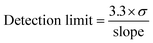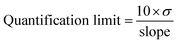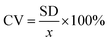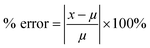Open Access Article
Open Access ArticleElectrochemical investigation of hydroxyapatite–lanthanum strontium cobalt ferrite composites (HA–LSCF) for SARS-CoV-2 aptasensors†
Yeni Wahyuni Hartati *a,
Melania Janisha Devi
*a,
Melania Janisha Devi a,
Irkham
a,
Irkham a,
Salsha Zulqaidaha,
Atiek Rostika Noviyanti
a,
Salsha Zulqaidaha,
Atiek Rostika Noviyanti a,
Siti Rochanib,
Seda Nur Topkaya
a,
Siti Rochanib,
Seda Nur Topkaya c and
Yasuaki Einaga
c and
Yasuaki Einaga d
d
aDepartment of Chemistry, Faculty of Mathematics and Natural Sciences, Universitas Padjadjaran, Indonesia. E-mail: yeni.w.hartati@unpad.ac.id
bMining Technology Research Center, National Research and Innovation Agency, Indonesia
cDepartment of Pharmacy, Izmir Katip Celebi University, Turkey
dDepartment of Chemistry, Keio University, 3-14-1 Hiyoshi, Yokohama 223-8522, Japan
First published on 5th July 2023
Abstract
The hydroxyapatite–lanthanum strontium cobalt ferrite (HA–LSCF) composite showed a good response on a screen-printed carbon electrode (SPCE) electrochemical aptasensor to detect SARS-CoV-2. SPCE/HA–LSCF with a thiolated aptamer has a strong affinity for the SARS-CoV-2 spike RBD protein. This occurs due to the binding of –SH to the HA-positive region. In the presence of LSCF, which is conductive, an increase in electron transfer from the redox system [Fe(CN)6]3−/4− occurs. The interaction of the aptamer with the RBD protein can be observed based on the decrease in the electron transfer process. As a result, the developed biosensor is highly sensitive to the SARS-CoV-2 spike RBD protein with a linear range of 0.125 to 2.0 ng mL−1, a detection limit of 0.012 ng mL−1, and a quantification limit of 0.040 ng mL−1. The analytical application of the aptasensor demonstrates its feasibility in the analysis of saliva or swab samples.
1 Introduction
Hydroxyapatite (HA) is a calcium phosphate mineral with the chemical formula Ca10(PO4)6(OH)2 (Ca/P = 1.67). HA has positive and negative charges on the surface to facilitate the adsorption process of molecules. In addition, HA has high affinity and good biocompatibility.1–6 Hexagonal cells are located in the outermost layers of HA and can bind other ions.7 The OH vacancies in the HA structure are essential as an electron trap. It is a recombination center for electrons and changes the surface connected to close interaction with biological living cells.8HA and its composites (with metals and their oxides) have been widely reported for applications to improve the separation performance of filter paper.9,10 HA-based materials for biosensor applications have also been widely developed with good biocompatibility properties. This includes HA nanoparticles modified graphite pencil electrodes applied for sequence-selective DNA hybridization monitored by differential pulse voltammetry (DPV) technique.11 Glassy carbon electrodes–HA is used for the detection of BK polyomavirus,12 cysteine,13 and folic acid.14 Furthermore, HA–reduced graphene oxide composites detect uric acid15 and dopamine.16 HA composite with ZnO and palladium nanoparticles modified on a carbon paste electrode (CPE) has also been developed to simultaneously determine arbutin (AT) and vitamin C (VC).17 HA modified with polypyrrole, and multi-walled carbon nanotubes (MWCNTs) on glassy carbon electrode for detecting Mycobacterium tuberculosis also showed good performance.18 However, HA has low conductivity in pure water.19 So to increase its conductivity properties, HA can be composited with other conductive materials.
The most popular and mixed conducting material for solid oxide fuel cells is lanthanum strontium cobalt ferrite (LSCF) perovskite. This is due to the material's excellent electrochemical performance and stability.20–23 In addition, HA functionalization with lanthanum oxide has shown good ionic and electronic conductivity and high thermal stability.24–27 In this work, we focus on composite of LSCF which only to increase of its electronic conductivity.
In 2020, the World Health Organization (WHO) declared coronavirus disease-19 (COVID-19) caused by the SARS-CoV-2 virus as a global pandemic. The standard gold method for detecting the presence of this virus is RT-PCR. However, electrochemical biosensors have been developed as an alternative method that is fast, easy, and simple to use for the detection of COVID-19. Molecular diagnosis is carried out by detecting SARS-CoV-2 biomarkers as targets, such as RNA, proteins, and antibodies.28 Antibody-based electrochemical biosensors (immunosensors) that have been developed to detect the COVID-19 antigen include graphene-modified gold electrodes,29 screen-printed carbon electrodes modified copper oxide nanocubes (SPCE/Cu2O NCs),30 SPCE–AuNP–ScFv,31 and paper-based biosensors.32 In addition, aptamer-based biosensors as recognition elements (aptasensors) have also been developed to detect COVID-19, including gold wire electrodes,33 GCE–Au,34 and SPCE–Au.35 Compared with antibodies, aptamer has several advantages, namely smaller size, thermal stability, high affinity, excellent sensitivity, easy synthesis, low toxicity, and easy chemical modification.36
This study developed a screen-printed carbon electrode based on HA–LSCF (SPCE/HA–LSCF) as a single-use, cost-effective, and sensitive aptasensor platform for electrochemical detection of SARS-CoV-2 S-RBD. Among the various aptamer immobilization techniques as bioreceptors,37 we immobilized to the electrode through the electrostatic interaction of the positive charge of HA with the thiolated aptamer. The SARS-CoV-2 S-RBD antigen was chosen as the antigen target molecule. Meanwhile, SPCE/HA–LSCF was developed and applied for the first time in an electrochemical aptasensor. In addition, DPV, electrochemical impedance spectroscopy (EIS), scanning electron microscope (SEM), and energy-dispervise X-ray spectroscopy (EDS) techniques examined the electrochemical and microscopic characterization of the electrodes. Optimization of the experimental parameters is discussed using the Box–Behnken experimental design. Under optimum conditions, variations in target concentration give changes in the peak current height of the K3[Fe(CN)6] redox system to produce quantitative electrochemical signals. HA and LSCF modifications provide a larger surface area and strengthen the detection of SARS-CoV-2 S-RBD. In addition, the aptasensor's precise analytical response, simple operation, and fast response time make it a suitable diagnostic device for monitoring COVID-19 in saliva samples or swabs.
2 Experimental
2.1 Materials
The hydroxyapatite–lanthanum strontium cobalt ferrite (HA–LSCF) composite was produced at the Laboratory of Inorganic Chemistry, Padjadjaran University. The phosphate-buffered saline (PBS) of pH 7.4 was obtained from Merck. Potassium ferricyanide K3[Fe(CN)6], potassium chloride (KCl), bovine serum albumin (BSA), and tris(2-carboxyethyl)phosphine (TCEP) were obtained from Sigma Aldrich in Singapore. In addition, an aptamer of thiol-5′-TTTTTCCAGCCACCGACCTTGTGCTTTGGGAGTGCTGGTCCAAGGGCGTTAAGGACA-3′ was obtained from Bioner in South Korea. Electrochemical measurements were performed using a Zimmer Peacock potentiostat, PSTrace 5.7 software, and SPCE (GSI Technologies, USA).2.2 HA–LSCF preparation
The HA–LSCF was prepared based on previous work, with modifications. HA was synthesized hydrothermally from chicken eggshells and diammonium hydrogen phosphate (DHP), (NH4)2HPO4. Chicken eggshells were crushed and calcined at 1000 °C for 5 hours to convert CaCO3 to CaO. CaO and DHP at a molar ratio of Ca/P = 1.67 were put into the autoclave and heated at 230 °C for 48 hours. The HA obtained was then dried at 110 °C for 2 hours. The purity of HA is 99.5%.25 The HA–LSCF composite was synthesized using the solid-state phase method. The mixture was crushed using an agate mortar and pestle for 5 minutes before being put into the cake and sintered at 730 °C for 5 hours. Furthermore, 0.1% w/v HA–LSCF was dispersed in demineralized water by ultrasonication for further use. The synthesized HA–LSCF was characterized using SEM-EDS.2.3 Assembly of the immunosensor
SPCE modified using HA–LSCF with direct assembly. 40 μL of HA–LSCF solution was dropped onto the electrode surface and incubated overnight at room temperature.38 25 μL of aptamer mixture (20 μL of aptamer-thiol and 5 μL of TCEP) was dropped in SPCE/HA–LSCF and incubated for 20 minutes at room temperature. Finally, 1% BSA was dropped on SPCE/HA–LSCF/aptamer to block non-specific bonds and incubated for 15 minutes at room temperature, then rinsed with PBS solution pH 7.4. Spike SARS-CoV-2 RBD protein with a specific concentration is dropped onto the electrode and incubated for 5 minutes at room temperature. At each assembly step, SPCE was electrochemically characterized by a potassium ferricyanide redox system (10 mM K3[Fe(CN)6] in 0.1 M KCl) using DPV over a potential range of −0.5 to +1.0 V at scan speed 0.008 V s−1 and EIS. SPCE surfaces were characterized using SEM-EDS. Fig. 1 illustrates the aptasensor for the detection of RBD protein S.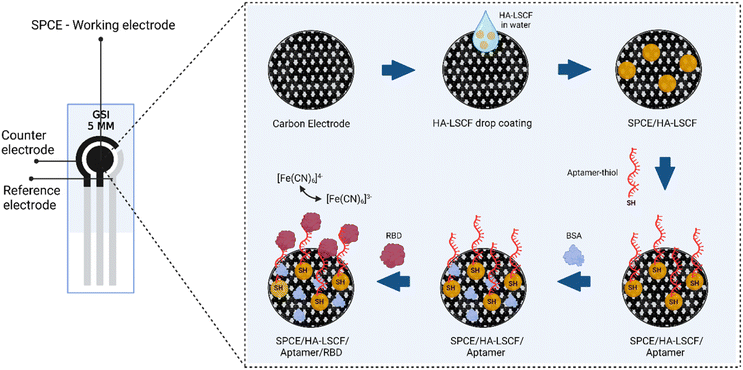 | ||
| Fig. 1 Illustration of HA–LSCF modified aptasensor for detection of RBD spike protein of SARS-CoV-2. | ||
2.4 Parameter optimization with the Box–Behnken experiment design
Factors such as aptamer concentration, aptamer incubation time and RBD spike protein were optimized in the experiment. Each factor was designed through 3 different levels, as shown in Table 1. The Minitab 19 program was used to process the experimental data.| Factor | Unit | Level | ||
|---|---|---|---|---|
| −1 | 0 | +1 | ||
| Aptamer concentration | μg mL−1 | 1.0 | 1.5 | 2.0 |
| Aptamer incubation time | min | 20 | 40 | 60 |
| RBD spike protein incubation time | min | 5 | 10 | 15 |
2.5 Determination of analytical parameters
The SARS-CoV-2 spike RBD protein with a concentration of 0.125, 0.25, 0.5, 1, and 2 ng mL−1 were tested on the aptasensor under optimal conditions. The electrochemical response was measured, and a curve was made between the changes in the average peak current (y) to the protein concentration (x).Limits of detection and quantification were determined by standard deviation of y-intercept of regression line. Limits of detection and quantification are calculated by entering the standard deviation and slope based on the equation:39
where σ is the standard deviation, and the slope is obtained from the regression equation y = bx + a.
The above procedure was used to determine precision and accuracy by measuring the RBD spike protein solution six times. The measurement results show the difference in the average peak current (ΔI) and the standard deviation. Precision is expressed by the coefficient of variation (CV), while accuracy is described by % E or percent relative error with the equation:39
| Precision = 100% − CV |
| Accuracy = 100% − % error |
2.6 Analysis of RBD spike protein in saliva samples
Saliva clinical samples from normal subjects were stored in 5 mL PBS pH 7.4. Then 1, 50, and 100 ng mL−1 standard RBD spike protein was added to a microtube. Electrochemical measurements were carried out with redox system of potassium ferricyanide (K3[Fe(CN)6] 10 mM in 0.1 M KCl) using DPV over a potential range of −0.5 to +1.0 V using a scanning speed of 0.008 V s−1.3 Results and discussion
3.1 Scanning electron microscope (SEM) characterization
The morphology of HA–LSCF is shown in Fig. 2(a). The synthesized HA–LSCF particles appear to form agglomerates. Pore size analysis was performed using Corel Draw. Pore size was calculated using the parallel dimension tools menu on a surface area of 191.44 μm2 with an average pore size of 0.27 μm. The pore size of a material can assists in biosensor binding. HA–LSCF semi-qualitative analysis with EDS is shown in Fig. 2(b). Semi-qualitative analysis was carried out to determine the elemental content contained in the HA–LSCF. The results showed that all HA–LSCF elements were detected from the EDS results.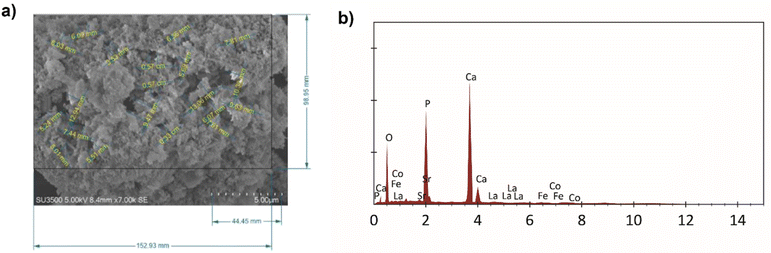 | ||
| Fig. 2 HA–LSCF characterization results using SEM (a) pore size of HA–LSCF, and (b) EDS result of HA–LSCF. | ||
Fig. 3(a) and (b) show the SPCE bare and SPCE/HA–LSCF SEM results. The SEM results of the SPCE/HA–LSCF surface were rougher than bare SPCE. This direct assembly modification method generates an uneven distribution of HA–LSCF across the electrode surface. This technique relies on physical adsorption, and another possibility is HA–LSCF as a suspension that does not dissolve well in distilled water. Fig. 3(d) shows the SPCE/HA–LSCF EDS results. All constituent elements of HA–LSCF were detected in the EDS results. The composition of the constituent elements of LSCF (La, Sr, Co, Fe) is less than the other elements (Ca, O, P, C) because of its small concentration at around 0.5%. Based on the results of SEM and EDS, it can be concluded that SPCE was successfully modified with the HA–LSCF in the surface.
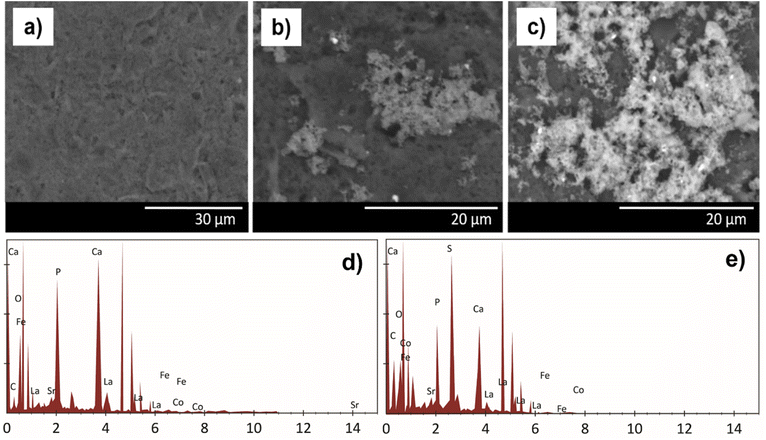 | ||
| Fig. 3 SPCE characterization results using SEM (a) SPCE bare, (b) SPCE/HA–LSCF, and (c) SPCE/HA–LSCF/aptamer. (d) EDS result of SPCE–HA–LSCF, and (e) SPCE/HA–LSCF/aptamer. | ||
Fig. 3(c) shows the surface morphology of SPCE/HA–LSCF/aptamer. Large molecules cover the electrodes on the SPCE/HA–LSCF/aptamer. Fig. 3(e) shows the SPCE EDS results after aptamer immobilization. The presence of sulphur element, which the contribution from the aptamer, indicates that the aptamer has been immobilized on the electrode surface. This is because sulfur, phosphate and carbon are aptamer elements.
3.2 Electrochemical characterization
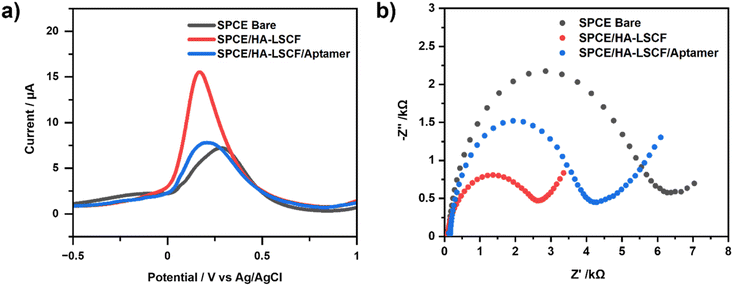
Fig. 4(a) shows the electrochemical characterization of SPCE using DPV with the redox compound [Fe(CN)6]3−/4−. The voltammogram shows an increase in current from 6.649 ± 0.076 μA to 14.389 ± 0.309 μA after modification with a current increase percentage of 216.40% from SPCE bare. The good electron conductivity of the HA–LSCF affects this current to increase the electron transfer at the electrodes. This indicates that HA–LSCF has been successfully modified on the SPCE surface. Meanwhile, the voltammogram shows that the peak current [Fe(CN)6]3−/4− at SPCE/HA–LSCF/aptamer decreased by 7.748 ± 0.082 μA, or the percentage decrease in peak current was 52.27%. This decrease in current is because the aptamer is a macromolecule that covers the surface of the electrode, thereby inhibiting the electron transfer of [Fe(CN)6]3−/4− species at the electrode. The negative charge of the aptamer can also contribute to the repulsive force against [Fe(CN)6]3−/4−, which causes a decreased current response after aptamer immobilization.40,41Fig. 4(b) shows the Nyquist plot of SPCE characterized using EIS. Bare SPCE shows greater resistance than SPCE/HA–LSCF. This indicates that the electron transfer resistance at the surface of the bare SPCE electrode is higher. The increase in resistance is inversely proportional to the increase in current according to Ohm's law. In the Nyquist plot, the half circle shows the magnitude of the charge transfer resistance (Rct). Therefore, the larger the diameter of the semicircle, the greater the charge transfer resistance.42 The Rct value is 5.436 ± 0.060 kΩ for SPCE bare and 2.268 ± 0.007 kΩ for SPCE/HA–LSCF. Meanwhile, the Rct value of the aptamer-immobilized SPCE increased to 3.887 ± 0.075 kΩ due to the inhibition of the electron transfer process [Fe(CN)6]3−/4−. The aptamer used to immobilize the electrodes is reacted with TCEP to produce free thiols. TCEP is a reducing agent that reduces disulfide bonds in the aptamer to produce free thiols.43 HA can attract a negatively charged aptamer because it has a positively charged side. Therefore, the aptamer can be immobilized on the surface of the HA-modified electrode. HA has a surface that can facilitate the adsorption of molecules. The positive surface of calcium ions (Ca2+) on HA can also bind negatively charged molecules.44
Furthermore, the empty electrode area that was not immobilized with aptamer was blocked with 1% BSA and incubated for 15 minutes. BSA was used as a blocking agent to prevent non-specific binding that interfered with the aptasensor reaction and its measurement. The electrodes were then rinsed using PBS pH 7.4 to remove species that were not bound to the electrodes, and the spike RBD protein was ready to be incubated on the electrode surface.
3.3 Optimization of Box–Behnken experiment design
Optimum experimental conditions were determined using the Box–Behnken experimental design. The RBD spike protein concentration used as a fixed variable was 1 ng mL−1. The independent variables chosen were aptamer concentration as factor 1 (X1), aptamer incubation time as factor 2 (X2), and RBD spike protein incubation time as factor 3 (X3). The experimental design with Box–Behnken has the advantage of having fewer trials but obtaining complete information. With three variables, there are only 15 trials. Each experiment was measured twice. The resulting response is processed using the Minitab 19 program. Optimum results are obtained for each factor with the maximum current.The regression equation obtained is as follows:
| y = −5.65 + 6.98X1 + 0.0701X2 + 0.181X3 + 1.462X12 − 0.00348X22 − 0.00108X32 + 0.02199X1X2 + 0.0977X1X3 − 0.000060X2X3 | (1) |
Description:
X1: aptamer concentration.
X2: aptamer incubation time.
X3: incubation time of RBD spike protein.
The regression equation shows the relationship between statistical and experimental data. Meanwhile, the positive and negative values in the regression equation indicate whether the factor influences the experimental response by increasing or decreasing the current. Optimum conditions for each factor were obtained at aptamer concentration of 1.0 μg mL−1, incubation times of aptamer and RBD spike protein were 20 minutes and 5 minutes, respectively.
The P value from the Analysis of Variance (ANOVA) test shows the significance of the factor on the response. P value < 0.05 indicates a significant effect on the response, while P value > 0.05 indicates an insignificant effect. The P values for each factor were aptamer concentration of 0.000, aptamer incubation time of 0.013, and protein S RBD incubation time of 0.439. Based on these data, the aptamer's concentration and incubation time significantly affected the current response. In contrast, the RBD protein S incubation time had no significant effect.
3.4 Aptasensor response against SARS-CoV-2 S-RBD
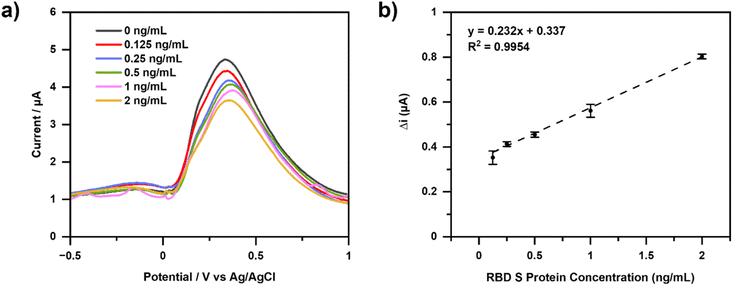
Calibration curves were made with variations in S-RBD concentrations which are 0, 0.125, 0.25, 0.5, 1.0, and 2.0 ng mL−1. DPV measurements were repeated three times under optimal conditions, and the results shown in Fig. 5. The interaction between the aptamer and RBD spike protein was characterized using DPV. As a result, there is a decrease in current when the RBD spike protein is added. This indicates that the S protein RBD is already bound to the aptamer, thereby blocking electron transfer and decreasing the current, which is proportional to the S protein RBD concentration.Aptamer binds specifically to the RBD spike protein. The ssDNA aptamer generated from SELEX was used with the RBD selection strategy based on the ACE2 competition carried out by Song et al. (2020). Hydrogen bonding occurs between the aptamer and the RBD spike protein. At the 3′ end of the aptamer, T42 and T43 can form hydrogen bonds with amino acids in the RBD spike protein, namely Thr500, Gln506, and Asn437. The binding between the aptamer and the RBD spike protein is almost similar to that of the RBD spike protein and ACE2, as well as the Thr500 amino acid.45
Fig. 5(a) shows the decrease in current resulting from measurements with increasing concentrations of RBD spike protein. Therefore, the concentration is directly proportional to the binding of the RBD spike protein to the aptamer. Electron transfer at the electrodes is impaired due to inhibition of the RBD spike protein, a large non-electroactive molecule.
Fig. 5(b) shows the calibration curve of the change in current for RBD spike protein concentration. The linear regression equation obtained is y = 0.232x + 0.337 with an R2 value of 0.9954. Limits of detection (LOD) is calculated as the analyte concentration giving a signal equal to the blank signal, yB, plus three standard deviations of the blank, sB. Furthermore, the limit of quantitation (LOQ) which is regarded as the lower limit for precise quantitative measurements, as opposed to qualitative detection. A value of yB +10sB has been suggested for this limit.46 The resulting LOD and LOQ values were 0.012 ng mL−1 and 0.040 ng mL−1, respectively (Table S1†). Precision and accuracy values were determined by six measurements of RBD spike protein with a concentration of 0.5 ng mL−1. The precision was found to be 98.54% (RSD = 1.46%) while the accuracy results are 97.53% (Table S2†).
Table 2 shows a comparison of this study with other SARS-CoV-2 detection aptasensors that have been reported. It was found that the detection limits in this study had higher and lower values than the others, but quantitative comparisons can be continued with standard PCR methods. The % recovery value was determined by storing saliva samples from normal subjects in 5 mL PBS pH 7.4 and adding 1, 50, and 100 ng mL−1 of RBD spike protein standard. The average concentration measured using 3 DPV measurements was 0.99, 49.71, and 99.68 ng mL−1 with a recovery value of 99.17%, 99.42%, and 99.71%, respectively.
| Method | Limit of detection | Linear range | Sample | Reference |
|---|---|---|---|---|
| Electrochemical aptamer based sensor using gold electrode | — | — | Salivary | 33 |
| Electrochemical aptamer based sensor using Shrinky-Dink electrode | 1 × 103 pg mL−1 | — | Salivary | 47 |
| Electrochemical aptasensor using SPCE–Cu2O for the spike RBD detection spike glycoprotein | 0.03 ± 0.01 × 10−3 pg mL−1 | 0.0001–1.2 × 106 pg mL−1 | Salivary | 30 |
| Electrochemical impedance spectroscopy-based aptasensor using a carbon nanofiber–gold for the RBD spike detection | 7.0 pM | 10–64 × 103 pM | Salivary | 48 |
| Electrochemical aptasensor for SARS-CoV-2 antigen detection based on aptamer-binding induced multiple hairpin assembly signal amplification | 9.79 × 10−3 pg mL−1 | 0.05–50 × 103 pg mL−1 | — | 49 |
| Nanoscale electrode–aptamer–SARS-CoV-2 imaging by photo-induced force microscopy | 1.30 pM (66 pg mL−1) | — | — | 34 |
| Folding-based electrochemical aptasensor | 7 × 103 pM | — | — | 50 |
| Optimization electrochemical aptasensor using SPCE–Au | — | 2.63 × 103 pg mL−1 | Salivary | 35 |
| HA–LaSCF for the RBD spike detection | 1.20 × 102 pg mL−1 | 0.125–2 × 103 pg mL−1 | — | This work |
4 Conclusions
HA–LSCF composite is a functional material that can be applied as a conductive adsorbent due to its unique structure. Electrochemical investigation for biosensor application, in this case, aptasensor,can also be categorized as very good. HA–LSCF was successfully immobilized on the SPCE surface. Due to the unique HA-based structure, the aptamer was successfully immobilized on the SPCE/HA–LSCF surface. This SPCE/HA–LSCF/aptamer can detect the SARS-CoV-2 spike RBD with a linear range of 0.125 to 2.0 ng mL−1 and a detection limit of 0.0120 ng mL−1. In addition, the developed HA–LSCF-based biosensor can be used as an alternative method for detecting the COVID-19 pandemic.Author contributions
Y. W. H. conceptualized, wrote the manuscript, funding acquisition the work. M. J. D., I., and S. Z., data curation, analysed data, methodology, software, visualization, and edited the manuscript. A. R. N., S. R., S. N. T., and Y. E. investigation, project administration, resources, and validation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Authors would like to thank Indonesian Ministry of Research, Technology and the National Innovation Agency for supporting this work with scheme no. 69/FI/P-KCOVID-19.2B3/IX/2020, and Academic Leadership Grant of Padjadjaran University No. 1549/UN6.3.1/PT.00/2023.References
- I. Cacciotti, A. Bianco, M. Lombardi and L. Montanaro, J. Eur. Ceram. Soc., 2009, 29, 2969–2978 CrossRef CAS.
- A. Fihri, C. Len, R. S. Varma and A. Solhy, Coord. Chem. Rev., 2017, 347, 48–76 CrossRef CAS.
- Y. W. Hartati, I. Irkham, S. Zulqaidah, R. S. Syafira, I. Kurnia, A. R. Noviyanti and S. N. Topkaya, Sens. Bio-Sens. Res., 2022, 38, 100542 CrossRef.
- M. Ibrahim, M. Labaki, J. M. Giraudon and J. F. Lamonier, J. Hazard. Mater., 2020, 383, 121139 CrossRef CAS PubMed.
- R. S. Syafira, M. J. Devi, S. Gaffar and Y. W. Hartati, Biointerface Res. Appl. Chem., 2023, 13, 499 CAS.
- S. Wang, Y. Lei, Y. Zhang, J. Tang, G. Shen and R. Yu, Anal. Biochem., 2010, 398, 191–197 CrossRef CAS PubMed.
- I. Gutowska, Z. Machoy and B. Machaliński, J. Biomed. Mater. Res., Part A, 2005, 75, 788–793 CrossRef PubMed.
- V. S. Bystrov, J. Coutinho, A. V. Bystrova, Y. D. Dekhtyar, R. C. Pullar, A. Poronin, E. Palcevskis, A. Dindune, B. Alkan, C. Durucan and E. V. Paramonova, J. Phys. D: Appl. Phys., 2015, 48, 195302 CrossRef.
- S. Pai, M. S. Kini and R. Selvaraj, Environ. Sci. Pollut. Res., 2021, 28, 11835–11849 CrossRef CAS PubMed.
- Q. Q. Zhang, Y. J. Zhu, J. Wu, Y. T. Shao and L. Y. Dong, J. Colloid Interface Sci., 2020, 575, 78–87 CrossRef CAS PubMed.
- A. Erdem and G. Congur, Electroanalysis, 2018, 30, 67–74 CrossRef CAS.
- A. Musolino, M. Raugi, R. Rizzo and M. Tucci, IEEE Trans. Magn., 2014, 50(12), 1–11 Search PubMed.
- N. Vladislavić, I. Š. Rončević, M. Buzuk, M. Buljac and I. Drventić, J. Solid State Electrochem., 2021, 25, 841–857 CrossRef.
- P. Kanchana, N. Lavanya and C. Sekar, Mater. Sci. Eng., C, 2014, 35, 85–91 CrossRef CAS PubMed.
- Y. Chen, W. Zhou, J. Ma, F. Ruan, X. Qi and Y. Cai, Microsc. Res. Tech., 2020, 83, 268–275 CrossRef CAS PubMed.
- T. V. Gopal, T. M. Reddy, P. Shaikshavali and G. Venkataprasad, Surf. Interfaces, 2021, 24, 101145 CrossRef CAS.
- S. A. Shahamirifard and M. Ghaedi, Biosens. Bioelectron., 2019, 141, 111474 CrossRef CAS PubMed.
- K. S. Rizi, B. Hatamluyi, M. Rezayi, Z. Meshkat, M. Sankian, K. Ghazvini, H. Farsiani and E. Aryan, Talanta, 2021, 226, 122099 CrossRef CAS PubMed.
- L. H. Huang, X. Y. Sun and J. M. Ouyang, Sci. Rep., 2019, 9, 18979 CrossRef CAS PubMed.
- H. Fan, M. Keane, P. Singh and M. Han, J. Power Sources, 2014, 268, 634–639 CrossRef CAS.
- G. D. Han, K. C. Neoh, K. Bae, H. J. Choi, S. W. Park, J. W. Son and J. H. Shim, J. Power Sources, 2016, 306, 503–509 CrossRef CAS.
- S. P. Jiang, Int. J. Hydrogen Energy, 2019, 44, 7448–7493 CrossRef CAS.
- M. Kim, D. H. Kim, G. D. Han, H. J. Choi, H. R. Choi and J. H. Shim, J. Alloys Compd., 2020, 843, 155806 CrossRef CAS.
- J. Mirzababaei and S. S. C. Chuang, Catalysts, 2014, 4, 146–161 CrossRef.
- A. R. Noviyanti, N. Akbar, Y. Deawati, E. E. Ernawati, Y. T. Malik, R. P. Fauzia and Risdiana, Heliyon, 2020, 6(4), e03655 CrossRef PubMed.
- P. La, M. Co, A. M. Couper, D. Pletcher, F. C. Walsh, J. W. Stevenson, I. Armstrong, R. D. Carneim, L. It Pederson and W. J. Weber, Electrochemical Methods, 1996, 143, 614 CrossRef.
- M. A. Peña and J. L. G. Fierro, Chem. Rev., 2001, 101, 1981–2017 CrossRef PubMed.
- F. Cui and H. S. Zhou, Biosens. Bioelectron., 2020, 165, 112349 CrossRef CAS PubMed.
- B. Mojsoska, S. Larsen, D. A. Olsen, J. S. Madsen, I. Brandslund and F. A. Alatraktchi, Sensors, 2021, 21, 1–11 CrossRef PubMed.
- Z. Rahmati, M. Roushani, H. Hosseini and H. Choobin, Microchim. Acta, 2021, 188, 105 CrossRef CAS PubMed.
- G. N. Fadhilah, M. Yusuf, A. K. Sari, T. R. Tohari, H. L. Wiraswati, S. Ekawardhani, L. Faridah, N. Fauziah, I. Anshori and Y. W. Hartati, ChemistrySelect, 2023, 8, e202203928 CrossRef CAS.
- A. Yakoh, U. Pimpitak, S. Rengpipat, N. Hirankarn, O. Chailapakul and S. Chaiyo, Biosens. Bioelectron., 2021, 176, 112912 CrossRef CAS PubMed.
- A. Idili, C. Parolo, R. Alvarez-Diduk and A. Merkoçi, ACS Sens., 2021, 6, 3093–3101 CrossRef CAS PubMed.
- J. C. Abrego-Martinez, M. Jafari, S. Chergui, C. Pavel, D. Che and M. Siaj, Biosens. Bioelectron., 2022, 195, 113595 CrossRef CAS PubMed.
- A. K. Sari, Y. W. Hartati, S. Gaffar, I. Anshori, D. Hidayat and H. L. Wiraswati, J. Electrochem. Sci. Eng., 2022, 12, 219–235 CrossRef CAS.
- D. H. Mohsin, M. S. Mashkour and F. Fatemi, Chem. Pap., 2021, 75, 279–295 CrossRef CAS.
- A. K. Sari, S. Gaffar and Y. W. Hartati, Anal. Bioanal. Electrochem., 2022, 14(1), 127–143 CAS.
- A. A. A. Khan, H. Ajab, A. Yaqub and M. A. Abdullah, E3S Web Conf., 2019, 76, 3–8 Search PubMed.
- P. Feldsine, C. Abeyta and W. H. Andrews, J. AOAC Int., 2002, 85(5), 1187–1200 CrossRef CAS PubMed.
- A. Lomae, P. Preechakasedkit, O. Hanpanich, T. Ozer, C. S. Henry, A. Maruyama, E. Pasomsub, A. Phuphuakrat, S. Rengpipat, T. Vilaivan, O. Chailapakul, N. Ruecha and N. Ngamrojanavanich, Talanta, 2023, 253, 123992 CrossRef CAS PubMed.
- Z. Liu, C. Wang, S. Zheng, X. Yang, H. Han, Y. Dai and R. Xiao, Nanomedicine, 2023, 47, 102624 CrossRef CAS PubMed.
- D. V. Ribeiro, C. A. C. Souza and J. C. C. Abrantes, Rev. IBRACON Estrut. Mater., 2015, 8, 529–546 CrossRef.
- L. Farzin, M. Shamsipur, L. Samandari and S. Sheibani, Microchim. Acta, 2018, 185, 1–9 CrossRef PubMed.
- K. Lin and J. Chang, Structure and properties of hydroxyapatite for biomedical applications, Hydroxiapatite (Hap) for Biomedical Applications, Elsevier Ltd., 2015, vol. 4214 Search PubMed.
- Y. Song, J. Song, X. Wei, M. Huang, M. Sun, L. Zhu, B. Lin, H. Shen, Z. Zhu and C. Yang, Anal. Chem., 2020, 92, 9895–9900 CrossRef CAS PubMed.
- J. Miller and J. C. Miller, Statistics and Chemometrics for Analytical Chemistry. Pearson Education. 2018 Search PubMed.
- J. A. Zakashansky, A. H. Imamura, D. F. Salgado, H. C. Romero Mercieca, R. F. L. Aguas, A. M. Lao, J. Pariser, N. Arroyo-Currás and M. Khine, Anal. Methods, 2021, 13, 874–883 RSC.
- M. A. Tabrizi and P. Acedo, Biosensors, 2022, 12(3), 142 CrossRef PubMed.
- J. Xue, Y. Li, J. Liu, Z. Zhang, R. Yu, Y. Huang, C. Li, A. Chen and J. Qiu, Talanta, 2022, 248, 123605 CrossRef CAS PubMed.
- F. Curti, S. Fortunati, W. Knoll, M. Giannetto, R. Corradini, A. Bertucci and M. Careri, ACS Appl. Mater. Interfaces, 2022, 14, 19204–19211 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed calculation of LoD, LoQ, RSD, precision, accuracy for aptasensor response against SARS-CoV-2 S-RBD. See DOI: https://doi.org/10.1039/d3ra01531a |
| This journal is © The Royal Society of Chemistry 2023 |