Open Access Article
Open Access ArticleRegulation of recombinant humanized collagen on HAP growth and its molecule simulation†
Zhilin Huang‡
 ab,
Chucheng Wang‡
ab,
Chucheng Wang‡ ab,
Xiaohui Chenab,
Shan Dingab,
Qi Xiangc,
Mo Xie
ab,
Xiaohui Chenab,
Shan Dingab,
Qi Xiangc,
Mo Xie ad,
Yadong Huang
ad,
Yadong Huang c and
Hong Li
c and
Hong Li *ab
*ab
aCollege of Chemistry and Materials Science, Jinan University, Guangzhou, 510632, China. E-mail: tlihong@jnu.edu.cn
bEngineering Research Center of Artificial Organs and Materials, Ministry of Education, Guangzhou, 510632, China
cInstitute of Biomedicine and Guangdong Provincial Key Laboratory of Bioengineering Medicine, Jinan University, Guangzhou, 510632, China
dGuangdong Provincial Key Laboratory of Functional Supramolecular Coordination Materials and Applications, Guangzhou, 510632, China
First published on 1st September 2023
Abstract
Hydroxyapatite (HAP) in natural bone is formed under the regulation of natural collagen I. Here, we report how recombinant humanized collagen I (rhCol I) regulates the growth of HAP nanocrystals in a long belt shape 100–150 nm in width and 200–300 nm in length. MD simulation results showed that the interactions between rhCol I and the (001), (100), and (211) planes of HAP mainly contributed to the electrostatic force and van der Waals forces via COO⋯Ca, –NH⋯Ca, CH⋯OPO3, and NH⋯OPO3 bonds, respectively. On the (001) plane, the interaction between –COO− and Ca was stronger than on the (100) and (211) planes, resulting in a large electrostatic force, which inhibited the growth of the (001) plane. The lowest energy of adsorption to the (211) plane resulted in the preferential growth of the (211) plane due to the weakest interaction with rhCol I. The detailed correlation between HAP and rhCol I could explain HAP growth under regulation by rhCol I. This study provides a reference for the bio-application of recombinant collagen.
1. Introduction
Hydroxyapatite (HAP, Ca10(PO4)6(OH)2)1 is one of the most important components of bone minerals, and synthesized HAP shows great promise in hard tissue restoration.2–5 Various synthesized HAP materials have been obtained and applied; however, these HAP materials, whose structure and biofunction are still far away from the natural bone apatite, still can not fully meet the requirements for the clinical treatment.6,7 Therefore, the synthesis of HAP is still a hot topic in the biomedical field.It is worth mentioning that bone is not monolithic but is essentially a nanohybrid, which constitutes HAP nanocrystals and collagen fibrils. HAP in natural bone is formed under the regulation of macromolecules via a process of biomineralization, in which collagen plays an important role.8,9 As we know, collagen may initiate and direct the growth of apatite crystals, providing a structural template for the directed nucleation and growth of apatite.10–12 Resembling bone formation in a biomimetic way, the HAP nanocrystals were prepared by regulating the nucleation and growth of calcium phosphate minerals by collagen in an aqueous solution.13 However, the use of natural collagen derived from animal tissue may raise many concerns related to the quality, purity, and possible immunological reactions,14 and also can lead to a variability in cross-linking to the crystal phase, or changes in the nanostructure, or even the bio-functions of the formed HAP. Therefore, scientists are trying to find a biomolecule that possesses collagen's partial or full functions in the biomedical field, such as amino acids, peptides, and other proteins.15
With the progress in genetic engineering, recombinant collagen with designated structural and functional characteristics has been developed and shown potential in the biomedical field. Recombinant collagen is generally prepared by transcribing a specific gene segment and is expressed in yeast, bacteria, or animal cells,16 with a consistent formulation output and high productivity. Considering the homology of the amino acid sequence, a human collagen gene that encodes a specific type of human collagen is mainly chosen to produce predictable and reliable recombinant collagen. In the early days, Cui et al. investigated the mineralization of recombinant human-like collagen, and found that it could regulate the deposition of HAP nanocrystals by the self-assembly of nano-fibrils of mineralized collagen.17 However, there was some differences between the amino acid sequences of recombinant human-like collagen and native type I collagen, because some modification was done in the recombinant collagen due to solubility issues. More recently, in March 2021, a standard guide for the definition of recombinant collagen was issued by the National Medical Products Administration of China (NMPA), and recombinant humanized collagen (rhCol I) was defined as a full-length or fragment of a functional amino acid sequence encoded by a specific type of human collagen gene, or a combination of the above functional fragments. RhCol I has the advantages of processability, being virus-free and water-soluble, and low immunogenicity, which we speculate mean it can be considered to regulate the formation of HAP in a biomimetic way.
In the study, we tried to explore the regulation of rhCol I on HAP growth. Although both collagen and noncollagenous proteins have been reported to contribute to the biomineralization of HAP,18,19 the collagen-assisted mineralization mechanism at the molecular level is still underdeveloped. Xue et al., using computational modeling, performed a detailed analysis of the Ca–P cluster growth process in the solution phase in the presence of human type I collagen, and revealed a nonclassical nucleation pathway for the HAP nucleation with the collagen I as the seed.20 In recent studies, molecular dynamics (MD) simulations have been employed as an efficient method to understand the interactions between collagen and the HAP face.21,22 The interaction sites,23–25 adsorption energies,26 substitution energies,27,28 and other information were obtained based on MD simulations. Even, the interfacial interactions between the HAP crystallographic surfaces and complex polymers (such as PE, PLA, and PA66) have been studied by calculating their binding energies.29 So far, and to the best of our knowledge, no study has yet analyzed the interaction between HAP and rhCol I. Herein, we employed MD simulation to study the interfacial interactions between HAP crystallographic surfaces and rhCol I. The study might help to explain the rhCol I dependence on HAP growth, and may provide support to develop novel biomaterials for tissue repair.
2. Experimental
2.1 Simulation and models
The adsorption energies due to the interaction of the collagen and HAP surfaces were calculated according to eqn (1):37
| Eads = Egra + Emolecule − Etot | (1) |
2.2 Synthesis of HAP crystals
Calcium chloride (CaCl2, purity ≥ 96 wt%), and dipotassium hydrogen phosphate (K2HPO4, purity ≥ 99 wt%) were purchased from Sigma-Aldrich. All chemicals were used without further purification. The solutions were prepared with ultrapure water (0.22 lS, 25 °C, MilliQ©, Millipore).First, 30 mM CaCl2 solution and 3 mM K2HPO4 solution as the stock liquids were prepared, and then appropriate amounts of the CaCl2 and K2HPO4 stock liquid were taken respectively and added into a centrifuge tube containing rhCol I solution. The final concentrations of CaCl2, K2HPO4, and rhCol I were 2.5 mM, 1.5 mM, and 200 μg mL−1 respectively. Finally, the mixed solution was placed in a constant temperature water bath at 37 °C for mineralization for 3 days, and the precipitate was taken out after centrifugation and washed several times using ultrapure water. As a control, the above reaction was carried out in the absence of rhCol I.
2.3 Characterization
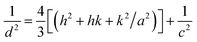 | (2) |
D = kλ/(β![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ) θ)
