Open Access Article
Open Access ArticleNaturally occurring phenylethanoids and phenylpropanoids: antimalarial potential†
Usama Ramadan Abdelmohsen *ab,
Soad A. L. Bayoumic,
Nesma M. Mohamedcd,
Yaser A. Mostafaef,
Che J. Ngwag,
Gabriele Pradelg and
Salwa F. Farag
*ab,
Soad A. L. Bayoumic,
Nesma M. Mohamedcd,
Yaser A. Mostafaef,
Che J. Ngwag,
Gabriele Pradelg and
Salwa F. Farag *ch
*ch
aDepartment of Pharmacognosy, Faculty of Pharmacy, Minia University, Minia 61519, Egypt. E-mail: usama.ramadan@mu.edu.eg
bDepartment of Pharmacognosy, Faculty of Pharmacy, Deraya University, 7 Universities Zone, 61111 New Minia City, Egypt
cDepartment of Pharmacognosy, Faculty of Pharmacy, Assiut University, Assiut 71526, Egypt. E-mail: farag_s@yahoo.com
dDepartment of Pharmacognosy, Faculty of Pharmacy, Badr University in Assiut, Assiut 77771, Egypt
ePharmaceutical Organic Chemistry Department, Faculty of Pharmacy, Assiut University, 71526 Assiut, Egypt
fPharmaceutical Chemistry Department, Faculty of Pharmacy, Badr University in Assiut, Assiut 77771, Egypt
gDivision of Cellular and Applied Infection Biology, Institute of Zoology, RWTH Aachen University, 52074 Aachen, Germany
hDepartment of Pharmacognosy, College of Pharmacy, Taif University, P.O. Box 11099, Taif 21944, Saudi Arabia
First published on 7th September 2023
Abstract
Malaria as an infectious disease is one of the world's most dangerous parasitic diseases. There is an urgent need for the development of new antimalarial drugs. Natural products are a very rich source of new bioactive compounds. Our research aims to shed light on the recent studies which demonstrated the antimalarial potential of phenylpropanoids as a major natural-products class. This study involves an in silico analysis of naturally-occurring phenylpropanoids and phenylethanoids which showed 25 compounds with moderate to strong binding affinity to various amino acid residues lining the active site; P. falciparum kinase (PfPK5), P. falciparum cytochrome bc1 complex (cyt bc1), and P. falciparum lysyl-tRNA synthetase (PfKRS1); of Plasmodium falciparum parasite, a unicellular protozoan which causes the most severe and life-threatening malaria. Furthermore, the study was augmented by the assessment of antiplasmodial activity of glandularin, a naturally occurring dibenzylbutyrolactolic lignan, against chloroquine-sensitive 3D7 strain of P. falciparum using SYBR green I-based fluorescence assay, which showed high antimalarial activity with IC50 value of 11.2 μM after 24 hours of incubation. Our results highlight phenylpropanoids and glandularin in particular as a promising chemical lead for development of antimalarial drugs.
Introduction
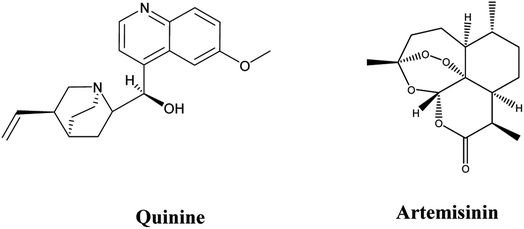
Vector-borne protozoan illnesses like trypanosomiasis, leishmaniasis, and malaria are well-known public health threats due to their high morbidity and mortality rates. They are widely distributed in tropical regions where the presence of poverty and conditions that are conducive to vectors responsible for the transmission of diseases are found. Numerous strategies have been applied to reduce malaria burden either by vector control or by using vaccines.1,2Quinine (Fig. 1) was the first drug, from natural source, used in treatment of malaria. It was used as the first-choice drug in treatment of malaria for around 100 years, until the emergence of resistant parasites. Later on, quinine was replaced by artemisinin (Fig. 1), isolated from Artemisia annua, which has been widely used in Chinese traditional medicine and has proven effectiveness against all multi-drug resistant P. falciparum strains. Unfortunately, high failure rates of artemisinin combinations therapy was detected in the Greater Mekong sub-regions in Southeast Asia. This was attributed to the emergence of artemisinin resistance. Recently, independent emergence of artemisinin resistance in East Africa (Rwanda and Uganda) has arisen.3
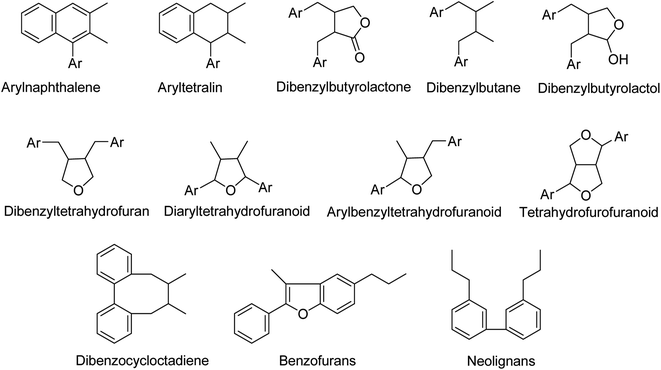
Secondary metabolites (specialized metabolites) are natural compounds with a variety of biological activities and chemical configurations.4–8 Among which are simple phenolics such as phenylethanoids and phenylpropanoids classed as (C6–C2) and C6–C3 compounds. Additionally, polyphenolics as lignans and neolignans constitute a significant class of secondary metabolites.9–11 In a more specific context, lignans are phenylpropanoid dimers linked by a C–C-bond between carbons 8 and 8′ in the side chain12–15 including many subtypes according to the nature and position of the linkage between the phenylpropane units (Fig. 2). Neolignans are a class of lignans that lack the β-β′ (also termed 8-8′) phenylpropane linkage that are characteristic of classical lignans.16–19
Different classes of phenylethanoids and phenylpropanoids are spread over various species of higher plants, however according to chemotaxonomy, the specific pattern of substitution of these classes may restrict their existence to certain genera and species.20,21 On the other hand, more than 200 lignans have been chemically and biologically characterized in different organs of about 70 plant families.19
