Open Access Article
Open Access ArticleEvaluation of the synergistic effect of chitosan metal ions (Cu2+/Co2+) in combination with antibiotics to counteract the effects on antibiotic resistant bacteria
Nouran A. Elbialy a,
Heba K. A. Elhakimb,
Mona Hassan Mohamedc and
Zainab Zakaria
a,
Heba K. A. Elhakimb,
Mona Hassan Mohamedc and
Zainab Zakaria *d
*d
aBiotechnology and Bimolecular Chemistry Department, Faculty of Science, Cairo University, Giza, Egypt
bBiochemistry Division, Faculty of Science, Cairo University, Giza, Egypt
cDepartment of Chemistry, Faculty of Science, Cairo University, Giza, Egypt
dResearch and Development Department, Faculty of Pharmacy, Heliopolis University, Cairo, Egypt. E-mail: zeinab.zakaria@hu.edu.eg
First published on 14th June 2023
Abstract
The effectiveness of antibiotics that save millions of lives is in danger due to the increasing rise of resistant bacteria around the world. We proposed chitosan–copper ions (CSNP–Cu2+) and chitosan–cobalt ion nanoparticles (CSNP–Co2+) as biodegradable nanoparticles loaded with metal ions synthesized via an ionic gelation method for treatment of antibiotic resistant bacteria. The nanoparticles were characterized using TEM, FT-IR, zeta potential and ICP-OES. The MIC was evaluated for the NPs in addition to evaluating the synergetic effect of the nanoparticles in combination with cefepime or penicillin for five different antibiotic resistant bacterial strains. In order to investigate the mode of action, MRSA, DSMZ 28766 and Escherichia coli E0157:H7 were selected for further evaluation of antibiotic resistant genes expression upon treatment with NPs. Finally, the cytotoxic activities were investigated using MCF7, HEPG2 and A549 and WI-38 cell lines. The results showed quasi spherical shape and mean particle size of 19.9 ± 5 nm, 21 ± 5 nm and 22.27 ± 5 for CSNP, CSNP–Cu2+ and CSNP–Co2+ respectively. FT-IR showed slight shifting of the hydroxyl and amine group's peaks of chitosan indicating the adsorption of metal ions. Both nanoparticles had antibacterial activity with MIC ranging between 125 and 62 μg ml−1 for the used standard bacterial strains. Moreover, the combination of each of the synthesized NP with either cefepime or penicillin not only showed a synergetic effect as antibacterial activity of each NP or antibiotics alone, but also decreased the fold of antibiotic resistance genes expression. The NPs showed potent cytotoxic activities for MCF-7, HepG2 and A549 cancer cell lines with lower cytotoxic values for the WI-38 normal cell line. The NPs' antibacterial activity may be due to penetration and rupture of the cell membrane and the outer membrane of Gram negative and Gram positive bacteria causing bacterial cell death, in addition to, penetration into the bacterial genes and blocking gene expression that is vital to bacterial growth. The fabricated nanoparticles can be an effective, affordable and biodegradable solution to challenge antibiotic resistant bacteria.
1 Introduction
One of the main threats to human health is the rise of pathogenic bacteria that are resistant to drugs. In the United States, antibiotic-resistant bacteria are responsible for almost 2 million cases of serious illness, including 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fatalities each year.1 The majority of multi-drug-resistant (MDR) infections necessitate protracted antibiotic therapy, often coupled with tissue debridement (i.e., surgical removal), which results in low patient compliance and high medical expenses. Antibiotic usage obviously promotes the emergence of resistance. Epidemiological studies have shown a link between the use of antibiotics and the establishment and spread of resistant bacterial species.2
000 fatalities each year.1 The majority of multi-drug-resistant (MDR) infections necessitate protracted antibiotic therapy, often coupled with tissue debridement (i.e., surgical removal), which results in low patient compliance and high medical expenses. Antibiotic usage obviously promotes the emergence of resistance. Epidemiological studies have shown a link between the use of antibiotics and the establishment and spread of resistant bacterial species.2
Many identified bacteria by the Centers for Disease Control and Prevention (CDC) as posing urgent, serious, and worrying risks already place a significant clinical and financial burden on the American health care system, patients, and their families.3 Due to improper antibiotic use, bacteria can become resistant to them especially in the middle east region.4 Bacteria produce β-lactamase and cleave β-lactam rings in antibiotics to cause β-lactamase resistance, which is one of several categories of antibiotic resistance.5 Studies have attempted β-lactam antibiotics and -lactamase inhibitors to solve this problem. Instances include clavulanic acid (amoxyclav), sulbactam (ampicillin/sulbactam), and tazobactam (piperacillin/tazobactam).6 Around the world, several types of antibiotic resistance genes (ARGs) are frequently discovered in livestock fertilizer are the fundamental cause of bacterial resistance. Re-potentiating the antibiotics by combining them with additional antibiotics or antibacterial substances, such as nanoparticles in synergistic ways is one technique to get around this issue.7 Most antibiotic resistance pathways are irrelevant for the nanoparticles (NPs). For instance, metallic components NPs that exhibit a potent capacity to prevent bacterial adherence and proliferation can be turned into antibacterial nanoparticles and nano-composites by part alloying, common factor, and heat processing.8–10 As a result, different types of nanoparticles synthesis have been used in order to get metal NPs, metal oxide NPs, nano-composites,11 magnetic NPs12 and biodegradable NPs for interesting new NP-based materials with antibacterial properties.13 Nanoparticles with polymer sizes between 10 and 1000 nm composed of natural or synthetic materials exhibit distinctive physical and chemical characteristics due to phenomena including the quantum size effect, micro size effect, surface effect, and macro-quantum tunnel effect.14 Chitosan (CS) has generated a significant interest due to its special combination of capabilities, including biocompatibility, biodegradability, metal complication, and antibacterial action.15 Consequently, CS has numerous existing and potential uses in numerous fields including combination to NPs.
CS has been shown to produce a number of complexes, including CS–silver, CS–zinc, and CS–manganese. It is also effective in vitro against a number of bacterial pathogens, including Escherichia coli, Salmonella choleraesuis, and Staphylococcus aureus.16,17 Pervious study investigated the synergic effect of a composites of tetracycline with copper oxide nanoparticles (CuONp) integrated in chitosan micro-particles (CsMp@CuONp) as antibacterial elements.18 Additionally, it was noted that the enhanced cytotoxic action of the CS–metal complex is mostly due to the strong affinity of metal ions for the cell membrane.19
The aim of the present study is to investigate the use of chitosan–metal ion NPs to reverse the action of antibiotic resistance using affordable and biodegradable method. Furthermore, this research evaluated the synergetic effect of synthesized nanoparticles on antibiotic resistance bacteria including Gram positive and Gram negative strains in combination with β-lactamase antibiotics. The antibiotic resistance gene expression due to the combination was evaluated in order to determine nanoparticles mode of action as well as investigating the cytotoxic activities on cancer cell lines.
2 Method
2.1. Nanoparticle synthesis
CSNP and nano-chitosan loaded with metal were purchased from NanoTech Egypt Company. Briefly, CSNP (Chitosan Egypt, Giza, Egypt) were prepared according to ionotropic gelation method between CS and sodium tripolyphosphate (TPP) (Sigma-Aldrich, St. Louis, MO, USA) in ratio 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 respectively. To obtain 1% CSNP solution, CS was dissolved in 1% acetic acid (v/v) (Dop Organik Kimya, Ankara, Turkey), while TPP was dissolve in 50 ml distilled water. The mixture was prepared by adding dropwise of TPP solution onto CS, then it was stirred for 40 min at room temperature.20 The suspension was centrifuged at 10
1 respectively. To obtain 1% CSNP solution, CS was dissolved in 1% acetic acid (v/v) (Dop Organik Kimya, Ankara, Turkey), while TPP was dissolve in 50 ml distilled water. The mixture was prepared by adding dropwise of TPP solution onto CS, then it was stirred for 40 min at room temperature.20 The suspension was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 12 min, and then the precipitate was washed three times with water. Finally, CSNP solution was freeze dried to be obtained in powder form.