| (3) |
In formula (3), D is the crystal diameter, k is the ratio constant of 0.89, λ is the wavelength of the X-ray of 0.15406 nm, β is the width of the diffraction peak, and θ is the corresponding angle of the diffraction peak.
3. Results and discussion
3.1 Synthesized HAP
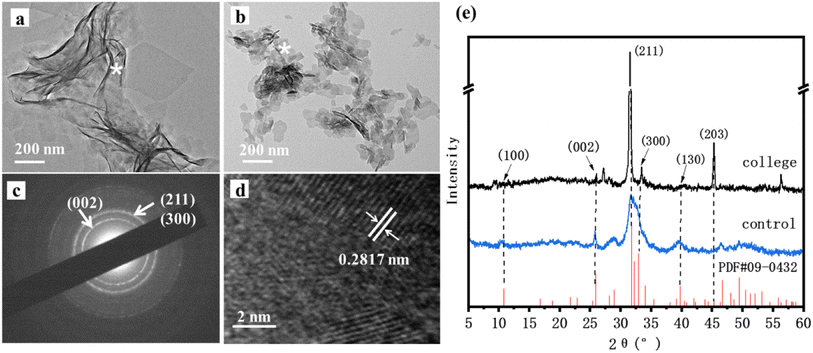
HAP crystals were observed via TEM as shown in Fig. 3a and b. It could be clearly seen that the synthesized HAP in the presence of the collagen had a belt-like structure, and these belts were 100–150 nm in width and 200–300 nm in length. However, it was found that plate-like HAP crystals 10–20 nm in width and 20–50 nm in length long formed in the control (Fig. 3b). In this case, we suppose that rhCol I acts as a soft template to guide and regulate HAP crystal growth with a long belt structure. The crystal structure of HAP in the presence of the rhCol I was further confirmed by selected area electron diffraction (SAED), as shown in Fig. 3c. The pattern of both HAP crystals showed a polycrystalline ring, and resolvable ring patterns ascribed to the (002), (211), and (300) planes of the HAP, respectively. The crystal structure was further evaluated by HRTEM lattice fringes (Fig. 3d). The lattice fringes of the synthesized HAP in the presence of the rhCol I showed a d-spacing of 0.2817 nm, corresponding to (211) planes.The XRD patterns shown in Fig. 3e gave all standard peaks matching with the JCPDS 09-0432 card of standard HAP, thus confirming the formation of the HAP phase in all the samples. The diffraction peak of the three main groups of HAP synthesized by rhCol I was at about 32°, corresponding to the (211), (112), and (300) crystal planes of HAP, and the peak shape was sharp and the intensity was high, indicating that the crystallinity of HAP was incisive. The diffraction peak intensity ratios of (211) to the (002) crystal plane in the standard HAP (JCPDS 09-0432) and blank control groups were 2.5 and 1.69, respectively, while the diffraction peak intensity ratio of the existing (211) to (002) crystal plane in the presence of rhCol I was 38.5, indicating that rhCol I made HAP grow preferentially along the (211) plane. The values for the HAP cell parameters and crystal size can be calculated by formulas (2) and (3), as shown in Table 1. The results show that the rhCol I regulated the crystal growth of HAP, which caused the cell parameters to increase the a value and decrease the c value. The crystal size increased to 50.300 nm according to Table 1.
| Sample | a (Å) | c (Å) | c/a | D (Å) |
|---|---|---|---|---|
| HAP (JCPDS 09-0432) | 9.418 | 6.884 | 0.731 | — |
| Blank control group | 9.473 | 6.852 | 0.723 | 216.200 |
| rhCol I | 9.523 | 6.820 | 0.716 | 503.000 |
Most studies suggest that the interaction between the collagen matrix and HAP crystals determines the eventual morphology of the HAP platelets in the bone.38,39 The (001) crystal plane is the most stable of all the crystal planes of HAP, and the interaction on the (001) crystal plane determines the morphology of HAP in the presence of proteins.40 However, HAP crystals in tooth enamel with high crystallinity have shown that matrix proteins only affect the growth of the (010) surface.41 This means that the interaction between the matrix and HAP also affects its preferential growth at different planes. Also, although most of the prior studies have shown the strong growth of (211) plane as the HAP crystal,42 no details about the interaction of HAP on the (211) plane have been reported. Herein, we tried to analyze the interaction between rhCol I and the (001) plane, (100) plane, and (211) plane using MD simulations.
3.2 Interaction between HAP and rhCol I
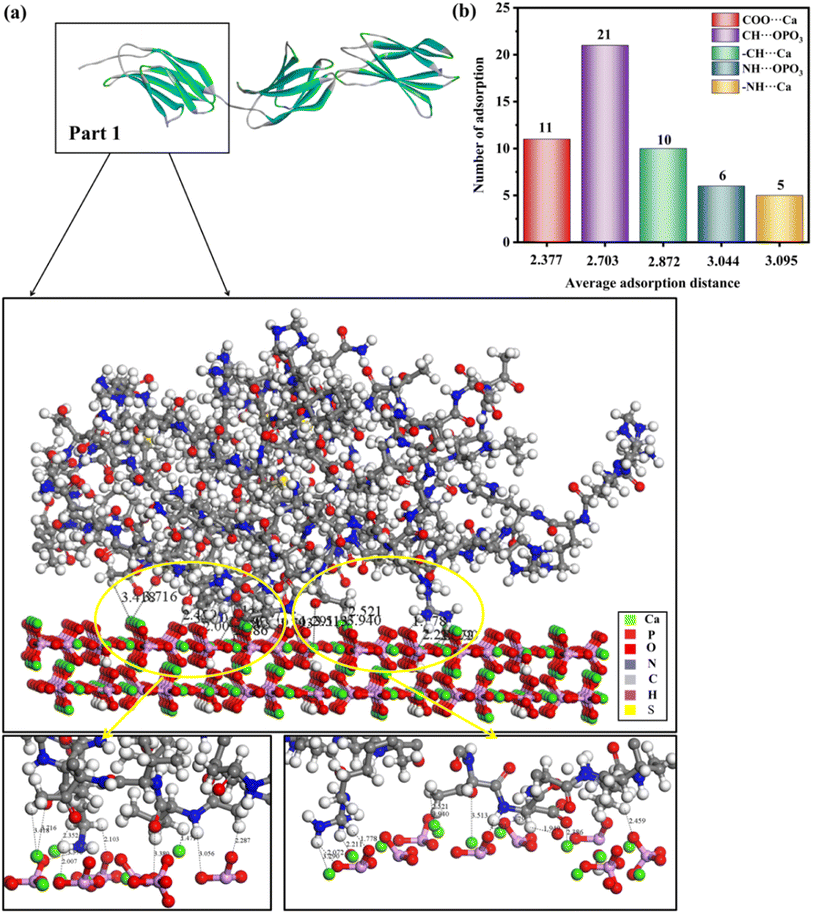
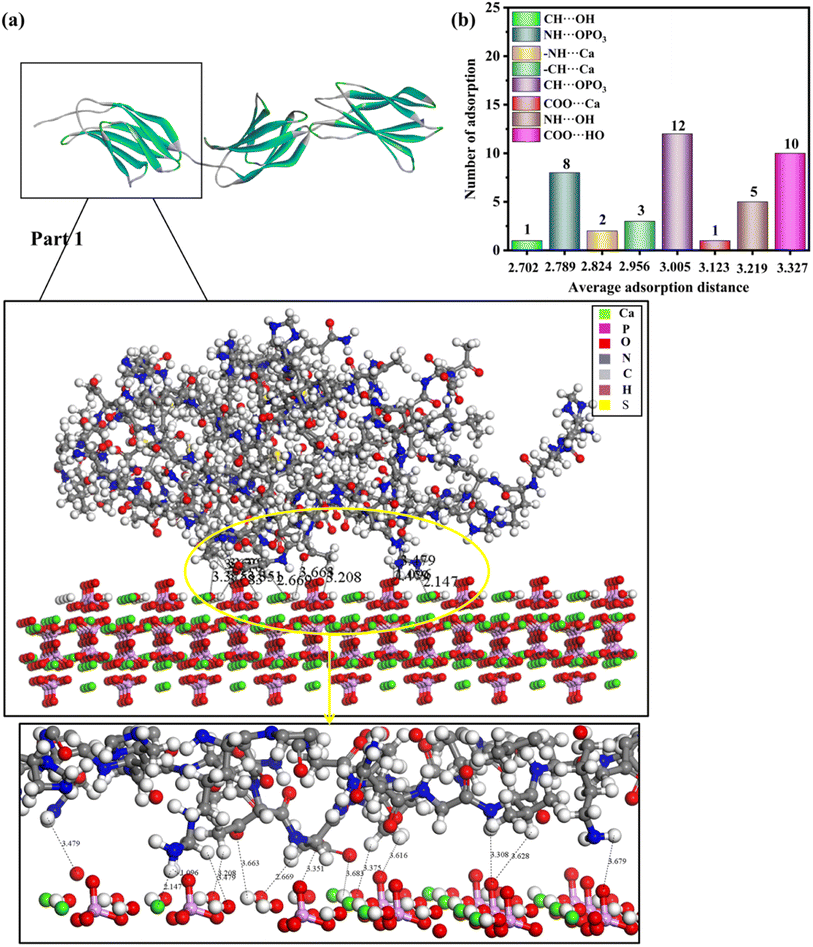
Generally, there are multiple steps for the interaction of biomolecules on the HAP surface: first of all, the molecule would spread to the HAP surface and then be adsorbed at the active site of the HAP surface, eventually overlapping the structure after adsorption. As studied by Xue11 and Almora-Barrios,13 type I collagen can maintain a stable structure quite well, so we assumed that the rhCol I can keep the same configuration, as shown in Fig. 2a. Detailed information on the active sites of rhCol I for the (001), (100), and (211) surfaces of HAP was obtained and is shown in Fig. 4–6 and Fig S1–S6,† respectively.Looking at 3 parts of rhCol I, Part 1 was adsorbed at the (001) surface, as shown in Fig. 4a, and Part 2 and Part 3 on the (001) crystal plane, as shown in Fig. S1 and S2,† respectively. The most common interaction site of full-length rhCol I on the HAP (001) surface is between the hydrogen atom of –CH2 groups with the oxygen atom of the phosphoric acid group (CH⋯OPO3 = 1.686–3.967 Å), and the average interaction distance was 2.703 Å, according to our analysis as shown in Fig. 4b. Also, the oxygen atom of the carboxylic acids and the hydrogen atom of the –CH2 group can interact strongly with surface calcium ions (COO⋯Ca = 1.496–3.513 Å, –CH⋯Ca = 1.471–3.910 Å), respectively. It has been reported that the hydrogen atoms of the amine group (>NH2) will migrate to the oxygen atoms of the phosphoric acid group on the surface, which will form hydrogen bonds,21 whose range for NH⋯OPO3 is 2.072–3.701 Å. In addition, the hydrogen atom of the amine group has a small amount of interaction with surface calcium ions (–NH⋯Ca = 2.007–3.655 Å).
As shown in Fig. 5a, S3 and S4,† the rhCol I molecule was adsorbed on the (100) surface of HAP. The shortest average adsorption distance was between the hydrogen atoms of –CH2 groups with the oxygens of hydroxide (CH⋯OH), and the average adsorption distance was 2.702 Å. The most favorable mode of adsorption for the rhCol I is through a lot of interactions between the hydrogen atoms of –CH2 groups with the oxygen atom of the phosphoric acid group (CH⋯OPO3 = 2.314–3.740 Å), because they have the largest number of interactions, and the adsorption distance was shorter with an average adsorption distance of 3.005 Å. Hydrogen bondings occur between the hydrogen atom of the amine group with the oxygen of the phosphoric acid group and the oxygens of hydroxide (NH⋯OPO3 = 1.096–3.842 Å, NH⋯OH = 3.089–3.320 Å), which are in accord with results in the recent computational study of Almora-Barrios et al.21,43 The hydrogen atoms of the amine group and the hydrogen atoms of the –CH2 group also have a small amount of interaction with the surface calcium ions (–NH⋯Ca = 2.147–3.501 Å, –CH⋯Ca = 1.747–3.879 Å). The interaction of the oxygen atoms of the carboxyl group with surface calcium ion (COO⋯Ca = 3.123 Å) was weaker than that on the (001) plane. However, the interaction between the oxygen atom of the carboxyl group and the hydrogen atom of the hydroxyl group (COO⋯HO = 2.560–3.967 Å) was stronger than that of the (001) plane.
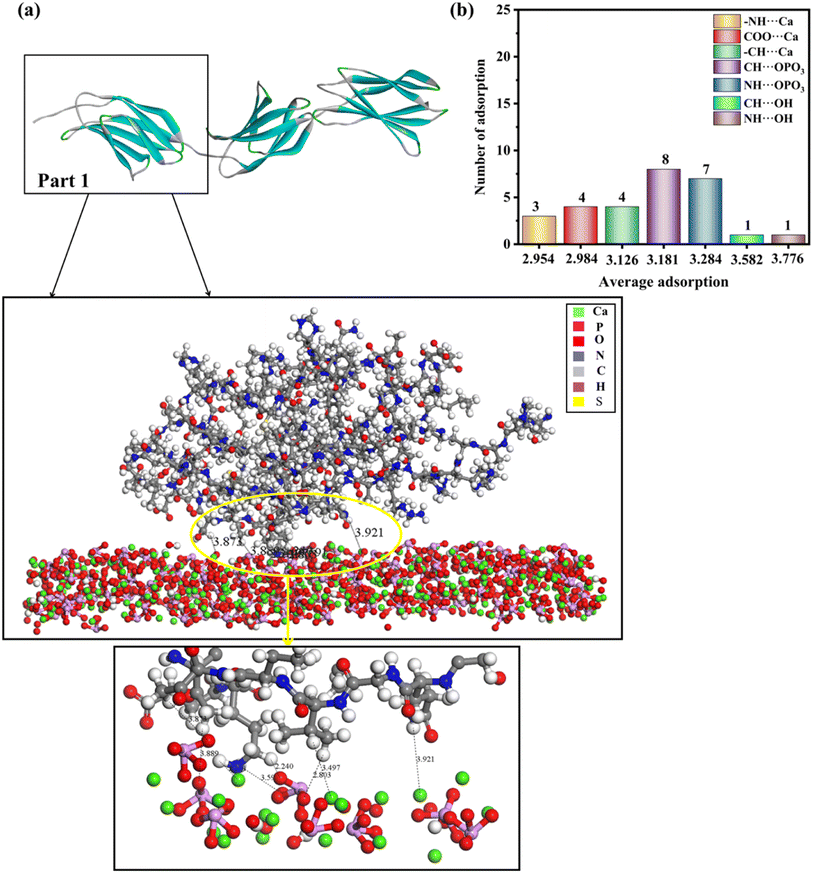
Fig. 6, S5 and S6† show the lowest-energy configuration on the (211) surface, where the shortest average adsorption distance was between the hydrogen atoms of the amine group with the calcium ions on the surface (–NH⋯Ca = 2.201–3.352 Å), and the average adsorption distance was 2.954 Å. In addition, the interaction with the surface occurs mainly via the hydrogen atom of the –CH2 group and the hydrogen atoms of the amine group with the oxygen of the phosphoric acid group (CH⋯OPO3 = 2.240–3.889 Å, NH⋯OPO3 = 2.326–3.951 Å). The oxygen atoms of the carboxyl group and the hydrogen atoms of the –CH2 group will have relatively strong interactions with the calcium ions on the surface (COO⋯Ca = 2.798–3.118 Å, –CH⋯Ca = 2.364–3.497 Å), and the average adsorption distances were 2.954, 2.984, and 3.126 Å, respectively. In addition, the hydroxyl group on the (211) surface interacted only slightly with hydrogen atoms of the amine group and hydrogen atoms of the –CH2 group, because the number of interactions between them was minimal and the adsorption distance was longer.
Based on the analysis above, the interaction between rhCol I and HAP depends mainly on electrostatic interaction energy and van der Waals forces. As shown in Fig. 4–6, the interactions between the rhCol I and (001), (100), and (211) planes of HAP were mainly via COO⋯Ca, –NH⋯Ca, CH⋯OPO3, and NH⋯OPO3, respectively. These interactions mean that the growth of HAP also was affected by the main chain group of CH2, which showed a crystal with a long belt shape. Relatively, the adsorption sites on the (001) plane were the most abundant (the total number of adsorption sites was 53), which means a strong interaction between the rhCol I and on the plane of HAP; meanwhile, the least was related to the adsorption sites on the (211) plane, which indicated a weak interaction between them.
Typically, almost all collagen peptides form multiple interactions with the HAP surface through –COO− and >NH2 groups,44,45 and the electrostatic interaction is the main driving force of collagen, peptides, amino acids, and other proteins, where their functional groups, such as carboxylic acids, can directly bond with surface Ca ions.46–48 For natural collagen, representing a triple-helical molecule consisting of two α1 chains and one α2 chain formed via chemical bonding and hydrogen bonding, free charged resides would be reserved. MD simulations indicated that these charged residues could allow the formation of CaP clusters.20 Combined with previous reports,49–51 it is argued that rhCol I has a stable molecular configuration, which may be formed via the interaction of electrostatic interaction; therefore, the electrostatic interaction and van der Waals forces are the main interactions between rhCol I and HAP crystals.
3.3 Absorption energy
A negative adsorption energy indicates that adsorption of a protein at the surface is thermodynamically favorable. Usually, if the adsorption distance on this surface is shorter, the interaction force will be stronger, leading to stronger adsorption on this surface, and then the adsorption energy will be higher. Based on the results of the MD simulations, we speculated that the adsorption energy of the (001) plane would be the highest and that of the (211) plane would be the weakest.The adsorption energy is a macroscopic representation of the interaction force between molecules. The adsorption energy of the rhCol I on the surface of HAP was calculated according to eqn (1), as shown in Fig. 7. The electrostatic interaction energy plays an important role in the adsorption process here because the electrostatic interaction energy (Eelec) was significantly greater than the van der Waals interaction energy (EvdW) and hydrogen bonding energy (EH-bond). The adsorption energy of the rhCol I on the (001) crystal plane of HAP was the highest (−1417.262 kcal mol−1); thus the adsorption was the strongest, indicating that rhCol I tends to be strongly adsorbed on the (001) surface. Then the inhibitory effect on the growth of the (001) surface had the strongest effect. However, the adsorption energy on the (211) plane was −419.841 kcal mol−1, and the adsorption energy was the lowest. Therefore, it can be inferred that the growth of the (211) plane was greater than the other two planes, resulting in the obvious preferential growth of (211), which is consistent with the results from the experimental and MD simulation studies.
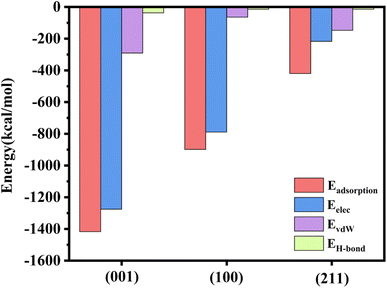 | ||
| Fig. 7 Value of adsorption energy, electrostatic energy, van der Waals energy, and hydrogen bonding energy of rhCol I on the crystal surface. | ||
Besides, ongoing work in our group will take into account the effect of water and carbonate on the adsorption of collagen at HAP surfaces, which clearly takes place in an aqueous environment. The presence and competition of surrounding water molecules and carbonate may well affect the adsorption of the collagen to the HAP surfaces and hence HAP growth.
4. Conclusions
Based on MD simulation, the (001), (100), and (211) planes of HAP crystal strongly interact with rhCol I mainly via COO⋯Ca, –NH⋯Ca, CH⋯OPO3, and NH⋯OPO3, respectively, which can be related to the electrostatic interaction and van der Waals forces. On the (001) plane, the interaction between –COO− and Ca was stronger than the other two crystal planes, resulting in a greater electrostatic force, which may result in a strong adsorption on the (001) plane, and further inhibit the growth of (001) of HAP. The free energy of adsorption of the rhCol I on the (001), (100), and (211) planes of HAP was in accord with the results from the MD simulation, respectively. Due to the lowest adsorption energy of (211) plane, preferential growth of the (211) plane was determined, which agreed with our experimental results. Here, MD simulations of rhCol I regulation of HAP crystal growth are presented, which may be helpful to synthesize HAP under regulation by novel biomolecules.Author contributions
Zhilin Huang: methodology, date curation, writing-original draft. Chucheng Wang: methodology, date curation, investigation. Xiaohui Chen: investigation, validation. Hong Li: methodology, resources, writing-review & editing, funding acquisition. Shan Ding, Qi Xiang, Mo Xie, Yadong Huang: methodology, resources.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (31971284), Guangdong Provincial Key Areas R&D Programs (2022B1111080007) and the Key Areas Research and Development Program of Guangzhou (202103030003).References
- S. Awasthi, S. K. Pandey and E. Arunan, J. Mater. Chem. B, 2021, 9, 228–249 RSC.
- Z. Zou, J. Xie and E. Macías-Sánchez, Cryst. Growth Des., 2020, 21, 414–423 CrossRef.
- T. W. Sun, Y. J. Zhu and F. Chen, RSC Adv., 2018, 8, 26218–26229 RSC.
- X. Zheng, P. Zhang and Z. Fu, RSC Adv., 2021, 11, 19041–19058 RSC.
- D. Luo, S. Lin and X. Li, Mater. Lett., 2011, 65, 2395–2397 CrossRef CAS.
- A. Hertz and I. J. Bruce, Nanomedicine, 2007, 2, 899–918 CrossRef CAS PubMed.
- Y. Khan, M. J. Yaszemski and A. G. Mikos, J. Bone Jt. Surg., 2008, 90, 36–42 CrossRef PubMed.
- F. Nudelman, K. Pieterse and A. George, Nat. Mater., 2010, 9, 1004–1009 CrossRef CAS PubMed.
- A.-W. Xu, Y. Maa and H. Cölfen, J. Mater. Chem., 2007, 17, 415–444 RSC.
- Y. Wang, T. Azaïs and M. Robin, Nat. Mater., 2012, 11, 724–733 CrossRef CAS PubMed.
- Z. Xue, X. Wang and D. Xu, Phys. Chem. Chem. Phys., 2022, 24, 11370–11381 RSC.
- L. Chen, R. Jacquet and E. Lowder, Bone, 2015, 71, 7–16 CrossRef CAS PubMed.
- N. Almora-Barrios and N. H. De Leeuw, Cryst. Growth Des., 2012, 12, 756–763 CrossRef CAS.
- T. Luo and K. L. Kiick, Eur. Polym. J., 2013, 49, 2998–3009 CrossRef CAS PubMed.
- W. J. Chung, K. Y. Kwon and J. Song, Langmuir, 2011, 27, 7620–7628 CrossRef CAS PubMed.
- T. Wang, J. Lew and J. Premkumar, Eng. Biol., 2017, 1, 18–23 CrossRef.
- Y. Zhai, F. Z. Cui and Y. Wang, Curr. Appl. Phys., 2005, 5, 429–432 CrossRef.
- A. L. Boskey, Bone Miner., 1989, 6, 111–123 CrossRef CAS PubMed.
- B. M. Oosterlaken, M. P. Vena and G. With, Adv. Mater., 2021, 33, 2004418 CrossRef CAS PubMed.
- Z. Xue, M. Yang and D. Xu, J. Phys. Chem. C, 2019, 123, 2533–2543 CrossRef CAS.
- C. Liao, Y. Xie and J. Zhou, RSC Adv., 2014, 4, 15759–15769 RSC.
- N. Almora-Barrios and N. H. de Leeuw, Langmuir, 2010, 26, 14535–14542 CrossRef CAS PubMed.
- J. Zeng, S. Yang and H. Yu, Langmuir, 2021, 37, 3410–3419 CrossRef CAS PubMed.
- J. W. Shen, T. Wu and Q. Wang, Biomaterials, 2008, 29, 513–532 CrossRef CAS PubMed.
- S. Shang, Q. Zhao and D. Zhang, Mater. Sci. Eng., C, 2019, 105, 110017 CrossRef CAS PubMed.
- S. E. Hoff, J. Liu and H. Heinz, J. Colloid Interface Sci., 2022, 605, 685–700 CrossRef CAS PubMed.
- X. Wang, L. Zhang and Z. Liu, RSC Adv., 2018, 8, 3716–3722 RSC.
- X. Wang, X. Yang and H. Chen, Langmuir, 2020, 36, 10822–10829 CrossRef CAS PubMed.
- H. Zhang, X. Lu and Y. Leng, Acta Biomater., 2009, 5, 1169–1181 CrossRef CAS PubMed.
- L. Veselinović, L. Karanović and Z. Stojanović, J. Appl. Crystallogr., 2010, 43, 320–327 CrossRef.
- Z. Du, H. Su and W. Wang, Nat. Protoc., 2021, 16, 5634–5651 CrossRef CAS PubMed.
- S. L. Mayo, B. D. Olafson and W. A. Goddard, J. Phys. Chem., 1990, 94, 8897–8909 CrossRef CAS.
- M. S. Oliveira, S. N. Costa and N. K. V. Monteiro, J. Braz. Chem. Soc., 2019, 30, 2367–2375 CAS.
- W. G. Knupp, M. S. Ribeiro and M. Mir, Appl. Surf. Sci., 2019, 495, 143493 CrossRef CAS.
- M. H. Ri, C. J. Yu and Y. M. Jang, J. Mater. Sci., 2016, 51, 3125–3135 CrossRef CAS.
- R. Gholizadeh and Y. Wang, Chem. Eng. Sci., 2018, 184, 62–71 CrossRef CAS.
- R. Bhowmik, K. S. Katti and D. Katti, Polymers, 2007, 48, 664–674 CrossRef CAS.
- Y. F. Xu, F. Nudelman and E. D. Eren, Nat. Commun., 2020, 11, 1–12 CrossRef PubMed.
- W. B. Tien, M. T. Chen and P. C. Yao, Mater. Sci. Eng., C, 2012, 32, 2096–2102 CrossRef CAS PubMed.
- J. W. Shen, T. Wu and Q. Wang, Biomaterials, 2008, 29, 513–532 CrossRef CAS PubMed.
- E. F. Bres and J. L. Hutchison, J. Biomed. Mater. Res., 2002, 63, 433–440 CrossRef CAS PubMed.
- Y. Zhai and F. Z. Cui, J. Cryst. Growth, 2006, 291, 202–206 CrossRef CAS.
- N. Almora-Barrios, K. F. Austen and N. H. de Leeuw, Langmuir, 2009, 25, 5018–5025 CrossRef CAS PubMed.
- H. Gu, Z. Xue and M. Wang, J. Phys. Chem. B, 2019, 123, 3372–3382 CrossRef CAS PubMed.
- K. Wang, M. H. Wang and Q. G. Wang, J. Eur. Ceram. Soc., 2017, 37, 2509–2520 CrossRef CAS.
- M. Cutini, M. Corno and D. Costa, J. Phys. Chem. C, 2017, 123, 7540–7550 CrossRef.
- W. Wang, Z. Xue and D. Xu, J. Phys. Chem. B, 2021, 125, 5078–5088 CrossRef CAS PubMed.
- Z. Wang, Z. Xu and W. Zhao, J. Mater. Chem. B, 2015, 3, 9157–9167 RSC.
- E. Troy, M. A. Tilbury and A. M. Power, Polymers, 2021, 13, 3321 CrossRef CAS PubMed.
- P. V. Azzopardi, J. O'Young and G. Lajoie, PLoS One, 2010, 5, e9330 CrossRef PubMed.
- Y. Xie, C. Liao and J. Zhou, Biophys. Chem., 2013, 179, 26–34 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra03810f |
| ‡ These authors contributed to equally to the work. |
| This journal is © The Royal Society of Chemistry 2023 |