Several studies reported that phenylethanoids possess antibacterial, anticancer, antidiabetic, anti-inflammatory, anti-obesity, antioxidant, antiviral, and neuroprotective properties.21 Whereas, plants rich in phenylpropanoids have been reported to be used traditionally for treatment of several skin inflammatory disorders as sores, wound healing purposes, sedative activity, and relief of general body exhaustion.15,22 Through the last few years, many research studies have revealed their anti-inflammatory, anti-atherosclerosis, anti-platelet-aggregation, antihypertension, antifatigue, analgesic, hepatoprotective and immunostimulant benefits.23–25 Furthermore, they are known for their potent antioxidant capacity,26–28 their capabilities to control diabetes,29,30 in addition to their notable effects as anticancer, nephroprotective, cardioprotective and neuroprotective agents.10,26,27 Moreover, they were reported to inhibit the growth of bacteria, fungus, and yeast in several investigations and also showed potent activity even against resistant microbes.10,14 Some studies also discussed their antiviral activity against different viruses as HIV-1, H1N1, SARS-CoV and hepatitis C viruses, and highlighted the possible mechanism of action of these compounds.10,17–19,24,31,32
In 2016, a study conducted by Hematpoor and co-workers reported the larvicidal and ovicidal activity against Aedes aegypti, Aedes albopictus and Culex quinquefasciatus of Piper sarmentosum phenylpropanoids.33
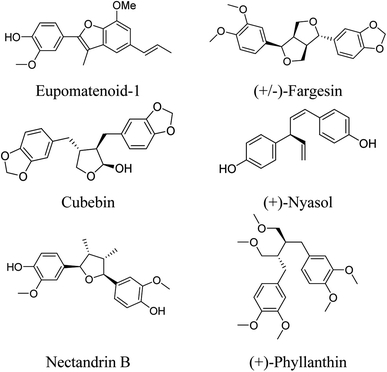
Only few studies reported their antiprotozoal activity for compacting vector-borne protozoan illnesses. Oketch-Rabah and his co-workers, in 1997, reported the antiprotozoal activity of Asparagus africanus Lam. (Liliaceae) roots. The lignan (+)-nyasol (Fig. 3) potently inhibits the growth of Leishmania major promastigotes, the IC50 being 12 μM, and moderately inhibits P. falciparum schizonts with the IC50 49 μM.34 Also, tetrahydrofuran lignans, isolated from Nectandra megapotamica (Lauraceae), showed in vitro activity against Leishmania donovani and P. falciparum.35
Furthermore, it had been reported that the rhizomes of Aristolochia elegans Mast (F. Aristolochiaceae) exhibited antiprotozoal activities against Entamoeba histolytica and Giardia lamblia due to eupomatenoid-1 which was very active (IC50 < 0.624 μg mL−1); in contrast, fargesin and cubebin (Fig. 3) were moderately active (IC50 < 275 μg mL−1).36
In 2017, Otero and co-workers demonstrated the antiprotozoal activity of some hybrids of caffeic acid against Trypanosoma cruzi and amastigotes of Leishmania (Viannia) panamensis protozoa.10,37 In 2020, Maia and co-workers had been reported the virtual screening and the in vitro assessment of the antileishmanial activity of lignans.38
Additionally, lignans isolated from Haplophyllum tuberculatum (Rutaceae) exhibited antiprotozoal activity against L. donovani amastigotes, P. falciparum, and Trypanosoma brucei rhodesiense bloodstream forms. Nectandrin B (Fig. 3) exhibited the highest activity against L. donovani (IC50 4.5 μM) and the highest selectivity index of 25.5.39
Lignans isolated from Holostylis reniformis Duch. (Aristolochiaceae) and Rhaphidophora decursiva possessed high antiplasmodial activity against P. falciparum.40–42 4-(1,3-dimethoxypropyl)phenol, a compound isolated from Alpinia galanga (L.) (Zingiberaceae), showed significant activity against L. major (IC50 = 27.8 ± 0.34 μg mL−1) and was selectively active against T. brucei gambisense and T. brucei rhodeisense with potent activity with IC50 23.66 ± 0.87 μg mL−1 and 26.85 ± 2.20 μg mL−1, respectively.43 Moreover, Komlaga and co-workers, reported isolation of a dibenzyl lignan, named as (+)-phyllanthin (Fig. 3), which displayed potent antiplasmodial activity against chloroquinine-resistant P. falciparum strains W2 and 3D7 (IC50 = 26.23 ± 3.47 & 5.65 ± 1.48 μM, respectively).44
According to previous studies, the mechanisms underlying the antimalarial activity of natural products are most likely through interfering with the parasite's haem detoxification, triggering oxidative stress and lipid peroxidation, inhibition of the enzymes involved in fatty acid, protein and calcium metabolism.45,46
Clearly, we highlighted the pivotal role of natural products (especially secondary metabolites) in compacting malaria as an epidemic disease. Hence, this study encompasses screening of the antimalarial activities of both phenylethanoids and phenylpropanoids, in addition to their derivatives via computational approaches using in silico molecular docking studies, aiming at discovery of novel scaffolds that afford safer and effective medicinal treatments to reduce the high malaria mortality and morbidity rates and to overcome the limitations of the conventional antimalarial drugs. Also, exploring antimalarial activity of glandularin isolated from isolated from the leaves of Glandularia × hybrida against 3D7 strains of P. falciparum, in addition to its binding interactions within crystal structure of three known P. falciparum active sites.