000g for 12 min, and then the precipitate was washed three times with water. Finally, CSNP solution was freeze dried to be obtained in powder form.
Chitosan nanoparticle was loaded to Cu2+ (CSNP–Cu2+) and Co2+ (CSNP–Co2+) via addition each of 5 ml metal ions solutions with concentration 120 μg ml−1. Copper sulfate and cobalt chloride (Loba Chemie Pvt. Ltd) were directly used as sources of each Cu2+ and Co2+, into nano-chitosan suspensions (0.3%, w/v) and stirring for 12 h at room temperature.16 Then, CSNP loaded with metal ions were purified as described for CSNP purification.
2.2. Characterization of nanoparticles
Fourier transform infra-red spectroscopy (FT-IR) spectra were obtained using the IRAffinity-1 spectrophotometer from Shimadzu, Japan. The spectrum was measured between 400 and 4000 cm−1. Focusing on nanoparticles, a transmission electron microscope (TEM) was used to assess the morphology and particle size distribution using a JOEL model 2100 from Japan with an 8000 kV accelerating voltage. CSNP loaded metal ions was analyzed using an inductively coupled plasma optical emission spectroscopy using ICP-OES (Ultima Expert, JOBIN YVON Technology) to quantify the amount of Cu2+ and Co2+ ions. Zetasizer Nano ZS, Malvern, UK was used to characterize the zeta potential of the synthetized nanoparticles.2.3. Antibacterial activity of CSNP–Cu2+ and CSNP–Co2+
Micro-dilution broth method was used according to the CLSI guidelines M100 (ref. 21) for determination of minimum inhibitory concentration (MIC) for each of the commercial antibiotic used (cefepime and penicillin), against nano-chitosan metal solutions, using five antibiotic resistant Gram positive and Gram negative bacterial strains. The used bacterial strains were MRSA DSMZ 28766, MRSA DSMZ 46320, Klebsiella pneumonia DSMZ 26371, ESBL2-1 Escherichia coli DSMZ 5923, and Escherichia coli E0157:H7 standard strain.Briefly, each of bacterial strain were cultured overnight at 37 °C and then the final bacterial concentration was adjusted to 2 × 106 colony-forming units (CFU ml−1) in nutrient broth (Difco). Two-fold serial dilutions were prepared of antibiotics to reach the final concentration of [78, 156.25, 312.5, 625, 1250, 2500, 5000, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg ml−1] meanwhile CSNP, CSNP–Cu2+ and CSNP–Co2+ solutions were also prepared in two fold serial dilution to give the final concentration of [7.8, 15.625, 31.25, 62.5, 125, 250, 500, 1000 μg ml−1]. In sterile 96-well plates, 100 μl of each dilution of antibiotics, CSNP, CSNP–Cu2+ and CSNP–Co2+ were placed into the well containing 100 μl of bacterial suspension. Triplicate samples were performed for each treatment, as well as medium and bacteria were used as negative and positive control, respectively. The MIC was determined using turbidity measurements for all wells after overnight incubation at 37 °C at 630 nm using a micro-plate reader (BioTek, USA).
000 μg ml−1] meanwhile CSNP, CSNP–Cu2+ and CSNP–Co2+ solutions were also prepared in two fold serial dilution to give the final concentration of [7.8, 15.625, 31.25, 62.5, 125, 250, 500, 1000 μg ml−1]. In sterile 96-well plates, 100 μl of each dilution of antibiotics, CSNP, CSNP–Cu2+ and CSNP–Co2+ were placed into the well containing 100 μl of bacterial suspension. Triplicate samples were performed for each treatment, as well as medium and bacteria were used as negative and positive control, respectively. The MIC was determined using turbidity measurements for all wells after overnight incubation at 37 °C at 630 nm using a micro-plate reader (BioTek, USA).
2.4. CSNP–Cu2+ and CSNP–Co2+ in combination of antibiotic: synergistic interaction between antibiotics and CS–metal ions NPs
The checkerboard experiment,22 in which the antibiotics [cefepime and penicillin] and CS metal loaded nanoparticles [CSNP–Cu2+ and CSNP–Co2+] were serially diluted in all drug combinations and was used to detect the synergistic interaction of the two medications. A 2-fold serial dilution from 5 mg ml−1 MIC to 0.039 mg ml−1 MIC was used to evaluate the fractional inhibitory concentration (FIC) of each antibiotic [cefepime and penicillin] against the concentration for the manufactured nanoparticles [CSNP–Cu2+ and CSNP–Co2+] varied from 0.25 mg ml−1 MIC to 0.0019 mg ml−1 MIC using the same bacterial strains used for MIC determination. Cefepime and penicillin concentrations were lowered vertically while CS–metal ions NPs were varied in descending order; a negative control with culture alone without any antimicrobial agent was used in the plates, and then kept at 37 °C overnight. The FIC was determined by dividing its combined MIC by its individual MIC as following equation: FIC = CNP/MICNP + CAb/MICAb, where MICNP and MICAb are the MICs of synthesized nanoparticles and antibiotic drug alone, respectively, and CAb and MICAb are the concentrations of the nanoparticles and antibiotic compounds in combination, respectively. The results were interpreted in accordance with the European Committee on Antimicrobial Susceptibility Testing's recommendations.22,232.5. Cytotoxicity evaluation of CSNP–Cu2+ and CSNP–Co2+
Normal human fetal lung fibroblast (WI-38), human breast carcinoma (MCF-7), hepatocellular carcinoma (HepG-2) and pulmonary epithelial cell carcinoma (A549) cell lines were purchased from The Holding Company for Biological Products & Vaccines (VACSERA, Cairo, Egypt, https://www.vacsera.com/). A549 and WI-38 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 μg streptomycin and 100 units penicillin, while HepG-2 and MCF-7 were maintained in RPMI medium supplemented with 10% fetal bovine serum, 100 μg streptomycin and 100 units penicillin. Temperature was adjusted at 37 °C in a humidified atmosphere consisting of 95% O2 and 5% CO2 for cell culture conditions.The synthesized CSNP–Cu2+ and CSNP–Co2+ ions were prepared in two fold serial dilution to a concentration range of [3.9, 7.8, 15.625, 31.25, 62.5, 125, 250, 500 μg ml−1] in sterile phosphate buffer saline (PBS). Cytotoxicity was measured for cell lines using MTT assay, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (Serva Electrophores, Germany). For cell monolayers, 100 μl of cell suspension aliquot (104 cells per well) were plated in 96-well tissue culture plate, then incubated at 37 °C for 24 h in a humidified incubator with 5% CO2 in order to allow attachment of cells to the plate. Negative control with no cells and positive control cell culture was included. Afterwards, cells were incubated for another 48 h at 37 °C with the prepared CSNP–Cu2+ and CSNP–Co2+ NP, then washed twice by PBS (Lonza Bioproducts, Belgium). The reduction of MTT to formazan was achieved by adding to each well 50 μl of 0.5 μg ml−1 MTT, and then incubated for 4 h. Finally, cells were treated with 50 μl of DMSO to solubilize the purple crystals of formazan. Absorbance of the cells was measured at 570 nm with microplate ELISA reader (BioTek, USA). Control and samples were assayed in quadrates for each concentration. The relative viability of cells was expressed as follows: % cell viability = (A570 of treated cells/A570 of positive control cells) × 100. The 50% inhibitory concentration (IC50) was estimated from graphic plots of the dose–response curve for each concentration using GraphPad Prism software (San Diego, CA, USA).
