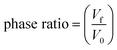Open Access Article
Open Access ArticleHexafluoroisopropanol-based deep eutectic solvents for high-performance DNA extraction†
Jia Xu ,
Yuan Yang,
Xiaonan Cai and
Han Xiao
,
Yuan Yang,
Xiaonan Cai and
Han Xiao *
*
Institute of Maternal and Child Health, Wuhan Children's Hospital (Wuhan Maternal and Child Healthcare Hospital), Tongji Medical College, Huazhong University of Science & Technology, Wuhan, 430016, China. E-mail: tjxiaohan@hust.edu.cn
First published on 24th March 2023
Abstract
In this study, hexafluoroisopropanol (HFIP)-based deep eutectic solvents (DESs) were developed and used for DNA extraction from human whole blood samples for the first time. HFIP-based DESs were prepared using HFIP and choline chloride (ChCl)/tetrabutylammonium chloride/cetyltrimethylammonium bromide as the hydrogen bond donor and acceptor, respectively. The two-phase system formation was promoted with different inorganic salts as the phase-forming component. According to the strong phase separation capability and high DNA extraction efficiency, DESs consisting of HFIP/ChCl-(NH4)2SO4, HFIP/ChCl-Na2SO4 and HFIP/ChCl-MgSO4 were then selected for application in DNA extraction. The factors that could have impacted the DNA extraction process, including molar ratio of DES, DES addition, salt species and addition, and sample pH, were systematically investigated via single-factor experimental analysis. Furthermore, we selectively examined bovine serum albumin and RNA to assess the specificity of the HFIP-based DESs for DNA extraction. Conclusively, 93.9%, 96.7% and 99.8% DNA could be extracted using the HFIP/ChCl-(NH4)2SO4, HFIP/ChCl-Na2SO4, and HFIP/ChCl-MgSO4 systems, respectively. Moreover, the developed systems were successfully used to extract DNA from human whole blood with satisfactory results. The DNA secondary structure was stable after DES extraction with the electrostatic interaction between DES and DNA as the main force driving DNA adsorption by DES.
Introduction
Nucleic acid purification and enrichment are crucial for conducting reliable research in genetic profiling, clinical trials, and food safety.1–3 Nucleic acid extraction is a complex process, as proteins and polysaccharides in cells and the environment disturb the purification of nucleic acids, and high concentrations of enzymes can also degrade nucleic acids.4 Conventional DNA purification methods (such as phenol-chloroform liquid–liquid extraction) require the extensive use of organic solvents, which are not environmentally friendly.5 Commercialized DNA extraction kits require repeated elution and centrifugation steps, which may result in the loss of DNA. Therefore, it is of great significance to develop new DNA purification and separation technologies that are economical, efficient, environmentally-friendly, and easy to operate.Aqueous two-phase systems (ATPSs) are mild, biocompatible, and environmentally-friendly liquid–liquid extraction, separation, and purification techniques that have been extensively used in biotechnological applications.6,7 Polymer/polymer, polymer/salt, and small molecular alcohol/salt constituents are commoly used in the development of ATPSs.8–10 These functions on the principle of spontaneous phase separation, which occurs between two distinct water-soluble compounds above a certain concentration. A high content of both phase components in water can provide a biocompatible environment for bioactive components, thus protecting their structural integrity and biological activity.11–13 The demand for environmentally friendly solvents led to the use of deep eutectic solvents (DESs) in extraction processes. Generally, DESs are typically prepared with two components, hydrogen bond donors (HBDs) and hydrogen bond acceptors (HBAs), with the aid of heating and stirring.14,15 The formed eutectic mixtures generally exhibit much lower melting points than those of their individual components. Compared with traditional organic solvents, DESs have low toxicity, and high thermal stability, are non-volatile, and can readily dissolve many organic and inorganic compounds.16 They have abundant functional groups, that can attract target compounds by hydrogen bonding and hydrophobic interactions.17–20 DESs have recently been used in analytical chemistry as effective extractants for the separation and preconcentration of analytes from a variety of complex matrices, such as biological fluid, food and environmental samples.21–23 DESs are excellent solvents for DNA.24 Mondal et al. studied the solubility and stability of DNA in bio-based DESs and found that DNA is soluble as well as chemically and structurally stable in DESs.24
Hexafluoroisopropanol (HFIP), a strong HBD (1.96), much larger than that of water (1.02), can aid in the preparation of DESs with a variety of HBAs.25–27 HFIP-based DESs have been successfully employed in the purification of pesticides, anthraquinones, and dyes25–27 and are also used as environmental reaction media.28 In this study, we synthesized HFIP-based DESs and used them for the extraction and quantification of DNA for the first time. Three types of HFIP-based DESs were directly synthesized and applied in the rapid and efficient extraction of DNA from spiked aqueous solution and human whole blood samples. To comprehensively understand the ability of DESs to extract DNA, the proportions of DESs components were explored in detail, including those of HBD species and inorganic salts. HFIP/ChCl DESs and three types of sulfates were then used to investigate the influencing factors and back-extraction performance. Bovine serum albumin (BSA) and RNA were used to assess the specificity of the HFIP-based DESs for DNA extraction. In addition, the HFIP/ChCl DES method was used to extract DNA from human whole blood samples to evaluate its practical applicability.