Experimental
Molecular docking simulations
In silico simulations of 25 molecules were performed using Molecular Operating Environment (MOE ® software) within crystal structure of three P. falciparum proteins; P. falciparum cell cycle regulator and non-human derived cyclin-dependent kinase (CDK) which called the P. falciparum kinase (PfPK5; PDB ID: 1V0P), P. falciparum cytochrome bc1 complex (cyt bc1; PDB ID: 4PD4), and P. falciparum lysyl-tRNA synthetase (PfKRS1; PDB ID: 6AGT) revealed from protein data bank (RCSB Protein data bank; https://www.rcsb.org/). Crystal structure of three proteins were validated by re-docking of co-crystallized ligands and their docking score and RMSD (Å) were in the acceptable range. Structure of 25 test molecules were drawn using Chem®Draw program and were energy minimized using MOE ligand preparation tool. Finally, docking protocol and visual inspection of obtained docking poses was done as reported (diterpenoids profile of the marine sponge Chelonaplysilla erecta and candidacy as potential antitumor drugs investigated by molecular docking and pharmacokinetic studies, Natural Product Research, https://doi.org/10.1080/14786419.2022.2063856), and data were listed in Table S3† and represented as schematic diagrams in Fig. 4, 5 and S3.†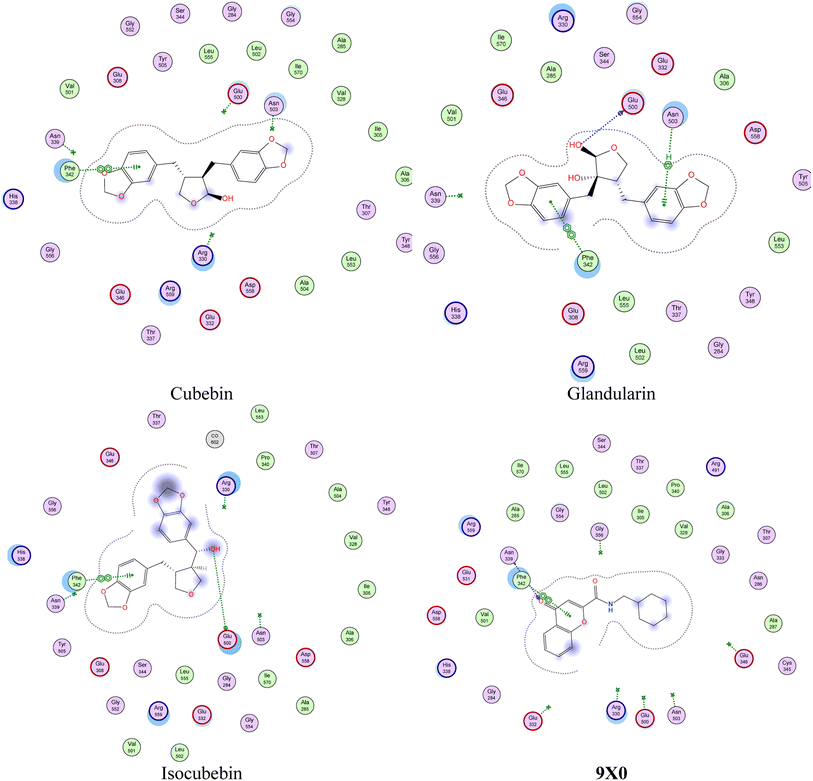 | ||
| Fig. 4 Presumptive mode of interaction of test molecules and 9X0 within active site of P. falciparum lysyl-tRNA synthetase (PfKRS1; PDB ID: 6AGT). | ||
Plant preparation and glandularin isolation
In a previous study, leaves of Glandularia × hybrida were extracted using ethanol and the ethanolic extract was further defatted using n-hexane. The defatted extract was subjected to a series of chromatographic procedures to isolate glandularin. Glandularin, a dibenzylbutyrolactolic lignin, was obtained as colorless residue, identified using different spectroscopic techniques, and the stereocenters were determined using X-ray crystallographic analysis. Glandularin has been previously characterized.47P. falciparum culture medium and maintenance
The parasites were cultured as described previously.48 In brief, parasites were grown RPMI 1640 medium supplemented with 5% pooled human sera A+, along with 11 mM glucose, 25 mM HEPES, and 29 μM hypoxanthine. Human sera were heat-deactivated, aliquot and stored frozen at −20 °C until use. Complete medium was prepared fresh just before use and pre-warmed at 37 °C.Synchronization of parasite culture
Sorbitol treatment or ring stage isolation is a strategy for synchronizing parasite culture. Sorbitol synchronization method is based on parasitized RBC membrane differential permeability. Although RBCs are impermeable to sorbitol, infected RBCs with mature stages have a permeable membrane due to parasite-induced structural changes. This feature is utilized to selectively destroy mature forms of the parasite using osmotic shock while leaving uninfected RBCs and RBCs parasitized by ring stages unaffected. Synchronization was done using the procedure as reported before.49 Primarily, Sorbitol synchronized parasites (100 μl) at 2% haematocrit and 1% parasitaemia were incubated under normal culture conditions in the absence or presence of increasing concentrations of the tested compound. Artemisinin was used as positive controls, while 0.4% (v/v) DMSO was used as the negative control. Individual samples were seeded in 96 well plates and incubated at 37 °C for 48 h.SYBR green-I fluorescence assay for parasitemia determination
The SYBR green-I-based fluorescence assay was performed to measure the growth inhibition effect of glandularin against 3D7 strains of P. falciparum as described by Johnson, et al. 2007.50,51 After 12, 24 and 48 h of incubation, 100 μl of SYBR green I lysis buffer was added to each well and mixed gently twice with a multi-channel pipette and incubated in dark at 37 °C for 1 h. Fluorescence was measured with a Victor fluorescence multi-well plate reader (PerkinElmer, Waltham, MA) with an excitation and emission wavelength of 485 and 530 nm, respectively. The fluorescence counts were plotted against the drug concentration and the 50% inhibitory concentration (IC50) was determined by analysis of dose response curves.Results were validated microscopically by examination of Giemsa-stained smears of glandularin treated parasite cultures. Blood films were stained with a 7.5% Giemsa solution (pH of 7.2) for 15 min. Parasitemia was counted using a bright field microscope, and the morphological changes were noted via imaging.
