Open Access Article
Open Access ArticleDesign, synthesis, anticancer and in silico assessment of 8-piperazinyl caffeinyl-triazolylmethyl hybrid conjugates†
Mohammad Navid Soltani Rad *ab,
Somayeh Behrouz
*ab,
Somayeh Behrouz *ab,
Kiana Shahbazkhani
*ab,
Kiana Shahbazkhani a,
Marzieh Behrouza,
Elham Zarenezhad
a,
Marzieh Behrouza,
Elham Zarenezhad c and
Ali Ghanbariasadd
c and
Ali Ghanbariasadd
aDepartment of Chemistry, Shiraz University of Technology, Shiraz 71555-313, Iran. E-mail: soltani@sutech.ac.ir; behrouz@sutech.ac.ir; Fax: +98 71 3735 4520; Tel: +98 71 3735 4500
bMedicinal Chemistry Research Laboratory, Novel Technology for Health Research Center, Shiraz University of Technology, Shiraz 71555-313, Iran
cNon-communicable Diseases Research Center, Fasa University of Medical Sciences, Fasa, Iran
dDepartment of Medical Biotechnology, School of Medicine, Fasa University of Medical Sciences, Fasa, Iran
First published on 18th August 2023
Abstract
In this paper, we have assessed the design, synthesis, characterization, anticancer properties, toxicity, and in silico study of 8-piperazinyl caffeinyl-triazolylmethyl derivatives as new caffeine hybrid conjugates. These compounds consist of four moieties comprising 8-caffeinyl, piperazinyl, 1,2,3-triazolyl, and alkyl substituents. The synthesis of these compounds was started by bromination of caffeine to attain 8-BC, SNAr reaction with piperazine to acquire 8-PC, N-propargylation of 8-PC and finally click Huisgen cycloaddition with diverse alkyl azides. These compounds were in vitro tested against two significant cancer cell lines comprising breast cancer MCF-7 (ATCC HTB-22) and melanoma cancer A-375 (ATCC CRL-1619) cell lines and activities compared with methotrexate (MTX) as a reference drug. Anticancer assessments indicated 12j (IC50 = 323 ± 2.6) and 12k (IC50 = 175 ± 3.2) were the most potent compounds against A-375 and MCF-7 cell growth, respectively and their activities were even stronger than MTX (IC50 = 418 ± 2 for A375 and IC50 = 343 ± 3.6 for MCF-7). Toxicities were determined by screening compounds against normal cell line HEK-293 (ATCC CRL-11268) and indicated that except 12i (IC50 = 371 ± 2.3), 12j (IC50 = 418 ± 2.4), and MTX (IC50 = 199 ± 2.4), all compounds are non-toxic. Docking was conducted for 12j and 12k and determined the strong binding affinities to B-RAF kinase and hDHFR enzymes, respectively. In silico pharmacokinetic and physiochemical profiles of tested compounds were investigated which indicated that most compounds obeyed Lipinski's rule of five (RO5). The DFT study on M06-2X/6-311G (d,p) was used to indicate HOMO, LUMO, MEP, and other parameters for a better understanding of 12j and 12k reactivity. Owing to anticancer properties, toxicity, and in silico data, 12j and 12k can be proposed for further research in the future.
1 Introduction
Cancer is a multifactorial disease that is influenced by various factors such as lifestyle, population, genetics, and surroundings.1 Based on statistics, cancer is one of the leading causes of mortality worldwide. The occurrence rate of cancer varies across 140 countries, and more than eleven million people are diagnosed with cancer every year.2 It is estimated that there will be 16 million new cases of cancer annually before 2020. Amongst cancer types, breast cancer is one of the deadliest and the most common cancers in women that affect and threaten the lives of women over the age of 50. About 2.3 million women in 2020 were diagnosed with breast cancer and 685![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths worldwide.3 There were 7.8 million women alive who were diagnosed with breast cancer in 2015–2020, making it the world's most prevalent cancer. Breast cancer occurs in many women of any race or nationality worldwide, poor or rich, and causes huge costs for individuals, societies, and governments.4 Melanoma is a type of skin cancer that can spread to other areas of the body. The main cause of melanoma is UV light, which comes from the sun and is used in sunbeds. Getting melanoma is widely dependent on factors such as age and having pale skin, a large number of moles, and a family history of skin cancer. At least 132
000 deaths worldwide.3 There were 7.8 million women alive who were diagnosed with breast cancer in 2015–2020, making it the world's most prevalent cancer. Breast cancer occurs in many women of any race or nationality worldwide, poor or rich, and causes huge costs for individuals, societies, and governments.4 Melanoma is a type of skin cancer that can spread to other areas of the body. The main cause of melanoma is UV light, which comes from the sun and is used in sunbeds. Getting melanoma is widely dependent on factors such as age and having pale skin, a large number of moles, and a family history of skin cancer. At least 132![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 malignant melanomas occur globally each year. There has been a significant increase in the incidence of skin cancers since the 1970s. A changing lifestyle and sun-seeking behavior are responsible for an extensive increment in skin cancers. Also, the depletion of the ozone layer, which provides a protective filter against UV radiation, aggravates the problem.5 Despite the development of chemotherapeutic agents in the remedy of all types of cancer, these compounds have been envisaged many shortcomings and problems like lack of selectivity and severe toxicity against normal cells, low efficacy, high prices, inconstancy, and so on. Hence, ongoing and restless efforts are underway to find potent drugs with minimal toxicity, high efficacy, and cheap expense of production.6
000 malignant melanomas occur globally each year. There has been a significant increase in the incidence of skin cancers since the 1970s. A changing lifestyle and sun-seeking behavior are responsible for an extensive increment in skin cancers. Also, the depletion of the ozone layer, which provides a protective filter against UV radiation, aggravates the problem.5 Despite the development of chemotherapeutic agents in the remedy of all types of cancer, these compounds have been envisaged many shortcomings and problems like lack of selectivity and severe toxicity against normal cells, low efficacy, high prices, inconstancy, and so on. Hence, ongoing and restless efforts are underway to find potent drugs with minimal toxicity, high efficacy, and cheap expense of production.6
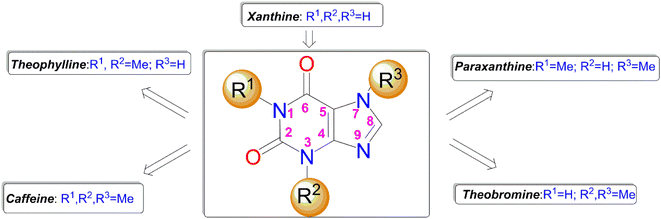
Over the years, naturally occurring compounds have aroused special attention from the pharmaceutical point of view, mainly because of their notable therapeutic properties.7 Among naturally occurring compounds methylxanthines comprising caffeine, theophylline, theobromine and paraxanthine (Fig. 1) are unique scaffolds with immense biological activities.8 They are ubiquitously found in many plants, seeds of coffee, cacao, and tea leaves.9 In particular, among methylxanthines, caffeine is one of the most well-known and widely used molecules and extensively exploited in the history life of human beings.10 Caffeine has gained particular attention in recent decades owing to its magnificent chemotherapeutic properties such as diuretic, vasodilator, analgesic, and so on.11 Unlike the most N-methyl xanthines, the caffeine can be solely modified through its C8 site with a few electrophiles or active species.12–15
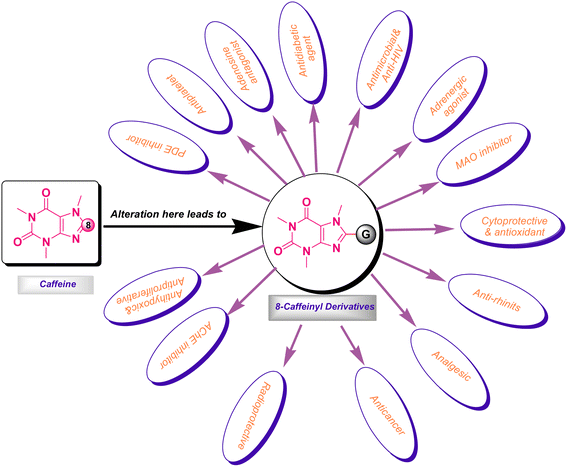
It is worth mentioning that the functionalization of caffeine C8 position creates remarkable biological profiles with numerous medicinal properties, which are summarized in Fig. 2.
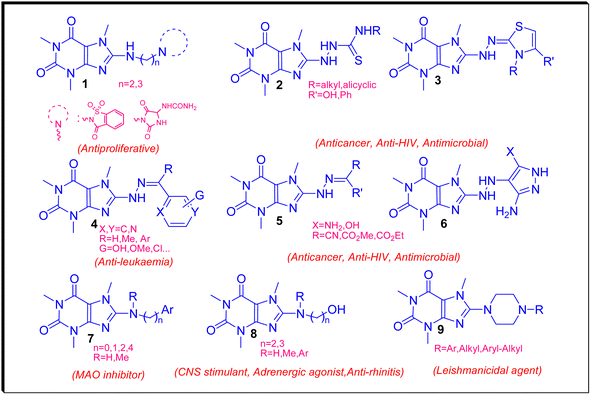
Recently, we have reviewed and categorized the C8-modified caffeine derivatives to C8–C, C8–O, C8–S, and C8–N on the basis of the atom's type linked to C8.16 Among C8-modified caffeine derivatives, the C8–N analogues are fabulous as they show considerable biological activities comprising anticancer, antimicrobial, antiviral and CNS stimulants.12,16–22 The structures of some C8–N derivatives 1–9 and their corresponding biological activities are demonstrated in Fig. 3.
Piperazine is an important heterocyclic core in medicinal chemistry because it is a versatile building block that can be used to synthesize a wide range of bioactive molecules.23 Perhaps, it is no exaggeration to claim that plenty of approved drugs involve piperazinyl moieties in their molecular frameworks.24 Parental piperazine is used as an anthelmintic or anti-parasitic drug through antagonizing of the parasite worm's GABA receptors that cause the paralysis of the worm muscles and subsequent excretion from the patient's intestines.25 One of the key reasons for its importance is its ability to act as a scaffold for the development of drugs that target many enzymes and/or receptors. Piperazine derivatives have been shown to exhibit a range of pharmacological activities including anxiolytic, sedative, antipsychotic, and antidepressant effects. These compounds work by interacting with various neurotransmitter systems in the brain including the dopamine, serotonin, and GABA systems. In addition to its use in CNS drugs, piperazine is also an important building block for the development of other types of drugs including anticancer, antihistamines, antivirals, and antibiotics. For example, ciprofloxacin, a fluoroquinolone antibiotic widely used to treat several bacterial infections, contains a piperazine ring in its structure. Overall, the unique properties and versatility of piperazine make it an important heterocyclic core in medicinal chemistry and drug discovery. Its ability to act as a scaffold for the development of drugs with a wide range of pharmacological activities makes it an essential tool for the design and synthesis of new therapeutics.26
1,2,3-Triazoles are versatile heterocyclic compounds that can be easily synthesized and modified, hence they have found particular attention in medicinal chemistry.27 Since 1,2,3-triazoles are well recognized by target proteins like enzymes or receptors, they have been shown to exhibit a range of biological activities including antimicrobial, antifungal, antitumor, antiviral, and anti-inflammatory properties.28 One of the key reasons for their importance is their ability to act as non-hydrolyzable bioisosteres of an amide bonds. For example, 1,2,3-triazoles can be used as surrogates for the amide bond in peptides, which can improve their stability and bioavailability.29 In addition, the ease and selectivity of 1,2,3-triazoles synthesis through ‘Click’ Huisgen azide–alkyne cycloaddition method provides an efficient tool to conjugate two or more pharmacophores in a target molecule. This has led to their widespread use in drug discovery and development, as well as in the synthesis of other bioactive molecules.
Hybrid molecules are important in medicinal chemistry because they can combine the beneficial properties of two or more different pharmacophores into a single compound.30 This can lead to improved therapeutic efficacy, reduced toxicity, and/or enhanced selectivity for a specific target. The design of hybrid molecules with a dual mode of action is an important technique extensively applied in drug discovery. For example, a hybrid molecule could be designed to combine an antimicrobial agent with an anti-inflammatory agent, leading to a compound that can kill bacteria and reduce inflammation, simultaneously. In addition, hybrid molecules can also be used to overcome drug resistance, which is a major challenge in the development of new therapeutics.31 Overall, the ability to combine the beneficial properties of different pharmacophores into a single hybrid molecule has the potential to significantly improve the efficacy and safety of drugs and to accelerate the development of drug discovery for a wide range of diseases.32
Recently, we reported the synthesis and biological assessments of 8-(4-alkylpiperazinyl) caffeine 10 and 8-caffeinyl-triazolylmethyl hybrid conjugates 11 (Fig. 4) exhibiting remarkable leishmanicidal and anticancer properties.16,22 In pursuit of our ongoing research on novel 8-caffeinyl derivatives and also inspired by the structure of compounds 10 and 11, hereby we have disclosed the synthesis, characterization, anticancer, and in silico investigation of new 8-piperazinyl caffeinyl-triazolylmethyl hybrid conjugates 12.
