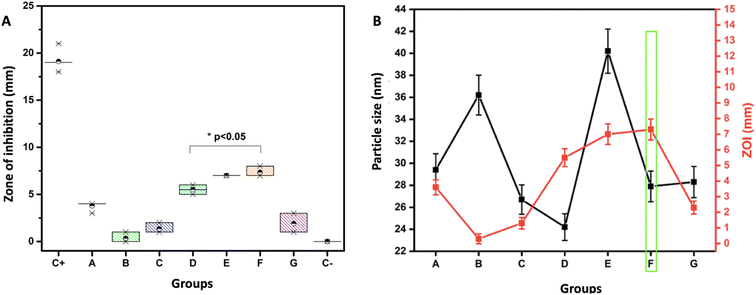
Dental caries continues to be a public health problem affecting more than 90% of the world's population, mainly developing countries. The Gram-positive bacteria S. mutans is the principal microorganism which grows due to its acidogenic and aciduric characteristics that contribute to the colonization and formation on the dental surfaces as well as the demineralization of hard dental tissues.61 So, based on this fact, we chose this bacterium as a model pathogen to evaluate the antibacterial activity of biosynthesized AgNPs in this study. The results of the agar diffusion experiment show the following zones of inhibition (ZOI): group A: 3.6 ± 0.5 mm, group B: 0.3 ± 0.5 mm, group C: 1.3 ± 0.5 mm, group D: 5.5 ± 0.5 mm, group E: 7.0 ± 0.5 mm, group F: 7.3 ± 0.5 mm and group G: 2.3 ± 0.5 mm, positive control 19.0 ± 0.5 mm, and negative control 0 mm, showing significant differences (p < 0.05) as shown in Fig. 5A. The ZOI of all groups are presented after deducting from the positive control values.AgNPs generally express antimicrobial behavior against many pathogens, including bacteria, fungi, and viruses.26,49,62 A high bactericidal activity against Gram-positive and Gram-negative bacteria is seen when the AgNP size is between 10–100 nm has been described,59 which coincides with our results where we found bacterial inhibition zones of up to 13.3 ± 0.5 mm in the Gram-positive bacteria studied with AgNPs between 24 to 40 nm. Enan et al. reported the ZOI of up to 18 ± 2.5 mm in S. mutans with AgNPs biosynthesized using Cupressus macrocarpa extract with a particle size of 13.5–25.8 nm.63 Al-Ansari et al. also reported a similar concept for 18.3 ± 0.5 nm, AgNPs synthesized with gum arabic with an average particle size of 8.41 nm,52 which coincides with the previous literature, a smaller particle size exhibits a more significant antimicrobial effect.63,64
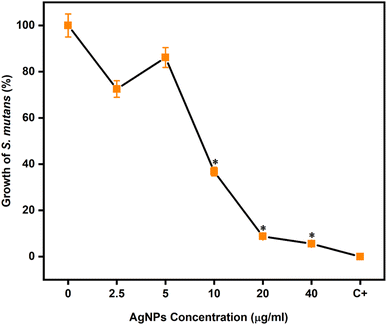
With the previously described results, group F was identified as the optimized condition and taken into consideration for further studies. The AgNPs are of spherical morphology and monodispersity (n = 3), with an average particle size of 28.5 ± 8.16 nm (n = 3), mean ZOI of 13.3 ± 0.5 mm against S. mutans (n = 9) (p < 0.05) (Fig. 5B). Based on these promising results, the microdilution broth method was adopted to determine the effect of AgNPs (group F) against microbial growth by incubating with different concentrations. The results are as follows for 2.5 μg mL−1 = 72.5 ± 3.6%, 5 μg mL−1 = 86.1 ± 4.3%, 10 μg mL−1 = 36.8 ± 1.8%, 20 μg mL−1 = 8.7 ± 0.4% & 40 μg mL−1 = 5.5 ± 0.2%. A MIC of 10 μg mL−1 was identified, and according to the Clinical and Laboratory Standards Institute (CLSI),39 the value is considered to be intermediate, and an MBC at 40 μg mL−1 is deemed sensitive (Fig. 6). The obtained MIC agrees with the results of Tavaf et al., who also report a MIC of 10 μg (ref. 65) and it is close to the range reported by Ma et al., of 50 μM (8.5 μg mL−1) for S. mutans.66 On the other hand, Espinosa-Cristóbal et al. found 16.7 μg mL−1 with an average size of 8.1 nm and 66.8 μg mL−1 of 20.1 nm,67 which concludes that the NPs size plays a vital role in exhibiting antimicrobial effect.63
The plausible mechanism behind the AgNPs bactericidal activity is due to their enhanced cell wall binding and the enormous generation of free radicals.68 The Ag+ ions released from the NPs against bacteria can interact with peptidoglycan cell membranes, causing cell lysis, preventing DNA replication, and disrupting protein synthesis.69
3.4 Immunohistochemistry analysis
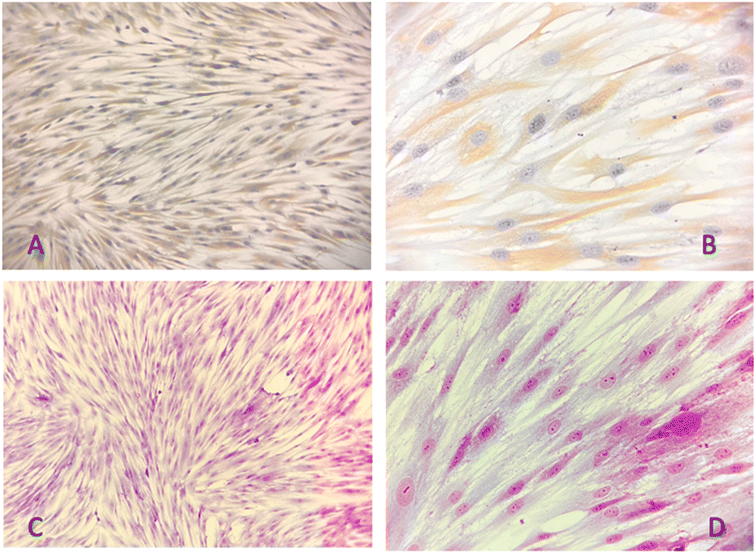
From immunohistochemistry, the photomicrographs examined show a confluence of HGF obtained from explants with in vitro growth on an electrocharged slide. Masson's trichrome staining indicates oval and some polygonal-shaped cells with basophilic staining, nuclei, and nucleoli evident, condensed chromatin arranged in a fibrillar stroma of connective tissue with strong positivity. The vimentin antibody shows strong immunopositivity (+++) with antibody uptake in the cytoplasm without colocalization (Fig. 7A–D). This confirms that the in vitro-grown cells were HGF which was further used for various studies.3.5 Cytotoxicity
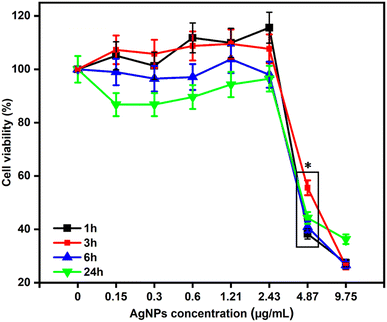
The MTT assay result showed a dosage and time-dependent effect when HGF was in contact with AgNPs at 24, which is shown in Fig. 8, where moderate cytotoxicity was seen. The CC50 of AgNPs was identified to be 4.5 μg mL−1 at the end of 24 h incubation. This result corroborates with the reported values of Huang et al. in other cell lines; as the concentration increases, moderate cytotoxicity can be identified.70 Yen et al. found that 10 nm AgNPs at a concentration greater than 25 μg mL−1 are considered cytotoxic; thus, it concludes that the particle size and concentration are directly proportional to the cytotoxic effect.71 The cellular uptake of AgNPs is through endocytosis, a time, dose, and energy-dependent process.72 Once internalized, they induce effects such as oxidative stress, cell membrane deterioration, cell cycle rest, inflammation, DNA damage, genotoxicity, and apoptosis.73 This explains the concept behind the moderate cytotoxicity of as-synthesized AgNPs by the Pelargonium x hortorum leaves extract in this study.3.6 ROS studies
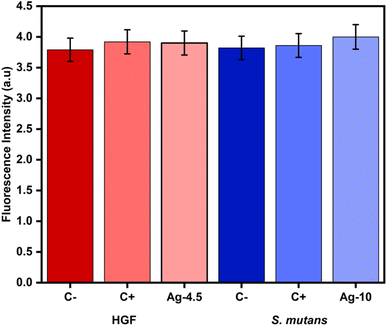
The release of metal ions is related to the generation of ROS in both prokaryotic and eukaryotic cells. Intracellular ROS are triggered as a cell defense mechanism and are eliminated through antioxidants to maintain cellular homeostasis. Excessive production of these antioxidants generates oxidative stress, which leads to alterations in DNA, proteins, and lipids, thereby causing cell death.74Some studies show that ROS formation is mainly because of NPs interactions with the bacteria. Fageria et al. found that biosynthesized protein-capped AgNPs at a concentration of 5, 10, and 40 μg mL−1 cause an increased ROS production in cancer cells relating to concentration-dependent cytotoxicity.75 The production of a higher level of an antioxidant enzyme could be a compensatory mechanism to cope with the increased level of oxidative stress due to ROS generated by treatment with AgNPs.75 Quinteros et al. found that AgNPs are related to the production of ROS with levels of oxidized proteins and lipids in Staphylococcus aureus and Escherichia coli. These results agree with those reported by Fegeria et al., where oxidative stress is related to one of the mechanisms by which AgNPs produce antimicrobial effects.76 Ghabban et al. found ROS production in S. mutans with concentrations of 3.5 and 5.3 μg mL−1 of AgNPs.77 The outcomes from our research differ from theirs, where 10 μg mL−1 in the prokaryotic cell did not lead to any ROS production.
Regarding eukaryotic cells, Gurunathan et al. reports an increase in ROS production in mouse embryonic fibroblast cells exposed to 5–20 μg mL−1 of AgNPs. Identifying a dose-dependent effect, lower concentration-lower ROS production,78 the results of this study show an immediate impact, with 4.5 μg mL−1 of AgNPs observing a slight increase in free radical production in the study groups, without statistically significant results (p > 0.05) which is represented in Fig. 9. Our results concur with Saliev et al., who identified lower ROS production in skin fibroblasts with AgNPs of spherical morphology compared to those of triangular morphology; with circular shapes, no significant increase in ROS formation was detected compared to the control group.79
3.7 Nuclei fluorescence assay
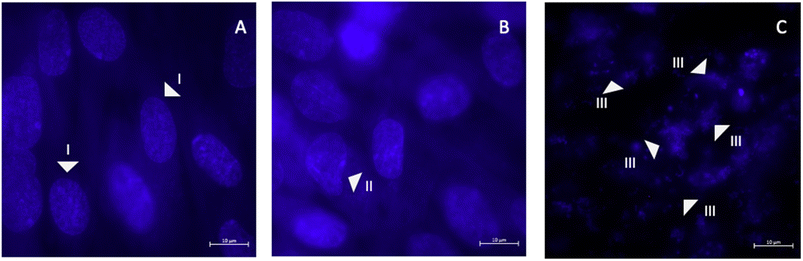
The impact of AgNPs on the HGF cell nuclear morphology was evaluated using DAPI staining and visualization through fluorescence microscopy. From the images, intact cell nuclei for the negative control (HGF) and AgNPs CC50 of 4.5 μg mL−1, and in the case of positive control (H2O2), a complete fragmentation was observed (Fig. 10A–C). The interaction of AgNPs with the HGF cell nucleus has resulted in negligible effect, thereby causing a shrunken nuclear membrane without causing any deformation, blebbing, destruction of the nuclear contents or apoptosis. It is prevalent when the Ag+ ion is reduced with chemicals, even at deficient concentrations. It may lead to cytoplasmic condensation, rounded cells, progressive nuclear shrinkage, and nuclear fragmentation, as the most identifiable nuclear alterations related to cell apoptosis were reported by certain literature. For instance, when we compared with other green synthesis using Terminalia chebula seed extract and synthesized nearly spherical polydisperse 30 to 150 nm sized AgNPs. The biocompatibility was tested with the McCoy fibroblast cell line, where 100 μg mL−1 (IC50 – 67.26 μg mL−1) showed high inhibition along with apoptosis and nuclear fragmentation causing apoptosis.80Considering this evaluation, very few studies have been carried out and determined its effect on biocompatibility when it comes to green synthesized-AgNPs. Therefore, most studies use AgNPs for anticancer studies, showing a significant impact by inducing apoptotic pathways and leading to the destruction of cancer cells.81–86 Thus, we conclude that the pelargonium-mediated AgNPs are highly biocompatible and can be used for diverse biomedical applications.