Results and discussion
The main problem facing malaria transmission areas is the development of drug resistant parasite strains. This enhances the importance of discovering new antimalarial agents with better activity.52 Our study aimed to spot the light on antimalarial active phenylethanoids and phenylpropanoids through in silico and in vitro assessments.Molecular docking simulations
A series of 261 naturally occurring phenylethanoids and phenylpropanoids from different plant families (Tables S1, S2 and Fig. S1†) including 27 representatives with previously reported antiprotozoal activity (Table S1 and Fig. S2†) were investigated for their antimalarial potential.Based on Gao et al. findings about the effect of quinone on ubiquitin in bc1 complex and inhibition of Qi site of P. falciparium cyt bc1,53 and Hoepfner et al. who utilized the reverse genomic approaches to identify P. falciparum lysyl-tRNA synthetase as the novel druggable drug target for both blood and liver stages for a compound identified from such phenotypic screening, we run two different virtual screening experiments, one through target predication simulations using Swiss Target Predication facility which gave insights about major target for such class of compounds as inhibitors of both kinases and secreted proteins54 and other one using in silico molecular docking screening within active site of 3 proteins; P. falciparum kinase, P. falciparum cytochrome bc1 complex, and P. falciparum lysyl-tRNA synthetase; isolated from P. falciparum malarial strain. As shown in Table S3,† most of selected compounds have moderate to strong docking score within active site of selected proteins, as shown in Fig. S4.†
Remarkably, some of these molecules were found to have a docking score higher with tighter binding interactions (in the form of H-donor and H-acceptor, in addition to hydrophobic interactions) with various amino acids lining active sites than did the co-crystallized ligands, as shown in Fig. S3 and S5.† Moreover, exploration of binding interactions of glandularin and its structurally-related compounds; cubebin and isocubebin; within one of the most popular proteins affecting P. falciparum activity; lysyl-tRNA synthetase (PfKRS1),55 as shown in Fig. 4, revealed interestingly that glandularin showed comparable binding energy to that of co-crystallized ligand; 9X0; with strong binding interactions with key amino acid residues lining PfKRS1 active site in the form of three stabilizing H-bonds and hydrophobic interactions indicating its possibility as antimalarial target, as shown in Fig. 5.
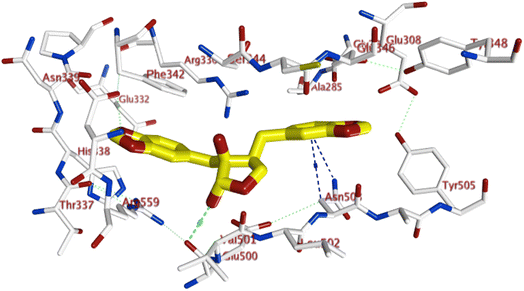 | ||
| Fig. 5 3D binding interactions of glandularin within 6AGT active site; showing H-bonding (red-dotted lines) and various hydrophobic interactions (blue-dotted lines). | ||
Interestingly, there isn't much data in available literature discussing the structural activity relationship of these compounds selected for molecular docking study and how their structures might affect their biological activity on target proteins. Hence, few of these well-characterized compounds were selected and checked for their affinity for the study's chosen target proteins correlated with their structures to get a good idea of their biological activity in upcoming in vitro tests.