2.6. Antibiotic resistant genes expression modulation by the effect of CSNP–Cu2+ and CSNP–Co2+ in combination with antibiotics
MRSA DSMZ 28766 was selected as one of the Gram positive bacterial strains and E. coli E0157:H7 was selected as one of the Gram negative bacterial strains for further evaluation of antibiotic resistant genes expression. Bacterial isolates were cultured in 10 ml aliquots in nutrient broth medium overnight at 37 °C containing CSNP–Cu2+, CSNP–Co2+ in combination with antibiotics at the corresponding sub-MIC concentrations; then, the cell pellets of each treatment were harvested by centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min and stored at −80 °C until RNA extraction.
000 rpm for 10 min and stored at −80 °C until RNA extraction.
RNA extraction was performed by, Total RNA Extraction Kit (easy-spin™ (DNA-free, iNtRON BIOTECHNOLOGY, USA)) according to manufacturer instructions. RNA was treated with DNase in order to remove any genomic DNA impurities by DNase I, RNase-free (Thermo Fisher Scientific, USA). RNA concentration was determined using the NanoDrop instrument (Nano-100, UK). The Maxime RT PreMix Kit (iNtRON BIOTECHNOLOGY, USA) was used to synthesize cDNA from 50 ng of RNA using a PCR thermocycler (A & E Lab, UK) according to manufacturer instructions.
Relative quantification of gene expression was carried out using the Cycle Threshold (CT) comparative method. Quantitative PCR amplification was done by Maxima SYBR Green qPCR Master Mix (2×) ROX solution (Thermo Scientific, USA) using thermo cycler Mx 3005 pro (Agilent Technologies). The thermal profile was: pre-treatment at 50 °C for 2 min, initial denaturation at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at specific annealing temperature for 30 s, and extension at 72 °C for 30 s. Samples were prepared in a 25 μl reaction mixture containing 12.5 μl Maxima SYBR Green qPCR Master Mix (2×), primer forward and reverse both were 1 pmol μl−1, template cDNA 50 ng, and RNase free water up to final volume 25 μl, all PCR reactions were performed in duplicate. The specific primers of genes are listed in Table 1. The fold of gene expression level were calculated with Ct value relative to 16s RNA gene using the equation, fold = 2−(ΔΔCt).
| Bacteria | Genes name | Primer sequence | Reference |
|---|---|---|---|
| MRSA DSMZ 46320 | MecA | F-ACTGATTAACCCAGTACAGATCCTTTC | 46 |
| R-TCCAAACTTTGTTTTTCGTGTCTTT | |||
| Blaz | F-CCTAAGGGCCAATCTGAACC | Designed | |
| R-ACACTCTTGGCGGTTTCACT | |||
| PBP-4 | F-AATGAGTTTGCCGGGTACAG | Designed | |
| R-CCATTAATGCATTCCCCATC | |||
| PBP-1 | F-AATGGCAATTTTGCATCACA | Designed | |
| R-CCACGTTTAGGCTGCTTCTC | |||
| 16S rRNA | F-GTGACAAACCGGAGGAAGGT | 47 | |
| R-ATCCGAACTGAGAACAACTTTATGG | |||
| E. coli E0157:H7 | blaTEM | F-GCATCTTACGGATGGCATGA | 48 |
| R-GTCCTCCGATCGTTGTCAGAA | |||
| blaCMY | F-GGCAAACAGTGGCAGGGTAT | 48 | |
| R-AATGCGGCTTTATCCCTAACG | |||
| blaSHV | F-TTCAAAGGCCGGCATTTTCA | Designed | |
| R-CAGCGGTAAATCGTGGAGTG | Designed | ||
| 16s rRNA | F-CATGCCGCGTGTATGAAGAA | 49 | |
| R-CGGGTAACGTCAATGAGCAAA |
2.7. Statistical analysis
All the data was expressed as mean ± standard deviation (SD). Statistical analysis was performed using Graphpad Prizm software version 6.1. Histograms were graphed using OriginPro 8.5. A value of p less than 0.05 was considered as statistically significant.3 Result
3.1. Characterization of CSNP–Cu2+ and CSNP–Co2+
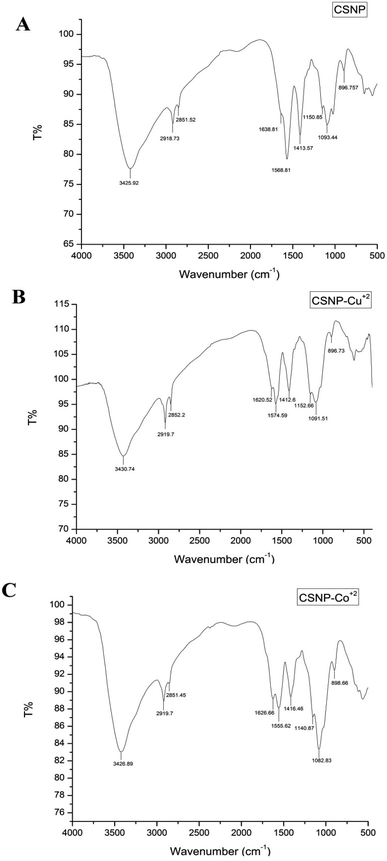
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, cross-linked between the phosphoric groups of TPP and ammonium ions of the CS within the nanoparticles that serves to enhance both the inter- and intra-molecular interaction in chitosan nanoparticles.24 The absorption peaks at 1093.44 and 896.757 cm−1 are recognized due to the anti-symmetric stretching vibration of C–O–C bridges and assigned to glucopyranose ring in CS matrix. As shown in Fig. 1B, the peaks at 1638.81 cm−1, 1568.81 cm−1 corresponding to the vibration of NH2 in amine groups shifts to 1620.52 and 1574.59 cm−1, respectively, indicating that NH2 took part in adsorption Cu2+. Also, the peak corresponding to the stretching vibration of OH-group shifts from 3425.92 cm−1 to 3430.74 cm−1 after Cu2+ sorption, indicating that –OH group also take part in sorption. FT-IR results in Fig. 1C illustrates that same shifting peaks related to hydroxyl slightly shifted to 3426.89 cm−1 and amine groups shifted to 1626.66 and 1555.62 cm−1 indicate that they were the two major functional groups responsible for adsorption of Co2+.25
O, cross-linked between the phosphoric groups of TPP and ammonium ions of the CS within the nanoparticles that serves to enhance both the inter- and intra-molecular interaction in chitosan nanoparticles.24 The absorption peaks at 1093.44 and 896.757 cm−1 are recognized due to the anti-symmetric stretching vibration of C–O–C bridges and assigned to glucopyranose ring in CS matrix. As shown in Fig. 1B, the peaks at 1638.81 cm−1, 1568.81 cm−1 corresponding to the vibration of NH2 in amine groups shifts to 1620.52 and 1574.59 cm−1, respectively, indicating that NH2 took part in adsorption Cu2+. Also, the peak corresponding to the stretching vibration of OH-group shifts from 3425.92 cm−1 to 3430.74 cm−1 after Cu2+ sorption, indicating that –OH group also take part in sorption. FT-IR results in Fig. 1C illustrates that same shifting peaks related to hydroxyl slightly shifted to 3426.89 cm−1 and amine groups shifted to 1626.66 and 1555.62 cm−1 indicate that they were the two major functional groups responsible for adsorption of Co2+.25