Materials and methods
Materials and reagents
HFIP was purchased from Aladdin Chemistry Co., Ltd. (Shanghai, China). Choline chloride (ChCl), tetrabutylammonium chloride (TBAC), cetyltrimethylammonium bromide (CTAB), salmon sperm DNA sodium salt, ribonucleic acid (yeast) (RNA) and BSA were purchased from Macklin (Shanghai, China). (NH4)2SO4, K2HPO4, KH2PO4, Na2CO3, Na2HPO4, Na2SO4, MgSO4, hydrochloric acid (HCl), ammonia solution (25–28%, w/v) and ethylenediaminetetraacetic acid disodium salt dehydrate (EDTA-Na2·2H2O) were purchased from Sinopharm Chemical Reagent, Co., Ltd. (Shanghai, China). Tris-hydrochloride (Tris-HCl) was purchased from Guangzhou Saiguo Biotech Co., Ltd (Guangzhou, China). All other chemicals used were of analytical reagent grade and were not further purified unless otherwise indicated. Deionized water (18.25 MΩ) was used in all experiments.Human whole blood samples from healthy volunteers were provided by the Wuhan Children's Hospital (China). Appropriate amounts of anticoagulant were added to the blood samples which were then stored at −20 °C until use.
Instruments
Agitation and extraction were accomplished using a UXI orbital shaker (Huxi, Shanghai, China). The DNA concentrations of the solution were determined using an ultraviolet-1600PC UV-Vis spectrophotometer (XIPU, Shanghai, China) with a 1.0 cm quartz cell. DNA amplification was performed using a T100™ thermal cycler (Bio-Rad Laboratories, USA). The polymerase chain reaction (PCR) products were analyzed using 1.5% agarose gel electrophoresis with ethidium bromide staining followed by visualization on ChemiDoc XRS+ Imaging System (Bio-Rad Laboratories, USA). Fourier-transform infrared (FT-IR) spectra were recorded using a 470FT-IR spectrometer (Thermo NICOLET, USA) at room temperature. The 1H–1H nuclear Overhauser spectroscopy (NOESY) and 1H nuclear magnetic resonance (1H NMR) spectra were obtained using an Avance NEO 400 MHz spectrometer (Bruker, Germany). Circular dichroism (CD) spectra were obtained using JASCO J-1500 (JASCO, Japan). Dynamic light scattering (DLS) was evaluated using a Zeta sizer Nano ZS90 (Malvern, England).DESs synthesis
Three kinds of DESs were synthesized based on ChCl, TBAC, and CTAB used as HBAs and HFIP as the HBD. HBA and HBD were placed in a screw-cap pressure tube at a designed molar ratio. The mixture was then heated at 80 °C for 2 h with constant magnetic stirring until a homogeneous and stable liquid formed. All the prepared DESs were stored at room temperature for usage.DNA partitioning
The phase-forming components were essential for DNA partitioning. Various two-phase systems consisting of 0.5 mL DES (HBA and HFIP molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) and 0.8 g inorganic salts were prepared in 5 mL of TE buffer (pH 7.0). The DNA concentration was 10 μg mL−1. The solution was then vigorously mixed using an orbital shaker and left to equilibrate for 10 min. After full phase separation, the phase behavior was recorded and the DNA concentration in the bottom phase was quantified using a UV-Vis spectrophotometer at 260 nm.
2) and 0.8 g inorganic salts were prepared in 5 mL of TE buffer (pH 7.0). The DNA concentration was 10 μg mL−1. The solution was then vigorously mixed using an orbital shaker and left to equilibrate for 10 min. After full phase separation, the phase behavior was recorded and the DNA concentration in the bottom phase was quantified using a UV-Vis spectrophotometer at 260 nm.
DNA extraction
To evaluate the extraction performance of the proposed HFIP/ChCl DES, several influencing factors, including the HBA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HBD molar ratio, DES addition, salt addition, and solution pH, were explored.
HBD molar ratio, DES addition, salt addition, and solution pH, were explored.
An appropriate amount of inorganic salt was dissolved in 5 mL TE buffer with a certain amount of DNA (10 μg mL−1). A certain volume of DES was then added. The mixture was mixed well and left to stand for 10 min to allow it to separate into two phases. The bottom phase was withdrawn using a microsyringe and diluted with methanol at four times the phase volume. The DNA concentration in the bottom phases was determined at 260 nm using a UV-Vis spectrophotometer. Additionally, a blank sample without DNA was analyzed in the same system to eliminate any background effects.
The DNA extraction efficiency (%) and phase ratio were calculated according to the following formulas:
where, C0 is the spiked DNA concentration and Cf is the DNA concentration in the DES-rich phase after extraction. V0 is the volume of the DNA sample and Vf is the volume of the DES-rich phase.
Sample preparation
A human whole blood sample (200 μL) was mixed with proteinase K (20 μL, 100 mg mL−1) and Triton X-100 (20 μL) and the mixture was incubated at 70 °C for 10 min. The mixture was then diluted to 1 mL in TE buffer (pH 8.0). After centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 2 min, the supernatant (900 μL) was mixed with the same volume of anhydrous methanol. After another 2 min centrifugation cycle, the supernatant (1 mL) was mixed with 4 mL of TE buffer (pH 8.0). Then, (NH4)2SO4 (1.4 g), Na2SO4 (0.6 g), or MgSO4 (1.2 g) was added to the solution and the mixture was stirred well. Finally, DES (0.7 mL) was added to the mixture. The system was allowed to stand for 10 min to allow the DNA to be completely partitioned between the two phases. Finally, 200 μL of the bottom phase was withdrawn using a microsyringe and transferred to a 1.5 mL vial with 400 μL of distilled water. The mixture was vortexed and incubated at 90 °C for 5 min and then centrifuged to achieve complete separation. The clear and transparent supernatant was directly used as the DNA template for PCR.
000 rpm for 2 min, the supernatant (900 μL) was mixed with the same volume of anhydrous methanol. After another 2 min centrifugation cycle, the supernatant (1 mL) was mixed with 4 mL of TE buffer (pH 8.0). Then, (NH4)2SO4 (1.4 g), Na2SO4 (0.6 g), or MgSO4 (1.2 g) was added to the solution and the mixture was stirred well. Finally, DES (0.7 mL) was added to the mixture. The system was allowed to stand for 10 min to allow the DNA to be completely partitioned between the two phases. Finally, 200 μL of the bottom phase was withdrawn using a microsyringe and transferred to a 1.5 mL vial with 400 μL of distilled water. The mixture was vortexed and incubated at 90 °C for 5 min and then centrifuged to achieve complete separation. The clear and transparent supernatant was directly used as the DNA template for PCR.
Results and discussion
The FT-IR spectra and 1H NMR results were used to confirm the formation of hydrogen bonds in the synthesized HFIP-based DESs. The FT-IR characterization of all the prepared HFIP-based DESs is shown in Fig. 1. The stretching vibration peaks of O–H in pure HFIP and ChCl were at 3424.98 cm−1 and 3293.82 cm−1, respectively, which shifted to a lower wavenumber of 3165.54 cm−1 at in ChCl/HFIP. This was due to the reduction in the force constant caused by the transfer of the electron cloud of the oxygen atom to the hydrogen bond. The shift in the –OH stretching vibration indicated the existence of hydrogen bonding between ChCl and HFIP. In addition, compared with the spectra of ChCl and HFIP, both characteristic peaks were distinguishable in the ChCl/HFIP spectra. Furthermore, no new peaks were detected, indicating that there was no chemical reaction during DES synthesis. A similar phenomenon was observed for FT-IR spectra of the other HFIP-based DESs, which confirmed the presence of hydrogen bonds in these DESs (Fig. 1b).The NOESY spectrum was shown in Fig. S1.† The results showed there was a strong hydrogen bonding between O–H in HFIP and O–H in ChCl. The 1H NMR results of all of the prepared HFIP-based DESs are shown in Fig. S2.† The 1H NMR of CTAB-HFIP was as follows: δ 4.62 (s, 1H), 3.47 (m, 2H), 3.38 (s, 9H), 1.76 (s, 2H), 1.37 (s, 26H), and 0.9 (t, 3H). The 1H NMR of ChCl-HFIP was as follows: δ 4.60 (s, 1H), 4.01 (dd, 2H), 3.52 (m, 2H), and 3.21 (d, 9H). The 1H NMR of TBAC-HFIP was as follows: δ 4.53 (s, 1H), 3.11 (m, 8H), 1.36 (m, 8H), 1.32 (m, 8H), and 0.94 (t, 12H). The characteristic peaks are consistent with those reported previously.27,29
DES selection
To evaluate the partitioning performance and ability of the established HFIP-based DESs (HFIP/ChCl, HFIP/TBAC, and HFIP/CTAB) to form phases, salmon sperm DNA was selected as a model molecule and seven types of inorganic salts ((NH4)2SO4, K2HPO4, KH2PO4, Na2CO3, Na2HPO4, MgSO4, and Na2SO4) were used as phase separation inducers. After phase separation to equilibrium, DNA was transferred to the bottom DES-rich phase, while the top phase contained mostly inorganic salts.The DNA partitioning results are summarized in Table S1.† The HFIP/CTAB-Na2CO3 and HFIP/CTAB-K2HPO4 systems formed a homogeneous phase and could not achieve phase separation. The HFIP/CTAB-Na2HPO4 system was divided into three phases; thus, it could not achieve two-phase separation. The other solutions, including all of the HFIP/ChCl and HFIP/TBAC systems, HFIP/CTAB-Na2SO4, HFIP/CTAB-KH2PO4, HFIP/CTAB-MgSO4, and HFIP/CTAB-(NH4)2SO4 achieved transparent liquid–liquid two-phase formation. In these two-phase systems, 50 μL of the bottom phase was extracted with 200 μL of water to support the extraction of DNA into the aqueous phase. After vortexing and centrifugation, the systems, including all of the HFIP/TBAC, HFIP/ChCl-(NH4)2SO4, HFIP/ChCl-MgSO4, HFIP/ChCl-K2HPO4, HFIP/ChCl-Na2CO3, HFIP/ChCl-Na2HPO4, HFIP/ChCl-Na2SO4, and HFIP/CTAB-MgSO4 systems formed two phases with a clear boundary. The DNA concentration in the top aqueous phase was then measured using a UV-Vis spectrophotometer at 260 nm. It is showed that, in all of the HFIP/TBAC systems, and the HFIP/ChCl-K2HPO4, HFIP/ChCl-Na2CO3, HFIP/ChCl-Na2HPO4 and the HFIP/CTAB-MgSO4 systems, almost no DNA can be detected in the bottom phase, indicating that there is no DNA transferring into the DES-rich phase. However, DNA recovered in the DES-rich phase was more than 50% in three systems including HFIP/ChCl-(NH4)2SO4, HFIP/ChCl-Na2SO4, and HFIP/ChCl-MgSO4 systems, thus these systems were used in subsequent experiments.
Effect of molar proportion of DES
Three types of DESs consisting of ChCl and HFIP with different molar ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5, 1
1.5, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, and 1
3, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) were formed to explore their influence on DNA extraction. The distribution of DNA was mainly related to the relative affinity of the two phases. It can be seen from the phase ratio results (red lines) that the volume of bottom DES-rich phase increased as the proportion of HFIP in DES increased, leading to the change of DNA content in the DES-rich phase. For HFIP/ChCl-(NH4)2SO4, the extraction efficiency of DNA in the bottom phase increased as molar ratio varied from 1
4) were formed to explore their influence on DNA extraction. The distribution of DNA was mainly related to the relative affinity of the two phases. It can be seen from the phase ratio results (red lines) that the volume of bottom DES-rich phase increased as the proportion of HFIP in DES increased, leading to the change of DNA content in the DES-rich phase. For HFIP/ChCl-(NH4)2SO4, the extraction efficiency of DNA in the bottom phase increased as molar ratio varied from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 to 1
1.5 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 and then decreased at the molar ratio of 1
3 and then decreased at the molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 (Fig. 2). The same phenomenon was observed in the HFIP/ChCl-Na2SO4 and HFIP/ChCl-MgSO4 systems. The optimal ChCl/HFIP molar ratio was selected at 1
4 (Fig. 2). The same phenomenon was observed in the HFIP/ChCl-Na2SO4 and HFIP/ChCl-MgSO4 systems. The optimal ChCl/HFIP molar ratio was selected at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.
3.
 | ||
| Fig. 2 The effects of molar ratio of DES (ChCl/HFIP) on extraction efficiency and phase ratio in HFIP/ChCl-(NH4)2SO4 system (a), HFIP/ChCl-Na2SO4 system (b), and HFIP/ChCl-MgSO4 system (c). | ||
Effect of inorganic salt concentration
The inorganic salts can be used as inducers for phase separation in two-phase systems, and can also affect the ionic strength and phase ratio. As shown in Fig. 3a, for HFIP/ChCl-(NH4)2SO4 system, the effects of the (NH4)2SO4 addition in the range of 0.6–2.0 g was investigated. The system with 0.6 g (NH4)2SO4 did not show separation into two phases. Regarding (NH4)2SO4, the DNA extraction efficiency increased as the concentration increased from 0.8 g to 1.4 g, while it exhibited a small decrease as the concentration exceeded 1.4 g. As for HFIP/ChCl-Na2SO4 system, the Na2SO4 concentration in the range of 0.4–1.6 g was investigated, 0.4 g Na2SO4 could not induce the phase separation. The increasing addition of Na2SO4 reduced the transfer of DNA into DES-rich phase; as a result, the extraction efficiency of DNA was decreased (Fig. 3b). Thus, 0.6 g of Na2SO4 was sufficient for DNA extraction. In the HFIP/ChCl-MgSO4 system, the MgSO4 addition in the range of 0.4–2.0 g was investigated; 0.4 g MgSO4 could not induce the phase separation. As shown in Fig. 3c, there was an increased change in the extraction of DNA when the concentration of MgSO4 increased from 0.6 g to 1.2 g. Thereafter, a significant decrease was observed; thus, the optimum amount of MgSO4 was 1.2 g. | ||
| Fig. 3 The effects of (NH4)2SO4 mass (a), Na2SO4 mass (b), and MgSO4 mass (c) on the extraction efficiency. | ||
Effect of DES addition
The amount of DES is an essential parameter for DNA extraction. In the absence of DES, sufficient interactions could not occur between DES and DNA, but excess DES increased the solution viscosity to block DNA dispersion. The effects of DES addition on the DNA extraction efficiency are illustrated in Fig. 4. In the HFIP/ChCl-(NH4)2SO4 and HFIP/ChCl-MgSO4 systems, the DNA extraction efficiency increased with the amount of DES. However, as the amount of DES further increased, there was no significant increase in the extraction efficiency. Therefore, 0.7 mL and 1 mL DES were selected as the most suitable amount for the HFIP/ChCl-(NH4)2SO4 and HFIP/ChCl-MgSO4 systems. For HFIP/ChCl-Na2SO4 system, excess DES decreased the DNA extraction efficiency; 0.7 mL of DES was selected as appropriate amounts for subsequent experiments. | ||
| Fig. 4 The effects of DES addition on extraction efficiency in HFIP/ChCl-(NH4)2SO4 system (a), HFIP/ChCl-Na2SO4 system (b), and HFIP/ChCl-MgSO4 system (c). | ||
Effect of sample pH
Considering that the solution pH can affect DES decomposition, and harsh pH conditions may compromise the structural integrity of DNA, the effect of pH on the extraction of DNA was investigated at pH values ranging from 2 to 9. TE buffer was used to adjust the solution pH. As shown in Fig. 5, the maximum peak areas were obtained at pH 8 for all of the studied systems; however, at other pH values, the extraction efficiency was lower. An alkaline medium was better for DNA extraction than an acidic medium in the HFIP/ChCl system, but the extraction efficiency decreased with a continuous increase in the pH. The extraction efficiency ranged from 73.8% to 93.9% for the HFIP/ChCl-(NH4)2SO4 system, 71.5% to 96.7% in the HFIP/ChCl-Na2SO4 system, and 48.6% to 99.8% in HFIP/ChCl-MgSO4 system as the sample pH ranged from 2 to 9, indicating that the DNA was preferentially enriched in the DES-rich phase. The bottom phase had a higher affinity toward DNA, promoting its distribution in the DES-rich phase.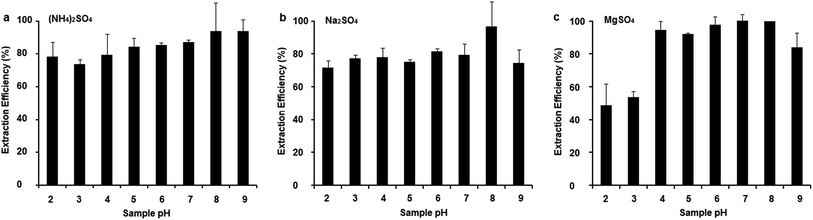 | ||
| Fig. 5 The effects of solution pH on extraction efficiency in HFIP/ChCl-(NH4)2SO4 system (a), HFIP/ChCl-Na2SO4 system (b), and HFIP/ChCl-MgSO4 system (c). | ||
Specificity
The purity of DNA directly determined the reliability and sensitivity of further downstream application. DNA, RNA, and protein contaminants must be separated. RNA (yeast) and BSA were selected as model interfering molecules, and 10 μg mL−1 RNA or 0.2 mg mL−1 BSA was added to the three prepared HFIP/ChCl systems under the optimal conditions. We investigated the RNA and BSA extraction efficiencies under different pH conditions by adding RNA or BSA instead of DNA, to the systems. After the complete separation of the systems, the RNA or BSA in the DES-rich phase was diluted and analyzed. As shown in Fig. S3 and S4,† when the pH ranged from 2 to 9, the BSA extraction efficiency was lower than 49.9%, 47.7%, and 73.9% for HFIP/ChCl-(NH4)2SO4, HFIP/ChCl-Na2SO4, and HFIP/ChCl-MgSO4 systems, respectively. When the pH value ranged from 2 to 9, the RNA extraction efficiency was lower than 26.3%, 12.4%, and 19.6% for HFIP/ChCl-(NH4)2SO4, HFIP/ChCl-Na2SO4, and HFIP/ChCl-MgSO4 system, respectively. Under the optimum condition of pH 8.0, the BSA and RNA extraction efficiencies were below 43%, and 20%, respectively. RNA and BSA tended to transfer to the top aqueous phase in the studied HFIP/ChCl systems. The results showed that the HFIP/ChCl system had good selectivity for DNA.Analysis of human blood samples
After the first extraction process, 93.9%, 96.7% and 99.8% of the DNA in the HFIP/ChCl-(NH4)2SO4, HFIP/ChCl-Na2SO4, and HFIP/ChCl-MgSO4 systems was extracted to the DES-rich phase under the optimum extraction conditions, respectively. The purification of DNA using DES-based extraction was intended to obtain high-purity DNA to meet the requirements of further downstream application. Therefore, the back-extraction process was carried out to back-extract the DNA to the aqueous phase. Herein, a human whole blood sample was selected for the separation process. After back-extraction, the recovered DNA in the aqueous phase was assayed by 1.5% agarose gel electrophoresis, as shown in Fig. S5.† The A260/A280 ratio of the recovered DNA was in the range of 1.64 to 1.89. The DNA recovered via back extraction was directly used as a template for PCR. A 274 bp TP53 gene fragment and a 335 bp EGFR gene fragment were amplified; the primer details are shown in Table S2.† The PCR mixtures (25 μL) consisted of 1 μL of recovered DNA, 0.5 μL each of the forward and reverse primers (10 mmol L−1), and 12.5 μL of Taq PCR Master Mix. The thermo cycling conditions were as follows: 10 min at 95 °C for denaturation, followed by 35 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 1 min. This was followed by a final holding step at 72 °C for 5 min. The analytical result is presented in Fig. S6.†In addition, we compared the proposed HFIP-based DESs extraction method with a commercial DNA extraction kit (Tiangen Biotech, Beijing, China). For 200 μL of human whole blood, approximately 40 μg, 30 μg and 70 μg of DNA was recovered in the HFIP/ChCl-(NH4)2SO4, HFIP/ChCl-Na2SO4 and HFIP/ChCl-MgSO4 systems under the optimal conditions, respectively. However, approximately 3.5 μg of DNA was recovered by the commercial kit. The proposed method takes 30 min to extract DNA from human whole blood and involves four centrifugation steps and two standing steps. The method of commercial DNA extraction kit requires about 31 min for total extraction and consists of six centrifugation steps and three standing steps. The HFIP-based DESs extraction method proposed is simpler than the method of commercial kit, and requires similar extraction time. The results indicated that the proposed method could be applied to effectively extract DNA from human whole blood samples.
Extraction mechanism
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N of the bases.24 After the DNA was extracted to the DES-rich phase, the symmetric stretching vibration band attributed to the PO2− groups at 1180 cm−1 was not affected, whereas the asymmetric band at 1257 cm−1 was found to have disappeared. These results indicated that an interaction occurred between ammonium chloride cations and the phosphate groups of DNA, which was caused by the electrostatic attraction between DNA and DES.
N of the bases.24 After the DNA was extracted to the DES-rich phase, the symmetric stretching vibration band attributed to the PO2− groups at 1180 cm−1 was not affected, whereas the asymmetric band at 1257 cm−1 was found to have disappeared. These results indicated that an interaction occurred between ammonium chloride cations and the phosphate groups of DNA, which was caused by the electrostatic attraction between DNA and DES.
Conclusion
In this study, HFIP-based DESs were prepared with various HBAs, including ChCl, TBAC, and CTAB. The prepared HFIP/ChCl DES could help extract DNA quickly and efficiently. Under the optimized conditions, the maximum extraction efficiency for DNA was found to be more than 93% in the HFIP/ChCl DES systems. Moreover, it was also found that the HFIP/ChCl DES systems were beneficial enriching DNA, other than RNA and BSA. To some extent, the above DES systems achieved selective DNA extraction. The proposed method was demonstrated with the extraction of DNA from a human whole blood sample. The DNA extracted using the HFIP/ChCl DES systems were suitable for PCR amplification. The extraction mechanism was also discussed. The results showed that the DNA secondary structure was stable after DES extraction and that the electrostatic interaction between DES and DNA was the main force driving DNA adsorption by DES. This approach provides a method for DNA extraction and a new direction for the development of DESs in the field of extraction and separation.Ethical statement
The authors state that all experiments were performed in compliance with the relevant laws and institutional guidelines. The human whole blood used in here was the remaining blood sample after routine clinical examination and the informed consent is exempted with the consent of the ethics committee of Wuhan Children's Hospital (Wuhan Maternal and Child Healthcare Hospital). This study has been approved by the ethics committee of this hospital (No. 2022R024-E03).Author contributions
Jia Xu: conceptualization, methodology, formal analysis, validation, writing-original draft. Yuan Yang: methodology, formal analysis, validation. Xiaonan Cai: methodology, formal analysis, validation. Han Xiao: project administration.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This research was supported by the National Natural Science Foundation of China (Nos. 81802109).References
- Y. Chen, Y. Liu, Y. Shi, J. Ping, J. Wu and H. Chen, TrAC, Trends Anal. Chem., 2020, 127, 115912 CrossRef CAS PubMed.
- M. N. Emaus, M. Varona, D. R. Eitzmann, S. Hsieh, V. R. Zeger and J. L. Anderson, TrAC, Trends Anal. Chem., 2020, 130, 115985 CrossRef CAS.
- A. G. Pérez, E. González-Martínez, C. R. D. Águila, D. A. González-Martínez, G. G. Ruiz, A. G. Artalejo and H. Yee-Madeira, Colloids Surf., A, 2020, 591, 124500 CrossRef.
- X. Wang, M. Liu, F. Peng and X. Ding, J. Chromatogr. A, 2021, 1659, 462626 CrossRef CAS PubMed.
- T. Li, M. D. Joshi, D. R. Ronning and J. L. Anderson, J. Chromatogr. A, 2013, 1272, 8–14 CrossRef CAS PubMed.
- K. K. Athira and R. L. Gardas, Fluid Phase Equilib., 2022, 558, 113463 CrossRef CAS.
- W. N. Phong, P. L. Show, Y. H. Chow and T. C. Ling, J. Biosci. Bioeng., 2018, 126(3), 273–281 CrossRef CAS PubMed.
- Y. Zhou, X. Yang, L. Bai, Z. Wu, J. Zhang, Z. Qin and J. Fan, J. Mol. Liq., 2022, 368, 120630 CrossRef CAS.
- X. H. Zhang, H. N. Cui, J. J. Zheng, X. D. Qing, K. L. Yang, Y. Q. Zhang, L. M. Ren, L. Y. Pan and X. L. Yin, Food Res. Int., 2023, 163, 112278 CrossRef CAS PubMed.
- Y. Chen, X. Liang and G. M. Kontogeorgis, Sep. Purif. Technol., 2023, 306, 122624 CrossRef CAS.
- K. Xu, Y. Wang, Y. Huang, N. Li and Q. Wen, Anal. Chim. Acta, 2015, 864, 9–20 CrossRef CAS PubMed.
- P. Xu, Y. Wang, J. Chen, X. Wei, W. Xu, R. Ni, J. Meng and Y. Zhou, Talanta, 2018, 189, 467–479 CrossRef CAS PubMed.
- F. Raji and A. Rahbar-Kelishami, Colloids Surf., A, 2021, 624, 126823 CrossRef CAS.
- A. M. Ramezani, R. Ahmadi and Y. Yamini, TrAC, Trends Anal. Chem., 2022, 149, 116566 CrossRef CAS.
- P. Janicka, M. Kaykhaii, J. Płotka-Wasylka and J. Gębicki, Green Chem., 2022, 24, 5035 RSC.
- Q. Zhang, K. O. Vigier, S. Royer and F. Jérôme, Chem. Soc. Rev., 2012, 41, 7108–7146 RSC.
- M. M. Contreras-Gámez, Á. Galán-Martín, N. Seixas, A. M. C. Lopes, A. Silvestre and E. Castro, Bioresour. Technol., 2023, 369, 128396 CrossRef PubMed.
- R. Rashid, S. M. Wani, S. Manzoor, F. A. Masoodi and M. M. Dar, Food Chem., 2023, 398, 133871 CrossRef CAS PubMed.
- O. Mokhodoeva, V. Maksimova, A. Shishov and V. Shkinev, Sep. Purif. Technol., 2023, 305, 122427 CrossRef CAS.
- F. Peng, M. Liu, X. Wang and X. Ding, Anal. Chim. Acta, 2021, 1181, 338899 CrossRef CAS PubMed.
- F. Shakirova, A. Shishov and A. Bulatov, Talanta, 2021, 234, 122660 CrossRef CAS PubMed.
- J. Cao, C. Wang, L. Shi, Y. Cheng, H. Hu, B. Zeng and F. Zhao, Food Chem., 2022, 383, 132586 CrossRef CAS PubMed.
- A. Duque, J. Grau, J. L. Benedé, R. M. Alonso, M. A. Campanero and A. Chisvert, Talanta, 2022, 243, 123378 CrossRef CAS PubMed.
- D. Mondal, M. Sharma, C. Mukesh, V. Gupta and K. Prasad, Chem. Commun., 2013, 49, 9606–9608 RSC.
- W. W. Deng, Y. Zong and Y. X. Xiao, ACS Sustainable Chem. Eng., 2017, 5, 4267–4275 CrossRef CAS.
- W. Deng, L. Yu, X. Li, J. Chen, X. Wang, Z. Deng and Y. Xiao, Food Chem., 2019, 274, 891–899 CrossRef CAS PubMed.
- K. Xu, P. Xu and Y. Wang, Talanta, 2020, 213, 120839 CrossRef CAS PubMed.
- L. Wang, D. Dai, Q. Chen and M. He, J. Fluorine Chem., 2014, 158, 44–47 CrossRef CAS.
- M. Nemati, M. R. A. Mogaddam, M. A. Farazajdeh, M. Tuzen and J. Khandaghi, J. Chromatogr. A, 2021, 1660, 462653 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ra00315a |
| This journal is © The Royal Society of Chemistry 2023 |