Firstly, by examining the docking score of the well-known polyphenols (feddeiphenol A and C, and surinamensin) within the active site of P. falciparum kinase (PfPK5; PDB ID: 1V0P) (Table S3†), we found that there is no big difference between the C-linked polynuclear compound, feddeiphenol A, and its ether-analog feddeiphenol C. This might be because the chelate-type intramolecular H-bond formed between etherial O-atom and neighbouring OH-group which impairs free rotation around the central atom and hence it does not significantly improve docking efficiency compared to non-etherial analog. However, when we worked on docking simulations of these compounds within P. falciparum lysyl-tRNA synthetase (PfKRS1; PDB ID: 6AGT) to examine the significance of the presence of additional methoxy group in one of surinamensin's aromatic rings, we found that it didn't significantly improve the docking profile of surinamensin compared to its dimethoxy-analog, feddeiphenol C, and by visually inspecting its docking poses, indeed none of the 3,4,5-OMe groups participate in any binding interactions. Moreover, the hydroxy propanone side chain of feddeiphenol C was found to participate in a strong H-bond interaction with ASN 339, which could be responsible for raising its docking score above the surinamensin one.
Secondly, examining caffeic acid derivatives with moderate to strong docking score (rosmarinic acid, ethyl rosmarinate, and shimobashiric acid B) (Table S3†), we found that the introduction of lipophilic group as ethyl group of ethyl ester derivative (ethyl rosmarinate) significantly improved docking score compared to free acids, rosmarinic acid and shimobashiric acid B within P. falciparum kinase (PfPK5; PDB ID: 1V0P) active site which was the opposite found within both P. falciparum cytochrome bc1 complex (cyt bc1; PDB ID: 4PD4), and P. falciparum lysyl-tRNA synthetase (PfKRS1; PDB ID: 6AGT) active sites. Additionally, by examining the docking pose of rosmarinic acid, we found that the carboxylic group has no participation in any binding interactions with amino acid residues lining the 1V0P active site. So, all these interesting results could be attributed to the addition of lipophilicity via a lipophilic alkyl group to these caffeic acid derivatives (as in ethyl rosmarinate and shimobashiric acid B). Finally, the presence of hydroxyl group either on tetrahydrofuran ring or the methylenic carbon improved the docking score of these alcoholic derivatives (isocubebin, cubebin, and glandularin) over non-alcoholic ones (hinokinin and savinin) through the formation of H-bond with amino acid residues lining all the three active sites of P. falciparum kinase (PfPK5; PDB ID: 1V0P), P. falciparum cytochrome bc1 complex (cyt bc1; PDB ID: 4PD4), and P. falciparum lysyl-tRNA synthetase (PfKRS1; PDB ID: 6AGT) proteins is a common finding among members of lignan's class.
In vitro antimalarial activity
The in silico docking study was further augmented by in vitro assessment of the glandularin for their antiplasmodial activity against chloroquine-sensitive 3D7 strain of P. falciparum using SYBR green I-based fluorescence assay and results were listed in Table 1. According to antimalarial activity scale set by Philippe and co-workers, the results showed moderate antimalarial activity of glandularin with an IC50 value of 11.2 μM after 24 hours of incubation56 (N.B. the positive control, artemisinin, showed IC50 of 0.02 μM at the same incubation time interval).| Time (h) | Glandularin IC50 values (μM) |
|---|---|
| 12 | 62.9 |
| 24 | 11.2 |
| 48 | 53.4 |
Remarkably, microscopical examination of Giemsa-stained blood smears of untreated (control) and glandularin-treated parasite cultures showed clearly the antimalarial effect of glandularin on infected cells, as shown in Fig. 6.
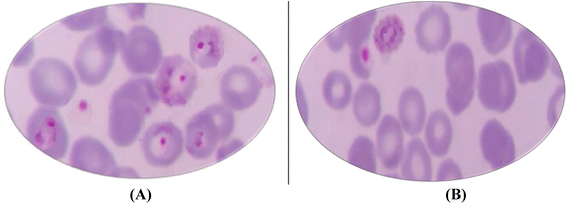 | ||
| Fig. 6 (A) Giemsa-stained thin blood smears showing numerous P. falciparum (ring stage) in the untreated control sample; (B) after treatment with Glandularin. | ||
Glandularin was of a special antimalarial concern in our study due to its antimalarial potential showed in high docking scores in the in silico study, together with promising antimalarial activity against 3D7 strain of P. falciparum in the in vitro study. Glandularin's structure is closely related to cubebin (8α-hydroxy cubebin). Cubebin and its analogues were assessed before for their antiprotozoal activities in many studies revealing their potent activities. Carlis and co-workers assessed cubebin, its oxidation product hinokinin, and dihydrocubebin for their in vitro anthelmintic activity against gastrointestinal nematodes using the egg hatch test, larval development test and L3 migration inhibition test.57 These compounds showed ovicidal activity with EC50 values of 150.00 μg mL−1, 186.70 μg mL−1 and 68.38 μg mL−1, respectively. In larval development test, cubebin showed an EC50 value of 14.89 μg mL−1, and an EC50 value of 30.75 μg mL−1 for hinokinin. On the other hand, hinokinin showed 100% inhibition for the larval development at all concentrations evaluated. In the L3 migration inhibition test, larval migration was inhibited by dihydrocubebin with a value of 100% in all concentrations evaluated, while cubebin and hinokinin exhibited EC50 values of 0.89 μg mL−1 and 0.34 μg mL−1, respectively. Additionally, cubebein and hinokinin, were reported for their antiprotozoal activity against T. cruzi, the primary cause of Chagas' diseases.58 The antiprotozoal activity was assessed in vivo using tissue morphometric analysis, showing significant decrease in parasitemia in the groups treated with cubebin and hinokinin orally. These compounds and their closely structural analogues are worthy to be further investigated for antiprotozoal activity including underlying mechanisms of their activities and studying of their structural activity relationships together with chemical transformations which may help in adjusting their activities.