3.2. Antibacterial activity of CSNP–Cu2+ and CSNP–Co2+
As represented in Table 2, MIC of cefepime was 2.5 mg ml−1 for MRSA DSMZ 28766 and Klebsiella pneumonia DSMZ 26371, while the MIC for both E. coli strains was 5 mg ml−1 and 1.25 mg ml−1 for MRSA DSMZ 46320. Moreover, penicillin MIC values against all tested bacterial strains were 5 mg ml−1, except for (MRSA) DSMZ 46320 the MIC was 2.5 mg ml−1 indicated that those strains are resistant to the used antibiotics as shown in Table 3.| Drug | MRSA DSMZ 28766 | MRSA DSMZ 46320 | Klebsiella pneumonia DSMZ 26371 | E. coli DSMZ 5923 | E. coli E0157:H7 |
|---|---|---|---|---|---|
| Cefepime | 2.5 | 1.25 | 2.5 | 5 | 5 |
| Penicillin | 5 | 2.5 | 5 | 5 | 5 |
| CSNP–Co2+ | 0.125 | 0.125 | 0.0625 | 0.125 | 0.125 |
| CSNP–Cu2+ | 0.0625 | 0.0625 | 0.125 | 0.0625 | 0.125 |
| Combination drug | MRSA DSMZ 28766 | MRSA DSMZ 46320 | Klebsiella pneumonia DSMZ 26371 | E. coli DSMZ 5923 | E. coli E0157:H7 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC NP (mg ml−1) | MIC antibiotic (mg ml−1) | FIC | MIC NP (mg ml−1) | MIC antibiotic (mg ml−1) | FIC | MIC NP (mg ml−1) | MIC antibiotic (mg ml−1) | FIC | MIC NP (mg ml−1) | MIC antibiotic (mg ml−1) | FIC | MIC NP (mg ml−1) | MIC antibiotic (mg ml−1) | FIC | |
| a The FIC index is interpreted as follows: FIC ≤ 0.5, synergy; 0.5 ≤ FIC ≤ 2, addition of effects; 2 ≤ FIC ≤ 4, indifference and for FIC > 4, antagonism. | |||||||||||||||
| CSNP–Co2+ + penicillin | 0.03125 | 0.625 | 0.37 | 0.0625 | 0.625 | 0.75 | 0.015625 | 0.3125 | 0.31 | 0.03125 | 0.625 | 0.37 | 0.015625 | 0.3125 | 0.31 |
| CSNP–Cu2+ + penicillin | 0.015625 | 0.3125 | 0.31 | 0.03125 | 0.3125 | 0.62 | 0.03125 | 0.625 | 0.375 | 0.015625 | 0.3125 | 0.31 | 0.015625 | 0.3125 | 0.31 |
| CSNP–Co2+ + cefepime | 0.03125 | 0.3125 | 0.37 | 0.015625 | 0.078125 | 0.18 | 0.03125 | 0.3125 | 0.625 | 0.015625 | 0.3125 | 0.18 | 0.007813 | 0.15625 | 0.15 |
| CSNP–Cu2+ + cefepime | 0.015625 | 0.15625 | 0.31 | 0.015625 | 0.078125 | 0.31 | 0.03125 | 0.3125 | 0.375 | 0.015625 | 0.3125 | 0.31 | 0.007813 | 0.15625 | 0.15 |
Synthesized nanoparticles showed antibacterial activity as shown in Table 3. The recorded results also clearly showed that the MIC of CSNP–Co2+ for MRSA DSMZ 28766, MRSA DSMZ 46320, and ESBL2-1 E. coli DSMZ 5923 was 0.125 mg ml−1 while for Klebsiella pneumonia DSMZ 26371 and E. coli E0157:H7 was 0.0625 mg ml−1. Also CSNP–Cu2+ MIC obtained for MRSA DSMZ 28766, MRSA DSMZ 46320, ESBL2-1 E. coli DSMZ 5923, and E. coli E0157:H7 was 0.0625 mg ml−1, while for Klebsiella pneumonia DSMZ 26371 was 0.125 mg ml−1.
3.3. CSNP–Cu2+ and CSNP–Co2+ in combination of antibiotic: synergistic interaction between antibiotics and CS–metal ions NPs
Upon the combination of each of the synthesized NP with either cefepime or penicillin, the MIC of each combination was lowered greatly as shown in Table 3. MIC of cefepime in combination with CSNP–Co2+ and CSNP–Cu2+ against MRSA DSMZ 28766, MRSA DSMZ 46320, Klebsiella pneumonia DSMZ 26371 and E. coli E0157:H7, reduced from 8 up to 32 folds, while the MIC for penicillin in combination with the synthesized nanoparticle lowered 8–16 fold.The calculated FIC for each combination revealed that the combination of each of the synthesized NP with either cefepime or penicillin had a synergetic effect as antibacterial activity of each NP or antibiotics alone, except for the combination of penicillin and both types of nanoparticles had an additive effect upon treatment with MRSA, DSMZ 46320, as well as the combination of cefepime and CSNP–Co2+ upon treatment with Klebsiella pneumonia DSMZ 26371. When the FIC index is less than 0.5, the combination works synergistically; when it is between 0.5 and 2, it behaves additively; when it is between 2 but and 4, it performs indifferently; and when it is greater than 4, it responds antagonistically.
3.4. Cytotoxicity evaluation of CSNP–Cu2+ and CSNP–Co2+
Results presented in Fig. 5 indicated that the synthesized nanoparticles had a potent cytotoxic effect for the malignant A549, MCF-7 and HepG-2 cancer cell lines with lower cytotoxicity to the nonmalignant WI-38 cell line. The recorded IC50 values of CSNP–Cu2+ were found to be 163.5 μg ml−1, 49.8 μg ml−1, 62.5 μg ml−1 and 52.4 μg ml−1 for WI-38, MCF7, HEPG2 and A549, respectively. While the measured IC50 values of CSNP–Co2+ for WI-38, MCF7, HEPG2 and A549 were 166.3 μg ml−1, 66.7 μg ml−1, 53.4 μg ml−1, and 61.8 μg ml−1, respectively. This indicates a targeting effect of CSNP–Cu2+ and CSNP–Co2+ to cancer cell line comparable to normal cell line.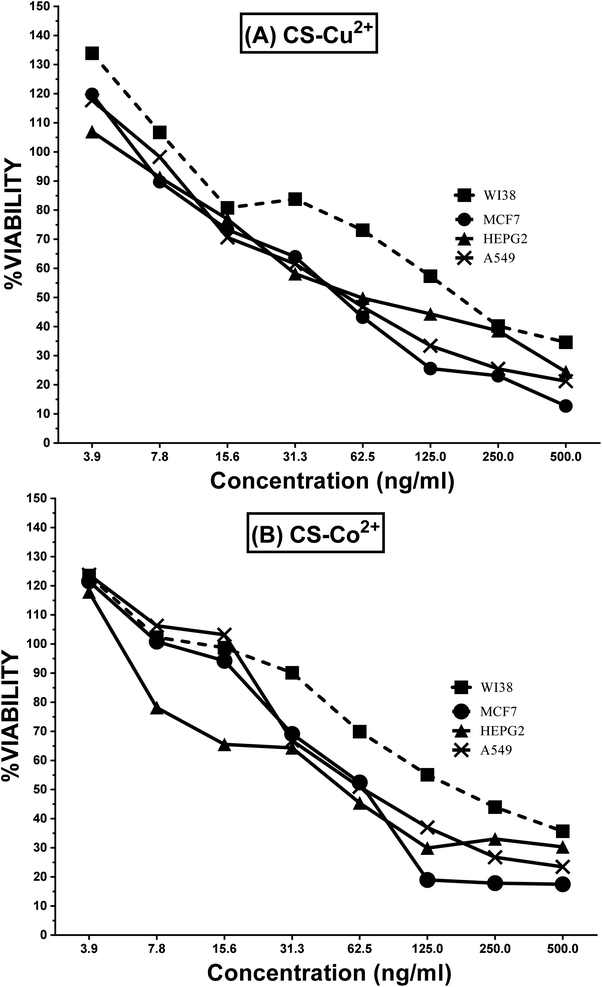 | ||
| Fig. 5 MTT cytotoxicity curve for mean% of viability ± standard error (SE) of (A) CSNP–Cu2+ and (B) CSNP–Co2+ for A549, MCF-7, HepG-2 and Hbf-4 cell line. | ||
3.5. Antibiotic resistant genes expression modulation by the effect of the of CSNP–Cu2+ and CSNP–Co2+ in combination with antibiotics
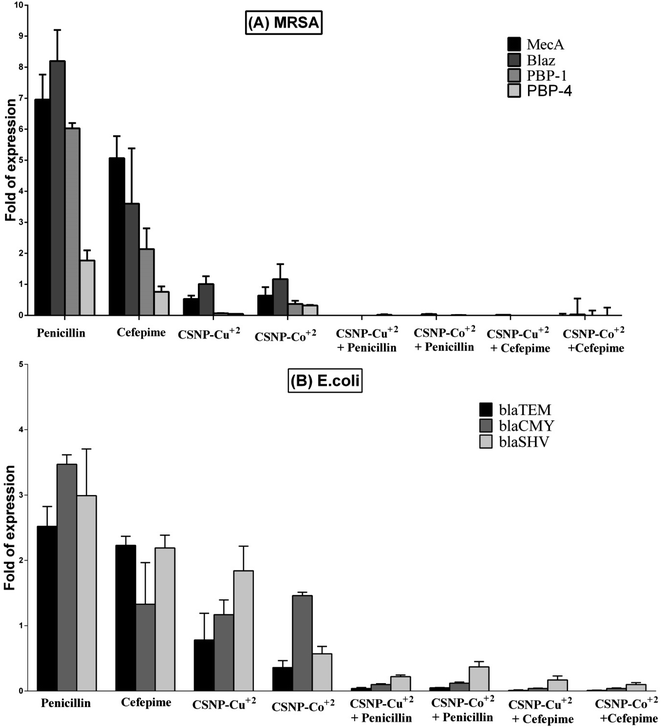