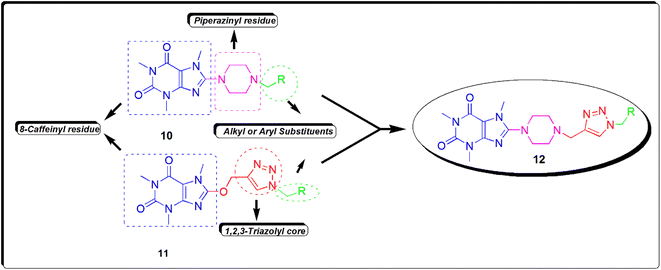 | ||
| Fig. 4 The structure of new 8-piperazinyl caffeinyl-triazolylmethyl hybrid conjugates 12 and comparing its structure with previously synthesized compounds 10 and 11. | ||
2 Results and discussion
2.1. Chemistry
The synthesis of 8-piperazinyl caffeinyl-triazolylmethyl hybrid conjugates 12 was performed due to Scheme 1.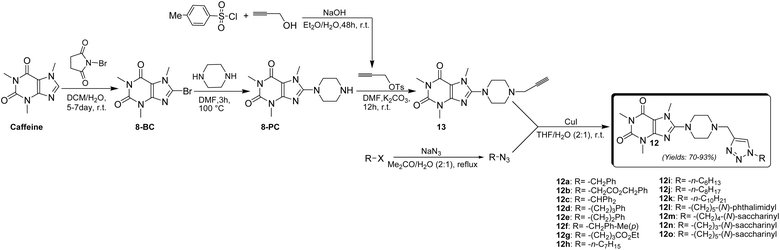 | ||
| Scheme 1 General synthetic pathway for preparing novel 8-piperazinyl caffeinyl-triazolylmethyl hybrid conjugates 12. | ||
Due to Scheme 1, this synthesis was initiated by 8-bromocaffeine (8-BC) preparation. The bromination of caffeine should be achieved to enhance the positive charge density on C(8) for the subsequent SNAr-type reaction. The 8-BC was synthesized due to standard procedure using NBS in a mixture of DCM and water at room temperature.33 Also, 8-BC can be synthesized using other methods;34,35 however, none of them were efficient in comparison with NBS. Applying the NBS method almost yielded the pure 8-BC, quantitatively. Having prepared 8-BC, the 8-piperazinyl caffeine (8-PC) was then acquired by a SNAr-type coupling reaction of piperazine with electrophilic C(8) of 8-BC in DMF at 100 °C.22 8-PC is an odorless white solid (m.p. = 165–167 °C) with apparent basic properties (pH ≈ 12.5 in 0.01 M aqueous solution at 25 °C). 8-PC is freely soluble in water, but it partially dissolves in solvents like DMF, DCM, or CHCl3. To afford alkyne 13, 8-PC was used for N-propargylation reaction with propargyl tosylate which was previously prepared by the reaction of TsCl and propargyl alcohol through the well-established method.36 The use of propargyl tosylate is preferable to propargyl bromide since propargyl tosylate is a safe and cheap analogue of expensive propargyl bromide. To this end, TsCl was used to react with an excess amount of propargyl alcohol in diethyl ether and an aqueous NaOH mixture at room temperature. This procedure almost affords a quantitative yield of propargyl tosylate (≈100%) as a yellow oil. Alkyne 13 was prepared as a pale yellow solid (m.p. = 140–142 °C) by coupling of 8-PC with propargyl tosylate using K2CO3 as a base in anhydrous DMF at ambient temperature for 24 h. To achieve the final products, the alkyl azides were prepared by reaction of diverse alkyl halides with NaN3 in acetone-water solution at reflux condition. Ultimately, the cuprous catalyzed ‘Click’ Huisgen cycloaddition reaction of alkyl azides with alkyne 13 in THF-H2O (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
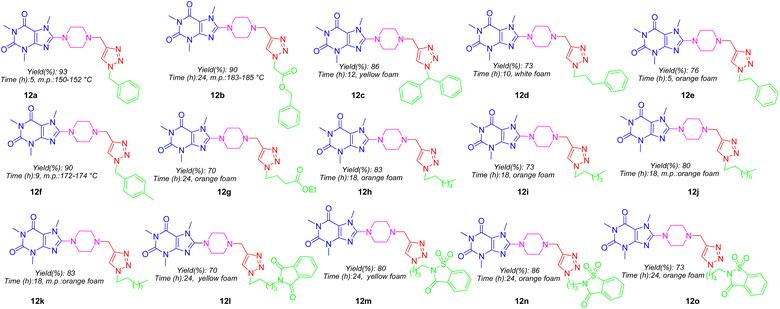
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) media at ambient temperature achieved final products 12a–12o in good yields which structures are depicted in Fig. 5. As can be seen in Fig. 5, the target compounds merely vary in alkyl residues. They are involved in aryl-alkyl residues (12a, 12c–12f), alkyl-esters (12b and 12g), normal alkyls (12h–12k), and alkyl-imides (12l–12o).
1) media at ambient temperature achieved final products 12a–12o in good yields which structures are depicted in Fig. 5. As can be seen in Fig. 5, the target compounds merely vary in alkyl residues. They are involved in aryl-alkyl residues (12a, 12c–12f), alkyl-esters (12b and 12g), normal alkyls (12h–12k), and alkyl-imides (12l–12o).
2.2. In vitro anticancer and cytotoxic activity
A-375 cell line is a known human melanoma cell line that was isolated from a tumor from a 54 years old woman with malignant melanoma. This cell line is very useful for pharmacology, immuno-oncology, and toxicology research.37 MCF-7 cell line is one of the most popular human breast cancer cell lines in the world, which was derived from a 69 years old woman with breast adenocarcinoma in 1973. Researchers are interested in the MCF-7 cell line since this cell maintains several characteristics similar to the breast epithelium. This cell line is often used for in vitro study models of breast cancer biology. Regarding various available variants, this cell line has a wide range of applications in the development of chemotherapeutic drugs and also understanding the drug resistance.38 HEK-293 is known as a normal cell line that is derived from human embryonic kidney cells in 1973. This cell line is commonly applied for basic medical research and also pharmacology.39 The IC50 values related to toxicity tests for all synthetic products are summarized in Table 1. According to the results obtained from the MTT assay, it was determined that 12j (IC50 = 323 ± 2.6) is the most potent compound; moreover, 12i (IC50 = 371 ± 2.5), and 12k (IC50 = 367 ± 3.4) exhibited remarkable inhibition effects on A-375 cell line growth and their toxicities are even stronger than MTX as a reference drug. Additionally, 12h (IC50 = 498 ± 2) also demonstrated marginal toxicity in comparison with MTX for inhibition of A-375 cell line growth. In the case of the MCF-7 cell line, 12k (IC50 = 175 ± 3.2) was the most potent agent and other compounds encompassing 12h (IC50 = 306 ± 1.8), 12i (IC50 = 260 ± 2.2), and 12j (IC50 = 245 ± 2) displayed more toxicities than MTX. Amongst tested compounds effective on both A-375 and MCF-7, 12k (IC50 = 175 ± 3.2) displayed more selectivity owing to its non-toxicity against HEK-293 (IC50 > 500). Apart from 12i, 12j, and MTX, all other compounds have no toxicity effect on HEK-293.| Compound | A-375 | MCF-7 | HEK-293 |
|---|---|---|---|
| 12a | >500 | >500 | >500 |
| 12b | >500 | >500 | >500 |
| 12c | >500 | >500 | >500 |
| 12d | >500 | >500 | >500 |
| 12e | >500 | >500 | >500 |
| 12f | >500 | >500 | >500 |
| 12g | >500 | >500 | >500 |
| 12h | 498 ± 2 | 306 ± 1.8 | >500 |
| 12i | 371 ± 2.5 | 260 ± 2.2 | 371 ± 2.3 |
| 12j | 323 ± 2.6 | 245 ± 2.3 | 418 ± 2.4 |
| 12k | 367 ± 3.4 | 175 ± 3.2 | >500 |
| 12l | >500 | >500 | >500 |
| 12m | >500 | >500 | >500 |
| 12n | >500 | >500 | >500 |
| 12o | >500 | >500 | >500 |
| MTX | 418 ± 2 | 343 ± 3.6 | 199 ± 2.4 |
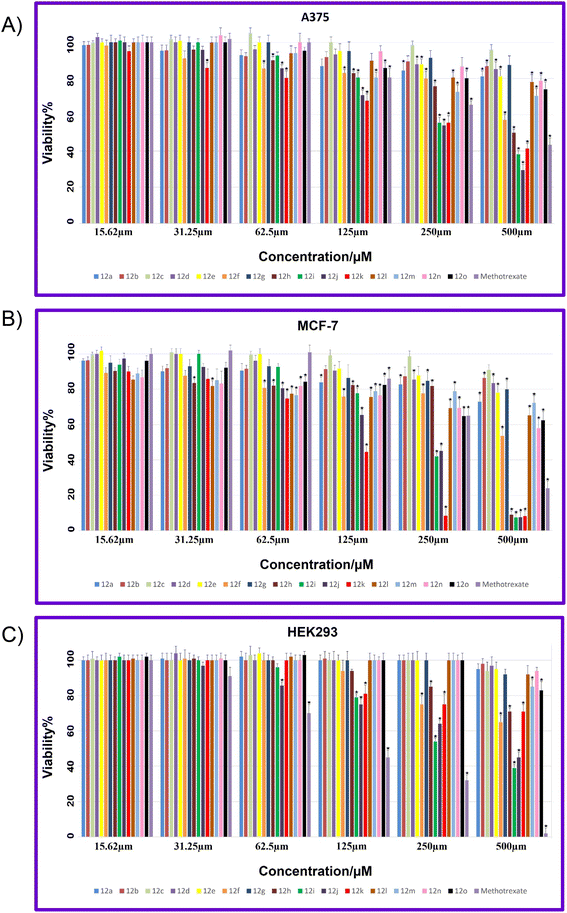
The viability percentages for examined cell lines at different concentrations (15.62, 31.25, 62.5, 125, 250, and 500, μM) of 12a–12o and MTX are shown in Fig. 6A–C. A p-value less than 0.05 (typically ≤0.05) is statistically significant that shown with stars in Fig. 6A–C. As it is indicated in Fig. 6A, for A375, the lowest viability values and hence the highest toxicity are related to 12j compared to all synthesized compounds. Also, 12i and 12k demonstrated lower viability and higher toxicity against A-375 compared to MTX as a reference drug. It is worth mentioning that 12f demonstrated a considerable decrease in viability at different concentrations while 12f displayed non-toxicity against the A375 cell line. In the case of MCF-7, the highest toxicity with the lowest viability are related to 12h, 12i, 12j, and 12k compared to all tested compounds (Fig. 1B). In general, all compounds showed lowest viability on MCF-7 cancer cells than A-375.
Fig. 6C showed the viability percentage for normal cell line HEK-293. As it is displayed, for HEK-293, the highest toxicity with the lowest viability was related to MTX at all examined concentrations. Two potent 12i and 12j involved the viability higher than MTX, in particular at higher concentrations that indicates the notable selectivity of 12i and 12j against HEK-293.
2.3. Structural activity relationship (SAR) analysis
As shown in Fig. 5, all title compounds are common in caffeinyl, piperazinyl, and 1,2,3-triazolyl methyl moieties, and their differences in the anticancer activity must be attributed to the substituents linked to the N1 atom of 1,2,3-triazolyl residue. In this context, compounds 12a–12o can be categorized into four groups. The first group involves simple aryl-alkyl substituents comprising 12a and 12c–12f in which due to Table 1, none of them displayed toxicity against both tested cell lines contrary to our expectations. 12b and 12g are the second group that includes alkyl-ester moieties without toxicity against cancer cell lines. The third group (12l–12o) consisting alkyl-imide moieties in which imides like phthalimide and saccharine are linked to ω-carbon of different alkyl chains. As indicated, none of them displayed toxicity against both cell lines. The fourth group contains normal alkyl chain substituents. Surprisingly, 12h–12k are the most potent compounds against both cancer cell lines. They only vary in the length of carbon chains. While 12j with octyl moiety is the most potent compound against A375; however, 12k with decanyl residue is the most potent agent against MCF-7. Hence, the presence of substituents particularly normal alkyl is critically essential to exhibit anticancer activity. Eventually, it can be determined that lipophilicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) and molecular topology are two main potential parameters that affect the anticancer trend in title compounds.
P) and molecular topology are two main potential parameters that affect the anticancer trend in title compounds.
2.4. Plausible mechanism of anticancer activity
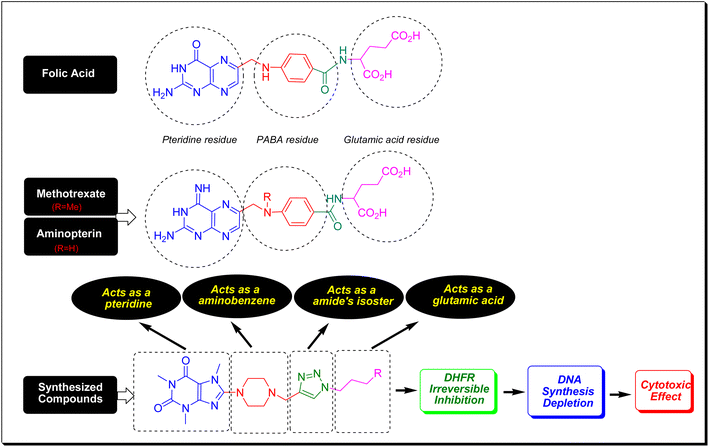
Folic acid or folacin is an aqueous vitamin so-called vitamin B9 which plays a crucial role in the biosynthesis of DNA, the modification of DNA and RNA, the synthesis of methionine, and diverse chemical reactions that happened in cellular metabolism. In fact, folic acid is a precursor for the biosynthesis of tetrahydrofolate (coenzyme F) through the catalytic action of dihydrofolate reductase (DHFR).40 By a one-carbon transfer metabolism, and the enzymatic role of thymidylate synthase (TS), coenzyme F transfers its methyl to deoxyuridine monophosphate (dUMP) and converts it to deoxythymidine monophosphate (dTMP). The dTMP is critically essential for DNA synthesis and cell replication.41 In this regard, the inhibition of DHFR prevents coenzyme F synthesis and hence blocks DNA synthesis and all biochemical events that pertained to DNA. Methotrexate (MTX) and aminopterin have a very close structural feature to folic acid (Fig. 7). Same as folic acid, MTX and aminopterin have been composed of three molecular fragments consisting of pteridine, p-aminobenzoic acid (PABA), and glutamic acid. They merely vary in pteridine core and this difference is responsible for the inhibition of DHFR.42 The key pharmacophoric site in folic acid, MTX, and aminopterin is the pteridine core which closely resembles xanthine alkaloids. As can be seen in Fig. 7, the compounds have conceptual similarities to folic acid, MTX, and aminopterin in which caffeinyl can be considered as a successor of pteridine, piperazinyl as aminobenzene, 1,2,3-triazolyl as a isostere of amide that can mimic the PABA-amide part and finally the alkyl substituents as a glutamic acid residue. Thus, owing to structural similarities of title compounds with MTX and aminopterin, it is proposed they potentially inhibit DHFR enzyme and confine the biosynthesis of DNA (Fig. 7).2.5. In silico studies
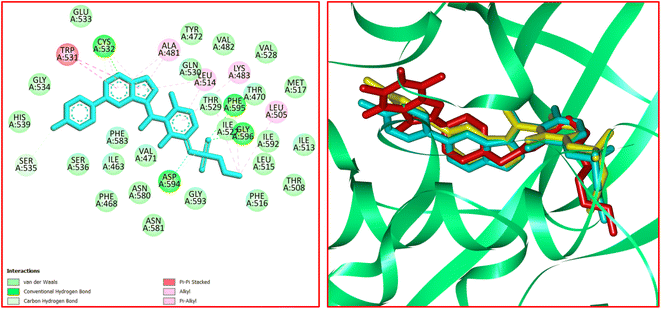
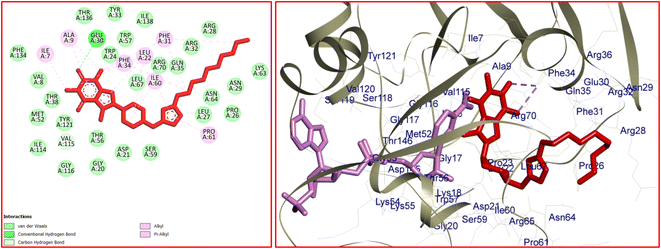
The significant interactions for stabilization of vemurafenib in the active site of B-RAF V600E kinase enzyme include π–alkyl, π–π*, van der Waals, hydrophobic, and hydrogen bond interactions.46 A π–π* stacking interaction was created between the heterocyclic ring of vemurafenib and Trp531. Four conventional hydrogen bonds were observed between vemurafenib and the residues of Cys532, Asp594, Phe595, and Gly596. Several carbon–hydrogen bonds were detected between vemurafenib and the residues of Cys532, Ser535, and Asp594. Additionally, the side chain of Ala481, Lys483, Leu514, Cys532, and Phe595 participated in π–alkyl interaction with the vemurafenib. The hydrophobic and van der Waals interactions of vemurafenib were detected with the side chains of Ile463, Thr470, Val471, Leu515, Phe516, Ile527, Gln530, Cys532, Gly534, Ser536, His539, Phe583, Ile592, and Gly593. The docked conformation of vemurafenib and the overlay view of conformations of co-crystallized vemurafenib, re-docked vemurafenib, and 12j in the active site of an enzyme is depicted in Fig. 8. Delightfully, 12j binds to the identical site similar to vemurafenib and exhibits robust interaction with the B-RAF V600E enzyme as illustrated in Fig. 8.
For 12j, the analysis of the docking protocol revealed that Phe583 is participated in both π–sigma and π–π interactions with piperazinyl residue and a five-membered ring of caffeine core, respectively. A conventional hydrogen bond was detected between Ser536 and O(6) of caffeine residue. In addition, three carbon–hydrogen bonds were observed between the 8-PC moiety of 12j and the residues of Cys532, Gly534, and Ser535. The residues of Ala481, Leu514, Trp531, Phe583, and Phe595 were involved in π–alkyl interactions with 12j. The hydrophobic and van der Waals interactions of 12j are similar to those observed for vemurafenib since both compounds were in the same binding pocket. The two-dimensional (2D) docked conformation of 12j and its three-dimensional (3D) docked conformation including hydrogen bonds are presented in Fig. 9. The ΔG values for vemurafenib and 12j were also calculated which are equal to −187.29 and −163.51 kcal mol−1, respectively. The total hydrogen bond energies for vemurafenib and 12j were −8.91 and −10.52 (kcal mol−1), respectively. These results indicated that hydrogen bonding interactions play a significant role in the stabilization of 12j in the binding site of the B-RAF V600E kinase enzyme.
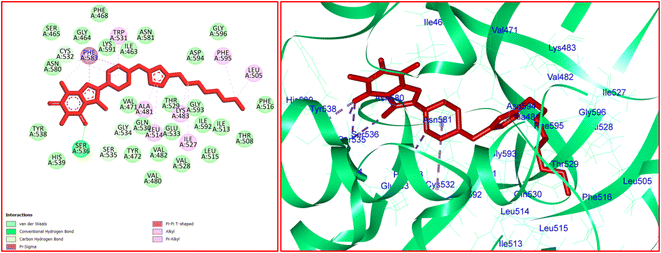 | ||
| Fig. 9 The 2D- [left] and 3D- [right] docked conformations of 12j in the active site of B-RAF V600E kinase. | ||
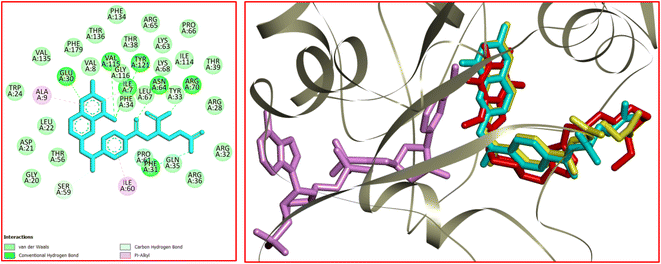
Dihydrofolate reductase (DHFR) plays a crucial role in the metabolism of folic acid and the biosynthesis of purines, thymidylate, and glycine.47 DHFR enzyme proves to be an important target to treat cancer. In this connection, the interactions, affinity, binding mode, and orientation of 12k were studied in the active site of human DHFR using MVD 6.0 software. The 3D structure of human DHFR (hDHFR) in complex with methotrexate (MTX) and nicotinamide adenine dinucleotide phosphate (NADPH) was received from RSCB (PDB code: 1U72). The analysis of the results was achieved using Discovery Studio 2021 software. The docking protocol was validated via re-docking of MTX in the active site of hDHFR. The RMSD value was equal to 0.71 Å which is desirable to continue the docking process. The analysis of the docking results revealed that H-bond interactions play a key role in the stabilization of MTX in the active site of hDHFR. Accordingly, seven strong hydrogen bonds were detected between MTX and the residues of Ile7, Glu30, Phe31, Asn64, Arg70, Val115, and Tyr121. The residue of Ser59 was involved in a carbon–hydrogen bond with the N–Me residue of MTX. Ile7 and Ala9 participated in π–alkyl interaction with the pteridine core of MTX. In addition, a π–alkyl interaction was formed between the residue of Ile60 and the aryl ring of MTX. The hydrophobic and van der Waals interactions of MTX were detected in a hydrophobic pocket consisting of the side chains of Ile7, Val8, Leu22, Phe31, Tyr33, Phe34, Gln35, Thr56, Ser59, pro61, Leu67, Lys68, Val115, and Gly116. The docked conformation of MTX and the overlay view of conformations of co-crystallized MTX, re-docked MTX, and 12k in the active site of an enzyme are depicted in Fig. 10. Delightfully, 12k and MTX bind to the same binding site and display potent interactions with the hDHFR enzyme as shown in Fig. 10.
In the case of 12k, the O(2) of caffeine core was involved in a conventional hydrogen bond interaction with Gln30. Two carbon–hydrogen bonds were detected between the N(3)–Me and N(7)–Me of caffeine core with Glu30 and Val115, respectively. In addition, four π–alkyl interactions were observed between 12k and the residues of Ala9, Leu22, Ile60, and Pro61. The van der Waals and hydrophobic interactions of 12k and MTX were similar as they were located in identical binding site. The 2D and 3D conformations of 12k are displayed in Fig. 11. The ΔG values for MTX and 12k were also calculated which are equal to −234.59 and −200.49 kcal mol−1, respectively. The total hydrogen bond energies for MTX and 12k were −7.15 and −3.61 (kcal mol−1), respectively. According to the results of the docking study, the stabilization of MTX and 12k in the binding site of the hDHFR enzyme could be facilitated by the strong hydrogen bond interactions.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) value ≤ 5. The in silico pharmacokinetic and physiochemical profiles of 12a–12o and MTX were evaluated using the preADMET online server,50 OSIRIS Property Explorer,51 and SwissADME online software.52 The outcomes of these assessments are presented in Tables 2 and 3.
P) value ≤ 5. The in silico pharmacokinetic and physiochemical profiles of 12a–12o and MTX were evaluated using the preADMET online server,50 OSIRIS Property Explorer,51 and SwissADME online software.52 The outcomes of these assessments are presented in Tables 2 and 3.
| Compd | Mwa | nRBb | nHBDc | nHBAd | c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P e P e |
TPSAf | Drug-likeness | Drug score |
|---|---|---|---|---|---|---|---|---|
a Molecular weight.b Number of rotatable bonds.c Number of hydrogen bond donors.d Number of hydrogen bond acceptors.e Logarithm of octanol–water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P).f Topological polar surface area (Å2).g Methotrexate. P).f Topological polar surface area (Å2).g Methotrexate. |
||||||||
| 12a | 449.51 | 5 | 0 | 6 | 0.51 | 95.63 | 2.80 | 0.78 |
| 12b | 507.54 | 8 | 0 | 8 | −0.02 | 121.90 | −10.72 | 0.36 |
| 12c | 525.60 | 6 | 0 | 6 | 2.30 | 95.63 | 3.00 | 0.44 |
| 12d | 477.56 | 7 | 0 | 6 | 1.30 | 95.63 | 1.95 | 0.70 |
| 12e | 463.54 | 6 | 0 | 6 | 0.85 | 95.63 | 3.10 | 0.76 |
| 12f | 463.54 | 5 | 0 | 6 | 0.85 | 95.63 | 1.57 | 0.71 |
| 12g | 473.53 | 9 | 0 | 8 | −0.13 | 121.90 | −11.69 | 0.39 |
| 12h | 457.57 | 9 | 0 | 6 | 1.68 | 95.63 | −12.12 | 0.38 |
| 12i | 443.55 | 8 | 0 | 6 | 1.22 | 95.63 | −6.98 | 0.40 |
| 12j | 471.60 | 10 | 0 | 6 | 2.13 | 95.63 | −14.99 | 0.37 |
| 12k | 499.65 | 12 | 0 | 6 | 3.04 | 95.63 | −14.99 | 0.33 |
| 12l | 574.63 | 9 | 0 | 8 | 1.19 | 133.00 | −10.66 | 0.30 |
| 12m | 596.66 | 8 | 0 | 9 | 0.53 | 158.40 | 2.49 | 0.58 |
| 12n | 582.63 | 7 | 0 | 9 | 0.07 | 158.40 | 3.63 | 0.61 |
| 12o | 610.69 | 9 | 0 | 9 | 0.98 | 158.40 | 0.61 | 0.48 |
| MTXg | 454.44 | 10 | 5 | 9 | 1.23 | 210.54 | −7.09 | 0.22 |
| Compd | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S S |
HIAa | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kpb Kpb |
P-gpc | BBBd | CYP2D6 | CYP3A4 | Carcino.e | hERG inhibitionf |
|---|---|---|---|---|---|---|---|---|---|
a Human intestinal absorption (%).b Skin permeability (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp, cm h−1).c P-glycoprotein.d Blood–brain barrier permeability (C. brain/C. blood).e Carcinogenicity (mouse).f Human ether-a-go-go related gene channel.g Methotrexate. S = substrate. WS = weak-substrate. NS = non-substrate. I = inhibitor. NI = non-inhibitor. NC = non-carcinogenic. Kp, cm h−1).c P-glycoprotein.d Blood–brain barrier permeability (C. brain/C. blood).e Carcinogenicity (mouse).f Human ether-a-go-go related gene channel.g Methotrexate. S = substrate. WS = weak-substrate. NS = non-substrate. I = inhibitor. NI = non-inhibitor. NC = non-carcinogenic. |
|||||||||
| a | −3.21 | 99.63 | −4.41 | I/S | 0.10 | NI/WS | NI/S | NC | Medium |
| b | −2.86 | 98.88 | −4.43 | I/S | 0.07 | NI/WS | NI/S | NC | Medium |
| c | −4.14 | 98.29 | −3.55 | I/S | 0.21 | NI/NS | I/S | NC | Medium |
| d | −3.06 | 99.55 | −4.26 | I/S | 0.12 | NI/WS | NI/S | NC | Medium |
| e | −2.69 | 99.61 | −4.35 | I/S | 0.11 | NI/WS | NI/S | NC | Medium |
| f | −2.76 | 99.61 | −4.44 | I/S | 0.11 | NI/WS | NI/S | NC | Medium |
| g | −1.78 | 95.54 | −4.72 | I/S | 0.06 | NI/WS | NI/S | NC | Medium |
| h | −3.62 | 99.28 | −4.48 | I/S | 0.07 | NI/WS | NI/WS | NC | Medium |
| i | −3.08 | 99.06 | −4.54 | I/S | 0.08 | NI/WS | NI/WS | NC | Medium |
| j | −3.97 | 99.44 | −4.42 | I/NS | 0.07 | NI/WS | NI/WS | NC | Medium |
| k | −4.68 | 99.62 | −4.26 | I/NS | 0.04 | NI/WS | NI/WS | NC | Medium |
| l | −3.26 | 99.88 | −4.70 | I/S | 0.08 | NI/WS | NI/S | NC | Medium |
| m | −3.10 | 98.10 | −3.88 | NI/S | 0.05 | NI/WS | NI/S | NC | Medium |
| n | −2.73 | 97.69 | −4.05 | NI/S | 0.05 | NI/WS | NI/S | NC | Medium |
| o | −3.47 | 98.47 | −3.71 | I/S | 0.06 | NI/WS | I/S | NC | Medium |
| MTXg | −1.19 | 36.61 | −4.63 | NI/S | 0.04 | NI/NS | I/WS | NC | High |
As depicted in Table 2, the molecular weights of MTX and the synthesized compounds except that of 12b, 12c, and 12l–12o are less than 500 Dalton. According to the obtained results, the number of rotatable bonds (nRB) for MTX and the synthesized compounds except that of 12k is between 5–10. In addition, MTX and 12a–12o as both hydrogen bond acceptors (nHBA) and hydrogen bond donors (nHBD) are obeyed from RO5. In RO5, the lipophilicity factor is a noteworthy property of a molecule that is specified by cLogP. The cLogP for MTX and 12a–12o are less than 5 which conforms to RO5. The total polar surface area (TPSA) is an adequate factor to determine the drug transport character. TPSA is calculated by the summation of surfaces of polar atoms in a molecule which is usually less than 140 Å2 for approved drugs. The TPSA contents of MTX and 12m–12o are higher than 140 Å2. Therefore, the synthesized compounds except that of 12m–12o might have suitable membrane permeability or oral absorption. The drug likenesses of synthesized derivatives were computed with OSIRIS Property Explorer software which found the negative values for 12b and 12g–12l as same as MTX. Based on the aforementioned factors, the software could predict another parameter known as the drug score. According to the obtained results, the maximum and minimum drug score values of title compounds belonged to 12a and 12l, respectively, which are higher than that of MTX in all cases. Overall, compounds 12j and 12k could be considered promising for extra research.
The pharmacokinetic profile of 12a–12o and MTX including absorption, distribution, metabolism, and toxicity properties were also measured (Table 3). The analyses were focused on the assessment of P-glycoprotein (P-gp) substrate and/or inhibitor, human intestinal absorption (HIA), aqueous solubility (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S), and skin permeability (log
S), and skin permeability (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp) to predict the absorption profile of the compounds. log
Kp) to predict the absorption profile of the compounds. log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S is >–4 for most of the traded drugs. As it was calculated, MTX and all compounds except 12c and 12k are soluble in water. The moderate water solubility was predicted for 12c and 12k. The HIA factor was calculated to be more than 95% for 12a–12o, while it is 36.61% for MTX. Thus, the intestinal absorption of 12a–12o would be ideal. Transdermal delivery is a significant issue in drug delivery. It is also famous for skin permeability which describes the importance of drug absorption through the skin. The computed data revealed that the transdermal delivery of 12a–12o and MTX are similar. The drug transporter P-glycoprotein (P-gp) is an essential carrier for excreting the substrates from the cells. While the substrates of P-gp could diminish the drug's bioavailability but the P-gp inhibitors could increase its bioavailability. Among compounds, 12j and 12K are non-substrates of P-gp that may have desirable bioavailability. While MTX, 12m, and 12n are P-gp non-inhibitors; however, other derivatives are inhibitors of P-gp which may exhibit appropriate bioavailability. The distribution of a drug between the brain (the most lipophilic) and the blood (the most hydrophilic) tissues is identified by a parameter which is known as blood–brain barrier (BBB) permeability. The data in Table 3 revealed that 12a–12o and MTX have low BBB permeability. Thus, these compounds may not cause neurotoxicity since they could not cross the blood–brain barrier. Among cytochrome P450 enzymes, CYP2D6 and CYP3A4 enzymes are remarkably important for the metabolism of drugs. All compounds are non-inhibitors of CYP2D6. Except MTX, all entries are substrates for CYP2D6. MTX and 12a–12o are substrates of CYP3A4 enzyme. All compounds except 12c and 12o are non-inhibitors of the CYP3A4. Therefore, the metabolism of the synthesized derivatives could be achieved in the liver. According to the computed data shown in Table 3, 12a–12o and MTX are non-carcinogenic in mice. Human ether-a-go-go related gene channel (hERG) inhibitors are potential factors that initiate lengthening the QT and cardiac side effects. It is predicted that MTX could display a high risk of hERG inhibition while 12a–12o showed the medium risk of hERG inhibitory activity. Given the predicted data shown in Tables 2 and 3 and also the biological assay results, 12j and 12k could be considered as drug candidates for further studies.