Conclusions
The study enclosed some of the reported naturally occurring phenylethanoids and phenylpropanoids with diverse activities. Additionally, docking simulations run on 25 selected phenylpropanoids for their antimalarial activities against P. falciparum within active site of 3 proteins; P. falciparum kinase, P. falciparum cytochrome bc1 complex, and P. falciparum lysyl-tRNA synthetase; isolated from P. falciparum malarial strain. The in silico study was in agreement with all the findings reported in the literature about their potential as antiplasmodial agents. Glandularin, as a dibenzylbutyrolactolic lignan, demonstrated an example for naturally occurring antimalarial drugs. Glandularin's docking score to co-crystallized ligand within active site of P. falciparum lysyl-tRNA synthetase revealed its high potential as promising antimalarial drug of natural source. This was more confirmed by the high antimalarial activity of glandularin that was exerted during its in vitro antimalarial assessment against chloroquine-sensitive 3D7 strain of P. falciparum. According to the reported data and the new findings in our study, plants based phenylpropanoids and phenylethanoids could be a new treasure and promising avenue for developing new antimalarial drugs that can be efficiently used against known drug-resistant P. falciparum strains.Conflicts of interest
The authors declare no conflicts of interest.References
- M. Ohashi, M. Amoa-bosompem, K. D. Kwofie, J. Agyapong, R. Adegle, M. M. Sakyiamah, F. Ayertey, K. B. Owusu, I. Tuffour, P. Atchoglo, N. H. Tung, T. Uto, F. Aboagye, A. A. Appiah, R. Appiah-opong, A. K. Nyarko, W. K. Anyan, I. Ayi, D. A. Boakye, K. A. Koram, D. Edoh, S. Yamaoka, Y. Shoyama and N. Ohta, Phyther. Res., 2018, 32, 1617–1630 CrossRef CAS PubMed
.
- D. Zofou, R. B. Nyasa, D. S. Nsagha, F. Ntie-kang, H. D. Meriki, J. C. N. Assob and V. Kuete, Infect. Dis. Poverty, 2014, 1–14 CrossRef PubMed
.
- L. Zhu, R. W. van der Pluijm, M. Kucharski, S. Nayak, J. Tripathi, N. J. White, N. P. J. Day, A. Faiz, A. P. Phyo, C. Amaratunga, D. Lek, E. A. Ashley, F. Nosten, F. Smithuis, H. Ginsburg, L. von Seidlein, K. Lin, M. Imwong, K. Chotivanich, M. Mayxay, M. Dhorda, H. C. Nguyen, T. N. T. Nguyen, O. Miotto, P. N. Newton, P. Jittamala, R. Tripura, S. Pukrittayakamee, T. J. Peto, T. T. Hien, A. M. Dondorp and Z. Bozdech, Commun. Biol., 2022, 5, 1–13 CrossRef PubMed
.
- S. O. Bruce, Secondary Metabolites from Natural Products, Secondary Metabolites, Trends and Reviews, 2016 Search PubMed
.
- U. R. Abdelmohsen, A. Albohy, B. S. Abdulrazik, S. A. L. Bayoumi, L. G. Malak, I. S. A. Khallaf, G. Bringmann and S. F. Farag, RSC Adv., 2021, 11, 16970–16979 RSC
.
- A. M. Ahmed, B. K. Mahmoud, N. Millán-Aguiñaga, U. R. Abdelmohsen and M. A. Fouad, RSC Adv., 2023, 13, 1339–1369 RSC
.
- O. H. Abdelhafez, J. R. Fahim, S. Y. Desoukey, M. S. Kamel and U. R. Abdelmohsen, Chem. Biodiversity, 2019, 16(6), e1800692 Search PubMed
.
- A. Sharma, B. Shahzad, A. Rehman, R. Bhardwaj and M. Landi, Molecules, 2019, 24, 1–22 Search PubMed
.
- I. F. G. Mera, D. E. G. Falconí and V. M. Córdova, Bionatura, 2019, 4(4), 1000–1009 Search PubMed
.
- A. K. Neelam and K. K. Sharma, Crit. Rev. Food Sci. Nutr., 2020, 60, 2655–2675 CrossRef PubMed
.
- W. Y. Huang, Y. Z. Cai and Y. Zhang, Nutr. Cancer, 2010, 62, 1–20 CAS
.
- M. Petersen, D. Strack and U. Matern, Annu. Plant Rev. Online, 2018, 2, 147–217 Search PubMed
.
- T. Vogt, Mol. Plant, 2010, 3, 2–20 CrossRef CAS PubMed
.
- F. Zálešák, D. J. Denis and J. Pospíšil, Pharmacol. Res., 2019, 146, 104284 CrossRef PubMed
.
- V. A. Kurkin, Chem. Nat. Compd., 2003, 39, 87–110 CrossRef
.
- R. G. Kamkumo, A. M. Ngoutane, L. R. Tchokouaha, P. V. Fokou, E. A. Madiesse, J. Legac, J. J. Kezetas, B. N. Lenta, F. F. Boyom, T. Dimo, W. F. Mbacham, J. Gut and P. J. Rosenthal, Malar. J., 2012, 11, 1–7 CrossRef PubMed
.
- R. B. Teponno, S. Kusari and M. Spiteller, Nat. Prod. Rep., 2016, 33, 1044–1092 RSC
.
- X. Y. Xu, D. Y. Wang, Y. P. Li, S. T. Deyrup and H. J. Zhang, Plant-derived lignans as potential antiviral agents: a systematic review, Springer Netherlands, 2022, vol. 21 Search PubMed
.