As MRSA DSMZ 28766 was chosen as one of the Gram positive bacterial strains and E. coli E0157:H7 was chosen as one of the Gram negative bacterial strains for further evaluation of antibiotic resistant genes expression, Fig. 6 illustrates that for both types of bacterial genes expression fold had the greatest expression fold value in the presence of penicillin and cefepime alone that was reflected by the antibiotics resistance and bacterial growth. In contrast the presence of the synthesized nanoparticle alone or in combination with both antibiotics affected on the fold of antibiotic gene expression for both type of bacterial strains. For MRSA DSMZ 28766, the expression value of MecA, Blaz, PBP-4 and PBP-1 genes were decrease almost 80% in the presence of nanoparticle in comparison to the presence of antibiotics. Meanwhile, the fold of expression of blaTEM, blaCMY, blaSHV genes for E. coli E0157:H7 decreased about 30% by the effect of nanoparticles comparable to the value of antibiotics. Moreover, the combination of antibiotics with the synthesized nanoparticles (CSNP–Co2+ + penicillin, CSNP–Cu2+ + penicillin, CSNP–Co2+ + cefepime and CSNP–Cu2+ + cefepime) have the greatest effect to decrease the fold of genes expression for both bacterial strains. However, the drug combination almost silenced the antibiotic resistance genes expression for Gram positive bacterial strain, MRSA DSMZ 28766, and to Gram negative bacterial strain, E. coli E0157:H7, which was reflected by antibiotics sensitivity and bacterial growth inhibition.4 Discussion
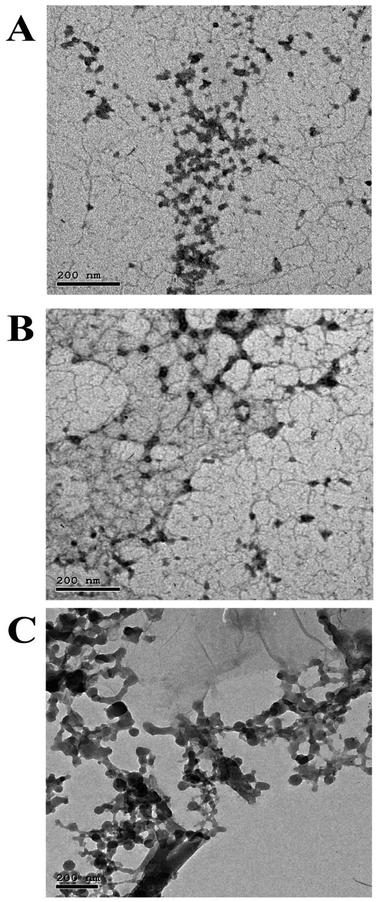
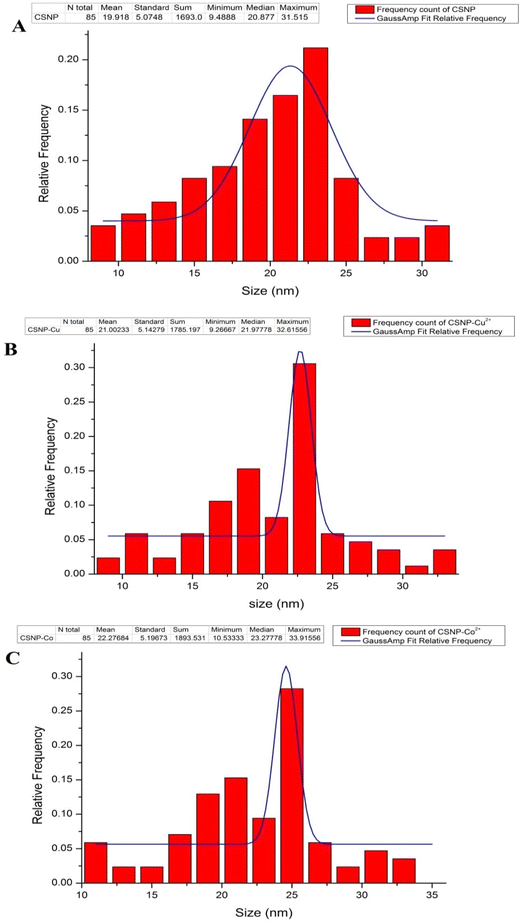
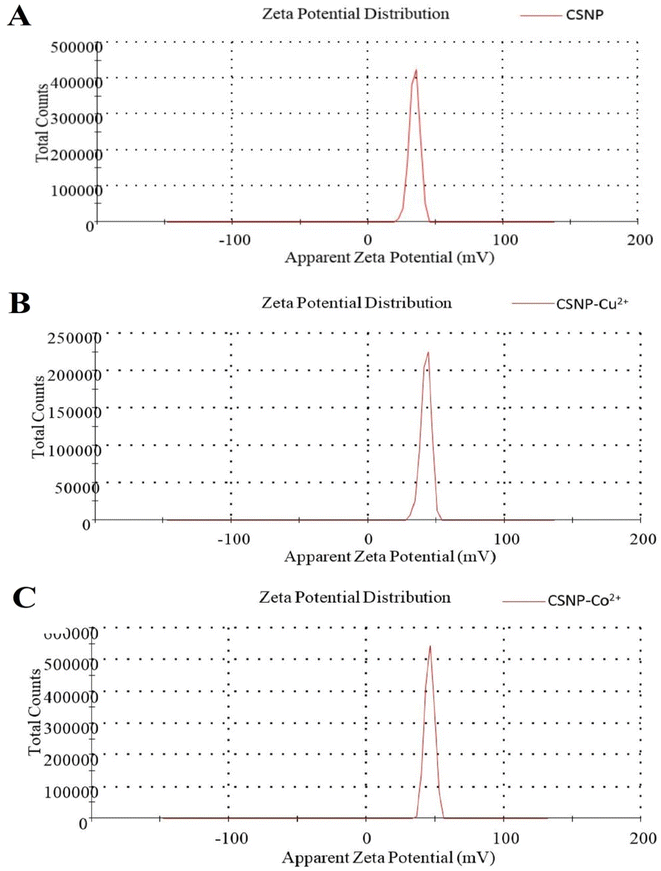
In the present study, a novel CSNP loaded with copper ions or cobalt ions with biological activities displaying potential uses in the medical, pharmaceutical, and industrial domains was effectively synthetized and characterized. Both CSNP–Cu2+ & CSNP–Co2+ were successfully synthesized via ionic gelation method, then characterized using FT-IR, TEM, ICP-OES and zeta potential to approve the alteration in size, shape and total charges of synthetized nanoparticles. Previous study indicated that metal ions could be effectively adsorbed on the CS surface at pH 6 (ref. 26) and the high affinity of the –NH2 or –OH groups of CSNP towards copper increases the stability of the metal ion.27 Throughout the synthesis steps of nanoparticles, the addition of TPP solution induces the formation of CSNP particles, which range in size from 9–33 nm and exhibit a zeta potential of 30 to 50 mV, depending on the mass ratio of CS to TPP or the molecular weight of CS. The particle size of synthesized nanoparticle was similar to each other as represented in Fig. 2 and 3. As the size distribution of the synthesized nanoparticles was taken along the diameter of the particles, it showed the mean particle size of 19.9 ± 5 nm for CSNP 21 ± 5 nm for CSNP–Cu2+ and 22.27 ± 5 for CSNP–Co2+. Also, zeta potential curve that represented in Fig. 4 indicated surface charge of +34.2, +42.2 and +45.8 mV for CSNP, CSNP–Cu2+ and CSNP–Co2+. Since our study focus on synthesis of metal ions loading onto chitosan nanoparticle, FT-IR & ICP-OES analyses were used to confirm the proper metal ions binding to CSNPs with quantification of metal ions concentrations. Meanwhile, FT-IR figures shows shifting peaks for –NH2 or –OH groups of CSNPs as illustrated in Fig. 4.In the present study, the synthesized nanoparticles had high antibacterial activity of, as indicated by MIC values, for tested antibiotic resistant bacteria, while those bacteria were resistant to penicillin and cefepime antibiotics. Moreover, the combination of each of the synthesized NP with either cefepime or penicillin had a synergetic effect as antibacterial activity of each NP or antibiotics alone, except for the combination of penicillin and both types of nanoparticles had an additive effect upon treatment with MRSA, as well as the combination of cefepime and CSNP–Co2+ upon treatment with Klebsiella pneumonia. Since our study focused on synthesized nanoparticle made from CS, many studies reported CSNP alone has demonstrated a wide spectrum of antimicrobial activity against Gram-positive and Gram-negative bacteria through different mechanisms.28 Positively charged CS can interact electrostatically with the negatively charged teichoic acid with peptidoglycans in Gram-positive bacteria, causing the cell membrane to rupture, internal components to leak out, and CS to enter the microbial cells. A previous work established that CS causes the leaking of proteins and other intracellular components.29 Also, CS's positive charges can disrupt the outer membrane (OM) in Gram-negative bacteria, allowing it to permeate the cell membrane and cause bacterial cell death. High negative charges from LPS can be countered by positive charges from CS.30 Previous research revealed that peptidoglycan hydrolysis can result in an improved electrostatic interaction, which is supported by measurements of the bacteria mixture's electric conductivity and the release of cytoplasmic β-galactosidase activities from E. coli into the growth media.31
Besides antibacterial activities of CS which has strong affinity towards metal ions because of the presence of numerous amine and hydroxyl groups, it can chelate with many metal ions. Indeed chelating metal ions properties can boost the antimicrobial activity of CS. The excellent antibacterial property of copper and cobalt for Gram positive and negative bacteria,32,33 as well as, cytotoxicity activity for multiple cancer cell lines have been reported previously.28 Previous study aimed to load different metal ions such as Ag+, Cu2+, Zn2+, Mn2+ or Fe2+ onto CSNPs, showed that antibacterial activity was significantly enhanced by the metal ions loaded, except for Fe2+. Especially for CSNP loaded Cu2+, the MIC for E. coli 25922, S. choleraesuis ATCC 50020 and S. aureus 25923 were 21–42 times lower than that of Cu2+.16 Another study focused on antibacterial behavior of CS-bivalent metal chelates (Co and Ni) against standard bacteria, Staphylococcus aureus ATCC 