Open Access Article
Open Access ArticleDevelopment of green fluorescent protein-based cAMP indicators for covering a wide range of cAMP concentrations†
Sohei Hiasaa,
Takeru Fujimoria,
Saki Aikib,
Hiroshi Ueda c,
Takashi Tsuboi*b and
Tetsuya Kitaguchi
c,
Takashi Tsuboi*b and
Tetsuya Kitaguchi *c
*c
aSchool of Life Science and Technology, Department of Life Science and Technology, Tokyo Institute of Technology, 4259 Nagatsuta-cho, Midori-ku, Yokohama-shi, Kanagawa 226-8501, Japan
bDepartment of Life Sciences, Graduate School of Arts and Sciences, The University of Tokyo, 3-8-1 Komaba, Meguro-ku, Tokyo 153-8902, Japan. E-mail: takatsuboi@bio.c.u-tokyo.ac.jp
cLaboratory for Chemistry and Life Science, Institute of Innovative Research, Tokyo Institute of Technology, 4259 Nagatsuta-cho, Midori-ku, Yokohama-shi, Kanagawa 226-8503, Japan. E-mail: kitaguct-gfp@umin.ac.jp
First published on 22nd May 2023
Abstract
There is a wide range in the concentration of intracellular cyclic adenosine 3′,5′-monophosphate (cAMP), which mediates specific effects as a second messenger in pathways affecting many physiological processes. Here, we developed green fluorescent cAMP indicators, named Green Falcan (Green fluorescent protein-based indicator visualizing cAMP dynamics) with various EC50 values (0.3, 1, 3, 10 μM) for covering the wide range of intracellular cAMP concentrations. The fluorescence intensity of Green Falcans increased in a cAMP dose-dependent manner, with a dynamic range of over 3-fold. Green Falcans showed a high specificity for cAMP over its structural analogues. When we expressed Green Falcans in HeLa cells, these indicators were applicable for visualization of cAMP dynamics in the low concentration range compared to the previously developed cAMP indicators, and visualized distinct kinetics of cAMP in various pathways with high spatiotemporal resolution in living cells. Furthermore, we demonstrated that Green Falcans are applicable to dual-color imaging with R-GECO, a red fluorescent Ca2+ indicator, in the cytoplasm and the nucleus. This study shows that Green Falcans open up a new avenue for understanding hierarchal and cooperative interactions with other molecules in various cAMP signaling pathways by multi-color imaging.
1. Introduction
Cyclic adenosine 3′,5′-monophosphate (cAMP) plays an important role as a second messenger with a wide range of concentrations from sub μM to μM in living cells.1–3 Many hormones or neurotransmitters stimulate G protein-coupled receptors in the cell membrane, leading to the activation of adenylate cyclase which catalyzes the conversion of ATP to cAMP.4 In living cells, cAMP binds to protein kinase A (PKA),5 exchange proteins directly activated by cAMP (Epac),6 or cyclic nucleotide-gated (CNG) ion channels,7 and causes many cellular functions such as hormone secretion,8 cell motility,9 and memory formation,10 etc.To date, many single-fluorescent protein (FP) based indicators have been developed as tools for detecting various intracellular signaling molecules (Ca2+, cAMP, cGMP, etc.).11–17 Since single-FP based indicators require single wavelength emission for detection, it is suitable for multicolor imaging to simultaneously visualize the hierarchal and cooperative relationship with other molecules in the same or distinct intracellular compartments, with high spatiotemporal resolution. They show increased and decreased FI in the presence of analytes, called turn-on type and turn-off types, respectively. A few turn-off type green fluorescent cAMP indicators have been produced,14,15,18 but distinguishing photobleach or the degradation of the indicator from analyte decrease is problematic. Several turn-on green fluorescent cAMP indicators had been developed, but their utility was limited by low dynamic range.18–20 Very recently, new cAMP indicators with higher dynamic range were developed and this limitation is being resolved,21 but it is not sufficient to monitor a wide range of cAMP concentrations in living cells (Table S1†).
Here, we utilized our previously described method of semi-rational molecular design and molecular evolution22 in developing indicators such as Green cGull,17 MaLionG,23 and Green Glifon,24 to generate powerful turn-on type green fluorescent cAMP indicators with expanded dynamic range and various half maximal effective concentration (EC50) employing PKA as a binding domain, named them Green Falcan 0.3, 1, 3, 10. We demonstrated that this useful method is not limited to cAMP indicators, but hopefully leads to the development of new indicators for a myriad of molecules.
2. Materials and methods
2.1. Chemicals
cAMP and cGMP were purchased from Sigma-Aldrich (St. Louis, MO, USA), forskolin from Tokyo Chemical Industry (Tokyo, Japan), 3-isobutyl-1-methylxanthine, 8-Br-cAMP and adenosine from Merck Millipore (Darmstadt, Germany), ATP, ADP and AMP from Oriental Yeast Co., Ltd. (Tokyo, Japan).2.2. Plasmid construction
The DNA fragment of GFP, restructured from circularly permuted (cp) GFP in G-GECO,13 a green fluorescent genetically-encoded Ca2+ indicator, was modified by PCR to contain KpnI and BglII restriction enzyme sites between N144 and S146 and cloned into the pRSET-A vector (Thermo Fisher Scientific, Massachusetts, USA). The cDNA for the mouse cAMP-dependent protein kinase (PKA) type I-beta regulatory subunit25 (NP_001240819.1) was obtained from adult mouse brain total RNA by RT-PCR. The region of the cDNA encoding the cAMP binding domain (amino acids 92–248) derived from site A, was ligated at the KpnI/BglII site of the GFP DNA. To improve the fluorescence intensity (FI), the N- and C-terminus linker lengths were optimized by inserting various lengths (N-terminus 0 to 9 amino acids; C-terminus 0 to 10 amino acids) of leucine zipper sequences26 between the GFP and the cAMP binding domain sequences. The plasmids encoding mutant indicators with different linker lengths at the N- and C-terminus of the cAMP binding domain were transformed into Escherichia coli JM109 (DE3) (Promega, Madison, USA), and these lysates were subjected to examine the FI change. The plasmid DNA of the mutant showing the maximum response was selected as a template for subsequent random mutagenesis. To further improve the FI change by introducing random mutations into the amino acid sequences around the linkers, PCR was performed using two degenerate primers containing NNK and MNN mixtures of bases. Plasmids carrying site-directed random mutations around linkers were transformed into JM109 (DE3), and around 50 colonies (empirically ∼15 kinds of amino acid coverage) were picked. The plasmid of the mutant with the maximum response was selected as the next template for the molecular evolution cycle of random mutagenesis, and this cycle continued until the response was more than 3-fold. Then, to adjust the half-maximal effective concentration (EC50) of Green Falcan by changing the affinity to cAMP, PCR was performed using two primers containing NNK and MNN designed to the region around R211 of PKA, which is important for cAMP binding.27 The mutants with various EC50 (1, 3, 10 μM) were selected by the dose-response curve using a four-parameter logistic curve fitting. For live-cell imaging in mammalian cells, Green Falcan DNA sequences were cloned into the pcDNA3.1 (−) vector (Thermo Fisher Scientific, Massachusetts, USA) (Fig. S2A–D†). To improve the solubility of the expressed proteins, the super acidic region of mouse amyloid β precursor protein (amino acids 190–286) was fused to the N-terminus of the indicators. For subcellular localization, the nuclear localization signal (nls; PKKKRKV) fused to the N-terminus of the indicators was derived from SV40. The cAMP indicator, cAMPr plasmid was obtained from Addgene (#99143, Massachusetts, USA), and was cloned into the pcDNA3.1 (+) vector for better expression.2.3. Protein production and purification
Green Falcans DNA in the pRSET-A vector were transformed into Escherichia coli JM109 (DE3) cells, cultured in 400 mL LB medium with 50 μg mL−1 ampicillin at 20 °C for 4 days, and harvested by centrifugation. The harvested cells were suspended in phosphate-buffered saline (PBS; pH 7.4) with 0.5% (v/v) Triton-X and 40 μg mL−1 lysozyme, and lysed by French press (Constant Systems Ltd, Daventry, UK). After centrifugation of the lysate, the supernatant containing the Green Falcans protein was purified by TALON® Metal Affinity Resin (TaKaRaBio, Shiga, Japan). After washing three times with 10 mM imidazole, the proteins were eluted from the resin with 300 mM imidazole. To remove the imidazole, the eluate was applied to PD-Miditrap G-25 (GE Healthcare, Chicago, USA) in 150 mM KCl, 50 mM HEPES-KOH (pH 7.4), 0.5% (v/v) Triton-X. The purified protein was stored at −80 °C.2.4. SDS-PAGE
The purified Green Falcan proteins (1 μg) were mixed with an equal volume of 2 × SDS loading buffer, and heated at 95 °C for 5 min. These were loaded in 10% acrylamide gel. Precision Plus Protein Unstained Standards (Bio-Rad, California, USA) was also loaded as a protein marker. After electrophoresis, the gel was dyed by CBB staining. For taking a picture, luminographI (ATTO, Tokyo, Japan) was used.2.5. In vitro spectroscopy
The excitation and emission spectra of Green Falcans were measured using a spectrophotometer (F2700, Hitachi, Tokyo, Japan). To examine the dose-response relationship, various concentrations of cAMP and cGMP (from 0.001 to 300 μM) were applied to 0.3 μM purified Green Falcan protein. Relative FI was calculated by dividing by the FI of the peak in the absence of cAMP. To obtain the EC50 value and dynamic range of the Green Falcans for cAMP and cGMP, a four-parameter logistic curve fitting (eqn (1)) was performed using QtiPlot (IONDEV SRL, Bucharest, Romania).
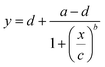 | (1) |
2.6. Cell culture and transfection
HeLa cells were cultured in Dulbecco's modified eagle medium (high glucose) with L-glutamine, sodium pyruvate, penicillin and streptomycin (Wako, Osaka, Japan), 10% (v/v) heat-inactivated fetal bovine serum (GE Healthcare, Chicago, USA). These cells were subcultured in 35 mm glass-bottomed dishes at 37 °C with 5% CO2 for live-cell imaging. Plasmids of each Green Falcans were transfected to these cells with Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific, Massachusetts, USA) according to the manufacturer's instruction. Then, HeLa cells were cultured at 37 °C overnight. After changing the medium, HeLa cells were cultured at 30 °C to 32 °C for at least 30 h to allow maturation of indicator chromophores.2.7. Live-cell imaging
Live-cell imaging was performed using a fluorescence inverted microscope (IX70, Olympus, Tokyo, Japan) equipped with an oil immersion ×40 objective lens (UApo/N 340, ×40, NA = 1.35, Olympus), a cooled CCD camera (Cool SNAP HQ2, Photometrics), a mercury lamp, an excitation filter of 460–480 nm, an emission filter of 495–540 nm and a dichroic mirror of 485 nm (U-MGFPHQ, Olympus). For dual-color imaging, the DA/FI/TR-3X3M-C Sedat filter set (OPTO-LINE, Inc., Saitama, Japan) is set by an excitation filter of 546–566 nm, an emission filter of 580.5–653.5 nm and triple band dichroic mirror of 403, 497 and 574 nm, was changed by using an HF110 high-speed filter wheel (Prior Scientific, Cambridge, UK). Fluorescence images were acquired every 5 s using Metafluor software (Molecular Devices, Sanjose, USA). The FI was measured by manually surrounding each cell with a region of interest (ROI) using ImageJ software (https://imagej.net/software/fiji/downloads).29 After subtracting the FI of a cell-free area, relative FI was calculated by dividing by the mean of the FI captured over 1 or 2 min before administering reagents. For statistical analysis, a Student's t-test was performed using R (https://www.r-project.org/)30 for Fig. 3F, S6B and S7C, F,† and one-way ANOVA was performed using EZR (https://www.jichi.ac.jp/saitama-sct/SaitamaHP.files/statmed.html)31 for Fig. 3C.3. Results and discussion
3.1. Generation of Green Falcans
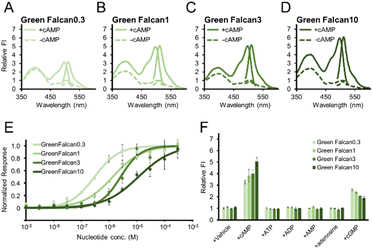
To generate turn-on type green fluorescent cAMP indicators with expanded dynamic range, we first constructed a prototype indicator by inserting the cAMP binding domain (amino acids 92–248) derived from site A of mouse cAMP-dependent protein kinase (PKA) type I-beta regulatory subunit27 (NP_001240819.1) into the vicinity of the chromophore of green fluorescent protein (GFP) (Fig. S1A†). When this prototype indicator was expressed in bacteria, the FI measured in the lysates increased slightly in the presence of cAMP. Since this response was insufficient for easy detection of minor changes in cAMP levels, we planned to improve the FI of prototype indicator by protein engineering. To begin, we selected the linker length between GFP and PKA (N-terminus: 4 amino acids, C-terminus: 3 amino acids) that displayed the largest FI change (1.3-fold) (Fig. S1B and C†). For further improvement of the dynamic range, we optimized the amino acid sequences around the N- and C-terminus linkers by site-directed random mutagenesis using PCR. After 40 cycles of random mutation, we obtained a green fluorescent cAMP indicator with more than 3-fold FI change in the presence of cAMP (Fig. S1D†). When the dose-response relationship to cAMP was examined, we found that the EC50, apparent Kd, of this indicator was 0.3 μM (Fig. S4E†). Generally, cAMP is known to function in a wide range of concentrations from sub μM to μM range in living cells.1 Therefore, to cover this range, we introduced random mutations around R211 of PKA, which is important for cAMP binding.27 Consequently, by the selection based on the response to several concentrations of cAMP, we succeeded in obtaining additional three green fluorescent cAMP indicators with various EC50 (1, 3, and 10 μM) (Fig. 1A, S4F–H†). The indicators, Green Falcan 0.3, 1, 3, and 10 were named after their EC50 value and abbreviation of green fluorescent protein-based indicator visualizing cAMP dynamics (Fig. 1B). Additionally, utilizing the same molecular design such as insertion of a ligand-binding domain into the vicinity of an FP chromophore and optimization of linker region to expand FI change, we used to develop our previous indicators for cAMP,14–16 cGMP,17 ATP,23 glucose,24 pyruvate33 and lactate33 demonstrates its versatility for generating genetically-encoded indicators based on fluorescent protein.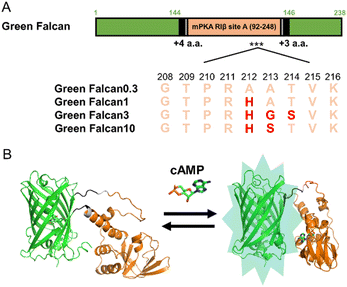 | ||
| Fig. 1 Schematic design of Green Falcan proteins. (A) Diagram for Green Falcan proteins. Red letters indicate mutated amino acids. (B) Schematic 3D structures of Green Falcan unbound (left) and bound (right) to cAMP. The schematic 3D structures were created with Alphafold2.32 | ||
3.2. Property of Green Falcans
To investigate the properties of the Green Falcans, we purified (Fig. S3†) and examined excitation and emission spectra for these indicators. All Green Falcans had two excitation peaks at around 400 nm and 498 nm with an emission peak at around 511 nm (Fig. 2A–D). The peaks in excitation at around 498 nm, and emission at around 511 nm, which are similar to the spectral properties of GFP,34 were increased more than 3-fold in the presence of cAMP. In the presence of cAMP, the absorption spectra of all Green Falcans showed a little decreased and increased peaks at around 400 nm and 500 nm, respectively (Fig. S4A–D†). It has been demonstrated that the GFP absorption peak observed at around 400 nm corresponds to the protonated state of the chromophore and that observed at 500 nm, to the deprotonated state.34 Thus, we think that a change in the population of the protonated/deprotonated state of chromophore contributes to the fluorescence intensity (FI) change. Interestingly, unlike other GFP-based indicators, Green Falcans have an excitation peak at around 400 nm, which increased in the presence of cAMP. Since the excitation peak derived from the protonated form of chromophore creates the fluorescence at around 511 nm by excited-state proton transfer,35,36 it seems that the proton transfer was also accelerated by the application of cAMP to induce FI change. However, the measurement with 400 nm excitation is not practical because the dynamic range with 400 nm excitation is lower than that with 480 nm, and the excitation at short wavelength causes cell damage.When the dose-response relationship of all Green Falcans was examined, we found that the EC50 value of Green Falcan 0.3, 1, 3, 10 for cAMP was 0.30, 1.4, 2.6 and 11 μM, and the dynamic range in the presence of cAMP was 3.9, 5.6, 4.7, 6.5-fold, respectively. While we found that the EC50 values of all Green Falcans for cGMP were higher than that for cAMP (Fig. 2E, S4E–H,† Table 1). However, since the EC50 values of Green Falcans for cAMP are about 10–100 times lower than that for cGMP, the detection of cAMP is still affected in the presence of cGMP. Therefore, the dose-response relationship for cAMP in the presence of cGMP (10 times EC50 for cAMP) was also examined. The responses of all Green Falcans for cAMP were smaller, while the EC50 values of all Green Falcans were similar to those without cGMP, suggesting that Green Falcans function efficiently in the presence of cGMP (Fig. S4I–L†). To examine the specificity of all Green Falcans for cAMP, we applied cAMP structural analogues (ATP, ADP, AMP, adenosine, cGMP) at 10 times the concentrations of the EC50 for each indicator. This demonstrated that all Green Falcans display high specificity for cAMP (Fig. 2F). The previously reported single-FP based indicators have shown pH sensitivity.14–17,23,24,33 Similarly, the FI of Green Falcans was increased depending on pH elevation (Fig. S4M–P†). These results suggest that by choosing the appropriate indicator, according to its EC50, for the expected intracellular cAMP range, these indicators can be used to specifically visualize a wide range of cAMP levels in living cells during a variety of physiological events. Although we have employed a cAMP binding domain from mouse Epac1 for developing cAMP indicators such as Flamindo,14 Flamindo2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 and Pink Flamindo,16 that from PKA was done in this study. Since introducing a mutation in the binding domain usually decreases the affinity for ligands, we recommend exploiting a binding domain with a higher affinity to develop indicators covering a wide range of ligand concentrations.
15 and Pink Flamindo,16 that from PKA was done in this study. Since introducing a mutation in the binding domain usually decreases the affinity for ligands, we recommend exploiting a binding domain with a higher affinity to develop indicators covering a wide range of ligand concentrations.
| Indicator name | EC50 (cAMP) (μM) | EC50 (cGMP) (μM) | Working range (cAMP) (μM) |
|---|---|---|---|
| Green Falcan 0.3 | 0.30 | 2.8 | 8.1 × 10−2–1.4 |
| Green Falcan 1 | 1.4 | 1.0 × 102 | 0.30–9.3 |
| Green Falcan 3 | 2.6 | 3.1 × 102 | 0.75–7.9 |
| Green Falcan 10 | 11 | 8.7 × 102 | 4.9–2.6 × 10−2 |
3.3. Applicability of Green Falcans to live-cell imaging
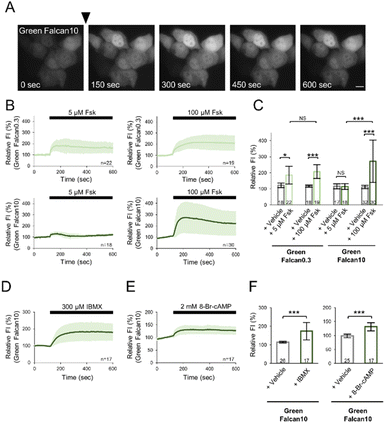
To investigate the applicability of Green Falcans to live-cell imaging, we expressed these indicators in HeLa cells, and monitored with several reagents increasing intracellular cAMP levels under a fluorescence microscope. Green Falcans were well expressed in HeLa cells, but had lower basal fluorescence intensity than HeLa cells expressing cAMPr, a previously developed cAMP indicator (Fig. S5A and B†). After the administration of 100 μM forskolin, an adenylate cyclase activator, the fluorescence intensity (FI) of these cells expressing each Green Falcan was significantly increased (Fig. 3A–C, S6A–C†), suggesting that all Green Falcans are able to be used in visualizing cAMP dynamics in living cells. Moreover, when the administration of 5 μM forskolin to HeLa cells expressing Green Falcan 0.3, 10, cAMPr or Pink Flamindo, previously developed cAMP indicators, the FI of Green Falcan 0.3 was significantly increased, but not of Green Falcan 10 and cAMPr (Fig. S7A–C†). The FI of Pink Flamindo was also increased, but the change was smaller than that of Green Falcan 0.3 (Fig. S7D–F†). In addition, when the dose-response relationship for forskolin in HeLa cells expressing Green Falcans, the FI of all Green Falcans by the administration of forskolin was increased in a dose dependent manner and Green Falcan 0.3 is most sensitive to forskolin (Fig. S6E–H†). These results suggest that a wide range of cAMP concentrations in a living cell is able to be monitored by choosing Green Falcan with a suitable working range. We further examined Green Falcan 10 carrying a similar EC50 with previous cAMP indicators in live-cell imaging. After the administration of 300 μM 3-isobutyl 1-methylxanthine (IBMX, phosphodiesterase inhibitor) or 2 mM 8-Br-cAMP (membrane-permeable cAMP analogue) to HeLa cells expressing Green Falcan 10, the FI of these cells was significantly increased (Fig. 3D–F, S6D†). Interestingly, the kinetics and amplitude of FI change after the administration of forskolin, IBMX and 8-Br-cAMP differed from each other, reflecting the distinct kinetics of various mechanisms for achieving cAMP increase such as production, inhibition of degradation, and membrane permeation. Therefore, these results suggest that the developed indicators are adaptable for detecting a spatial and temporal change of cAMP dynamics in various physiological phenomena.3.4. Dual-color imaging with Green Falcan and R-GECO
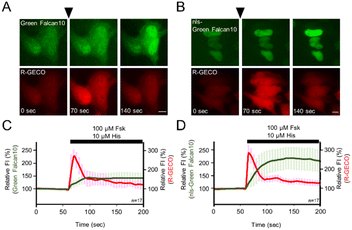
Many single-FP based indicators have been developed for visualizing with other signaling molecules. Since single-FP based indicators require single wavelength emission for detection, they are suitable for multicolor imaging. To demonstrate the advantage of single-FP indicators, we expressed both Green Falcan 10 and R-GECO,13 a red fluorescent Ca2+ indicator, in HeLa cells. After the administration of 100 μM forskolin and 10 μM histamine which causes Ca2+ elevation by stimulation of the histamine H1 receptor, the fluorescence intensity (FI) of both Green Falcan 10 and R-GECO was increased (Fig. 4A and C). We next expressed nuclear localization signal (nls)-Green Falcan 10 and R-GECO in HeLa cells to examine the applicability for visualization of dynamics of different molecules in a distinct intracellular compartment. After the administration of 100 μM forskolin and 10 μM histamine, the FI of both nls-Green Falcan 10 and R-GECO was increased (Fig. 4B and D). These results suggest that Green Falcans are applicable to dual-color imaging with other indicators, and to further hierarchical and cooperative analysis by multi-color imaging. Interestingly, the FI increase in the cytoplasm by the administration of forskolin and histamine was a little smaller than in the nucleus and was also smaller than by the administration of forskolin alone. These results could be ascribed to the accelerated degradation of cAMP by phosphodiesterase37 in the cytoplasm due to calcium elevation.4. Conclusion
In this study, we developed green fluorescent cAMP indicators with various EC50 by protein engineering, named Green Falcan 0.3, 1, 3 and 10. These indicators were able to detect cAMP dynamics in the cytoplasm and nucleus of living cells with high spatiotemporal resolution after the administration of several reagents and allowed dual-color imaging with R-GECO. Moreover, Green Falcan 0.3 was applicable to visualize a low concentration range of cAMP dynamics compared to previously developed cAMP indicators. Therefore, we expect that the Green Falcans will visualize unknown phenomena to unravel new physiological mechanism related to cAMP signaling in many organisms.38,39Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Dr Cong Quan Vu at Nano Life Science Institute, Kanazawa University for creating the schematic 3D structures of Green Falcan, and the Biomaterials Analysis Division, Tokyo Institute of Technology for DNA sequencing. This work was supported by The Japan Society for the Promotion of Science (JSPS), Grant-in-aid for Scientific Research (KAKENHI) (JP18H04832 and JP22H05176 to T. K., JP20H00575, JP20H04121, JP20H04765, and JP20H04836 to T. T.), Iketani Science and Technology Foundation (to T. K.), and the Cooperative Research Program of “Network Joint Research Center for Materials and Devices”.Notes and references
- S. R. Adams, A. T. Harootunian, Y. J. Buechler, S. S. Taylor and R. Y. Tsien, Nature, 1991, 349, 694–697 CrossRef CAS PubMed.
- A. Koschinski and M. Zaccolo, Sci. Rep., 2017, 7, 14090 CrossRef PubMed.
- S. Börner, F. Schwede, A. Schlipp, F. Berisha, D. Calebiro, M. J. Lohse and V. O. Nikolaev, Nat. Protoc., 2011, 6, 427–438 CrossRef PubMed.
- D. Willoughby and D. M. F. Cooper, Physiol. Rev., 2007, 87, 965–1010 CrossRef CAS PubMed.
- J. B. Shabb, Chem. Rev., 2001, 101, 2381–2411 CrossRef CAS PubMed.
- M. Grandoch, S. S. Roscioni and M. Schmidt, Br. J. Pharmacol., 2010, 159, 265–284 CrossRef CAS PubMed.
- U. B. Kaupp and R. Seifert, Physiol. Rev., 2002, 82, 769–824 CrossRef CAS PubMed.
- G. Tolhurst, Y. Zheng, H. E. Parker, A. M. Habib, F. Reimann and F. M. Gribble, Endocrinol., 2011, 152, 405–413 CrossRef CAS PubMed.
- A. Nakajima, S. Ishihara, D. Imoto and S. Sawai, Nat. Commun., 2014, 5, 5367 CrossRef CAS PubMed.
- E. R. Kandel, Mol. Brain, 2012, 5, 14 CrossRef CAS PubMed.
- T. Nagai, A. Sawano, P. Eun Sun and A. Miyawaki, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 3197–3202 CrossRef CAS PubMed.
- J. Nakai, M. Ohkura and K. Imoto, Nat. Biotechnol., 2001, 19, 137–141 CrossRef CAS PubMed.
- Y. Zhao, S. Araki, J. Wu, T. Teramoto, Y. F. Chang, M. Nakano, A. S. Abdelfattah, M. Fujiwara, T. Ishihara, T. Nagai and R. E. Campbell, Science, 2011, 333, 1888–1891 CrossRef CAS PubMed.
- T. Kitaguchi, M. Oya, Y. Wada, T. Tsuboi and A. Miyawaki, Biochem. J., 2013, 450, 363–373 CrossRef PubMed.
- H. Odaka, S. Arai, T. Inoue and T. Kitaguchi, PLoS One, 2014, 9, e100252 CrossRef PubMed.
- K. Harada, M. Ito, X. Wang, M. Tanaka, D. Wongso, A. Konno, H. Hirai, H. Hirase, T. Tsuboi and T. Kitaguchi, Sci. Rep., 2017, 7, 7351 CrossRef PubMed.
- S. Matsuda, K. Harada, M. Ito, M. Takizawa, D. Wongso, T. Tsuboi and T. Kitaguchi, ACS Sens., 2017, 2, 46–51 CrossRef CAS PubMed.
- P. H. Tewson, S. Martinka, N. C. Shaner, T. E. Hughes and A. M. Quinn, J. Biomol. Screening, 2016, 21, 298–305 CrossRef CAS PubMed.
- C. R. Hackley, E. O. Mazzoni and J. Blau, Sci. Signaling, 2018, 11, eaah3738 CrossRef PubMed.
- S. Kawata, Y. Mukai, Y. Nishimura, T. Takahashi and N. Saitoh, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2122618119 CrossRef CAS PubMed.
- L. Wang, C. Wu, W. Peng, Z. Zhou, J. Zeng, X. Li, Y. Yang, S. Yu, Y. Zou, M. Huang, C. Liu, Y. Chen, Y. Li, P. Ti, W. Liu, Y. Gao, W. Zheng, H. Zhong, S. Gao, Z. Lu, P. G. Ren, H. L. Ng, J. He, S. Chen, M. Xu, Y. Li and J. Chu, Nat. Commun., 2022, 13, 5363 CrossRef CAS PubMed.
- M. Mita, D. Wongso, H. Ueda, T. Tsuboi and T. Kitaguchi, Methods Mol. Biol., 2021, 2274, 89–100 CrossRef CAS PubMed.
- S. Arai, R. Kriszt, K. Harada, L. S. Looi, S. Matsuda, D. Wongso, S. Suo, S. Ishiura, Y. H. Tseng, M. Raghunath, T. Ito, T. Tsuboi and T. Kitaguchi, Angew. Chem., Int. Ed., 2018, 57, 10873–10878 CrossRef CAS PubMed.
- M. Mita, M. Ito, K. Harada, I. Sugawara, H. Ueda, T. Tsuboi and T. Kitaguchi, Anal. Chem., 2019, 91, 4821–4830 CrossRef CAS PubMed.
- C. H. Clegg, G. G. Cadd and G. S. McKnight, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 3703–3707 CrossRef CAS PubMed.
- I. Ghosh, A. D. Hamilton and L. Regan, J. Am. Chem. Soc., 2000, 122, 5658–5659 CrossRef CAS.
- R. A. Steinberg, M. M. Symcox, S. Sollid and D. Øgreid, J. Biol. Chem., 1996, 271, 27630–27636 CrossRef CAS PubMed.
- J. L. Sebaugh and P. D. McCray, Pharm. Stat., 2003, 2, 167–174 CrossRef.
- W. S. Rasband, ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA, 2018, https://imagej.nih.gov/ij/ Search PubMed.
- R Core Team, R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria, 2022, https://www.R-project.org/ Search PubMed.
- Y. Kanda, Bone Marrow Transplant., 2013, 48, 452–458 CrossRef CAS PubMed . https://www.jichi.ac.jp/saitama-sct/SaitamaHP.files/statmed.html.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Nature, 2021, 596, 583–589 CrossRef CAS PubMed.
- K. Harada, T. Chihara, Y. Hayasaka, M. Mita, M. Takizawa, K. Ishida, M. Arai, S. Tsuno, M. Matsumoto, T. Ishihara, H. Ueda, T. Kitaguchi and T. Tsuboi, Sci. Rep., 2020, 10, 19562 CrossRef CAS PubMed.
- R. Heim, D. C. Prasher and R. Y. Tsien, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 12501–12504 CrossRef CAS PubMed.
- D. Stoner-Ma, A. A. Jaye, K. L. Ronayne, J. Nappa, S. R. Meech and P. J. Tonge, J. Am. Chem. Soc., 2008, 130, 1227–1235 CrossRef CAS PubMed.
- K. Brejc, T. K. Sixma, P. A. Kitts, S. R. Kain, R. Y. Tsien, M. Ormö and S. J. Remington, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 2306–2311 CrossRef CAS PubMed.
- T. A. Goraya and D. M. F. Cooper, Cell. Signal., 2005, 17, 789–797 CrossRef CAS PubMed.
- I. Uno, K. Matsumoto and T. Ishikawa, J. Biol. Chem., 1983, 258, 3539–3542 CrossRef CAS PubMed.
- P. Milanesio, A. Arce-Rodríguez, A. Muñoz, B. Calles and V. de Lorenzo, Environ. Microbiol., 2011, 13, 324–339 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra01390a |
| This journal is © The Royal Society of Chemistry 2023 |