- Q. Cui, R. Du, M. Liu and L. Rong, Molecules, 2020, 25(1), 183 CrossRef CAS PubMed
.
- A. Latif, Y. Du, S. R. Dalal, M. L. Fernández-Murga, E. F. Merino, M. B. Casserac, M. Goetze and D. G. I. Kingstona, Chem. Biodivers., 2017, 14, 371–390 CrossRef PubMed
.
- L. Wu, M. I. Georgiev, H. Cao, L. Nahar, H. R. El-Seedi, S. D. Sarker, J. Xiao and B. Lu, Med. Res. Rev., 2020, 40, 2605–2649 CrossRef CAS PubMed
.
- M. Gálvez, C. Martin-cordero and M. J. Ayuso, DOI:10.1016/S1572-5995(06)80037-2.
- V. A. Kurkin, Adv. Biol. Chem., 2013, 03, 26–28 CrossRef CAS
.
- J. Pan, C. Yuan, C. Lin, Z. Jia and R. Zheng, Pharmazie, 2003, 58, 767–775 CAS
.
- R. de C. Sá, L. N. Andrade, R. dos R. B. de Oliveira and D. P. de Sousa, Molecules, 2014, 19, 1459–1480 CrossRef PubMed
.
- D. K. Maurya, T. Paul and A. Devasagayam, Food Chem. Toxicol., 2010, 48, 3369–3373 CrossRef CAS PubMed
.
- R. Ilijeva and G. Buchbauer, Nat. Prod. Commun., 2016, 11, 1619–1629 CrossRef PubMed
.
- J. Kyselka, D. Rabiej, M. Dragoun, F. Kreps, Z. Burčová, I. Němečková, J. Smolová, M. Bjelková, A. Szydłowska-czerniak, Š. Schmidt, L. Šarman and V. Filip, Eur. Food Res. Technol., 2017, 243, 1633–1644 CrossRef CAS
.
- A. Narasimhan, M. Chinnaiyan and B. Karundevi, Eur. J. Pharmacol., 2015, 761, 391–397 CrossRef CAS PubMed
.
- A. Meeprom, C. B. Chan, W. Sompong and S. Adisakwattana, Biomed. Pharmacother., 2018, 101, 777–785 CrossRef CAS PubMed
.
- R. S. Jansi, A. Khusro, P. Agastian, A. Alfarhan and N. A. Al-dhabi, Sci. Total Environ., 2021, 759, 143539 CrossRef PubMed
.
- R. M. Perez G, Pharm. Biol., 2003, 41, 107–157 CrossRef CAS
.
- A. Hematpoor, S. Y. Liew, W. L. Chong, M. S. Azirun, V. S. Lee and K. Awang, PLoS One, 2016, 11, 1–27 CrossRef PubMed
.
- H. A. Oketch-Rabah, S. F. Dossaji, S. B. Christensen, K. Frydenvang, E. Lemmich, C. Cornett, C. E. Olsen, M. Chen, A. Kharazmi and T. Theander, J. Nat. Prod., 1997, 60, 1017–1022 CrossRef CAS PubMed
.
- A. da Silva Filho, E. S. Costa, W. R. Cunha, M. L. A. e Silva and J. K. N. P. Dhammika Nanayakkara Bastos, Phyther. Res., 2008, 22, 544–549 CrossRef PubMed
.
- A. Jiménez-Arellanes, R. León-Díaz, M. Meckes, A. Tapia, G. M. Molina-Salinas, J. Luna-Herrera and L. Yépez-Mulia, Evid. Based Complementary Altern. Med., 2012, 2012, 593403 Search PubMed
.
- E. Otero, E. García, G. Palacios, L. M. Yepes, M. Carda, R. Agut, I. D. Vélez, W. I. Cardona and S. M. Robledo, Eur. J. Med. Chem., 2017, 141, 73–83 CrossRef CAS PubMed
.
- M. dos Santos Maia, J. P. R. Silva, T. A. de Lima Nunes, J. M. S. de Sousa, G. C. S. Rodrigues, A. F. M. Monteiro, J. F. Tavares, K. A. da Franca Rodrigues, F. J. B. Mendonça-Junior, L. Scotti and M. T. Scotti, Molecules, 2020, 25, 1–34 CrossRef PubMed
.
- A. B. Mahmoud, O. Danton, M. Kaiser, S. Han, A. Moreno, S. A. Algaffar, S. Khalid, W. K. Oh, M. Hamburger and P. Mäser, Molecules, 2020, 25, 1–15 Search PubMed
.
- X. N. Li, J. X. Pu, X. Du, L. M. Yang, H. M. An, C. Lei, F. He, X. Luo, Y. T. Zheng, Y. Lu, W. L. Xiao and H. D. Sun, J. Nat. Prod., 2009, 72, 1133–1141 CrossRef CAS PubMed
.
- H. J. Zhang, P. A. Tamez, V. D. Hoang, G. T. Tan, N. V. Hung, L. T. Xuan, L. M. Huong, N. M. Cuong, D. T. Thao, D. D. Soejarto, H. H. S. Fong and J. M. Pezzuto, J. Nat. Prod., 2001, 64, 772–777 CrossRef CAS PubMed
.
- L. Mamede, A. Ledoux, O. Jansen and M. Frédérich, Planta Med., 2020, 86, 585–618 CrossRef CAS PubMed
.
- M. I. Sulistyowaty, N. H. Uyen, K. Suganuma, B. Y. A. Chitama, K. Yahata, O. Kaneko, S. Sugimoto, Y. Yamano, S. Kawakami, H. Otsuka and K. Matsunami, Molecules, 2021, 26, 1–12 CrossRef PubMed
.
- G. Komlaga, G. Genta-jouve, S. Cojean, R. A. Dickson, M. L. K. Mensah, P. M. Loiseau, P. Champy and M. A. Beniddir, Tetrahedron Lett., 2017, 58, 3754–3756 CrossRef CAS
.
- N. Tajuddeen and F. R. Van Heerden, Malar. J., 2019, 18, 1–62 CrossRef PubMed
.
- M. L. López, R. Vommaro, M. Zalis, W. de Souza, S. Blair and C. Segura, Parasitol. Int., 2010, 59, 217–225 CrossRef PubMed
.
- N. M. Mohamed, M. A. M. Ahmed, S. I. Khan, F. R. Fronczek, A. F. Mohammed and S. A. Ross, Phytochemistry, 2022, 195, 113054 CrossRef CAS PubMed
.
- W. Trager and J. B. Jensen, Int. J. Parasitol., 1997, 27, 989–1006 CrossRef CAS PubMed
.
- M. Roncalés, J. Vidal, P. A. Torres and E. Herreros, Open J. Epidemiol., 2015, 05, 71–80 CrossRef
.
- J. D. Johnson, R. A. Dennull, L. Gerena, M. Lopez-Sanchez, N. E. Roncal and N. C. Waters, Antimicrob. Agents Chemother., 2007, 51, 1926–1933 CrossRef CAS PubMed
.
- M. Leidenberger, C. Voigtländer, N. Simon and B. Kappes, in Cell Viability Assays, Methods in Molecular Biology, ed. O. Gilbert and D. Friedrich, Humana Press, New York, NY, 2017, vol. 1601, pp. 97–110 Search PubMed
.
- P. Chaniad, M. Mungthin, A. Payaka, P. Viriyavejakul and C. Punsawad, BMC Complementary Med. Ther., 2021, 21, 1–10 CrossRef PubMed
.
- X. Gao, X. Wen, L. Esser, B. Quinn, L. Yu, C. A. Yu and D. Xia, Biochemistry, 2003, 42, 9067–9080 CrossRef CAS PubMed
.
- D. Hoepfner, C. W. McNamara, C. S. Lim, C. Studer, R. Riedl, T. Aust, S. L. McCormack, D. M. Plouffe, S. Meister, S. Schuierer, U. Plikat, N. Hartmann, F. Staedtler, S. Cotesta, E. K. Schmitt, F. Petersen, F. Supek, R. J. Glynne, J. A. Tallarico, J. A. Porter, M. C. Fishman, C. Bodenreider, T. T. Diagana, N. R. Movva and E. A. Winzeler, Cell Host Microbe, 2012, 11, 654–663 CrossRef CAS PubMed
.
- B. Baragaña, B. Forte, R. Choi, S. N. Hewitt, J. A. Bueren-Calabuig, J. P. Pisco, C. Peet, D. M. Dranow, D. A. Robinson, C. Jansen, N. R. Norcross, S. Vinayak, M. Anderson, C. F. Brooks, C. A. Cooper, S. Damerow, M. Delves, K. Dowers, J. Duffy, T. E. Edwards, I. Hallyburton, B. G. Horst, M. A. Hulverson, L. Ferguson, M. B. Jiménez-Díaz, R. S. Jumani, D. D. Lorimer, M. S. Love, S. Maher, H. Matthews, C. W. McNamara, P. Miller, S. O'Neill, K. K. Ojo, M. Osuna-Cabello, E. Pinto, J. Post, J. Riley, M. Rottmann, L. M. Sanz, P. Scullion, A. Sharma, S. M. Shepherd, Y. Shishikura, F. R. C. Simeons, E. E. Stebbins, L. Stojanovski, U. Straschil, F. K. Tamaki, J. Tamjar, L. S. Torrie, A. Vantaux, B. Witkowski, S. Wittlin, M. Yogavel, F. Zuccotto, I. Angulo-Barturen, R. Sinden, J. Baum, F. J. Gamo, P. Mäser, D. E. Kyle, E. A. Winzeler, P. J. Myler, P. G. Wyatt, D. Floyd, D. Matthews, A. Sharma, B. Striepen, C. D. Huston, D. W. Gray, A. H. Fairlamb, A. V. Pisliakov, C. Walpole, K. D. Read, W. C. Van Voorhis and I. H. Gilbert, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 7015–7020 CrossRef PubMed
.
- G. Philippe, L. Angenot, P. De Mol, E. Goffin, M. P. Hayette, M. Tits and M. Frédérich, J. Ethnopharmacol., 2005, 97, 535–539 CrossRef PubMed
.
- M. S. de P. Carlis, A. Féboli, A. C. de Laurentiz, R. da S. Filardi, A. H. P. de Oliveria, M. L. A. e Silva, L. A. dos Anjos, L. G. Magalhães and R. da S. de Laurentiz, Vet. Parasitol., 2019, 275, 108932 CrossRef PubMed
.
- V. R. Esperandim, D. da Silva Ferreira, K. C. S. Rezende, W. R. Cunha, J. Saraiva, J. K. Bastos, M. L. A. e Silva and S. de Albuquerque, Exp. Parasitol., 2013, 133, 442–446 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra04242a |
| This journal is © The Royal Society of Chemistry 2023 |