4533, S. faecalis ATCC 8043 and Escherichia coli ATCC 25923. They indicated that the inhibitory effects of the chelates were dependent not only on the property of the coordinated metal ion, but also on the molar ratio of the metal ion.34 Also Badawy et al., investigated antibacterial activity of synthesized CS and CS–AgNPs for E. coli and S. typhimurium in vitro which revealed that E. coli was more susceptible to CS–AgNPs than S. typhimurium and CS–AgNPs have more influence with increasing silver concentrations.35 A recent study discussed the synergetic effect between nanoparticle and antibiotics by Z. Assadi et al. which indicated antibacterial activity of CuONp and a synergistic effect of tetracycline/CuONp and tetracycline/CsMp@CuONp were investigated for MDR coagulase negative Staphylococcus, MDR Pseudomonas p41, MDR Pseudomonas p21, Staphylococcus aureus ATCC 6538 and Pseudomonas aeruginosa ATCC 9027. The average FIC of tetracycline with CuONp was 0.85 and for tetracycline with CsMp@CuONp reduced to 0.44 which indicated the additive and synergic effect respectively.18
Till now, there is no enough studies had been conducted to explore the antimicrobial activities of CSNP loaded with metal ions combination with certain antibiotics against MDR bacteria or antibiotic resistance bacteria. In contrast, few studies have previously been done to investigate the antibacterial properties of either antibiotic, metal ions in combination with beta-lactamase antibiotics or the combination of CSNPs with specific antibiotics when used against clinical infections of MDR.36,37 A study conducted metal–antibiotic complex using Ag+, Zn2+ and Cu2+ which was proven to be successful improving the activity of the antibiotic against β-lactamase-producing bacteria.37 Lamia Benhalima and Sandra Amri et al., showed a synergistic effects for antibiotic complexes with Ag+ and Cu2+. Furthermore, 25 different bacterial clinical isolates (16 Enterobacteriaceae, 5 Staphylococci, and 4 Pseudomonas), were tested for susceptibility to copper sulfate and fifty two percent of isolates were very susceptible to copper sulfate, with MICs ranging from 100 to 200 μg ml−1.38 Besides copper ions, cobalt metal ions form an important group of antimicrobial agent which have different active target from most bacteriostatic polymer.34,39 Moreover, a study focused on the chelation of CS with Co2+ and subsequent growth inhibitory studies against standard bacteria proved the effects of the metal coordination to CS with ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 inhibit the bacterial growth of Staphylococcus aureus ATCC 4533, S. faecalis ATCC 8043 and Escherichia coli ATCC 25923 with MIC value 125, 82.5 and 82.5 μg ml−1, respectively.34
3 inhibit the bacterial growth of Staphylococcus aureus ATCC 4533, S. faecalis ATCC 8043 and Escherichia coli ATCC 25923 with MIC value 125, 82.5 and 82.5 μg ml−1, respectively.34
In order to understand the mechanism of nanoparticles mode of action at molecular level, we studied the synthesized nanoparticles and their combination with antibiotics on resistant genes expression (MecA, Blaz, PBP-4 and PBP-1, for MRSA strain) and (blaTEM, blaCMY and blaSHV, for E. coli strain). The nanoparticles were effective alone in decreasing the fold of genes expression in comparison to antibiotics alone. Moreover, the combinations (CSNP–Co2+ + penicillin, CSNP–Cu2+ + penicillin, CSNP–Co2+ + cefepime and CSNP–Cu2+ + cefepime) have the greatest effect to decrease the fold of genes expression for both bacterial strains. However, the combination almost silenced the fold of MecA, Blaz, PBP-4 and PBP-1 genes expression for Gram positive bacterial strain, MRSA DSMZ 28766, in addition to blaTEM, blaCMY and blaSHV genes expression for Gram negative bacterial strain, E. coli E0157:H7. This findings support previous suggestion that the synthesized nanoparticles may penetrate the bacterial cell membrane, causing disturbance in resistant bacterial genes expressions and block gene expression vital to bacterial growth. In addition to gene expression silencing, also CS may electrostatically bind to peptidoglycans in Gram-positive bacteria, causing the cell membrane to rupture, while CS's positive charges can disrupt the outer membrane in Gram-negative bacteria, allowing it to permeate the cell membrane and cause bacterial cell death. Recently, the synergistic antibacterial potential of conventional β-lactam antibiotics combined with N-alkylaminated CSNPs against multidrug-resistant pathogen with blaCTX-M gene was investigate by Kaur et al. who showed that the developed nano-formulation resensitized the studied E. coli MDR strain to ampicillin and piperacillin by causing a 1000–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold decrease in their MIC values (5000–50
000-fold decrease in their MIC values (5000–50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mg l−1 to 5 mg l−1). Also, the combination of CS with cefoxitin and ceftazidime demonstrated a comparatively lower synergistic inhibitory effect owing to the higher susceptibility (MIC value = 0.5–5 mg l−1) to these antibiotics.40
000 mg l−1 to 5 mg l−1). Also, the combination of CS with cefoxitin and ceftazidime demonstrated a comparatively lower synergistic inhibitory effect owing to the higher susceptibility (MIC value = 0.5–5 mg l−1) to these antibiotics.40