S is >–4 for most of the traded drugs. As it was calculated, MTX and all compounds except 12c and 12k are soluble in water. The moderate water solubility was predicted for 12c and 12k. The HIA factor was calculated to be more than 95% for 12a–12o, while it is 36.61% for MTX. Thus, the intestinal absorption of 12a–12o would be ideal. Transdermal delivery is a significant issue in drug delivery. It is also famous for skin permeability which describes the importance of drug absorption through the skin. The computed data revealed that the transdermal delivery of 12a–12o and MTX are similar. The drug transporter P-glycoprotein (P-gp) is an essential carrier for excreting the substrates from the cells. While the substrates of P-gp could diminish the drug's bioavailability but the P-gp inhibitors could increase its bioavailability. Among compounds, 12j and 12K are non-substrates of P-gp that may have desirable bioavailability. While MTX, 12m, and 12n are P-gp non-inhibitors; however, other derivatives are inhibitors of P-gp which may exhibit appropriate bioavailability. The distribution of a drug between the brain (the most lipophilic) and the blood (the most hydrophilic) tissues is identified by a parameter which is known as blood–brain barrier (BBB) permeability. The data in Table 3 revealed that 12a–12o and MTX have low BBB permeability. Thus, these compounds may not cause neurotoxicity since they could not cross the blood–brain barrier. Among cytochrome P450 enzymes, CYP2D6 and CYP3A4 enzymes are remarkably important for the metabolism of drugs. All compounds are non-inhibitors of CYP2D6. Except MTX, all entries are substrates for CYP2D6. MTX and 12a–12o are substrates of CYP3A4 enzyme. All compounds except 12c and 12o are non-inhibitors of the CYP3A4. Therefore, the metabolism of the synthesized derivatives could be achieved in the liver. According to the computed data shown in Table 3, 12a–12o and MTX are non-carcinogenic in mice. Human ether-a-go-go related gene channel (hERG) inhibitors are potential factors that initiate lengthening the QT and cardiac side effects. It is predicted that MTX could display a high risk of hERG inhibition while 12a–12o showed the medium risk of hERG inhibitory activity. Given the predicted data shown in Tables 2 and 3 and also the biological assay results, 12j and 12k could be considered as drug candidates for further studies.
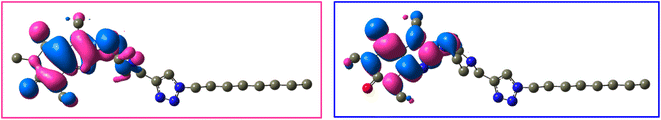
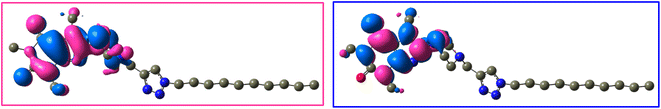
The frontier molecular orbitals (LUMO and HOMO) of 12j and 12k with the isovalue of 0.03 are shown in Fig. 12 and 13, respectively. The HOMO and LUMO energy levels of 12j are −6.866 and 0.352 ev, and those of 12k are −6.867 and 0.351 ev, respectively. On both molecules, HOMO and LUMO often distribute on the caffeine core. While the molecular orbitals lie on both carbonyl groups of caffeinyl residue in LUMO; however, one of its carbonyl groups is involved in HOMO. Also, the piperazinyl ring exhibits a significant contribution in LUMO compared to HOMO. As can be seen in Fig. 12 and 13, the triazole ring and alkyl chain have no contribution to HOMO or LUMO.
For both 12j and 12k, the total energy (Etot), enthalpy (H), Gibbs free energy (G), and molecular entropy (S) were calculated (Table 4). The Etot, H, and G were corrected by considering the zero-point energy (ZPE) calculation. The reactivity descriptors are based on the frontier orbitals and the electrical gap (E-gap) which are about 7.21 ev for 12j and 12k. The DFT analysis computed the reactivity characteristics such as softness (σ), hardness (η), electron affinity (A), and ionization energy (I). As depicted in Table 4, the reactivity descriptors for 12j and 12k have similar values which could be ascribed to the slight difference in their alkyl-chain length.
| Descriptor | 12j | 12k |
|---|---|---|
| ZPE | 16.81 | 18.37 |
| Etot | −41935.62 | −44073.06 |
| H | −41934.67 | −44072.04 |
| G | −41937.54 | −44075.09 |
| S | 926.76 | 988.94 |
| E-gap | 7.219 | 7.219 |
| σ | 0.277 | 0.277 |
| η | 3.609 | 3.610 |
| A | −0.352 | −0.351 |
| I | 6.866 | 6.867 |
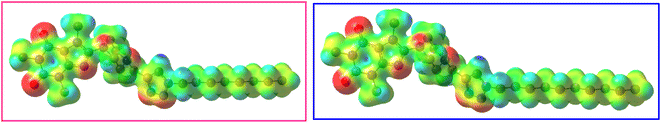
The molecular electrostatic potential (MEP) is a notable parameter to determine which regions of a molecule are susceptible to nucleophilic or electrophilic interaction with the surface of receptors or enzymes.54 In this regard, MEP of 12j and 12k were obtained using the DFT method at M06-2X/6-311G(d,p) level of theory (Fig. 14). The MEP is mapped on the total density surfaces of 12j and 12k with an isosurface value of 0.02. In Fig. 14, the regions with the positive electrostatic potentials are illustrated in blue and the atoms carrying the negative electrostatic potentials are presented in red.
3 Conclusion
In conclusion, it was explained the design, synthesis, and characterization of the new 8-caffeinyl-triazolylmethyl hybrid conjugates. These compounds were in vitro tested against two significant cancer cell lines comprising breast cancer MCF-7 (ATCC HTB-22) and melanoma cancer A-375 (ATCC CRL-1619) and their anticancer activity was compared with methotrexate (MTX) as a reference drug. Results have determined that 12i (IC50 = 371 ± 2.5), 12j (IC50 = 323 ± 2.6) and 12k (IC50 = 367 ± 3.4) displayed apparent inhibition effects on A-375 cell line growth and their toxicities are even stronger than MTX (IC50 = 418 ± 2). In the same circumstances, 12h (IC50 = 306 ± 1.8), 12i (IC50 = 260 ± 2.2), 12j (IC50 = 245 ± 2), and 12k (IC50 = 175 ± 3.2) demonstrated potent activity against MCF-7 cell growth and their activity are even stronger than MTX (IC50 = 343 ± 3.6). To recognize the toxicity, compounds were screened against normal cell line HEK-293 (ATCC CRL-11268) in which specified that except 12i (IC50 = 371 ± 2.3), 12j (IC50 = 418 ± 2.4), and MTX (IC50 = 199 ± 2.4), all compounds are non-toxic. The viability assessments have indicated that the lowest viability values were related to 12i–12k (for A375), 12h–12k (for MCF7), and MTX (for HEK-293) at various concentrations. Docking results have determined that 12j and 12k have strong binding affinities to B-RAF kinase and hDHFR enzymes, respectively. The in silico pharmacokinetic and physiochemical profiles of tested compounds were assessed using preADMET online server, OSIRIS Property Explorer, and SwissADME online software which indicated that most compounds obeyed Lipinski's rule of five (RO5). The DFT study on M06-2X/6-311G (d,p) determined that HOMO and LUMO often distribute on the caffeine core of 12j and 12k and like pteridine moiety in MTX is the main residue for interaction with the active site of the enzyme. Ultimately, regarding anticancer and in silico data, 12j and 12k can be considered as future drug candidates.4 Experimental
4.1. Materials and cell lines
Cancer cell lines including breast cancer line MCF-7 (ATCC HTB-22), melanoma cell line A-375 (ATCC CRL-1619), and normal cell line HEK-293 (ATCC CRL-11268) supplied from Pasteur Institute of Iran. Fetal bovine serum (FBS) was purchased from Gibco (USA). Phosphate-buffered saline (PBS) tablets, tetrazolium salt and 3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyltetrazo-lium bromide (MTT) were obtained from Sigma-Aldrich (USA). Dimethyl sulfoxide (DMSO), trypsin, penicillin or streptomycin, and Dulbecco's Modified Eagle's Media (DMEM) cell culture medium were supplied from Shellmax (China).4.2. Evaluation of anticancer activity
MTT assay was used for evaluating the anticancer activity of title compounds and MTX.55 All compounds were dissolved in PBS solution containing 0.5% DMSO; required dilution was prepared using the same solvent.The cell lines containing (MCF-7, A-375, and HEK-293) were cultured in culture flasks in DMEM complete medium (containing FBS 10% and penicillin-streptomycin 1%) and incubated at 37 °C under CO2 (5%) and air (95%), separately. Trypsin was applied for separation of various cell lines; then, they were seeded in 96-well plates and incubated for 48 h. The culture media was discarded, and a 75 μL complete fresh medium was added to each well. After adding the sample's desired amount, concentrations were set at 15.62, 31.25, 62.5, 125, 250, and 500 μM. After incubation (48 h, 37 °C, air 95%, CO2 5%), the plates' content was discarded, and washed with 100 μL PBS. Then, 100 μL MTT solution (0.5 mg.mL−1) was added to each well and was incubated for another 4 h. Next, 100 μL per well DMSO was added to dissolve formed formazan crystals. In each plate, six wells were considered the control group in each plate, filled with PBS solution containing 0.5% DMSO (25 μL) and DMEM containing the cells (75 μL). Finally, the absorbance (A) of each well was measured at 570 nm using a plate reader (ELISA Plate Reader). The cell viability at each concentration was calculated by eqn (1).
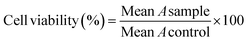 | (1) |
4.3. Molecular docking studies procedure
The molecular docking study was performed using Molegro Virtual Docker (MVD 6.0) software with its default parameters.44 To do the molecular docking study, the crystal structures of B-RAF V600E in complex with vemurafenib (PDB code: 3OG7) and human DHFR in complex with MTX and NADPH (PDB code: 1U72) were selected and obtained from the RCSB (http://www.rcsb.org). The structure of the enzyme was modified by eliminating all water molecules and ligands, compensating the missing hydrogen atoms, and defining the correct bond and atom types. In the case of B-RAF V600E, chain B was removed and the docking study was continued with importing of chain A to the software. In the case of hDHFR, the docking study was achieved in the presence of NADPH. Subsequently, the docking study was carried out based on 40 independent runs. The active site of the target enzyme was considered to include the residues of amino acids in a 7 Å radius around the inhibitor ligand. The structures of vemurafenib, methotrexate, 12j, and 12k were optimized using the DFT method at M06-2X/6-311G(d,p) level of theory with Gaussian 09 program package.56 Firstly, the docking process was endorsed via re-docking of vemurafenib and methotrexate in the active site of B-RAF V600E and XXX, respectively. Then, the study was extended by importing the optimized structure of the related ligands 12j and 12k to the software. All interactions were visualized and evaluated on the basis of the docking results using the Discovery Studio 2021 visualizer.4.4. Chemistry general
All chemicals were purchased from Merck and/or other chemical vendors and straightforwardly applied without further purifications. The reactions were monitored by TLC using SILG/UV 254 silica-gel plates. The purifications by a short column chromatography were done on the silica gel 60 (0.063–0.200 mm, 70–230 mesh; ASTM). The melting points are measured on the Electrothermal IA 9000 in open capillary tubes and they are uncorrected. The PerkinElmer 240-B micro-analyzer, Shimadzu GC/MS-QP 1000-EX apparatus (m/z; rel.%), and the Shimadzu FT-IR-8300 spectrophotometer were used to obtain the elemental analyses, GC/MS, and IR spectra, respectively. 1H- and 13C-NMR spectrum was recorded on the Brüker Avance-DPX-300 spectrometer operating at 300/75 MHz, respectively. Chemical shifts are given in δ relative to tetramethylsilane (TMS) as an internal standard and coupling constants J are given in Hz. The abbreviations used for 1H-NMR signals are s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, br = broad and etc.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (30 mL) in a round bottom flask (100 mL) was refluxed for 3 h (TLC control). Afterward, acetone was evaporated at reduced pressure, and then, the crude product was dissolved in CHCl3 (150 mL) and washed with H2O (2 × 150 mL). The organic layer was dried (Na2SO4, 10 g) and concentrated to afford the crude product which was purified by column chromatography eluting with EtOAc-n-hexane.57
1 (30 mL) in a round bottom flask (100 mL) was refluxed for 3 h (TLC control). Afterward, acetone was evaporated at reduced pressure, and then, the crude product was dissolved in CHCl3 (150 mL) and washed with H2O (2 × 150 mL). The organic layer was dried (Na2SO4, 10 g) and concentrated to afford the crude product which was purified by column chromatography eluting with EtOAc-n-hexane.57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): 0.15, 1H NMR (CDCl3, 400 MHz): δppm = 2.58–2.61 (m, 4H, 2NCH2), 2.89 (s, 1H, CH), 3.38 (s, 3H, N(1)–CH3), 3.50–3.52 (complex, 7H, 2NCH2, N(3)–CH3), 3.57 (s, 2H, NCH2), 3.72 (s, 3H, N(7)–CH3). 13C NMR (CDCl3, 100 MHz): δppm = 28.19, 29.90, 32.41, 44.74, 50.07, 52.60, 76.09, 79.11, 107.10, 149.70, 152.29, 155.42, 166.52. IR (KBr): 3215, 2948, 2190, 1697, 1601, 1518 cm−1. MS (EI): m/z (%) = 316 (18.4) [M+]. Anal. calc. for C15H20N6O2: C, 56.95; H, 6.37; N, 26.56; found: C, 56.80; H, 6.49; N, 26.73.
1): 0.15, 1H NMR (CDCl3, 400 MHz): δppm = 2.58–2.61 (m, 4H, 2NCH2), 2.89 (s, 1H, CH), 3.38 (s, 3H, N(1)–CH3), 3.50–3.52 (complex, 7H, 2NCH2, N(3)–CH3), 3.57 (s, 2H, NCH2), 3.72 (s, 3H, N(7)–CH3). 13C NMR (CDCl3, 100 MHz): δppm = 28.19, 29.90, 32.41, 44.74, 50.07, 52.60, 76.09, 79.11, 107.10, 149.70, 152.29, 155.42, 166.52. IR (KBr): 3215, 2948, 2190, 1697, 1601, 1518 cm−1. MS (EI): m/z (%) = 316 (18.4) [M+]. Anal. calc. for C15H20N6O2: C, 56.95; H, 6.37; N, 26.56; found: C, 56.80; H, 6.49; N, 26.73.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 V/V, 30 mL). The reaction mixture was stirred at room temperature till TLC indicated no further progress of the reaction (Fig. 5). Afterward, the solvent was evaporated and the remaining was dissolved in CHCl3 (200 mL) and washed with water (2 × 100 mL). The organic layer was dried on Na2SO4 (10 g) and evaporated in vacuo. The product was purified by recrystallization and/or short column chromatography on silica gel eluting with proper solvents as described below.
2 V/V, 30 mL). The reaction mixture was stirred at room temperature till TLC indicated no further progress of the reaction (Fig. 5). Afterward, the solvent was evaporated and the remaining was dissolved in CHCl3 (200 mL) and washed with water (2 × 100 mL). The organic layer was dried on Na2SO4 (10 g) and evaporated in vacuo. The product was purified by recrystallization and/or short column chromatography on silica gel eluting with proper solvents as described below.4.4.7.1 8-(4-((1-Benzyl-1H-1,2,3-triazol-4-yl)methyl)piperazin-1-yl)-1,3,7-trimethyl-3,7-dihydro-1H-purine-2,6-dione (12a). Recrystallization with EtOAc/MeOH (1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) afforded the pure product as white solid (4.17 g, 93%); m.p.: 150–152 °C, Rf (EtOAc/MeOH, 4
3) afforded the pure product as white solid (4.17 g, 93%); m.p.: 150–152 °C, Rf (EtOAc/MeOH, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): 0.53, 1H NMR (CDCl3, 400 MHz): δppm = 2.64 (t, J = 4.4 Hz, 4H, 2NCH2), 3.26 (t, J = 4.4 Hz, 4H, 2NCH2), 3.36 (s, 3H, N(1)–CH3), 3.49 (s, 3H, N(3)–CH3), 3.71 (complex, 5H, N(7)–CH3, NCH2C
1): 0.53, 1H NMR (CDCl3, 400 MHz): δppm = 2.64 (t, J = 4.4 Hz, 4H, 2NCH2), 3.26 (t, J = 4.4 Hz, 4H, 2NCH2), 3.36 (s, 3H, N(1)–CH3), 3.49 (s, 3H, N(3)–CH3), 3.71 (complex, 5H, N(7)–CH3, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 5.52 (s, 2H, NCH2Ph), 7.26–7.28 (m, 3H, aryl), 7.36–7.37 (m, 2H, aryl), 7.42 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 27.73, 29.68, 32.57, 49.47, 52.25, 53.26, 54.17, 105.34, 122.58, 128.08, 128.78, 129.13, 134.59, 144.48, 147.41, 151.72, 154.90, 156.24. IR (KBr): 3056, 2983, 2851, 1697, 1658, 1610, 1523, 1468, 1146 cm−1. MS (EI): m/z (%) = 449 (23.7) [M+]. Anal. calc. for C22H27N9O2: C, 58.78; H, 6.05; N, 28.04; found: C, 58.97; H, 6.23; N, 28.19.
C), 5.52 (s, 2H, NCH2Ph), 7.26–7.28 (m, 3H, aryl), 7.36–7.37 (m, 2H, aryl), 7.42 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 27.73, 29.68, 32.57, 49.47, 52.25, 53.26, 54.17, 105.34, 122.58, 128.08, 128.78, 129.13, 134.59, 144.48, 147.41, 151.72, 154.90, 156.24. IR (KBr): 3056, 2983, 2851, 1697, 1658, 1610, 1523, 1468, 1146 cm−1. MS (EI): m/z (%) = 449 (23.7) [M+]. Anal. calc. for C22H27N9O2: C, 58.78; H, 6.05; N, 28.04; found: C, 58.97; H, 6.23; N, 28.19.
4.4.7.2 Benzyl 2-(4-((4-(1,3,7-trimethyl-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-8-yl)piperazin-1-yl)methyl)-1H-1,2,3-triazol-1-yl)acetate (12b). Recrystallization with EtOAc afforded the pure product as white solid (4.56 g, 90%); m.p.: 183–185 °C, Rf (EtOAc/MeOH, 4
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): 0.54, 1H NMR (CDCl3, 400 MHz): δppm = 2.67 (br s, 4H, 2NCH2), 3.29 (br s, 4H, 2NCH2), 3.39 (s, 3H, N(1)–CH3), 3.53 (s, 3H, N(3)–CH3), 3.73 (s, 3H, N(7)–CH3), 3.79 (s, 2H, NCH2C
1): 0.54, 1H NMR (CDCl3, 400 MHz): δppm = 2.67 (br s, 4H, 2NCH2), 3.29 (br s, 4H, 2NCH2), 3.39 (s, 3H, N(1)–CH3), 3.53 (s, 3H, N(3)–CH3), 3.73 (s, 3H, N(7)–CH3), 3.79 (s, 2H, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 5.23 (s, 2H, NCH2CO2), 5.24 (s, 2H, CH2Ph), 7.34–7.38 (m, 5H, aryl), 7.65 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 27.75, 29.70, 32.59, 49.53, 50.84, 52.19, 53.19, 68.08, 105.37, 124.11, 128.59, 128.77, 128.90, 134.44, 144.64, 147.45, 151.76, 154.95, 156.28, 166.19. IR (KBr): 3070, 2989, 2846, 1715, 1698, 1663, 1607, 1527, 1454, 1142 cm−1. MS (EI): m/z (%) = 527 (25.9) [M+]. Anal. calc. for C24H29N9O4: C, 56.79; H, 5.76; N, 24.84; found: C, 56.60; H, 5.98; N, 25.01.
C), 5.23 (s, 2H, NCH2CO2), 5.24 (s, 2H, CH2Ph), 7.34–7.38 (m, 5H, aryl), 7.65 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 27.75, 29.70, 32.59, 49.53, 50.84, 52.19, 53.19, 68.08, 105.37, 124.11, 128.59, 128.77, 128.90, 134.44, 144.64, 147.45, 151.76, 154.95, 156.28, 166.19. IR (KBr): 3070, 2989, 2846, 1715, 1698, 1663, 1607, 1527, 1454, 1142 cm−1. MS (EI): m/z (%) = 527 (25.9) [M+]. Anal. calc. for C24H29N9O4: C, 56.79; H, 5.76; N, 24.84; found: C, 56.60; H, 5.98; N, 25.01.
4.4.7.3 8-(4-((1-Benzhydryl-1H-1,2,3-triazol-4-yl)methyl)piperazin-1-yl)-1,3,7-trimethyl-3,7-dihydro-1H-purine-2,6-dione (12c). Column chromatography on silica gel eluted with EtOAc afforded the pure product as pale yellow foam (4.51 g, 86%), Rf (EtOAc/MeOH, 4
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): 0.77, 1H NMR (DMSO-d6, 400 MHz): δppm = 2.56 (br s, 4H, 2NCH2), 3.14 (s, 3H, N(1)–CH3), 3.21 (s, 4H, 2NCH2), 3.29 (s, 3H, N(3)–CH3), 3.39 (s, 2H, NCH2C
1): 0.77, 1H NMR (DMSO-d6, 400 MHz): δppm = 2.56 (br s, 4H, 2NCH2), 3.14 (s, 3H, N(1)–CH3), 3.21 (s, 4H, 2NCH2), 3.29 (s, 3H, N(3)–CH3), 3.39 (s, 2H, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 3.59 (s, 3H, N(7)–CH3), 3.66 (s, 1H, NCH), 7.22–7.41 (m, 10H, aryl), 8.04 (s, 1H, C(5)–H of triazole). 13C NMR (DMSO-d6, 100 MHz): δppm = 27.25, 29.24, 32.30, 48.93, 51.60, 52.28, 66.61, 104.15, 124.09, 127.95, 128.12, 128.68, 138.85, 142.90, 146.58, 150.72, 153.64, 155.63. IR (KBr): 3028, 2991, 2838, 1695, 1650, 1608, 1520, 1475, 1127 cm−1. MS (EI): m/z (%) = 525 (29.1) [M+]. Anal. calc. for C28H31N9O2: C, 63.98; H, 5.94; N, 23.98; found: C, 64.12; H, 6.09; N, 23.80.
C), 3.59 (s, 3H, N(7)–CH3), 3.66 (s, 1H, NCH), 7.22–7.41 (m, 10H, aryl), 8.04 (s, 1H, C(5)–H of triazole). 13C NMR (DMSO-d6, 100 MHz): δppm = 27.25, 29.24, 32.30, 48.93, 51.60, 52.28, 66.61, 104.15, 124.09, 127.95, 128.12, 128.68, 138.85, 142.90, 146.58, 150.72, 153.64, 155.63. IR (KBr): 3028, 2991, 2838, 1695, 1650, 1608, 1520, 1475, 1127 cm−1. MS (EI): m/z (%) = 525 (29.1) [M+]. Anal. calc. for C28H31N9O2: C, 63.98; H, 5.94; N, 23.98; found: C, 64.12; H, 6.09; N, 23.80.
4.4.7.4 1,3,7-Trimethyl-8-(4-((1-(3-phenylpropyl)-1H-1,2,3-triazol-4-yl)methyl)piperazin-1-yl)-3,7-dihydro-1H-purine-2,6-dione (12d). Column chromatography on silica gel eluted with EtOAc afforded the pure product as white foam (3.57 g, 75%), Rf (EtOAc/MeOH, 4
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): 0.48, 1H NMR (CDCl3, 400 MHz): δppm = 2.19 (quint, J = 7.2 Hz, 2H, NCH2CH2), 2.56–2.60 (complex, 6H, 2NCH2, CH2Ph), 3.22 (br s, 4H, 2NCH2), 3.29 (s, 3H, N(1)–CH3), 3.43 (s, 3H, N(3)–CH3), 3.64–3.68 (complex, 5H, N(7)–CH3, NCH2C
1): 0.48, 1H NMR (CDCl3, 400 MHz): δppm = 2.19 (quint, J = 7.2 Hz, 2H, NCH2CH2), 2.56–2.60 (complex, 6H, 2NCH2, CH2Ph), 3.22 (br s, 4H, 2NCH2), 3.29 (s, 3H, N(1)–CH3), 3.43 (s, 3H, N(3)–CH3), 3.64–3.68 (complex, 5H, N(7)–CH3, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 4.29 (t, J = 7.2 Hz, 2H, NCH2CH2), 7.08–7.24 (m, 5H, aryl), 7.42 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 14.19, 27.74, 29.69, 31.67, 32.52, 32.57, 49.48, 52.21, 53.22, 105.36, 126.39, 128.16, 128.39, 128.63, 128.72, 140.07, 147.42, 151.73, 154.92, 156.24. IR (KBr): 3064, 2953, 2870, 1690, 1647, 1612, 1519, 1462, 1136 cm−1. MS (EI): m/z (%) = 477 (27.3) [M+]. Anal. calc. for C24H31N9O2: C, 60.36; H, 6.54; N, 26.40; found: C, 60.51; H, 6.72; N, 26.61.
C), 4.29 (t, J = 7.2 Hz, 2H, NCH2CH2), 7.08–7.24 (m, 5H, aryl), 7.42 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 14.19, 27.74, 29.69, 31.67, 32.52, 32.57, 49.48, 52.21, 53.22, 105.36, 126.39, 128.16, 128.39, 128.63, 128.72, 140.07, 147.42, 151.73, 154.92, 156.24. IR (KBr): 3064, 2953, 2870, 1690, 1647, 1612, 1519, 1462, 1136 cm−1. MS (EI): m/z (%) = 477 (27.3) [M+]. Anal. calc. for C24H31N9O2: C, 60.36; H, 6.54; N, 26.40; found: C, 60.51; H, 6.72; N, 26.61.
4.4.7.5 1,3,7-Trimethyl-8-(4-((1-phenethyl-1H-1,2,3-triazol-4-yl)methyl)piperazin-1-yl)-3,7-dihydro-1H-purine-2,6-dione (12e). Column chromatography on silica gel eluted with n-hexane/EtOAc (1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) afforded the pure product as orange foam (3.56 g, 77%), Rf (EtOAc/MeOH, 4
1) afforded the pure product as orange foam (3.56 g, 77%), Rf (EtOAc/MeOH, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): 0.47, 1H NMR (CDCl3, 400 MHz): δppm = 2.17 (br s, 2H, CH2Ph), 2.66 (br s, 4H, 2NCH2), 3.28 (br s, 4H, 2NCH2), 3.38 (s, 3H, N(1)–CH3), 3.51 (s, 3H, N(3)–CH3), 3.72–3.80 (complex, 5H, NCH2C
1): 0.47, 1H NMR (CDCl3, 400 MHz): δppm = 2.17 (br s, 2H, CH2Ph), 2.66 (br s, 4H, 2NCH2), 3.28 (br s, 4H, 2NCH2), 3.38 (s, 3H, N(1)–CH3), 3.51 (s, 3H, N(3)–CH3), 3.72–3.80 (complex, 5H, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, N(7)–CH3), 5.49 (br s, 2H, NCH2CH2), 7.18 (br s, 5H, aryl), 7.41 (br s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 13.07, 21.17, 27.75, 29.70, 30.96, 32.59, 49.50, 52.27, 105.36, 125.69, 128.19, 129.79, 131.50, 132.98, 138.78, 147.43, 151.75, 154.93, 156.28. IR (KBr): 3100, 2948, 2859, 1695, 1647, 1604, 1532, 1458, 1130 cm−1. MS (EI): m/z (%) = 463 (24.7) [M+]. Anal. calc. for C23H29N9O2: C, 59.60; H, 6.31; N, 27.20; found: C, 59.46; H, 6.12; N, 27.04.
C, N(7)–CH3), 5.49 (br s, 2H, NCH2CH2), 7.18 (br s, 5H, aryl), 7.41 (br s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 13.07, 21.17, 27.75, 29.70, 30.96, 32.59, 49.50, 52.27, 105.36, 125.69, 128.19, 129.79, 131.50, 132.98, 138.78, 147.43, 151.75, 154.93, 156.28. IR (KBr): 3100, 2948, 2859, 1695, 1647, 1604, 1532, 1458, 1130 cm−1. MS (EI): m/z (%) = 463 (24.7) [M+]. Anal. calc. for C23H29N9O2: C, 59.60; H, 6.31; N, 27.20; found: C, 59.46; H, 6.12; N, 27.04.
4.4.7.6 1,3,7-Trimethyl-8-(4-((1-(4-methylbenzyl)-1H-1,2,3-triazol-4-yl)methyl)piperazin-1-yl)-3,7-dihydro-1H-purine-2,6-dione (12f). Recrystallization with EtOAc afforded the pure product as pale orange solid (4.21 g, 91%), m.p.: 172–174 °C, Rf (EtOAc/MeOH, 4
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): 0.55, 1H NMR (CDCl3, 400 MHz): δppm = 2.17 (s, 3H, CH3Ph), 2.61 (br s, 4H, 2NCH2), 3.21–3.27 (m, 4H, 2NCH2), 3.38 (s, 3H, N(1)–CH3), 3.52 (s, 3H, N(3)–CH3), 3.70–3.73 (complex, 5H, NCH2C
1): 0.55, 1H NMR (CDCl3, 400 MHz): δppm = 2.17 (s, 3H, CH3Ph), 2.61 (br s, 4H, 2NCH2), 3.21–3.27 (m, 4H, 2NCH2), 3.38 (s, 3H, N(1)–CH3), 3.52 (s, 3H, N(3)–CH3), 3.70–3.73 (complex, 5H, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, N(7)–CH3), 4.61 (s, 2H, NCH2Ph), 7.08 (d, J = 6.8 Hz, 2H, aryl), 7.23–7.29 (complex, 3H, aryl, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 21.65, 27.75, 29.71, 30.95, 32.60, 36.70, 49.53, 52.10, 105.37, 127.07, 128.71, 128.78, 131.06, 132.73, 137.05, 147.44, 151.75, 154.94, 156.29. IR (KBr): 3039, 2961, 2874, 1696, 1664, 1612, 1540, 1449, 1132 cm−1. MS (EI): m/z (%) = 463 (22.6) [M+]. Anal. calc. for C23H29N9O2: C, 59.60; H, 6.31; N, 27.20; found: C, 59.78; H, 6.50; N, 27.06.
C, N(7)–CH3), 4.61 (s, 2H, NCH2Ph), 7.08 (d, J = 6.8 Hz, 2H, aryl), 7.23–7.29 (complex, 3H, aryl, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 21.65, 27.75, 29.71, 30.95, 32.60, 36.70, 49.53, 52.10, 105.37, 127.07, 128.71, 128.78, 131.06, 132.73, 137.05, 147.44, 151.75, 154.94, 156.29. IR (KBr): 3039, 2961, 2874, 1696, 1664, 1612, 1540, 1449, 1132 cm−1. MS (EI): m/z (%) = 463 (22.6) [M+]. Anal. calc. for C23H29N9O2: C, 59.60; H, 6.31; N, 27.20; found: C, 59.78; H, 6.50; N, 27.06.
4.4.7.7 Ethyl 4-(4-((4-(1,3,7-trimethyl-2,6-dioxo-2,3,6,7-tetrahydro-1H-purin-8-yl)piperazin-1-yl)methyl)-1H-1,2,3-triazol-1-yl)butanoate (12g). Column chromatography on silica gel eluted with n-hexane/EtOAc (1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) afforded the pure product as orange foam (3.40 g, 72%), Rf (EtOAc/MeOH, 4
5) afforded the pure product as orange foam (3.40 g, 72%), Rf (EtOAc/MeOH, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): 0.46, 1H NMR (CDCl3, 400 MHz): δppm = 1.14 (t, J = 6.0 Hz, 3H, OCH2CH3), 2.17 (q, J = 6.8 Hz, 2H, CH2CH2CO2), 2.28 (t, J = 7.2 Hz, 2H, CH2CO2), 2.61 (br s, 4H, 2NCH2), 3.22 (br s, 7H, 2NCH2, N(1)–CH3), 3.38 (s, 3H, N(3)–CH3), 3.63 (s, 3H, N(7)–CH3), 3.67 (s, 2H, NCH2C
1): 0.46, 1H NMR (CDCl3, 400 MHz): δppm = 1.14 (t, J = 6.0 Hz, 3H, OCH2CH3), 2.17 (q, J = 6.8 Hz, 2H, CH2CH2CO2), 2.28 (t, J = 7.2 Hz, 2H, CH2CO2), 2.61 (br s, 4H, 2NCH2), 3.22 (br s, 7H, 2NCH2, N(1)–CH3), 3.38 (s, 3H, N(3)–CH3), 3.63 (s, 3H, N(7)–CH3), 3.67 (s, 2H, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 4.02 (q, J = 6.0 Hz, 2H, OCH2), 4.38 (t, J = 6.8 Hz, 2H, NCH2CH2), 7.56 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 14.05, 25.30, 27.49, 29.45, 30.56, 32.40, 49.07, 49.21, 51.98, 52.93, 60.48, 104.98, 122.96, 143.65, 147.12, 151.38, 154.49, 155.99, 172.08. IR (KBr): 3050, 2945, 2831, 1718, 1699, 1653, 1605, 1530, 1455, 1127 cm−1. MS (EI): m/z (%) = 473 (21.3) [M+]. Anal. calc. for C21H31N9O4: C, 53.26; H, 6.60; N, 26.62; found: C, 53.50; H, 6.81; N, 26.80.
C), 4.02 (q, J = 6.0 Hz, 2H, OCH2), 4.38 (t, J = 6.8 Hz, 2H, NCH2CH2), 7.56 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 14.05, 25.30, 27.49, 29.45, 30.56, 32.40, 49.07, 49.21, 51.98, 52.93, 60.48, 104.98, 122.96, 143.65, 147.12, 151.38, 154.49, 155.99, 172.08. IR (KBr): 3050, 2945, 2831, 1718, 1699, 1653, 1605, 1530, 1455, 1127 cm−1. MS (EI): m/z (%) = 473 (21.3) [M+]. Anal. calc. for C21H31N9O4: C, 53.26; H, 6.60; N, 26.62; found: C, 53.50; H, 6.81; N, 26.80.
4.4.7.8 8-(4-((1-Heptyl-1H-1,2,3-triazol-4-yl)methyl)piperazin-1-yl)-1,3,7-trimethyl-3,7-dihydro-1H-purine-2,6-dione (12h). Column chromatography on silica gel eluted with n-hexane/EtOAc (1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) afforded the pure product as orange foam (3.97 g, 87%), Rf (EtOAc/MeOH, 4
4) afforded the pure product as orange foam (3.97 g, 87%), Rf (EtOAc/MeOH, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) = 0.40, 1H NMR (CDCl3, 400 MHz): δppm = 0.78 (t, J = 7.2 Hz, 3H, CH2CH3), 1.14–1.24 (m, 10H, 5CH2), 2.61 (br s, 4H, 2NCH2), 3.23–3.26 (complex, 7H, 2NCH2, N(1)–CH3), 3.40 (s, 3H, N(3)–CH3), 3.64–3.67 (complex, 5H, NCH2C
1) = 0.40, 1H NMR (CDCl3, 400 MHz): δppm = 0.78 (t, J = 7.2 Hz, 3H, CH2CH3), 1.14–1.24 (m, 10H, 5CH2), 2.61 (br s, 4H, 2NCH2), 3.23–3.26 (complex, 7H, 2NCH2, N(1)–CH3), 3.40 (s, 3H, N(3)–CH3), 3.64–3.67 (complex, 5H, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, N(7)–CH3), 4.28 (t, J = 7.2 Hz, 2H, NCH2CH2), 7.48 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 13.90, 14.08, 22.36, 26.31, 27.56, 28.50, 29.52, 30.15, 31.42, 32.45, 49.39, 52.12, 60.21, 105.13, 122.51, 147.26, 151.53, 154.67, 156.14, 170.91. IR (KBr): 3085, 2970, 2848, 1694, 1650, 1603, 1520, 1475, 1142 cm−1. MS (EI): m/z (%) = 457 (27.1) [M+]. Anal. calc. for C22H35N9O2: C, 57.75; H, 7.71; N, 27.55; found: C, 57.60; H, 7.93; N, 27.71.
C, N(7)–CH3), 4.28 (t, J = 7.2 Hz, 2H, NCH2CH2), 7.48 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 13.90, 14.08, 22.36, 26.31, 27.56, 28.50, 29.52, 30.15, 31.42, 32.45, 49.39, 52.12, 60.21, 105.13, 122.51, 147.26, 151.53, 154.67, 156.14, 170.91. IR (KBr): 3085, 2970, 2848, 1694, 1650, 1603, 1520, 1475, 1142 cm−1. MS (EI): m/z (%) = 457 (27.1) [M+]. Anal. calc. for C22H35N9O2: C, 57.75; H, 7.71; N, 27.55; found: C, 57.60; H, 7.93; N, 27.71.
4.4.7.9 8-(4-((1-Hexyl-1H-1,2,3-triazol-4-yl)methyl)piperazin-1-yl)-1,3,7-trimethyl-3,7-dihydro-1H-purine-2,6-dione (12i). Column chromatography on silica gel eluted with n-hexane/EtOAc (1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) afforded the pure product as orange foam (3.67 g, 83%), Rf (EtOAc/MeOH, 4
2) afforded the pure product as orange foam (3.67 g, 83%), Rf (EtOAc/MeOH, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): 0.60, 1H NMR (CDCl3, 400 MHz): δppm = 0.78 (t, J = 6.8 Hz, 3H, CH2CH3), 1.15–1.20 (m, 8H, 4CH2), 2.61 (br s, 4H, 2NCH2), 3.23–3.26 (complex, 7H, 2NCH2, N(1)–CH3), 3.41 (s, 3H, N(3)–CH3), 3.64–3.71 (complex, 5H, NCH2C
1): 0.60, 1H NMR (CDCl3, 400 MHz): δppm = 0.78 (t, J = 6.8 Hz, 3H, CH2CH3), 1.15–1.20 (m, 8H, 4CH2), 2.61 (br s, 4H, 2NCH2), 3.23–3.26 (complex, 7H, 2NCH2, N(1)–CH3), 3.41 (s, 3H, N(3)–CH3), 3.64–3.71 (complex, 5H, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, N(7)–CH3), 4.28 (t, J = 7.2 Hz, 2H, NCH2CH2), 7.48 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 14.09, 22.46, 26.37, 27.58, 28.81, 28.92, 29.53, 30.17, 31.56, 32.46, 49.35, 52.11, 105.16, 122.53, 147.27, 151.55, 154.69, 156.12, 170.94. IR (KBr): 3074, 2935, 2840, 1696, 1651, 1610, 1528, 1457, 1150 cm−1. MS (EI): m/z (%) = 443 (21.6) [M+]. Anal. calc. for C21H33N9O2: C, 56.87; H, 7.50; N, 28.42; found: C, 57.03; H, 7.69; N, 28.62.
C, N(7)–CH3), 4.28 (t, J = 7.2 Hz, 2H, NCH2CH2), 7.48 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 14.09, 22.46, 26.37, 27.58, 28.81, 28.92, 29.53, 30.17, 31.56, 32.46, 49.35, 52.11, 105.16, 122.53, 147.27, 151.55, 154.69, 156.12, 170.94. IR (KBr): 3074, 2935, 2840, 1696, 1651, 1610, 1528, 1457, 1150 cm−1. MS (EI): m/z (%) = 443 (21.6) [M+]. Anal. calc. for C21H33N9O2: C, 56.87; H, 7.50; N, 28.42; found: C, 57.03; H, 7.69; N, 28.62.
4.4.7.10 1,3,7-Trimethyl-8-(4-((1-octyl-1H-1,2,3-triazol-4-yl)methyl)piperazin-1-yl)-3,7-dihydro-1H-purine-2,6-dione (12j). Column chromatography on silica gel eluted with n-hexane/EtOAc (1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) afforded the pure product as orange foam (3.81 g, 81%), Rf (EtOAc/MeOH, 4
6) afforded the pure product as orange foam (3.81 g, 81%), Rf (EtOAc/MeOH, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): 0.38, 1H NMR (CDCl3, 400 MHz): δppm = 0.79 (t, J = 7.2 Hz, 3H, CH2CH3), 1.17–1.24 (m, 12H, 6CH2), 2.61 (s, 4H, 2NCH2), 3.23–3.29 (complex, 7H, 2NCH2, N(1)–CH3), 3.43 (s, 3H, N(3)–CH3), 3.65–3.69 (complex, 5H, NCH2C
1): 0.38, 1H NMR (CDCl3, 400 MHz): δppm = 0.79 (t, J = 7.2 Hz, 3H, CH2CH3), 1.17–1.24 (m, 12H, 6CH2), 2.61 (s, 4H, 2NCH2), 3.23–3.29 (complex, 7H, 2NCH2, N(1)–CH3), 3.43 (s, 3H, N(3)–CH3), 3.65–3.69 (complex, 5H, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, N(7)–CH3), 4.28 (t, J = 7.2 Hz, 2H, NCH2CH2), 7.45 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 14.04, 22.55, 26.45, 27.70, 28.89, 29.00, 29.65, 30.24, 31.63, 32.55, 49.43, 50.36, 52.20, 53.22, 105.29, 122.44, 147.38, 151.68, 154.84, 156.21, 163.08. IR (KBr): 3040, 2980, 2829, 1698, 1647, 1612, 1518, 1438, 1156 cm−1. MS (EI): m/z (%) = 471 (25.7) [M+]. Anal. calc. for C23H37N9O2: C, 58.58; H, 7.91; N, 26.73; found: C, 58.39; H, 7.70; N, 26.56.
C, N(7)–CH3), 4.28 (t, J = 7.2 Hz, 2H, NCH2CH2), 7.45 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 14.04, 22.55, 26.45, 27.70, 28.89, 29.00, 29.65, 30.24, 31.63, 32.55, 49.43, 50.36, 52.20, 53.22, 105.29, 122.44, 147.38, 151.68, 154.84, 156.21, 163.08. IR (KBr): 3040, 2980, 2829, 1698, 1647, 1612, 1518, 1438, 1156 cm−1. MS (EI): m/z (%) = 471 (25.7) [M+]. Anal. calc. for C23H37N9O2: C, 58.58; H, 7.91; N, 26.73; found: C, 58.39; H, 7.70; N, 26.56.
4.4.7.11 8-(4-((1-Decyl-1H-1,2,3-triazol-4-yl)methyl)piperazin-1-yl)-1,3,7-trimethyl-3,7-dihydro-1H-purine-2,6-dione (12k). Column chromatography on silica gel eluted with n-hexane/EtOAc (1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) afforded the pure product as orange foam (3.99 g, 80%), Rf (EtOAc/MeOH, 4
6) afforded the pure product as orange foam (3.99 g, 80%), Rf (EtOAc/MeOH, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): 0.40, 1H NMR (CDCl3, 400 MHz): δppm = 0.80 (t, J = 7.2 Hz, 3H, CH2CH3), 1.17–1.24 (m, 16H, 8CH2), 2.63 (br s, 4H, 2NCH2), 3.25–3.30 (complex, 7H, 2NCH2, N(1)–CH3), 3.44 (s, 3H, N(3)–CH3), 3.66–3.72 (complex, 5H, NCH2C
1): 0.40, 1H NMR (CDCl3, 400 MHz): δppm = 0.80 (t, J = 7.2 Hz, 3H, CH2CH3), 1.17–1.24 (m, 16H, 8CH2), 2.63 (br s, 4H, 2NCH2), 3.25–3.30 (complex, 7H, 2NCH2, N(1)–CH3), 3.44 (s, 3H, N(3)–CH3), 3.66–3.72 (complex, 5H, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, N(7)–CH3), 4.29 (br s, 2H, NCH2CH2), 7.46 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 14.10, 14.19, 21.06, 22.64, 26.51, 27.74, 28.96, 29.24, 29.37, 29.44, 29.69, 31.83, 32.58, 60.39, 105.37, 124.29, 147.41, 151.74, 154.92, 156.23, 170.88. IR (KBr): 3068, 2971, 2850, 1699, 1642, 1607, 1531, 1472, 1159 cm−1. MS (EI): m/z (%) = 499 (26.7) [M+]. Anal. calc. for C25H41N9O2: C, 60.10; H, 8.27; N, 25.23; found: C, 60.28; H, 8.46; N, 25.42.
C, N(7)–CH3), 4.29 (br s, 2H, NCH2CH2), 7.46 (s, 1H, C(5)–H of triazole). 13C NMR (CDCl3, 100 MHz): δppm = 14.10, 14.19, 21.06, 22.64, 26.51, 27.74, 28.96, 29.24, 29.37, 29.44, 29.69, 31.83, 32.58, 60.39, 105.37, 124.29, 147.41, 151.74, 154.92, 156.23, 170.88. IR (KBr): 3068, 2971, 2850, 1699, 1642, 1607, 1531, 1472, 1159 cm−1. MS (EI): m/z (%) = 499 (26.7) [M+]. Anal. calc. for C25H41N9O2: C, 60.10; H, 8.27; N, 25.23; found: C, 60.28; H, 8.46; N, 25.42.
4.4.7.12 8-(4-((1-(5-(1,3-Dioxoisoindolin-2-yl)pentyl)-1H-1,2,3-triazol-4-yl)methyl)piperazin-1-yl)-1,3,7-trimethyl-3,7-dihydro-1H-purine-2,6-dione (12l). Column chromatography on silica gel eluted with n-hexane/EtOAc (1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) afforded the pure product as yellow foam (4.47 g, 78%), Rf (EtOAc/MeOH, 4
6) afforded the pure product as yellow foam (4.47 g, 78%), Rf (EtOAc/MeOH, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): 0.35, 1H NMR (CDCl3, 400 MHz): δppm = 1.18–1.23 (m, 6H, 3CH2), 1.83 (t, J = 6.8 Hz, 2H, CONCH2), 2.65 (br s, 4H, 2NCH2), 3.26–3.28 (complex, 7H, 2NCH2, N(1)–CH3), 3.40 (s, 3H, N(3)–CH3), 3.63–3.67 (complex, 5H, NCH2C
1): 0.35, 1H NMR (CDCl3, 400 MHz): δppm = 1.18–1.23 (m, 6H, 3CH2), 1.83 (t, J = 6.8 Hz, 2H, CONCH2), 2.65 (br s, 4H, 2NCH2), 3.26–3.28 (complex, 7H, 2NCH2, N(1)–CH3), 3.40 (s, 3H, N(3)–CH3), 3.63–3.67 (complex, 5H, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, N(7)–CH3), 4.27 (t, J = 6.8 Hz, 2H, NCH2CH2), 7.47 (s, 1H, C(5)–H of triazole), 7.78 (br s, 4H, aryl). 13C NMR (CDCl3, 100 MHz): δppm = 20.65, 23.97, 27.34, 28.91, 30.97, 33.02, 41.24, 49.13, 51.70, 53.26, 54.47, 106.38, 123.46, 127.35, 130.58, 132.91, 143.75, 149.41, 151.62, 155.96, 164.39, 168.54. IR (KBr): 3100, 2960, 2847, 1750, 1693, 1652, 1609, 1535, 1460, 1149 cm−1. MS (EI): m/z (%) = 574 (29.4) [M+]. Anal. calc. for C28H34N10O4: C, 58.52; H, 5.96; N, 24.37; found: C, 58.75; H, 6.15; N, 24.52.
C, N(7)–CH3), 4.27 (t, J = 6.8 Hz, 2H, NCH2CH2), 7.47 (s, 1H, C(5)–H of triazole), 7.78 (br s, 4H, aryl). 13C NMR (CDCl3, 100 MHz): δppm = 20.65, 23.97, 27.34, 28.91, 30.97, 33.02, 41.24, 49.13, 51.70, 53.26, 54.47, 106.38, 123.46, 127.35, 130.58, 132.91, 143.75, 149.41, 151.62, 155.96, 164.39, 168.54. IR (KBr): 3100, 2960, 2847, 1750, 1693, 1652, 1609, 1535, 1460, 1149 cm−1. MS (EI): m/z (%) = 574 (29.4) [M+]. Anal. calc. for C28H34N10O4: C, 58.52; H, 5.96; N, 24.37; found: C, 58.75; H, 6.15; N, 24.52.
4.4.7.13 8-(4-((1-(4-(1,1-Dioxido-3-oxobenzo[d]isothiazol-2(3H)-yl)butyl)-1H-1,2,3-triazol-4-yl)methyl)piperazin-1-yl)-1,3,7-trimethyl-3,7-dihydro-1H-purine-2,6-dione (12m). Column chromatography on silica gel eluted with n-hexane/EtOAc (1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) afforded the pure product as yellow foam (4.06 g, 82%), Rf (EtOAc/MeOH, 4
6) afforded the pure product as yellow foam (4.06 g, 82%), Rf (EtOAc/MeOH, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): 0.30, 1H NMR (CDCl3, 400 MHz): δppm = 1.31–1.41 (m, 2H, CH2), 1.53–1.60 (m, 2H, CH2), 1.87 (t, J = 7.6 Hz, 2H, SO2NCH2), 2.63 (br s, 4H, 2NCH2), 3.20–3.28 (complex, 7H, 2NCH2, N(1)–CH3), 3.43 (s, 3H, N(3)–CH3), 3.65–3.70 (complex, 5H, NCH2C
1): 0.30, 1H NMR (CDCl3, 400 MHz): δppm = 1.31–1.41 (m, 2H, CH2), 1.53–1.60 (m, 2H, CH2), 1.87 (t, J = 7.6 Hz, 2H, SO2NCH2), 2.63 (br s, 4H, 2NCH2), 3.20–3.28 (complex, 7H, 2NCH2, N(1)–CH3), 3.43 (s, 3H, N(3)–CH3), 3.65–3.70 (complex, 5H, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, N(7)–CH3), 4.30 (t, J = 6.8 Hz, 2H, NCH2CH2), 7.49 (s, 1H, C(5)–H of triazole), 7.77–7.85 (m, 2H, aryl), 7.97–7.99 (m, 2H, aryl). 13C NMR (CDCl3, 100 MHz): δppm = 21.65, 24.12, 26.65, 29.70, 33.16, 43.56, 49.53, 52.19, 53.45, 56.01, 106.02, 123.32, 126.32, 127.54, 128.77, 131.49, 133.38, 140.49, 143.75, 149.76, 151.10, 156.28, 163.91, 169.07. IR (KBr): 3045, 2996, 2857, 1740, 1695, 1652, 1609, 1520, 1470, 1367, 1152 cm−1. MS (EI): m/z (%) = 596 (27.9) [M+]. Anal. calc. for C26H32N10O5S: C, 52.34; H, 5.41; N, 23.48; found: C, 52.18; H, 5.19; N, 23.33.
C, N(7)–CH3), 4.30 (t, J = 6.8 Hz, 2H, NCH2CH2), 7.49 (s, 1H, C(5)–H of triazole), 7.77–7.85 (m, 2H, aryl), 7.97–7.99 (m, 2H, aryl). 13C NMR (CDCl3, 100 MHz): δppm = 21.65, 24.12, 26.65, 29.70, 33.16, 43.56, 49.53, 52.19, 53.45, 56.01, 106.02, 123.32, 126.32, 127.54, 128.77, 131.49, 133.38, 140.49, 143.75, 149.76, 151.10, 156.28, 163.91, 169.07. IR (KBr): 3045, 2996, 2857, 1740, 1695, 1652, 1609, 1520, 1470, 1367, 1152 cm−1. MS (EI): m/z (%) = 596 (27.9) [M+]. Anal. calc. for C26H32N10O5S: C, 52.34; H, 5.41; N, 23.48; found: C, 52.18; H, 5.19; N, 23.33.
4.4.7.14 8-(4-((1-(3-(1,1-Dioxido-3-oxobenzo[d]isothiazol-2(3H)-yl)propyl)-1H-1,2,3-triazol-4-yl)methyl)piperazin-1-yl)-1,3,7-trimethyl-3,7-dihydro-1H-purine-2,6-dione (12n). Column chromatography on silica gel eluted with n-hexane/EtOAc (1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) afforded the pure product as orange foam (4.88 g, 84%), Rf (EtOAc/MeOH, 4
6) afforded the pure product as orange foam (4.88 g, 84%), Rf (EtOAc/MeOH, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): 0.39, 1H NMR (CDCl3, 400 MHz): δppm = 1.16–1.19 (m, 2H, NCH2CH2), 2.44 (t, J = 6.4 Hz, 2H, SO2NCH2), 2.63 (br s, 4H, 2NCH2), 3.25–2.29 (complex, 7H, 2NCH2, N(1)–CH3), 3.43 (s, 3H, N(3)–CH3), 3.64–3.65 (complex, 5H, N(7)–CH3, NCH2C
1): 0.39, 1H NMR (CDCl3, 400 MHz): δppm = 1.16–1.19 (m, 2H, NCH2CH2), 2.44 (t, J = 6.4 Hz, 2H, SO2NCH2), 2.63 (br s, 4H, 2NCH2), 3.25–2.29 (complex, 7H, 2NCH2, N(1)–CH3), 3.43 (s, 3H, N(3)–CH3), 3.64–3.65 (complex, 5H, N(7)–CH3, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 4.44 (t, J = 6.4 Hz, 2H, NCH2CH2), 7.65 (s, 1H, C(5)–H of triazole), 7.81–8.01 (m, 4H, aryl). 13C NMR (CDCl3, 100 MHz): δppm = 21.24, 25.58, 27.43, 31.67, 41.36, 48.24, 50.12, 52.21, 55.17, 106.43, 122.67, 126.73, 127.55, 128.39, 131.23, 132.60, 140.78, 144.39, 150.40, 152.46, 156.85, 164.61, 169.79. IR (KBr): 3038, 2990, 2851, 1743, 1699, 1656, 1610, 1528, 1462, 1364, 1159 cm−1. MS (EI): m/z (%) = 582 (26.3) [M+]. Anal. calc. for C25H30N10O5S: C, 51.54; H, 5.19; N, 24.04; found: C, 51.75; H, 5.38; N, 24.23.
C), 4.44 (t, J = 6.4 Hz, 2H, NCH2CH2), 7.65 (s, 1H, C(5)–H of triazole), 7.81–8.01 (m, 4H, aryl). 13C NMR (CDCl3, 100 MHz): δppm = 21.24, 25.58, 27.43, 31.67, 41.36, 48.24, 50.12, 52.21, 55.17, 106.43, 122.67, 126.73, 127.55, 128.39, 131.23, 132.60, 140.78, 144.39, 150.40, 152.46, 156.85, 164.61, 169.79. IR (KBr): 3038, 2990, 2851, 1743, 1699, 1656, 1610, 1528, 1462, 1364, 1159 cm−1. MS (EI): m/z (%) = 582 (26.3) [M+]. Anal. calc. for C25H30N10O5S: C, 51.54; H, 5.19; N, 24.04; found: C, 51.75; H, 5.38; N, 24.23.
4.4.7.15 8-(4-((1-(5-(1,1-Dioxido-3-oxobenzo[d]isothiazol-2(3H)-yl)pentyl)-1H-1,2,3-triazol-4-yl)methyl)piperazin-1-yl)-1,3,7-trimethyl-3,7-dihydro-1H-purine-2,6-dione (12o). Column chromatography on silica gel eluted with n-hexane/EtOAc (1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6) afforded the pure product as orange foam (4.81 g, 79%), Rf (EtOAc/MeOH, 4
6) afforded the pure product as orange foam (4.81 g, 79%), Rf (EtOAc/MeOH, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1): 0.33, 1H NMR (CDCl3, 400 MHz): δppm = 1.15–1.18 (m, 2H, CH2), 1.32–1.37 (m, 2H, CH2), 1.51–1.58 (m, 2H, NCH2C
1): 0.33, 1H NMR (CDCl3, 400 MHz): δppm = 1.15–1.18 (m, 2H, CH2), 1.32–1.37 (m, 2H, CH2), 1.51–1.58 (m, 2H, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1.85–1.92 (m, 2H, SO2NCH2), 2.63 (br s, 4H, 2NCH2), 3.18–3.26 (complex, 7H, 2NCH2, N(1)–CH3), 3.41 (s, 3H, N(3)–CH3), 3.64–3.70 (complex, 5H, NCH2C
C), 1.85–1.92 (m, 2H, SO2NCH2), 2.63 (br s, 4H, 2NCH2), 3.18–3.26 (complex, 7H, 2NCH2, N(1)–CH3), 3.41 (s, 3H, N(3)–CH3), 3.64–3.70 (complex, 5H, NCH2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, N(7)–CH3), 4.31 (t, J = 6.4 Hz, 2H, NCH2CH2), 7.31 (s, 1H, C(5)–H of triazole), 7.80–7.88 (m, 4H, aryl). 13C NMR (CDCl3, 100 MHz): δppm = 19.35, 22.91, 25.19, 28.46, 29.71, 32.95, 45.71, 49.53, 50.96, 52.71, 55.27, 106.81, 123.54, 126.64, 127.78, 129.04, 132.84, 134.21, 140.74, 144.19, 149.12, 151.75, 153.95, 163.37, 168.76. IR (KBr): 3050, 2986, 2844, 1741, 1697, 1648, 1611, 1529, 1459, 1364, 1150 cm−1. MS (EI): m/z (%) = 610 (29.7) [M+]. Anal. calc. for C27H34N10O5S: C, 53.10; H, 5.61; N, 22.94; found: C, 52.92; H, 5.42; N, 22.76.
C, N(7)–CH3), 4.31 (t, J = 6.4 Hz, 2H, NCH2CH2), 7.31 (s, 1H, C(5)–H of triazole), 7.80–7.88 (m, 4H, aryl). 13C NMR (CDCl3, 100 MHz): δppm = 19.35, 22.91, 25.19, 28.46, 29.71, 32.95, 45.71, 49.53, 50.96, 52.71, 55.27, 106.81, 123.54, 126.64, 127.78, 129.04, 132.84, 134.21, 140.74, 144.19, 149.12, 151.75, 153.95, 163.37, 168.76. IR (KBr): 3050, 2986, 2844, 1741, 1697, 1648, 1611, 1529, 1459, 1364, 1150 cm−1. MS (EI): m/z (%) = 610 (29.7) [M+]. Anal. calc. for C27H34N10O5S: C, 53.10; H, 5.61; N, 22.94; found: C, 52.92; H, 5.42; N, 22.76.
Conflicts of interest
The authors have declared no conflict of interest.Acknowledgements
The authors wish to thank the Shiraz University of Technology research council for partial support of this work.References
- C. de Martel, D. Georges, F. Bray, J. Ferlay and G. M. Clifford, Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis, Lancet Global Health, 2020, 8, e180–e190, DOI:10.1016/S2214-109X(19)30488-7.
- https://www.who.int/news-room/fact-sheets/detail/cancer, accessed 1 July 2023.
- https://www.who.int/news-room/fact-sheets/detail/breast-cancer, accessed 1 July 2023.
- S. Łukasiewicz, M. Czeczelewski, A. Forma, J. Baj, R. Sitarz and A. Stanisławek, Breast Cancer—Epidemiology, Risk Factors, Classification, Prognostic Markers, and Current Treatment Strategies—An Updated Review, Cancers, 2021, 13, 4287, DOI:10.3390/cancers13174287.
- P. Kumar and T. N. Patel, Melanoma therapeutics: a literature review, J. Biomed. Res., 2022, 36, 77–97, DOI:10.7555/JBR.36.20210163.
- V. Prasad, K. De Jesús and S. Mailankody, The high price of anticancer drugs: origins, implications, barriers, solutions, Nat. Rev. Clin. Oncol., 2017, 14, 381–390, DOI:10.1038/nrclinonc.2017.31.
- K. Dzobo, The role of natural products as sources of therapeutic agents for innovative drug discovery, Comprehensive Pharm., 2022, vol. 2, pp. 408–422, DOI:10.1016/B978-0-12-820472-6.00041-4.
- B. G. Katzung, Basic & Clinical Pharmacology, Lange Medical Publications, McGraw-Hill Companies, 14 edn, 2018 Search PubMed.
- R. Franco, A. Oñatibia-Astibia and E. Martínez-Pinilla, Health Benefits of Methylxanthines in Cacao and Chocolate, Nutrients, 2013, 5, 4159–4173, DOI:10.3390/nu5104159.
- T. M. McLellan, J. A. Caldwell b and H. R. Lieberman, A review of caffeine's effects on cognitive, physical and occupational performance, Neurosci. Biobehav. Rev., 2016, 71, 294–312, DOI:10.1016/j.neubiorev.2016.09.001.
- N. Singh, A. K. Shreshtha, M. S. Thakur and S. Patra, Xanthine scaffold: scope and potential in drug development, Heliyon, 2018, 4, e00829, DOI:10.1016/j.heliyon.2018.
- J. Klosa, Caffeno-3-alkanolamines, US Pat., 3094531, 1963 Search PubMed.
- B. R. K. Shyamlal, M. Mathur, D. K. Yadav and S. Chaudhary, Microwave-assisted modified synthesis of C8-analogues of naturally occurring methylxanthines: Synthesis, biological evaluation and their practical applications, Fitoterapia, 2020, 143, 104533, DOI:10.1016/j.fitote.2020.104533.
- T. Zheng, H. Sun, F. Lu, K. Harms and X. Li, Cobalt induced C–H bond activation and C8-arylation of caffeine with aryl bromides, Inorg. Chem. Commun., 2013, 30, 139–142, DOI:10.1016/j.inoche.2013.01.025.
- W. Jiang, J. Zhuge, J. Li, G. Histand and D. Lin, Direct sulfenylation of the purine C8–H bond with thiophenols, J. Org. Chem., 2020, 85, 2415–2425, DOI:10.1021/acs.joc.9b03115.
- M. N. Soltani Rad, S. Behrouz, S. Aghajani, M. Behrouz, E. Zarenezhad and A. Ghanbariasad, Design, synthesis, anticancer and in silico assessment of 8-caffeinyl-triazolylmethoxy hybrid conjugate, RSC Adv., 2023, 13, 3056–3070, 10.1039/d2ra07683.
- F. A. Ashour, S. M. Rida, S. A. M. El-Hawash, M. M. ElSemary and M. H. Badr, Synthesis, anticancer, anti-HIV-1, and antimicrobial activity of some tricyclic triazino and triazolo[4,3-e]purine derivatives, Med. Chem. Res., 2012, 21, 1107–1119, DOI:10.1007/s00044-011-9612-6.
- S. M. Rida, F. A. Ashour, S. A. M. El-Hawash, M. M. El-Semary and M. H. Badr, Synthesis of some novel substituted purine derivatives as potential anticancer, anti-HIV-1 and antimicrobial agents, Arch. Pharm. Chem. Life Sci., 2007, 340, 185–194, DOI:10.1002/ardp.200600118.
- H. P. Booysen, C. Moraal, G. Terre'Blanche, A. Petzer, J. J. Bergh and J. P. Petzer, Thio- and aminocaffeine analogues as inhibitors of human monoamine oxidase, Bioorg. Med. Chem., 2011, 19, 7507–7518, DOI:10.1016/j.bmc.2011.10.036.
- R. Kaplánek, M. Jakubek, J. Rak, Z. Kejík, M. Havlík, B. Dolenský, I. Frydrych, M. Hajdúch, M. Kolář, K. Bogdanová, J. Králová, P. Džubák and V. Král, Caffeine-hydrazones as anticancer agents with pronounced selectivity toward T-lymphoblastic leukaemia cells, Bioorg. Chem., 2015, 60, 19–29, DOI:10.1016/j.bioorg.2015.03.003.
- M. Georgieva, J. Mitkov, G. Momekov, B. Zlatkov, P. Peikov and A. Zlatkov, Synthesis, structural and spectral analysis of some 8- substituted derivatives of 1,3,7-trimethylxanthine with antiprolipherative activity, World J. Pharm. Pharm. Sci., 2014, 3, 60–83 CAS.
- M. N. Soltani Rad, S. Behrouz, K. Zokaei, M. Behrouz and A. G. E. Zarenezhad, Synthesis of Some Novel 8-(4-Alkylpiperazinyl) Caffeine Derivatives as Potent Anti-Leishmania Agents, Bioorg. Chem., 2022, 128, 106062, DOI:10.1016/j.bioorg.2022.106062.
- M. Shaquiquzzaman, G. Verma, A. Marella, M. Akhter, W. Akhtar, M. F. Khan, S. Tasneem and M. Mumtaz Alam, Piperazine scaffold: A remarkable tool in generation of diverse pharmacological agents, Eur. J. Med. Chem., 2015, 102, 487–529, DOI:10.1016/j.ejmech.2015.07.026.
- A. Kleeman, J. Engel, B. Kutscher and D. Reichert, Pharmaceutical Substances, Thieme, Stuttgart, 3rd edn, 1999 Search PubMed.
- R. J. Martin, Modes of action of anthelmintic drugs, Vet. J., 1997, 154, 11–34, DOI:10.1016/S1090-0233(05)80005-X.
- A. K. Rathi, R. Syed, H.-S. Shin and R. V. Patel, Piperazine derivatives for therapeutic use: a patent review (2010-present), Expert Opin. Ther. Pat., 2016, 26, 777–797, DOI:10.1080/13543776.2016.1189902.
- J. E. Moses and A. D. Moorhouse, The growing applications of click chemistry, Chem. Soc. Rev., 2007, 36, 1249–1262, 10.1039/B613014N.
- E. Bonandi, M. S. Christodoulou, G. Fumagalli, D. Perdicchia, G. Rastelli and D. Passarella, The 1,2,3-triazole ring as a bioisostere in medicinal chemistry, Drug Discovery Today, 2017, 22, 1572–1581, DOI:10.1016/j.drudis.2017.05.014.
- S. G. Agalave, S. R. Maujan and V. S. Pore, Click chemistry: 1,2,3-triazoles as pharmacophores, Chem.–Asian J., 2011, 6, 2696–2718, DOI:10.1002/asia.201100432.
- B. Meunier, Hybrid molecules with a dual mode of action: Dream or reality?, Acc. Chem. Res., 2008, 41, 69–77, DOI:10.1021/ar7000843.
- W. Cardona-G, A. F. Yepes and A. Herrera-R, Hybrid Molecules: Promising compounds for the development of new treatments against leishmaniasis and chagas disease, Curr. Med. Chem., 2018, 25, 3637–3679, DOI:10.2174/0929867325666180309111428.
- I. A. Shagufta, Therapeutic significance of molecular hybrids for breast cancer research and treatment, RSC Med. Chem., 2023, 14, 218–238, 10.1039/D2MD00356B.
- M. N. Soltani Rad and S. Maghsoudi, Two-step three-component process for one-pot synthesis of 8-alkylmercaptocaffeine derivatives, RSC Adv., 2016, 6, 70335–70342, 10.1039/c6ra17814f.
- J. Mitkov, M. Georgieva and A. Zlatkov, Development of an optimized synthetic approach for synthesis of caffeine-8-thioglycolic acid and its ester derivatives, Pharmacia, 2012, 1–4, 17–23 Search PubMed.
- J. Mitkov, L. Nikolova, I. Nikolova, N. Danchev and A. Zlatkov, Synthesis and brain antihypoxic activity of some aminoalcoholic derivatives of caffeine-8-thioglycolic acid, C. R. Acad. Bulg. Sci., 2010, 63, 1075–1082 CAS.
- S. Behrouz, M. N. Soltani Rad, B. Taghavi Shahraki, M. Fathalipour, M. Behrouz and H. Mirkhani, Design, synthesis, and in silico studies of novel eugenyloxy propanol azole derivatives having potent antinociceptive activity and evaluation of their β-adrenoceptor blocking property, Mol. Diversity, 2019, 23, 147–164, DOI:10.1007/s11030-018-9867-7.
- E. Caputo, L. Maiorana, V. Vasta, F. M. Pezzino, S. Sunkara, K. Wynne, G. Elia, F. M. Marincola, J. A. McCubrey, M. Libra, S. Travali and M. Kane, Characterization of human melanoma cell lines and melanocytes by proteome analysis, Cell Cycle, 2011, 10, 2924–2936, DOI:10.4161/cc.10.17.17068.
- Ş. Comşa, A. M. Cimpean and M. Raica, The story of MCF-7 breast cancer cell line: 40 years of experience in research, Anticancer Res., 2015, 35, 3147–3154 Search PubMed.
- Y.-C. Lin, M. Boone, L. Meuris, I. Lemmens, N. Van Roy, A. Soete, J. Reumers, M. Moisse, S. Plaisance, R. Drmanac, J. Chen, F. Speleman, D. Lambrechts, Y. Van de Peer, J. Tavernier and N. Callewaert, Genome dynamics of the human embryonic kidney 293 lineage in response to cell biology manipulations, Nat. Commun., 2014, 5, 4767, DOI:10.1038/ncomms5767.
- https://en.wikipedia.org/wiki/Folate#cite_note-PKIN2020Folate-8, accessed 7 July 2023.
- B. Chabner, and D. L. Longo, Cancer Chemotherapy and Biotherapy Principles and Practice, Oxford University press, Wolters Kluwer Health/Lippincott Williams & Wilkins, 5th edn, 2011 Search PubMed.
- L. Genestier, R. Paillot, L. Quemeneur, K. Izeradjene and J. P. Revillard, Mechanisms of action of methotrexate, Immunopharmacol, 2000, 47, 247–257, DOI:10.1016/s0162-3109(00)00189-2.
- I. Puzanov, R. K. Amaravadi, G. A. McArthur, K. T. Flaherty, P. B. Chapman, J. A. Sosman, A. Ribas, M. Shackleton, P. Hwu, B. Chmielowski, K. B. Nolop, P. S. Lin and K. B. Kim, Long-term outcome in BRAF(V600E) melanoma patients treated with vemurafenib: Patterns of disease progression and clinical management of limited progression, Eur. J. Cancer, 2015, 51, 1435–1443, DOI:10.1016/j.ejca.2015.04.010.
- Molegro Virtual Docker, version 6.0.0, CLC Bio, 8200, Aarhus N, Denmark, 2012 Search PubMed.
- G. Marcou and D. Rognan, Optimizing fragment and scaffold docking by use of molecular interaction fingerprints, J. Chem. Inf. Model., 2007, 47, 195–207, DOI:10.1021/ci600342e.
- A. B. Umar, A. Uzairu, G. A. Shallangwa and S. Uba, Design of potential anti-melanoma agents against SK-MEL-5 cell line using QSAR modeling and molecular docking methods, SN Appl. Sci., 2020, 2, 815, DOI:10.1007/s42452-020-2620-8.
- V. Cody, J. R. Luft and W. Pangborn, Understanding the role of Leu22 variants in methotrexate resistance: comparison of wild-type and Leu22Arg variant mouse and human dihydrofolate reductase ternary crystal complexes with methotrexate and NADPH, Acta Crystallogr., 2005, D61, 147–155, DOI:10.1107/S0907444904030422.
- R. J. Bienstock, Library Design, Search Methods, and Applications of Fragment-Based Drug Design, A.C.S., Washington, 2011 Search PubMed.
- C. A. Lipinski, F. Lombardo, B. W. Dominy and P. J. Feeney, Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings, Adv. Drug Delivery Rev., 2001, 46, 3–25, DOI:10.1016/S0169-409X(96)00423-1.
- PreADMET online server, https://preadmet.webservice.bmdrc.org/adme, accessed 27 May 2023 Search PubMed.
- OSIRIS Property Explorer, https://www.organic-chemistry.org/prog/peo, accessed 27 May 2023 Search PubMed.
- SwissADME online software, http://www.swissadme.ch/index.php, accessed 27 May 2023 Search PubMed.
- Y. Zhao and D. G. Truhlar, The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals, Theor. Chem. Acc., 2008, 120, 215–241, DOI:10.1007/s00214-007-0310-x.
- G. L. Patrick, An Introduction to Medicinal Chemistry, Oxford University Press, New York, 5nd edn, 2013 Search PubMed.
- T. Mosmann, Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays, J. Immunol. Methods, 1983, 65, 55–63, DOI:10.1016/0022-1759(83)90303-90304.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, et al., Gaussian 09, Revision A.01Gaussian, Inc., Wallingford CT, 2009 Search PubMed.
- M. N. Soltani Rad, S. Behrouz, J. Mohammadtaghi-Nezhad, E. Zarenezhad and M. Agholi, Silica-tethered cuprous acetophenone thiosemicarbazone (STCATSC) as a novel hybrid nano-catalyst for highly efficient synthesis of new 1,2,3-triazolyl-based metronidazole hybrid analogues having potent antigiardial activity, Appl. Organomet. Chem., 2019, 33, e4799, DOI:10.1002/aoc.4799.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra04817a |
| This journal is © The Royal Society of Chemistry 2023 |