Further studies will be carried out to understand the in-depth antibacterial mechanisms at a molecular level and anti-inflammatory behavior with regard to the Pelargonium x hortorum leaves extract-derived AgNPs in culture with cells and focused on the proteomic profile to understand the molecular antimicrobial mechanisms.
4. Conclusions
In conclusion, the results confirmed that the biosynthesis of AgNPs mediated with Pelargonium x hortorum produced a nearly spherical morphology with a well-defined shape and high stability. Based on the metabolomics analysis, the presence of flavonoids, lignans, phenyl propanoinds, among other classes of compounds, could be responsible for the green biosynthesis of the AgNPs as reducing and stabilizing agents. Specifically, group F, incorporating 1200 μL (AgNO3) precursor for 30 min synthesis, produced an average particle size of 27.9 nm and was considered an optimal condition. The AgNPs obtained raised an antimicrobial effect on S. mutans, showing an intermediate MIC and a sensitive MBC according to CLSI standards. A CC50 of 4.5 μg mL−1 was identified in the HGF cell line. AgNPs in contact with HGF and S. mutans did not induce reactive oxygen species (ROS). Finally, the AgNPs did not show any nuclear fragmentation by DAPI staining. Based on the studies mentioned above, we conclude pelargonium mediated AgNPs can be best suited for diverse biomedical applications.Author contributions
C. A. L.-A.: investigation, methodology, conceptualization, formal analysis, writing – original draft, R. G.-C.: investigation, supervision, methodology, writing – review & editing, R. M.: supervision, software, conceptualization, methodology, writing – review & editing, M. F.: methodology, conceptualization, investigation, writing –review & editing, software, M. C. A.-A.: writing –review & editing, G. H-P.: writing –review & editing, A. P.-G.: visualization, writing – review & editing, L. S. A.-T.: supervision, validation, resources, methodology, project administration, writing – review & editing.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The authors thank the financial support by UNAM-DGAPA-PAPIIT: IN211922, IT200922 & TA200123 and PAPIME: PE203622. Also acknowledges the technical assistance of Ing. Nydia Hernandez Rios (Fluorescence microscopy), Ing. Ma. Lourdes Palma Tirado (TEM analysis), INB-Juriquilla, M. C. B. Mata Ortega (FTIR analysis), CULagos, Dr Manuel Rangel-Grimaldo (LC-MS/MS), UNCG, and Mariano Jacome (GNPS), FQ, UNAM.References
- K. D. Jandt and D. C. Watts, Nanotechnology in dentistry: Present and future perspectives on dental nanomaterials, Dent. Mater., 2020, 36(11), 1365–1378, DOI:10.1016/j.dental.2020.08.006.
- S. J. P. Begum, S. Pratibha, J. M. Rawat, D. Venugopal, P. Sahu, A. Gowda, K. A. Qureshi and M. Jaremko, Recent advances in green synthesis, characterization, and applications of bioactive metallic nanoparticles, Pharmaceuticals, 2022, 15(4), 455, DOI:10.3390/ph15040455.
- P. Singh, Y. J. Kim, H. Singh, C. Wang, K. H. Hwang, M. E. Farh and D. C. Yang, Biosynthesis, characterization, and antimicrobial applications of silver nanoparticles, Int. J. Nanomed., 2015, 10, 2567–2577, DOI:10.2147/IJN.S72313.
- M. C. Roco, C. A. Mirkin and M. C. Hersam, Nanotechnology research directions for societal needs in 2020: summary of international study, J. Nanopart. Res., 2011, 13(3), 897–919, DOI:10.1007/s11051-011-0275-5.
- R. Khursheed, K. Dua, S. Vishwas, M. Gulati, N. K. Jha, G. M. Aldhafeeri, F. G. Alanazi, B. H. Goh, G. Gupta, K. R. Paudel, P. M. Hansbro, D. K. Chellappan and S. K. Singh, Biomedical applications of metallic nanoparticles in cancer: Current status and future perspectives, Biomed. Pharmacother., 2022, 150, 112951, DOI:10.1016/j.biopha.2022.112951.
- P. Singh, H. Singh, Y. J. Kim, R. Mathiyalagan, C. Wang and D. C. Yang, Extracellular synthesis of silver and gold nanoparticles by Sporosarcina koreensis DC4 and their biological applications, Enzyme Microb. Technol., 2016, 86, 75–83, DOI:10.1016/j.enzmictec.2016.02.005.
- O. Goldie, R. Manisekaran, J. Pravin and V. Subramanian, Inorganic nanoflotillas as engineered particles for drug and gene delivery, Eng. Nanobiomater., 2016, 429–483, DOI:10.1016/b978-0-323-41532-3.00014-2.
- S. Nakamura, M. Sato, Y. Sato, N. Ando, T. Takayama, M. Fujita and M. Ishihara, Synthesis and Application of Silver Nanoparticles (AgNPs) for the Prevention of Infection in Healthcare Workers, Int. J. Mol. Sci., 2019, 20(15), 3620, DOI:10.3390/ijms20153620.
- P. Sreedevi, P. S. Reddy and K. V. Suryanarayana Rao, Effect of magnetic field and radiation on heat transfer analysis of nanofluid inside a square cavity filled with silver nanoparticles: Tiwari–Das model, Alexandria Eng. J., 2022, 61(2), 1529–1541, DOI:10.1016/j.aej.2021.06.055.
- C. L. Mallucci, M. D. Jenkinson, E. J. Conroy, J. C. Hartley, M. Brown, T. Moitt, J. Dalton, T. Kearns, M. J. Griffiths, G. Culeddu, T. Solomon, D. Hughes and C. Gamble, BASICS study collaborators. Silver-impregnated, antibiotic-impregnated or non-impregnated ventriculoperitoneal shunts to prevent shunt infection: the BASICS three-arm RCT, Health Technol. Assess., 2020, 24(17), 1–114, DOI:10.3310/hta24170.
- G. Mancuso, A. Midiri, E. Gerace and C. Biondo, Bacterial Antibiotic Resistance: The Most Critical Pathogens, Pathogens, 2021, 10(10), 1310, DOI:10.3390/pathogens10101310.
- M. F. Varela, J. Stephen, M. Lekshmi, M. Ojha, N. Wenzel, L. M. Sanford, A. J. Hernandez, A. Parvathi and S. H. Kumar, Bacterial Resistance to Antimicrobial Agents, Antibiotics, 2021, 10(5), 593, DOI:10.3390/antibiotics10050593.
- L. M. Stabryla, K. A. Johnston, N. A Diemler, V. S. Cooper, J. E. Millstone, S. J. Haig and L. M Gilbertson, Role of bacterial motility in differential resistance mechanisms of silver nanoparticles and silver ions, Nat. Nanotechnol., 2021, 16, 996–1003, DOI:10.1038/s41565-021-00929-w.
- M. A. Huq, Green Synthesis of Silver Nanoparticles Using Pseudoduganella eburnea MAHUQ-39 and Their Antimicrobial Mechanisms Investigation against Drug Resistant Human Pathogens, Int. J. Mol. Sci., 2020, 21, 1510, DOI:10.3390/ijms21041510.
- S. Raja, V. Ramesh and V. Thivaharan, Green biosynthesis of silver nanoparticles using Calliandra haematocephala leaves extract, their antibacterial activity and hydrogen peroxide sensing capability, Arabian J. Chem., 2017, 10(2), 253–261, DOI:10.1016/j.arabjc.2015.06.023.
- A. K. Mittal, Y. Chisti and U. C. Banerjee, Synthesis of metallic nanoparticles using plant extracts, Biotechnol. Adv., 2013, 31(2), 346–356, DOI:10.1016/j.biotechadv.2013.01.003.
- N. Tripathi and M. K. Goshisht, Recent advances and mechanistic insights into antibacterial activity, antibiofilm activity, and cytotoxicity of silver nanoparticles, ACS Appl. Bio Mater., 2022, 5(4), 1391–1463, DOI:10.1021/acsabm.2c00014.
- C. Vanlalveni, S. Lallianrawna, A. Biswas, M. Selvaraj, B. Changmai and S. L. Rokhum, Green synthesis of silver nanoparticles using plant extracts and their antimicrobial activities: a review of recent literature, RSC Adv., 2021, 11, 2804–2837, 10.1039/D0RA09941D.
- A. Roy, O. Bulut, S. Some, A. K. Mandal and M. D. Yilmaz, Green synthesis of silver nanoparticles: biomolecule-nanoparticle organizations targeting antimicrobial activity, RSC Adv., 2019, 9(5), 2673–2702, 10.1039/C8RA08982E.
- T. R. Anju, S. Parvathy, M. V. Veettil, J. Rosemary, T. H. Ansalna, M. M. Shahzabanu and S. Devika, Green synthesis of silver nanoparticles from Aloe vera extract and its antimicrobial activity, Mater. Today, 2021, 43, 3956–3960, DOI:10.1016/j.matpr.2021.02.665.
- M. Behravan, A. Hossein Panahi, A. Naghizadeh, M. Ziaee, R. Mahdavi and A. Mirzapour, Facile green synthesis of silver nanoparticles using Berberis vulgaris and root aqueous extract and its antibacterial activity, Int. J. Biol. Macromol., 2019, 24, 148–154, DOI:10.1016/j.ijbiomac.2018.11.101.
- S. Pirtarighat, M. Ghannadnia and S. Baghshahi, Green synthesis of silver nanoparticles using the plant extract of Salvia spinosa grown in vitro and their antibacterial activity assessment, J. Nanostruct. Chem., 2019, 9(1), 1–9, DOI:10.1007/s40097-018-0291-4.
- S. Hamedi and S. A. Shojaosadati, Rapid and green synthesis of silver nanoparticles using Diospyros lotus extract: Evaluation of their biological and catalytic activities, Polyhedron, 2019, 171, 172–180, DOI:10.1016/j.poly.2019.07.010.
- R. D. Rivera-Rangel, M. P. González-Muñoz, M. Avila-Rodriguez, T. A. Razo-Lazcano and C. Solans, Green synthesis of silver nanoparticles in oil-in-water microemulsion and nano-emulsion using geranium aqueous extract as a reducing agent, Colloids Surf., A, 2018, 536, 60–67, DOI:10.1016/j.colsurfa.2017.07.051.
- R. R. Thanighai Arassu, B. Nambikkairaj and D. R. Ramya, Pelargonium graveolens plant essential oil mediated green synthesis of Silver Nano particles and its antifungal activity against human pathogenic fungi, J. Pharmacogn. Phytochem., 2018, 7(6), 1778–1784 Search PubMed.
- L. S. Acosta-Torres, I. Mendieta, R. E. Nuñez-Anita, M. Cajero-Juárez and V. Castaño, Cytocompatible antifungal acrylic resin containing silver nanoparticles for dentures, Int. J. Nanomed., 2012, 7, 4777–4786, DOI:10.2147/IJN.S32391.
- A. G. Mahabadi, A. Mirzakhani, A. Azizi, S. Chavoshi and S. Khaghani, Extracts of Pelargonium hortorum: A natural and efficient fluid for fast and eco-friendly biosynthesis of CeO2 nanoparticles for antioxidant and photocatalytic applications, Inorg. Chem. Commun., 2021, 127, 108553, DOI:10.1016/j.inoche.2021.108553.
- M. Mohammadlou, H. Jafarizadeh-Malmiri and H. Maghsoudi, Hydrothermal green synthesis of silver nanoparticles using Pelargonium/Geranium extract and evaluation of their antifungal activity, Green Process. Synth., 2017, 6(1), 31–42, DOI:10.1515/gps-2016-0075.
- T. Pluskal, S. Castillo, A. Villar-Briones and M. Orešič, MZmine 2: Modular Framework for Processing, Visualizing, and Analyzing Mass Spectrometry-Based Molecular Profile Data, BMC Bioinf., 2010, 11, 395, DOI:10.1186/1471-2105-11-395.
- T. El-Elimat, M. Figueroa, B. M. Ehrmann, N. B. Cech, C. J. Pearce and N. H. Oberlies, High-Resolution MS, MS/MS, and UV Database of Fungal Secondary Metabolites as a Dereplication Protocol for Bioactive Natural Products, J. Nat. Prod., 2013, 76, 1709–1716, DOI:10.1021/np4004307.
- R. Schmid, S. Heuckeroth, A. Korf, A. Smirnov, O. Myers, T. S. Dyrlund, R. Bushuiev, K. J. Murray, N. Hoffmann, M. Lu, A. Sarvepalli, Z. Zhang, M. Fleischauer, K. Dührkop, M. Wesner, S. J. Hoogstra, E. Rudt, O. Mokshyna, C. Brungs, K. Ponomarov, L. Mutabdžija, T. Damiani, C. J. Pudney, M. Earll, P. O. Helmer, T. R. Fallon, T. Schulze, A. Rivas-Ubach, A. Bilbao, H. Richter, L. F. Nothias, M. Wang, M. Orešič, J. K. Weng, S. Böcker, A. Jeibmann, H. Hayen, U. Karst, P. C. Dorrestein, D. Petras, X. Du and T. Pluskal, Integrative Analysis of Multimodal Mass Spectrometry Data in MZmine 3, Nat. Biotechnol., 2023, 41, 447–449, DOI:10.1038/s41587-023-01690-2.
- O. D. Myers, S. J. Sumner, S. Li, S. Barnes and X. Du, One Step Forward for Reducing False Positive and False Negative Compound Identifications from Mass Spectrometry Metabolomics Data: New Algorithms for Constructing Extracted Ion Chromatograms and Detecting Chromatographic Peaks, Anal. Chem., 2017, 89, 8696–8703, DOI:10.1021/acs.analchem.7b00947.
- M. Wang, J. P. Carver, V. V. Phelan, L. M. Sanchez, N. Garg, Y. Peng, D. X. Nguyen, J. D. Watrous, C. A. Kapono, T. Luzzatto-Knaan, C. Porto, A. Bouslimani, A. Melnik, V. Meehan, M. J. Liu, W. Crüsemann, M. Boudreau, P. Esquenazi, E. Sandoval, M. Calderón and P. L. Kastritis, Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking, Nat. Biotechnol., 2016, 34, 828–837, DOI:10.1038/nbt.3597.
- H. Horai, M. Arita, S. Kanaya, Y. Nihei, T. Ikeda, K. Suwa, Y. Ojima, K. Tanaka, S. Tanaka, K. Aoshima, Y. Oda, Y. Kakazu, M. Kusano, T. Tohge, F. Matsuda, Y. Sawada, M. Y. Hirai, H. Nakanishi, K. Ikeda and T. Nishioka, MassBank: a Public Repository for Sharing Mass Spectral Data for Life Sciences, Int. J. Mater. Sci., 2010, 45, 703–714, DOI:10.1002/jms.1777.
- H. Mohimani, A. Gurevich, A. Shlemov, A. Mikheenko, A. Korobeynikov, L. Cao, E. Shcherbin, L. Nothias, P. C. Dorrestein and P. A. Pevzne, Dereplication of Microbial Metabolites through Database Search of Mass Spectra, Nat. Commun., 2018, 9, 4035, DOI:10.1038/s41467-018-06082-8.
- P. Shannon, A. Markiel, O. Ozier, N. S. Baliga, J. M. Wang, D. Ramage, N. Amin, B. Schwikowski and T. Ideker, Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks, Genome Res., 2003, 13, 2498–2504, DOI:10.1101/gr.1239303.
- L. W. Sumner, A. Amberg, D. Barrett, M. H. Beale, R. Beger, C. A. Daykin, T. W.-M. Fan, O. Fiehn, R. Goodacre, J. L. Griffin, T. Hankemeier, N. Hardy, J. Harnly, R. Higashi, J. Kopka, A. N. Lane, J. C. Lindon, P. Marriott, A. W. Nicholls, M. D. Reily, J. J. Thaden and M. R. Viant, Proposed Minimum Reporting Standards for Chemical Analysis. Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI), Metabolomics, 2007, 3, 211–221, DOI:10.1007/s11306-007-0082-2.
- P. Serrano-Díaz, D. Williams, J. Vega-Arreguin, R. Manisekaran, J. Twigg, D. Morse, R. Garcia-Contreras, M. C. Arenas-Arrocena and L. Acosta-Torres, Geranium -mediated synthesis of silver nanoparticles and their transcriptomic effects on Candida albicans, Green Process. Synth., 2023, 12(1), 20228105, DOI:10.1515/gps-2022-8105.
- CLSI, Performance Standards for Antimicrobial Susceptibility Testing, CLSI supplement M100, Clinical and Laboratory Standards Institute, 31st edn, 2021 Search PubMed.
- K. Erikson, H. Tuominen, M. Vakkala, J. H. Liisanantti, T. Karttunen, H. Syrjälä and T. I. Ala-Kokko, Brain tight junction protein expression in sepsis in an autopsy series, Crit Care, 2020, 24, 385, DOI:10.1186/s13054-020-03101-3.
- M. T. Lis-Balch, A chemotaxonomic reappraisal of the Section Ciconium Pelargonium (Geraniaceae), S. Afr. J. Bot., 1996, 62(5), 277–279 CrossRef.
- M. T. Lis-Balch, A chemotaxonomic study of the Pelargonium (Geraniaceae) species and their modern cultivars, J. Hortic. Sci., 1997, 72, 791–795, DOI:10.1080/14620316.1997.11515572.
- M. Mohammadlou, H. Jafarizadeh-Malmiri and H. Maghsoudi, Hydrothermal green synthesis of silver nanoparticles using Pelargonium/Geranium leaves extract and evaluation of their antifungal activity, Green Process. Synth., 2017, 6, 31–42, DOI:10.1515/gps-2016-0075.
- A. Ganbarianzade Mahabadi, A. Mirzakhani, A. Azizi, S. Chavoshi and Sh. Khaghani, Extracts of Pelargonium hortorum: A natural and efficient fluid for fast and eco-friendly biosynthesis of CeO2 nanoparticles for antioxidant and photocatalytic applications, Inorg. Chem. Commun., 2021, 127, 108553, DOI:10.1016/j.inoche.2021.108553.
- D. S. Walters, L. R. Minard, R. Craig and R. O. Mumma, Geranium defensive agents. III Structural Determination and Biosynthetic Considerations of Anacardic Acids of Geranium, J. Chem. Ecol., 1988, 14(3), 743–751, DOI:10.1007/BF01018769.
- F. M. Abdel Bar, M. A. Alossaimi, E. Elekhnawy, M. Abdullah Abulrahman Alzeer, A. Abo Kamer, E. Moglad and M. H. ElNaggar, Anti-Quorum Sensing and Anti-Biofilm Activity of Pelargonium x hortorum Root Extract against Pseudomonas aeruginosa: Combinatorial Effect of Catechin and Gallic Acid, Molecules, 2022, 27, 7841, DOI:10.3390/molecules27227841.
- A. Venter, H. Fisher, G. I. Stafford and K. Gyebi Duodu, Pigmented flower extracts of plant species from the Geraniaceae and Lamiaceae families as natural food colourants: anthocyanin composition, thermal and oxidative stability, Int. J. Food Sci. Technol., 2022, 57, 4347–4355, DOI:10.1111/ijfs.15761.
- A. Zaki, M. N. Aziz, R. Ahmad, I. Ahamad, M. S. Ali, D. Yasin, B. Afzal, S. M. Ali, A. Chopra, V. Hadda, P. Srivastava, R. Kumar and T. Farma, Synthesis, purification and characterization of Plectonema derived AgNPs with elucidation of the role of protein in nanoparticle stabilization, RSC Adv., 2022, 12, 2497–2510, 10.1039/D1RA08396A.
- X. Wei, F. Cheng, Y. Yao, X. Yi, B. Wei, H. Li, Y. Wu and J. He, Facile synthesis of carbon dots and silver nanoparticles (CDs/AgNPs) composite for antibacterial application, RSC Adv., 2021, 11, 18417–18422, 10.1039/D1RA02600C.
- D. Garibo, H. A. Borbón-Nuñez, J. N. D. de León, E. García Mendoza, I. Estrada, Y. Toledano-Magaña, H. Tiznado, M. Ovalle-Marroquin, A. G. Soto-Ramos, A. Blanco, J. A. Rodríguez, O. A. Romo, L. A. Chávez-Almazán and A. Susarrey-Arce, Green synthesis of silver nanoparticles using Lysiloma acapulcensis exhibit high-antimicrobial activity, Sci. Rep., 2020, 10(1), 12805, DOI:10.1038/s41598-020-69606-7.
- M. M. Al-Ansari, P. Dhasarathan, A. J. A. Ranjitsingh and L. A. Al-Humaid, Ganoderma lucidum inspired silver nanoparticles and its biomedical applications with special reference to drug resistant Escherichia coli isolates from CAUTI, Saudi J. Biol. Sci., 2020, 27(11), 2993–3002, DOI:10.1016/j.sjbs.2020.09.008.
- M. M. Al-Ansari, N. D. Al-Dahmash and A. J. A. Ranjitsingh, Synthesis of silver nanoparticles using gum Arabic: Evaluation of its inhibitory action on Streptococcus mutans causing dental caries and endocarditis, J. Infect. Public Health, 2021, 14(3), 324–330, DOI:10.1016/j.jiph.2020.12.016.
- P. Banerjee, M. Satapathy, A. Mukhopahayay and P. Das, Leaf extract mediated green synthesis of silver nanoparticles from widely available Indian plants: synthesis, characterization, antimicrobial property and toxicity analysis, Bioresour. Bioprocess., 2014, 1, 1–10, DOI:10.1186/s40643-014-0003-y.
- A. Gour and N. K. Jain, Advances in green synthesis of nanoparticles, Artif. Cells, Nanomed., Biotechnol., 2019, 47(1), 844–851, DOI:10.1080/21691401.2019.1577878.
- A. A. Alqahtani, M. A. El Raey, E. Abdelsalam, A. M. Ibrahim, O. Alqahtani, Z. A. Torky and H. G. Attia, The Biosynthesized Zinc Oxide Nanoparticles' Antiviral Activity in Combination with Pelargonium zonale Extract against the Human Corona 229E Virus, Molecules, 2022, 27(23), 8362, DOI:10.3390/molecules27238362.
- B. Fardsadegh, H. Vaghari, R. Mohammad-Jafari, Y. Naijian and H. Jafarizadeh-Malmiri, Biosynthesis, characterization and antimicrobial activities assessment of fabricated selenium nanoparticles using Pelargonium zonale leaf extract, Green Process. Synth., 2019, 8(1), 191–198, DOI:10.1515/gps-2018-0060.
- M. Franco-Romano, M. L. A. Gil, J. M. Palacios-Santander, J. J. Delgado-Jaén, I. Naranjo-Rodríguez, J. L. Hidalgo-Hidalgo de Cisneros and L. M Cubillana- Aguilera, Sonosynthesis of gold nanoparticles from a geranium leaf extract, Ultrason. Sonochem., 2014, 21(4), 1570–1577, DOI:10.1016/j.ultsonch.2014.01.017.
- N. Liaqat, N. Jaha, K. Ur-Rahman, T. Anwar and H. Qureshi, Green synthesized silver nanoparticles: Optimization, characterization, antimicrobial activity, and cytotoxicity study by hemolysis assay, Front. Chem., 2022, 29(10), 952006, DOI:10.3389/fchem.2022.952006.
- D. S. Basavarajappa, R. S. Kumar, A. I. Almansour, B. Chakraborty, M. P. Bhat, S. K. Nagaraja, H. Hiremath, K. Perumal and S. Nayaka, Biofunctionalized silver nanoparticles synthesized from Passiflora vitifolia leaf extract and evaluation of its antimicrobial, antioxidant and anticancer activities, Biochem. Eng. J., 2022, 187, 108517, DOI:10.1016/j.bej.2022.108517.
- Y. Khane, K. Benouis, S. Albukhaty, G. M. Sulaiman, M. M. Abomughaid, A. Al Ali, D. Aouf, F. Fenniche, S. Khane, W. Chaibi, A. Henni, H. D. Bouras and N. Dizge, Green Synthesis of Silver Nanoparticles Using Aqueous Citrus limon Zest Extract: Characterization and Evaluation of Their Antioxidant and Antimicrobial Properties, Nanomaterials, 2022, 12, 1–20, DOI:10.3390/nano12122013.
- P. L. L. Freire, A. J. R. Albuquerque, F. C. Sampaio, A. Galembeck, M. A. P. Flores, T. C. M Stamford and A. Rosenblatt. AgNPs, The new allies against S. mutans biofilm - A pilot clinical trial and microbiological assay, Braz. Dent. J., 2017, 28(4), 417–422, DOI:10.1590/0103-6440201600994.
- O. Bilek, T. Fialova, A. Otahal, V. Adam, K. Smerkova and Z. Fohlerova, Antibacterial activity of AgNPs-TiO2 nanotubes: influence of different nanoparticle stabilizers, RSC Adv., 2020, 10, 44601–44610, 10.1039/D0RA07305A.
- E. T. Enan, A. A. Ashour, S. Basha, N. H. Felemban and S. M. F. Gad El-Rab, Antimicrobial activity of biosynthesized silver nanoparticles, amoxicillin, and glass-ionomer cement against Streptococcus mutans and Staphylococcus aureus, Nanotechnology, 2021, 32, 215101, DOI:10.1088/1361-6528/abe577.
- S. Wei, Y. Wang, Z. Tang, H. Xu, Z. Wang, T. Yang and T. Zou, A novel green synthesis of silver nanoparticles by the residues of Chinese herbal medicine and their biological activities, RSC Adv., 2021, 11, 1411–1419, 10.1039/D0RA08287B.
- Z. Tavaf, M. Tabatabaei, A. Khalafi-Nezhad and F. Panahi, Evaluation of antibacterial, antibofilm and antioxidant activities of synthesized silver nanoparticles (AgNPs) and casein peptide fragments against Streptococcus mutans, Eur. J. Integr. Med., 2017, 12, 163–171, DOI:10.1016/j.eujim.2017.05.011.
- X. Ma, J. Lang, P. Chen and R. Yang, Silver nanoparticles as an effective antimicrobial against otitis Media pathogens, AIChE J., 2021, 67, 1–14, DOI:10.1002/aic.17468.
- L. F. Espinosa-Cristóbal, N. López-Ruiz, D. Cabada-Tarín, S. Y. Reyes-López, A. Zaragoza-Contreras, D. Constandse-Cortéz, A. Donohué-Cornejo, K. Tovar-Carrillo, J. C. Cuevas-González and T. Kobayashi, Antiadherence and antimicrobial properties of silver nanoparticles against streptococcus mutans on brackets and wires used for orthodontic treatments, J. Nanomater., 2018, 1–11, DOI:10.1155/2018/9248527.
- M. Oves, M. Aslam, M. A. Rauf, S. Qayyum S, H. A. Qari, M. S. Khan, M. Z. Alam, S. Tabrez, A. Pugazhendhi and I. M. I. Ismail, Antimicrobial and anticancer activities of silver nanoparticles synthesized from the root hair extract of Phoenix dactylifera, Mater. Sci. Eng., C, 2018, 89, 429–443, DOI:10.1016/j.msec.2018.03.035.
- V. T. Noronha, A. J. Paula, G. Durán, A. Galembeck, K. Cogo-Müller, M. Franz-Montan and N. Durán, Silver nanoparticles in dentistry, Dent. Mater., 2017, 33(10), 1110–1126, DOI:10.1016/j.dental.2017.07.002.
- Y. Huang, X. Lü, R. Chen and Y. Chen, Comparative study of the effects of gold and silver nanoparticles on the metabolism of human dermal fibroblasts, Regener. Biomater., 2020, 221–232, DOI:10.1093/rb/rbz051.
- H. J. Yen, S. H. Hsu and C. L. Tsai, Cytotoxicity and immunological response of gold and silver nanoparticles of different sizes, Small, 2009, 5(13), 1553–1561, DOI:10.1002/smll.200900126.
- E. M. Luther, Y. Koehler, J. Diendorf, M. Epple and R. Dringen, Accumulation of silver nanoparticles by cultured primary brain astrocytes, Nanotechnology, 2011, 22, 375101, DOI:10.1088/0957-4484/22/37/375101.
- X. Jiang, R. Foldbjerg, T. Miclaus, L. Wang, R. Singh, Y. Hayashi, D. Sutherland, C. Chen, H. Autrup and C. Beer, Multi-platform genotoxicity analysis of silver nanoparticles in the model cell line CHO-K1, Toxicol. Lett., 2013, 222(1), 55–63, DOI:10.1016/j.toxlet.2013.07.011.
- T. Ameh, M. Gibb, D. Stevens, S. H. Pradhan, E. Braswell and C. M. Sayes, Silver and copper nanoparticles induce oxidative stress in bacteria and mammalian cells, Nanomaterials, 2022, 12(14), 2402, DOI:10.3390/nano12142402.
- L. Fageria, V. Pareek, R. V. Dilip, A. Bhargava, S. S. Pasha, I. R. Laskar, H. Saini, S. Dash, R. Chowdhury and J. Panwar, Biosynthesized Protein-Capped Silver Nanoparticles Induce ROS-Dependent Proapoptotic Signals and Prosurvival Autophagy in Cancer Cells, ACS Omega, 2017, 2(4), 1489–1504, DOI:10.1021/acsomega.7b00045.
- M. A. Quinteros, C. A. Viviana, R. Onnainty, V. S. Mary, M. G. Theumer, G. E. Granero, M. G. Paraje and P. L. Páez, Biosynthesized silver nanoparticles: Decoding their mechanism of action in Staphylococcus aureus and Escherichia coli, Int. J. Biochem. Cell Biol., 2018, 104, 87–93, DOI:10.1016/j.biocel.2018.09.006.
- H. Ghabban, S. F. Alnomasy, H. Almohammed, O. M. Al Idriss, S. Rabea and Y. Eltahir, Antibacterial, cytotoxic, and cellular mechanisms of green synthesized silver nanoparticles against some cariogenic bacteria (Streptococcus mutans and Actinomyces viscosus), J. Nanomater., 2022, 1–8, DOI:10.1155/2022/9721736.
- S. Gurunathan, M. Qasim, C. Park, H. Yoo, D. Y. Choi, H. Song, C. Park, J. H. Kim and K. Hong, Cytotoxicity and Transcriptomic Analysis of Silver Nanoparticles in Mouse Embryonic Fibroblast Cells, Int. J. Mol. Sci., 2018, 19(11), 3618, DOI:10.3390/ijms19113618.
- T. Saliev, D. M. Baiskhanova, A. Akhmetova, D. A. Begimbetova, M. Akishev, G. Kulsharova, A. Molkenov, T. Nurgozhin, T. Alekseyeva and S. Mikhalovsky, Impact of electromagnetic fields on in vitro toxicity of silver and graphene nanoparticles, Electromagn. Biol. Med., 2019, 38(1), 21–23, DOI:10.1080/15368378.2018.1534740.
- N. Muthusamy, P. Kanniah, P. Vijayakumar, U. Murugan, D. S. Raj and U. Sankaran, Green-Inspired Fabrication of Silver Nanoparticles and Examine its Potential In Vitro Cytotoxic and Antibacterial Activities, J. Inorg. Organomet. Polym., 2021, 31, 4693–4709, DOI:10.1007/s10904-021-02082-2.
- A. Baker, S. Iram, A. Syed, A. M. Elgorban, A. H. Bahkali, K. Ahmad, M. Sajid Khan and J. Kim, Fruit Derived Potentially Bioactive Bioengineered Silver Nanoparticles, Int. J. Nanomed., 2021, 18(16), 7711–7726, DOI:10.2147/IJN.S330763.
- I. Ullah, A. T. Khalil, M. Ali, J. Iqbal, W. Ali, S. Alarifi and Z. K. Shinwari, Green-Synthesized Silver Nanoparticles Induced Apoptotic Cell Death in MCF-7 Breast Cancer Cells by Generating Reactive Oxygen Species and Activating Caspase 3 and 9 Enzyme Activities, Oxid. Med. Cell. Longevity, 2020, 1–14, DOI:10.1155/2020/1215395P.
- Gupta, N. Rai, A. Verma, D. Saikia, S. P. Singh, R. Kumar, S. K. Singh, D. Kumar and V. Gautam, Green-Based Approach to Synthesize Silver Nanoparticles Using the Fungal Endophyte Penicillium oxalicum and Their Antimicrobial, Antioxidant, and In Vitro Anticancer Potential, ACS Omega, 2022, 7(50), 46653–46673, DOI:10.1021/acsomega.2c05605.
- M. Dadashpour, A. Firouzi-Amandi, M. Pourhassan-Moghaddam, M. J. Maleki, N. Soozangar, F. Jeddi, M. Nouri, N. Zarghami and Y. Pilehvar-Soltanahmadi, Biomimetic synthesis of silver nanoparticles using Matricaria chamomilla extract and their potential anticancer activity against human lung cancer cells, Mater. Sci. Eng., C, 2018, 1(92), 902–912, DOI:10.1016/j.msec.2018.07.053.
- S. Devanesan, M. Jayamala, M. S. AlSalhi, S. Umamaheshwari and A. J. A. Ranjitsingh, Antimicrobial and anticancer properties of Carica papaya leaves derived di-methyl flubendazole mediated silver nanoparticles, J. Infect. Public Health, 2021, 14(5), 577–587, DOI:10.1016/j.jiph.2021.02.004.
- J. Mittal, U. Pal, L. Sharma, A. K. Verma, M. Ghosh and M. M. Sharma, Unveiling the cytotoxicity of phytosynthesised silver nanoparticles using Tinospora cordifolia leaves against human lung adenocarcinoma A549 cell line, IET Nanobiotechnol., 2020, 14(3), 230–238, DOI:10.1049/iet-nbt.2019.0335.
| This journal is © The Royal Society of Chemistry 2023 |