Furthermore, when evaluating the cytotoxic activities of CSNP–Cu2+ & CSNP–Co2+, recorded IC50 values of CSNP–Cu2+ were found to be 163.5 μg ml−1, 49.8 μg ml−1, 62.5 μg ml−1 and 52.4 μg ml−1 for WI-38, MCF7, HEPG2 and A549, respectively. While the measured IC50 values of CSNP–Co2+ for WI-38, MCF7, HEPG2 and A549 were 166.3 μg ml−1, 66.7 μg ml−1, 53.4 μg ml−1, 61.8 μg ml−1, respectively. This indicating a potent cytotoxic activity for cancer cell lines compared to a lower cytotoxicity for normal cell line. As an anticancer agent, CS, CS derivatives, or CSNPs have demonstrated their cytotoxic potential. The main mechanism of antibacterial effect of CS is linked to disruptions of the cell cycle's normal operation, interference with the biological system's key tenets of protein or enzyme synthesis, disruption of the hormonal pathway to biosynthesis, and inhibition of the proliferation of cancer cells.41 The interaction between CS and metal complexes and free radical scavenging behavior is what gives these compounds their cytotoxic properties. Complexes with various copper to CS ratios were evaluated in vitro with 293 cells and HeLa cells as possible anticancer agents by Yong Zheng and his group.42 The compound showed more anticancer activity and less toxicity than other copper–CS complexes evaluated at a ratio of 0.11 mol copper for one CS residue. Cytotoxicity (IC50) of the complex was calculated to be 48 ± 4 and 34 ± 2 μmol l−1 for HeLa cells and 293 cell lines, respectively. This research also shown that the copper–CS compound reduced tumour cell growth by arresting the cell cycle in 293 cells at the S phase.42 It has been established that copper nanoparticles loaded with CS are biocompatible compound for the execution of the enhanced retention and permeation (EPR) effect to be preferentially accumulated in cancer cells in vivo. Their remarkable anticancer effects have been shown by maximum damage and apoptotic body formation in cancer cells. The selective uptake and accumulation of tiny nanoparticles (<200 nm) by cancer cells has been the focus of recent advancements in research. A greater expression of caspase 3 in an experimental setting has demonstrated enhanced apoptotic activity caused by an increase in caspase 3/7 activity.42,43 Chattopadhyay et al. developed CS-based delivery of cobalt oxide nanoparticles (CS–CoO NPs) to human leukemic cells and investigate their specific induction of apoptosis and CS–CoO NPs exhibit toxicity toward the Jurkat cell line in a dose-dependent fashion. It was observed from their experiment that NPs kill Jurkat cells in proportions of 3.66, 7.25, 15.16, 21.81, 28.84, and 46.84%, respectively, at doses of 1, 5, 10, 25, 50, and 100 μg ml−1. But CS–CoO NPs at a dose of 200 μg ml−1 kill Jurkat cells significantly (p = 0.05), in a proportion of 67.58%.44 In addition, Ambika, et al., studied the synthesis, characterization and anticancer activity of CS–cobalt nanoparticles upon the addition of noni fruit extract as a reducing agent using A549 cell line and the LD50 value was 97.83 μg ml−1.45
5 Conclusion
Chitosan–copper ions (CSNP–Cu2+) and chitosan–cobalt ions (CSNP–Co2+) biodegradable nanoparticles was synthesized by ionic gelation method to challenge antibiotic resistant bacteria. The synthesized nanoparticles showed synergetic antibacterial activities against five antibiotic resistant Gram positive and Gram negative bacterial strains in combination with cefepime and penicillin antibiotics. The antibiotics with NPs inhibited antibiotic resistant genes expressions of MRSA and E. coli strains suggesting that the NPs antibacterial activity is due to: (a) CS may electrostatically bind to peptidoglycans in Gram-positive bacteria, causing the cell membrane to rupture, while CS's positive charges can disrupt the outer membrane (OM) in Gram-negative bacteria, allowing it to permeate the cell membrane and cause bacterial cell death, (b) NPs penetration into the bacterial genetic content and blocking some gene expression vital to bacterial growth. Moreover, the NPs showed potent cytotoxic activities against MCF-7, HepG2 and A549 cancer cell lines with lower cytotoxic values against WI-38 normal cell line. Finally, CSNPs loaded with Cu2+ and Co2+ metal ions can be an effective, affordable and biodegradable solution to fight antibiotic resistant bacteria. Further investigations could be explored for various biomedical applications of the synthesized CSNP–Cu2+ and chitosan–cobalt ions CSNP–Co2+.Conflicts of interest
The authors report there are no competing interests to declare.References
- A. Gupta, S. Mumtaz, C.-H. Li, I. Hussain and V. M. Rotello, Chem. Soc. Rev., 2019, 48, 415–427 RSC.
- A. F. Read and R. J. Woods, Evol. Med. Public Health, 2014, 2014, 147 CrossRef PubMed.
- B. D. Lushniak, Public Health Rep., 2014, 129, 314–316 CrossRef PubMed.
- R. C. Arul Selvaraj, M. Rajendran and H. P. Nagaiah, Molecules, 2019, 24, 3055 CrossRef PubMed.
- M. Bassetti, M. Merelli, C. Temperoni and A. Astilean, Ann. Clin. Microbiol. Antimicrob., 2013, 12, 1–15 CrossRef PubMed.
- B. Pilmis, V. Jullien, A. Tabah, J.-R. Zahar and C. Brun-Buisson, Ann. Intensive Care, 2017, 7, 113 CrossRef PubMed.
- H. Yilmaz Atay, Antibacterial Activity of Chitosan-Based Systems, Drug Delivery and Biomedical Applications, 2020, pp. 457–489 Search PubMed.
- M. A. Mahdi, S. R. Yousefi, L. S. Jasim and M. Salavati-Niasari, Int. J. Hydrogen Energy, 2022, 47, 14319–14330 CrossRef CAS.
- S. R. Yousefi, H. A. Alshamsi, O. Amiri and M. Salavati-Niasari, J. Mol. Liq., 2021, 337, 116405 CrossRef CAS.
- S. R. Yousefi, M. Ghanbari, O. Amiri, Z. Marzhoseyni, P. Mehdizadeh, M. Hajizadeh-Oghaz and M. Salavati-Niasari, J. Am. Ceram. Soc., 2021, 104, 2952–2965 CrossRef CAS.
- P. Mehdizadeh, M. Jamdar, M. A. Mahdi, W. K. Abdulsahib, L. S. Jasim, S. Raheleh Yousefi and M. Salavati-Niasari, Arabian J. Chem., 2023, 16, 104579 CrossRef CAS.
- S. R. Yousefi, D. Ghanbari, M. Salavati-Niasari and M. Hassanpour, J. Mater. Sci.: Mater. Electron., 2016, 27, 1244–1253 CrossRef CAS.
- M. Chen, J. P. Liu and S. Sun, J. Am. Chem. Soc., 2004, 126, 8394–8395 CrossRef CAS PubMed.
- L. Wang, C. Hu and L. Shao, Int. J. Nanomed., 2017, 12, 1227–1249 CrossRef CAS PubMed.
- I. Aranaz, A. R. Alcántara, M. C. Civera, C. Arias, B. Elorza, A. Heras Caballero and N. Acosta, Polymers, 2021, 13, 3256 CrossRef CAS PubMed.
- W.-L. Du, S.-S. Niu, Y.-L. Xu, Z.-R. Xu and C.-L. Fan, Carbohydr. Polym., 2009, 75, 385–389 CrossRef CAS.
- X. Wang, Y. Du and H. Liu, Carbohydr. Polym., 2004, 56, 21–26 CrossRef CAS.
- Z. Assadi, G. Emtiazi and A. Zarrabi, J. Drug Delivery Sci. Technol., 2018, 44, 65–70 CrossRef CAS.
- J. Ai, W. Liao and Z.-L. Ren, RSC Adv., 2017, 7, 15971–15977 RSC.
- N. H. Hoang, T. Le Thanh, R. Sangpueak, J. Treekoon, C. Saengchan, W. Thepbandit, N. K. Papathoti, A. Kamkaew and N. Buensanteai, Polymers, 2022, 14, 662 CrossRef CAS PubMed.
- M. P. Weinstein and J. S. Lewis 2nd, J. Clin. Microbiol., 2020, 58, 018644 CrossRef PubMed.
- P. Bellio, L. Fagnani, L. Nazzicone and G. Celenza, MethodsX, 2021, 8, 101543 CrossRef CAS PubMed.
- S. K. Pillai, R. C. Moellering and G. M. Eliopoulos, Antibiotics in Laboratory Medicine, 2005, vol. 5, pp. 365–440 Search PubMed.
- N. A. S. Rozman, W. Y. Tong, C. R. Leong, M. R. Anuar, S. Karim, S. K. Ong, F. A. M. Yusof, W.-N. Tan, B. Sulaiman and M. L. Ooi, Sci. Rep., 2020, 10(1), 3307 CrossRef CAS PubMed.
- G. Neeraj, S. Krishnan, P. S. Kumar, K. Ravi and V. V. Kumar, J. Mol. Liq., 2015, 214, 335–346 CrossRef.
- M.-Q. Wang, Y.-J. Du, C. Wang, W.-J. Tao, Y.-D. He and H. Li, Biol. Trace Elem. Res., 2012, 149, 184–189 CrossRef CAS PubMed.
- M. Abomosallam, M. Elalfy, Z. Zheng, K. Nagata and M. Suzuki, Nanotechnol. Environ. Eng., 2022, 7, 35–47 CrossRef CAS.
- A. Chrzanowska, A. Drzewiecka-Antonik, K. Dobrzyńska, J. Stefańska, P. Pietrzyk, M. Struga and A. Bielenica, Int. J. Mol. Sci., 2021, 22, 11415 CrossRef CAS PubMed.
- P. Sahariah and M. Másson, Biomacromolecules, 2017, 18, 3846–3868 CrossRef CAS PubMed.
- D. Yan, Y. Li, Y. Liu, N. Li, X. Zhang and C. Yan, Molecules, 2021, 26(23), 7136 CrossRef CAS PubMed.
- P. Feng, Y. Luo, C. Ke, H. Qiu, W. Wang, Y. Zhu, R. Hou, L. Xu and S. Wu, Front. Bioeng. Biotechnol., 2021, 9, 650598 CrossRef PubMed.
- H. A. Anwar, C. H. Aldam, S. Visuvanathan and A. J. Hart, J. Bone Joint Surg. Br., 2007, 89, 1655–1659 CrossRef CAS PubMed.
- E. L. Chang, C. Simmers and D. A. Knight, Pharmaceuticals, 2010, 3, 1711–1728 CrossRef CAS PubMed.
- S. Adewuyi, K. T. Kareem, A. O. Atayese, S. A. Amolegbe and C. A. Akinremi, Int. J. Biol. Macromol., 2011, 48, 301–303 CrossRef CAS PubMed.
- M. E. I. Badawy, T. M. R. Lotfy and S. M. S. Shawir, Bull. Natl. Res. Cent., 2019, 43, 83 CrossRef.
- T. V. Nguyen, T. T. H. Nguyen, S.-L. Wang, T. P. K. Vo and A. D. Nguyen, Res. Chem. Intermed., 2017, 43, 3527–3537 CrossRef CAS.
- J. S. Möhler, T. Kolmar, K. Synnatschke, M. Hergert, L. A. Wilson, S. Ramu, A. G. Elliott, M. A. T. Blaskovich, H. E. Sidjabat, D. L. Paterson, G. Schenk, M. A. Cooper and Z. M. Ziora, J. Inorg. Biochem., 2017, 167, 134–141 CrossRef PubMed.
- L. Benhalima, S. Amri, M. Bensouilah and R. Ouzrout, Pak. J. Med. Sci., 2019, 35, 1322–1328 Search PubMed.
- M. O. Agwara, P. T. Ndifon, N. B. Ndosiri, A. G. Paboudam, D. M. Yufanyi and A. Mohamadou, Bull. Chem. Soc. Ethiop., 2010, 24(3), 383–389 CrossRef CAS.
- M. Kaur, Y. Cohen, E. Poverenov and E. Eltzov, Int. J. Biol. Macromol., 2022, 223, 1107–1114 CrossRef CAS PubMed.
- H. S. Adhikari and P. N. Yadav, Int. J. Biomater., 2018, 2018, 2952085 Search PubMed.
- A. J. Huh and Y. J. Kwon, J. Controlled Release, 2011, 156, 128–145 CrossRef CAS PubMed.
- P. V. Baptista, M. P. McCusker, A. Carvalho, D. A. Ferreira, N. M. Mohan, M. Martins and A. R. Fernandes, Front. Microbiol., 2018, 9, 1–26 CrossRef PubMed.
- S. Chattopadhyay, S. K. Dash, S. Kar Mahapatra, S. Tripathy, T. Ghosh, B. Das, D. Das, P. Pramanik and S. Roy, J. Biol. Inorg. Chem., 2014, 19, 399–414 CrossRef CAS PubMed.
- K. Ambika, S. Helen and U. Ramesh, Int. J. Res. Anal. Rev., 2018, 5(4), 348–351 Search PubMed.
- D. R. Long, J. Mead, J. M. Hendricks, M. E. Hardy and J. M. Voyich, Antimicrob. Agents Chemother., 2013, 57, 241–247 CrossRef CAS PubMed.
- J. Meng, H. Wang, Z. Hou, T. Chen, J. Fu, X. Ma, G. He, X. Xue, M. Jia and X. Luo, Antimicrob. Agents Chemother., 2009, 53, 2871–2878 CrossRef CAS PubMed.
- N. Roschanski, J. Fischer, B. Guerra and U. Roesler, PLoS One, 2014, 9, e100956 CrossRef PubMed.
- M. Smati, O. Clermont, F. Le Gal, O. Schichmanoff, F. Jauréguy, A. Eddi, E. Denamur, B. Picard and C. Group, Appl. Environ. Microbiol., 2013, 79, 5005–5012 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |