Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Strategies in the optimization of DNA hybridization conditions and its role in electrochemical detection of dengue virus (DENV) using response surface methodology (RSM)
Jahwarhar Izuan Abdul Rashid *a,
Nor Azah Yusofb,
Jaafar Abdullah
*a,
Nor Azah Yusofb,
Jaafar Abdullah b and
Rafidah Hanim Shomiad @ Shuebc
b and
Rafidah Hanim Shomiad @ Shuebc
aDepartment of Chemistry and Biology, Centre for Defence Foundation Studies, National Defence University of Malaysia, Sungai Besi Camp, 57000, Kuala Lumpur, Malaysia. E-mail: jahwarhar@upnm.edu.my
bDepartment of Chemistry, Faculty of Science, Universiti Putra Malaysia, Serdang, Selangor 43400, Malaysia
cInstitute for Research in Molecular Medicine (INFORMM), Universiti Sains Malaysia, 16150 Kubang Kerian, Kelantan, Malaysia
First published on 16th June 2023
Abstract
In recent years, limited research has been conducted on enhancing DNA hybridization-based biosensor approaches using statistical models. This study explores the application of response surface methodology (RSM) to improve the performance of a DNA hybridization biosensor for dengue virus (DENV) detection. The biosensor is based on silicon nanowires decorated with gold nanoparticles (SiNWs/AuNPs) and utilizes methylene blue as a redox indicator. The DNA hybridization process between the immobilized DNA probe and the target DENV gene was monitored using differential pulse voltammetry (DPV) based on the reduction of methylene blue. Fourier-transform infrared spectroscopy (FTIR) and electrochemical impedance spectroscopy (EIS) were employed to confirm successful DNA hybridization events on the modified screen-printed gold electrode (SPGE) surface. Several parameters, including pH buffer, NaCl concentration, temperature, and hybridization time, were simultaneously optimized, with NaCl concentration having the most significant impact on DNA hybridization events. This study enhances the understanding of the role of each parameter in influencing DNA hybridization detection in electrochemical biosensors. The optimized biosensor demonstrated the ability to detect complementary oligonucleotide and amplified DENV gene concentrations as low as 0.0891 ng µL−1 (10 pM) and 2.8 ng µL−1, respectively. The developed biosensor shows promise for rapid clinical diagnosis of dengue virus infection.
1. Introduction
In recent years, electrochemical biosensors have emerged as a promising platform for developing point-of-care (POC) devices for the rapid diagnosis of infectious diseases. These devices offer advantages such as ease of miniaturization, simplicity, versatility, and cost-effectiveness. Moreover, previous research has demonstrated that electrochemical approaches can achieve femtomolar or attomolar concentrations, which is beneficial for the clinical diagnosis of infectious diseases.1,2 Coupling POC devices with miniaturized electrochemical transducers has transformed the detection landscape, enabling real-time, rapid detection of various infectious diseases, including COVID-19,3 dengue virus (DENV),4 Zika virus,5 Influenza,6 Hepatitis,7 human immunodeficiency (HIV),8 Salmonella9 and Tuberculosis.10Electrochemical biosensors rely on changes in measurable redox current due to specific interactions between immobilized biological recognition elements and analytes, such as ssDNA/RNA-ssDNA, aptamer-antigens/proteins, antibodies-antigens, and whole cells-antigen/proteins. Among these, DNA hybridization, involving the interactions between ssDNA/RNA and its complementary target sequence, is widely used in electrochemical sensing for detecting specific DNA sequences. High accessibility of complementary targets to DNA probe-modified electrode surfaces for DNA hybridization plays a vital role in enhancing electrochemical DNA hybridization detection.
In addition to depending on the improvement of the optimal conditions of the hybridization process, the sensing layer for immobilization and hybridization also needs to be considered.
A good sensing layer can enhance immobilization and DNA hybridization, influencing the biosensor's electrochemical signal.
Numerous studies have improved biosensor performance by using nanomaterials such as multiwalled carbon nanotubes (MWCNTs),11 zinc oxide nanoparticles,11 gold nanoparticles,12 carbon dots Fe3O4,13 silica nanoparticles,14 Au–Pt bimetallic nanoparticles/graphene oxide15 and etc. as a sensing layer in DNA biosensors fabrication.
In the past few years, our group has focused on electrode modification using hybrid nanomaterials consisting of silicon nanowires (SiNWs) and gold nanoparticles (AuNPs). We found that SiNWs/AuNPs nanocomposites could discriminate electrochemical signals with and without the presence of dengue virus gene, making them suitable for use as sensing materials.3,15–17 Despite limited research on SiNWs in electrochemical DNA detection, their unique features, such as strong conductivity, high biocompatibility, and high surface-to-volume ratio, make them promising for this application. Although we have successfully enhanced DNA probe immobilization on SiNWs/AuNPs modified electrodes, there is room for improvement in biosensor sensitivity and performance. This includes enhancing the DNA hybridization process on the modified electrode.
Optimizing DNA hybridization conditions is crucial for achieving high sensitivity and selectivity in DNA sensors.16 Hence, the optimization of DNA hybridization condition is needed. These parameters include pH, ionic strength, hybridization temperature, and time. Careful control of these parameters can optimize the sensitivity and specificity of the biosensor, enabling accurate detection of target DNA sequences in a sample. Although there have been numerous studies looking into and highlighting the optimum value of the DNA hybridization parameters, most rely on the one-factor-at-a-time method, (changing one parameter at a time while maintaining the other parameters at a constant level), which has drawbacks. This approach cannot evaluate the effects and interactions of parameters on the response, and there is limited understanding of the role these parameters play in response behavior. Thus, one of promising strategies to overcome the above issue is the application of statistical modelling design known as response surface methodology (RSM) in the field of biosensor to optimize their performance.17,18 RSM involves using statistical techniques in the design of experiments, development of models, evaluation of key variables, and prediction of responses under optimal conditions. One of the unique aspects of using RSM is that it allows researchers to identify the combination of factors that will optimize the sensitivity, selectivity, and stability of the biosensor. This approach also enables researchers to identify the most important factors, as well as reduce the number of experiments required to identify the optimal combination of factors.19 Regarding practical application value, our previous study shows that the developed biosensor, which relies on the DNA hybridization principle, has the potential to act as a diagnostic platform for various infectious diseases. This allows for quicker detection and timely, suitable treatment. Additionally, the RSM optimization method employed in this research can be applied to numerous biosensing applications, thus contributing to the broader field of electrochemical biosensor development and optimization. To our knowledge, there is limited research focusing on optimizing DNA hybridization conditions for DNA electrochemical sensing applications. The relationship between parameters in DNA hybridization conditions and their impact on DNA biosensor performance is still poorly understood. Therefore, this study aims to utilize RSM to explore the behaviour of parameters in DNA hybridization conditions, shedding light on their influence on the efficiency and specificity of DNA hybridization. We applied the RSM approach to optimize DNA hybridization conditions such as pH buffer, NaCl concentration, temperature, and hybridization time, aiming to enhance the electrochemical current signal for detecting dengue virus, which was used as the main analyte in this work.
2. Experimental procedure
2.1 Reagents and apparatus
Silicon nanowires suspension with average diameter of 150 nm and average length of 20 µm, gold chloroauric acid salt (HAuCl4·4H2O), potassium ferricyanide (III) [K3Fe(CN)6], sodium citrate (Na3C6H5O7), (3-aminopropyl) triethoxysilane (APTES), 3-3′-dithiopropionic acid (DTPA), hydrogen peroxide (H2O2) (30% w/w in H2O) and ammonium hydroxide (NH4OH) (30% w/w), were purchased from Sigma-Aldrich (USA). Methylene blue (MB) was purchased from R&M Chemicals (Essex, UK). The oligonucleotides sequences of DENV which is based on study of Callahan et al.20 were purchased from First BASE Laboratories Sdn Bhd, Selangor, Malaysia. For DNA hybridization studies, the TE buffer (0.01 M Tris–HCl; pH 8.0 and 0.001 M EDTA) was used to dilute oligonucleotide stock solutions and remove unbound of dsDNA oligonucleotides. For electrochemical measurement, methylene blue (MB) solution (R&M Chemicals, UK) was prepared as a redox indicator containing 50 M of supporting electrolyte.The oligonucleotides sequence of dengue virus used in this study; thiolated probe DNA (5′ SH-(CH2)6-AAC AGC ATA TTG ACG CTG GGA GAG ACC-3); complementary target DNA (5′-GGT CTC TCC CAG CGT CAA TAT GCT GTT-3); one-base mismatch (5′-GGT CT![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) TCC CAG CGT CAA TAT GCT GTT-3′); three-base mismatch (5′-GGT CT
TCC CAG CGT CAA TAT GCT GTT-3′); three-base mismatch (5′-GGT CT![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) TCC C
TCC C![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) G CGT CAA TAT GC
G CGT CAA TAT GC![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) GTT-3′) and non-complementary (5′-TTC TGT GTT AGT ATC TGG GCC ATG TCC-3′). The calculated ΔG for the hybridization between the sample DNA and the probe is approximately −37.8 kcal mol−1. This negative value indicates that the hybridization process is thermodynamically favorable and spontaneous. Screen printed gold electrode (SPGE) (Dropsens, Spain) based-three electrode system; silver counter electrode and gold electrodes as working and counter electrode, respectively. µAUTOLAB (III) potentiostat (Eco Chemie, Utrecht, The Netherlands) connected to the computer was used to conduct the electrochemical analysis, which was were operated using (GPES) software version 4.9 (Eco Chemie, Netherlands).
GTT-3′) and non-complementary (5′-TTC TGT GTT AGT ATC TGG GCC ATG TCC-3′). The calculated ΔG for the hybridization between the sample DNA and the probe is approximately −37.8 kcal mol−1. This negative value indicates that the hybridization process is thermodynamically favorable and spontaneous. Screen printed gold electrode (SPGE) (Dropsens, Spain) based-three electrode system; silver counter electrode and gold electrodes as working and counter electrode, respectively. µAUTOLAB (III) potentiostat (Eco Chemie, Utrecht, The Netherlands) connected to the computer was used to conduct the electrochemical analysis, which was were operated using (GPES) software version 4.9 (Eco Chemie, Netherlands).
2.2 Fabrication of the SPGE surface, DNA immobilization, and hybridization
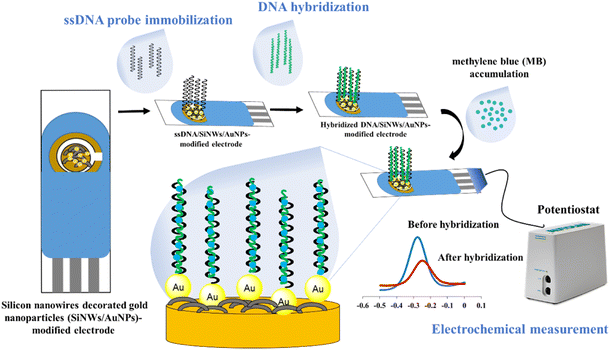
The fabrication of the SPGE surface using SiNWs/AuNPs nanocomposites for our developed DNA biosensor was based on our previous work21 as shown in Fig. 1. The working gold electrode surface was drop-casted with 6 µL of SiNWs suspension in 0.5% APTES solution, and dried for 3 hours at room temperature. It was then rinsed with ethyl-ethanol and cured for 30 minutes at 100 °C. The SiNW-modified SPGE was decorated with gold nanoparticles suspension as described in our previous work.2 To construct the biorecognition interface for the target dengue virus gene on the fabricated electrode surface, 10 µL of 5 µM thiolated ssDNA oligonucleotide probe was drop-casted onto the surface and left for 10 hours. The excess immobilized thiolated ssDNA probe was gently rinsed three times with TE buffer before hybridization. For the hybridization process, 10 µL of target DNA was directly drop-casted onto the surface of the thiolated ssDNA probe-modified electrode and incubated for 120 min at 40 °C. The hybridized dsDNA-modified electrode was rinsed with TE buffer and dried with N2 gas to remove unbound hybridized dsDNA. The same procedure was employed for the hybridization between immobilized ssDNA probe and other different ssDNA sequences and genomic dengue virus gene from real samples used in this study. The preparation of genomic dengue virus genes from real samples has been described in our previous work.42.3 Electrochemical measurement with methylene blue (MB) as a redox indicator
The hybridization was monitored by immersing the hybridized-modified electrode in 50 µM MB containing 50 mM Tris–HCl at pH 7.6 for 20 min, followed by the measurement of cyclic voltammetry (CV) and different pulse voltammetry (DPV) in 50 mM Tris–HCl at pH 7.6 in the potential ranging from −0.5 to 0 V. At the potential ranges of −0.5 to 0 V, step potential 0.005 V, modulation amplitude of 0.5 V with the interval time of 0.64 s at room temperature. The calculation of hybridization efficiency (%) is calculated based on our previous work2.4 Optimization of DNA hybridization conditions using central composite design (CCD)
Four independent variables, including pH hybridization buffer (6–8), NaCl concentration (1–1.5 M), hybridization temperature (40–45 °C), and incubation time (10–40 min), were applied to the RSM method to determine the optimum condition of the DNA hybridization process on the surface of the fabricated electrode. A total of 30 experiments were suggested according to the central composite design (CCD) using the statistical package, Design-Expert software version 6.0 (Stat-Ease Inc., Minneapolis, USA), with different combinations of parameter values, and hybridization efficiency was taken as the RSM response.All the experimental data of hybridization efficiency were fitted to the second order polynomial equation below:
| Y = X0 + X1A + X2B + X3C + X4D + X5A2 + X6B2 + X7C2 + X8D2 + X9AB + X10AC + X11AD + X11BC + X12BD + X13CD | (1) |
3. Result and discussion
3.1 Characterization of different modified SPGE
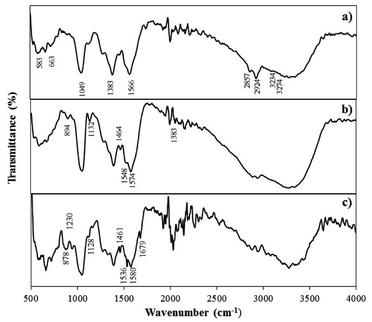
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N vibration of adenine), thymine (C2
N vibration of adenine), thymine (C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and guanine (C
O) and guanine (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretch of guanine) indicating the successful immobilization of the ssDNA probe on the electrode surface.28 Similar FTIR peaks were obtained for the ssDNA/AuNPs/SiNWs-modified electrodes after the introduction of complementary DNA target, confirming the presence of DNA immobilization and hybridization (Fig. 2c).
O stretch of guanine) indicating the successful immobilization of the ssDNA probe on the electrode surface.28 Similar FTIR peaks were obtained for the ssDNA/AuNPs/SiNWs-modified electrodes after the introduction of complementary DNA target, confirming the presence of DNA immobilization and hybridization (Fig. 2c).
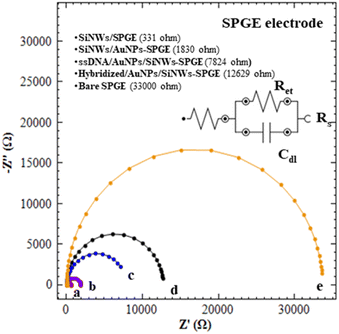
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Ω, (curve e). The Ret value for bare SPGE are dramatically decreased to 555 Ω and 331 Ω after the modification with SiNWs (curve a). This finding confirmed that the SiNWs may play an important role to facilitate electron transfer and thus improving the conductivity of bare electrodes. Curve b demonstrates that the Ret values of SiNWs/AuNPs-SPGE reach 1830 Ω, which is higher than curve a, primarily due to the modification of the SiNWs surface with APTES solution. This modification facilitates the formation of a self-assembled monolayer (SAM) and subsequent AuNPs decoration. Although the formation of SAM is crucial for AuNPs immobilization, it also influences the electron transfer process. As compared to curve c, the Ret values of curve d that kept increased suggested that the ssDNA (DNA probe) was successfully immobilized on SiNWs/AuNPs-SPGE. A further increase in Ret values of 12
000 Ω, (curve e). The Ret value for bare SPGE are dramatically decreased to 555 Ω and 331 Ω after the modification with SiNWs (curve a). This finding confirmed that the SiNWs may play an important role to facilitate electron transfer and thus improving the conductivity of bare electrodes. Curve b demonstrates that the Ret values of SiNWs/AuNPs-SPGE reach 1830 Ω, which is higher than curve a, primarily due to the modification of the SiNWs surface with APTES solution. This modification facilitates the formation of a self-assembled monolayer (SAM) and subsequent AuNPs decoration. Although the formation of SAM is crucial for AuNPs immobilization, it also influences the electron transfer process. As compared to curve c, the Ret values of curve d that kept increased suggested that the ssDNA (DNA probe) was successfully immobilized on SiNWs/AuNPs-SPGE. A further increase in Ret values of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 629 Ω (hybridized SiNWs/AuNPs-SPGE) was obtained. These increases of Ret values were due to the introduction of target DNA for hybrid DNA formation (curve d).
629 Ω (hybridized SiNWs/AuNPs-SPGE) was obtained. These increases of Ret values were due to the introduction of target DNA for hybrid DNA formation (curve d).
3.2 The optimization of DNA hybridization parameters using response surface methodology (RSM)
| Run | pH buffer | NaCl concentration (M) | Temperature (°C) | Hybridization time (min) | Hybridization efficiency (%) | |
|---|---|---|---|---|---|---|
| Experimental value | Predicted value | |||||
| 1 | 7 | 0.5 | 42.5 | 12.5 | 52.67 | 52.92 |
| 2 | 6 | 0.5 | 40 | 20 | 43.03 | 45.35 |
| 3 | 8 | 0.75 | 42.5 | 12.5 | 58.04 | 59.14 |
| 4 | 8 | 0.5 | 45 | 5 | 43.44 | 42.18 |
| 5 | 6 | 1 | 45 | 20 | 45.12 | 43.73 |
| 6 | 6 | 0.75 | 42.5 | 12.5 | 55.67 | 56.47 |
| 7 | 6 | 0.5 | 45 | 20 | 47.05 | 44.45 |
| 8 | 8 | 1 | 40 | 5 | 43.34 | 44.34 |
| 9 | 6 | 0.5 | 45 | 5 | 39.04 | 40.51 |
| 10 | 6 | 1 | 40 | 20 | 43.67 | 43.62 |
| 11 | 6 | 1 | 40 | 5 | 39.65 | 37.68 |
| 12 | 8 | 1 | 45 | 5 | 52.67 | 51.45 |
| 13 | 6 | 1 | 45 | 5 | 45.3 | 46.29 |
| 14 | 7 | 0.75 | 42.5 | 12.5 | 61.86 | 60.19 |
| 15 | 8 | 0.5 | 40 | 5 | 36.34 | 36.07 |
| 16 | 8 | 1 | 40 | 20 | 48.45 | 47.29 |
| 17 | 7 | 0.75 | 42.5 | 12.5 | 59.78 | 60.19 |
| 18 | 7 | 0.75 | 42.5 | 12.5 | 62.34 | 60.19 |
| 19 | 8 | 0.5 | 45 | 20 | 40.41 | 43.12 |
| 20 | 7 | 0.75 | 40 | 12.5 | 53.34 | 56.25 |
| 21 | 7 | 0.75 | 42.5 | 20 | 55.05 | 55.03 |
| 22 | 8 | 1 | 45 | 20 | 46.34 | 45.9 |
| 23 | 8 | 0.5 | 40 | 20 | 47.9 | 45.51 |
| 24 | 7 | 1 | 42.5 | 12.5 | 55.34 | 56.7 |
| 25 | 7 | 0.75 | 42.5 | 12.5 | 63.76 | 60.15 |
| 26 | 7 | 0.75 | 45 | 12.5 | 60.12 | 60.15 |
| 27 | 7 | 0.75 | 42.5 | 5 | 50.34 | 51.58 |
| 28 | 7 | 0.75 | 45 | 12.5 | 61.35 | 59.36 |
| 29 | 7 | 0.75 | 42.5 | 12.5 | 58.05 | 60.15 |
| 30 | 6 | 0.5 | 40 | 5 | 33.65 | 32.90 |
Table 2 presents the analysis of variance (ANOVA) of the quadratic model fitted to our experimental data. The model is highly significant, with a low probability value (<0.0001) for the fabricated electrode and a high F-value of 23.87, indicating that the quadratic model fits well and is adequate for our experimental data. A non-significant lack of fit F value of 1.56 was obtained in this study. The estimated regression coefficient, R2 of 0.957 and adjusted R2 of 0.9169 were obtained. The R2 values are close to one, indicating a good correlation between experimental data and predicted response. In this study, the low coefficient of variation (CV) of 4.92 indicates good reliability for both models. The adequate precision for the developed model in this study was found to be 15.71, which can be used to navigate the design space.30
| Sources | Sum of squares | df | Mean square | F-Value | P-Value |
|---|---|---|---|---|---|
| Model | 60.19 | 14 | 144.17 | 23.87 | <0.0001 |
| A | 1.3 | 1 | 32 | 5.3 | 0.0361 |
| B | 1.89 | 1 | 64.22 | 10.63 | 0.0053 |
| C | 1.56 | 1 | 43.22 | 7.21 | 0.017 |
| D | 1.72 | 1 | 53.39 | 8.84 | 0.0095 |
| A2 | −2.39 | 1 | 14.75 | 2.44 | 0.139 |
| B2 | −5.39 | 1 | 75.16 | 12.44 | 0.003 |
| C2 | −2.39 | 1 | 14.75 | 2.44 | 0.139 |
| D2 | −6.89 | 1 | 122.85 | 20.34 | 0.0004 |
| AB | 0.88 | 1 | 12.85 | 2.03 | 0.1749 |
| AC | −0.38 | 1 | 2.25 | 0.37 | 0.5508 |
| AD | −0.75 | 1 | 9 | 1.49 | 0.2411 |
| BC | 0.25 | 1 | 1 | 0.17 | 0.6899 |
| BD | −1.62 | 1 | 42.25 | 6.99 | 0.0184 |
| CD | −2.12 | 1 | 72.25 | 11.96 | 0.0035 |
| Residual | — | 15 | 6.04 | ||
| Lack of fit | — | 10 | 6.86 | 1.56 | 0.3258 |
| Pure error | — | 5 | 4.4 | ||
| Cor total | — | 29 | |||
| R2 = 0.957 | CV = 4.92 | Adeq precision = 15.705 | |||
| R2 adjusted = 0.9169 | |||||
| PRESS = 490.05 | |||||
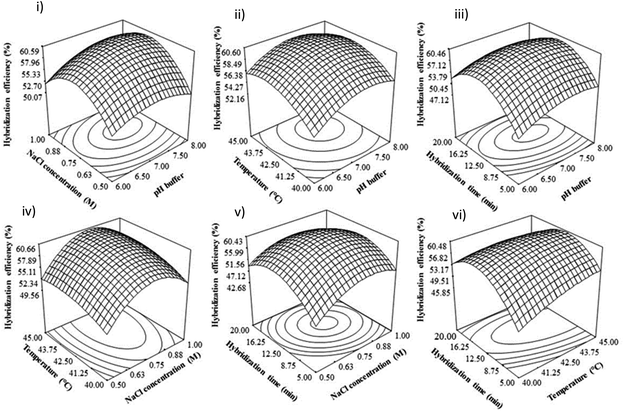
In Table 2, all the linear coefficient terms (A: pH buffer, B: NaCl concentration, C: hybridization temperature, D: hybridization time), quadratic coefficient terms (A2, B2), and interaction coefficient terms (BC, BD, CD) were statistically significant (P < 0.05). From this ANOVA analysis, there is a mutual interaction between NaCl concentration and hybridization temperature, NaCl concentration, and hybridization time, as well as hybridization temperature and time.
All of the experimental data for fabricated electrodes was fitted into the second-order full polynomial equation by applying multiple regression analysis as follows:
| Hybridiation efficiency (%) = 60.19 + 1.33A + 1.89B + 1.56C − 1.72D − 2.39A2 − 5.39B2 − 2.39C2 − 6.89D2 − 0.38AC − 0.75AD + 0.25BC − 1.62BD − 2.12CD | (2) |
3.3 The effect of hybridization parameters
In alkaline conditions, the hydrogen bond between DNA strands is disrupted, causing the double helix DNA to denature, separating from each other, and forming a single-stranded DNA coil.35–37 This can be attributed to the excess of OH− ions in alkaline solutions, resulting in the deprotonation of guanine and thymine bases and breaking the bonding of double helix DNA, which can influence the DNA hybridization rate.38,39 Besides that, Wang47 found that the electrode sensor would likely be easily damaged under highly acidic or alkaline solutions. Therefore, in this study, the DNA hybridization reaction was suppressed at extreme pH (low pH and high pH), and neutral pH is suitable for duplex DNA formation.
3.4 Optimization and verification of developed model
An optimal condition for DNA dengue virus detection signals to obtain the maximum DNA hybridization efficiency was determined using Derringer's desired function method in Design Expert 6.06 software. The optimal values of the studied parameters were suggested as pH 7.8, 1.45 M of NaCl concentration, 45 °C, and 10 minutes of hybridization time. Triplicate experiments (n = 3) were conducted to validate these optimal conditions, and a DNA hybridization efficiency of 62% (RSD 3.71%) was obtained. These experimental values were in agreement with the predicted values under the suggested optimal conditions. Thus, this optimized condition for DNA hybridization was used for real sample detection.3.5 The selectivity and sensitivity studies of our developed biosensor
Under optimized condition, the sensitivity of the developed DNA sensor was evaluated by the hybridization of probe DNA with different concentrations of complementary DNA target (Fig. 5a and b). Generally, the MB peak current of developed DNA sensor were decreased linearly with increasing in concentration of the synthetic complementary target. Fig. 5b, demonstrate the linear relationship where the MB peak current are decreased with increasing in concentration of the synthetic complementary DNA target following the equations below, respectively:| MB peak current (µA) = −0.0302 × ln(DNA target concentration (ng µL−1)) + 0 |
A cut-off value of 0.89 µA was calculated based on the triplicate analysis (n = 3) of the hybridization process between the DNA probe and non-complementary sequences. Consequently, the MB peak current values below the cut-off were interpreted as positive, while those above it were considered negative. Therefore, the lowest concentration of complementary DNA target that yielded an MB peak current below the cut-off value determined the limit of detection (LOD) for our developed DNA sensor. Fig. 5a and c show that the developed DNA sensors were able to detect complementary oligonucleotide dengue virus concentrations as low as 0.0891 ng µL−1 (10 pM), respectively. Fig. 5b displays the comparison of calibration curves for the level of detection before and after optimization by the developed DNA sensors. The limit of detection for the developed DNA sensor was significantly improved from 0.00891 ng µL−1 (non-optimized) to 0.0000891 ng µL−1 under optimized conditions (Fig. 5b). Our developed DNA sensor exhibited a detection limit that can reach the clinical level of RNA dengue virus concentration in blood, which is approximately 10 pM to 10 fM.77,78 The selectivity studies for our developed DNA sensor were investigated by hybridizing our developed biosensor with different kinds of synthetic DNA target sequences.
At a concentration of 89.1 ng µL−1 (10 nM), various synthetic DNA sequences, such as non-complementary, single-base mismatch, three-base mismatch, and complementary DNA sequences, were tested with our developed biosensor (Fig. 5c). As shown in Fig. 5c, the MB peak current for the non-complementary sequence detection is nearly equal to the MB current obtained for the background signal using a DNA probe without target DNA sequences, indicating that no DNA hybridization occurred. A decrease in MB peak current was observed upon the hybridization of complementary DNA targets (curve e), single-base mismatch (curve d), and three-base mismatch sequences (curve c) due to the inaccessibility of MB binding to the DNA surface, as previously described. The developed DNA biosensor demonstrated a relatively clear signal difference between single-base mismatch and complementary target DNA. This indicates that our DNA biosensor is capable of discriminating between single-base mismatch and complementary target DNA in dengue detection. Generally, the MB peak current generated by developed DNA sensor after hybridization increased in the following order: DNA probe without target > non-complementary DNA > three bases-mismatch > one base-mismatch > complementary sequences. This finding concludes that our newly developed DNA sensor demonstrates good selectivity, as it is able to discriminate between complementary, three base-mismatch, and non-complementary sequence detection.
3.6 The analytical performance of developed sensor on real sample detection
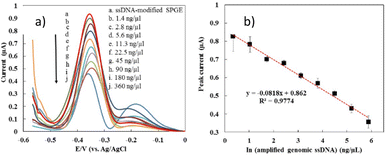
The optimal conditions obtained from the optimization process using the RSM approach were applied to detect actual samples from dengue virus samples. Fig. 6 demonstrates the DPV response of the developed biosensor after hybridization with the genomic ssDNA dengue virus at various concentrations, ranging from 1.4 ng µL−1 to 360 ng µL−1. It was shown that the MB current decreased with the increase in genomic ssDNA concentration. The detection limit of the developed sensor was estimated based on a cut-off value of 0.89 µA, where the peak current below the cut-off value is interpreted as positive and negative for those above it. The detection limit of our developed sensor was estimated as 2.8 ng µL−1. Fig. 6b exhibits that our developed biosensor displayed a linear relationship between the MB peak current and the natural logarithm of the genomic ssDNA concentration (ng µL−1), as stated in the equation below:| MB current (µA) = −0.08 × ln(genomic ssDNA concentration (ng µL−1)) + 0.86 | (3) |
In the previous work, the detection limit of pure synthetic oligonucleotide was 0.000891 ng µL−1, which was lower than the obtained detection limit of genomic DNA. As described before, the condition of real samples is very different from the pure synthetic oligonucleotide due to the presence of many impurities of biological molecules that can hinder the hybridization efficiency. In addition, the length of genomic ssDNA is longer and more bulky than the pure synthetic oligonucleotide which could affect the MB binding affinities to the DNA surface and MB current signal.79 The reproducibility of the fabricated electrode on detection of genomic ssDNA (5 ng µL−1) from serum spiked dengue virus were investigated showed a good reproducibility for nine measurements, where a RSD value of 9.34% and 8.23% were obtained respectively.
4. Conclusion
We employed a statistical modelling-based approach called RSM to enhance the sensitivity of our developed biosensor. This approach allowed us to better understand the role and behaviour of different parameters and their impact on the DNA hybridization process. Under optimized conditions, our developed biosensors were able to detect real genomic dengue sequences with a detection limit of 2.8 ng µL−1. Our approach, which combines sample preparation with electrochemical detection, is more specific, sensitive, and rapid than traditional methods such as gel electrophoresis visualization and ELISA assays, and is easier to use in practical settings like hospitals and laboratories.Conflicts of interest
No conflict of interest.Acknowledgements
This research was support by the Ministry of Science, Technology and Innovation (MOSTI) with grant number NND/ND/(1)/TD11-008 under the National Nanotechnology Directorate (NND) and National Defense University of Malaysia (NDUM). The author would also like to acknowledge the financial support provided by Malaysia for Fundamental Research Grant Scheme (FRGS) under Grant No. FRGS/1/2020/SSKK0/UPNM/02/1.References
- E. Safitri, L. Y. Heng, M. Ahmad, L. L. Tan, N. Nazaruddin, K. Suhud, C. P. Chiang and M. Iqhrammullah, Electrochemical DNA Biosensor Based on Mercaptopropionic Acid-Capped ZnS Quantum Dots for Determination of the Gender of Arowana Fish, Biosensors, 2022, 12(8), 650 CrossRef CAS PubMed.
- J. I. Abd Rashid, N. A. Yusof, J. Abdullah, U. Hashim and R. Hajian, A novel disposable biosensor based on SiNWs/AuNPs modified-screen printed electrode for dengue virus DNA oligomer detection, IEEE Sens. J., 2015, 15(8), 4420–4427 CAS.
- F. Jiang, Z. Xiao, T. Wang, J. Wang, L. Bie, L. Saleh, K. Frey, L. Zhang and J. Wang, Rapid and sensitive multiplex detection of COVID-19 antigens and antibody using electrochemical immunosensor-/aptasensor-enabled biochips, Chem. Commun., 2022, 58(52), 7285–7288 RSC.
- J. I. A. Rashid, N. A. Yusof, J. Abdullah and R. H. Shomiad, Strategies for the preparation of non-amplified and amplified genomic dengue gene samples for electrochemical DNA biosensing applications, RSC Adv., 2022, 12(1), 1–10 RSC.
- S. Mao, L. Fu, C. Yin, X. Liu and H. Karimi-Maleh, The role of electrochemical biosensors in SARS-CoV-2 detection: a bibliometrics-based analysis and review, RSC Adv., 2022, 12(35), 22592–22607 RSC.
- J. A. Park, J. Kim, S. M. Kim, H. Sohn, C. Park, T.-H. Kim, J.-H. Lee, M.-H. Lee and T. Lee, Fabrication of electrochemical influenza virus (H1N1) biosensor composed of multifunctional DNA four-way junction and molybdenum disulfide hybrid material, Materials, 2021, 14(2), 343 CrossRef CAS PubMed.
- W.-I. Lee, A. Subramanian, S. Mueller, K. Levon, C.-Y. Nam and M. H. Rafailovich, Potentiometric Biosensors Based on Molecular-Imprinted Self-Assembled Monolayer Films for Rapid Detection of Influenza A Virus and SARS-CoV-2 Spike Protein, ACS Appl. Nano Mater., 2022, 5(4), 5045–5055 CrossRef CAS PubMed.
- J. L. Gogola, G. Martins, A. Gevaerd, L. Blanes, J. Cardoso, F. K. Marchini, C. E. Banks, M. F. Bergamini and L. H. Marcolino-Junior, Label-free aptasensor for p24-HIV protein detection based on graphene quantum dots as an electrochemical signal amplifier, Anal. Chim. Acta, 2021, 1166, 338548 CrossRef CAS PubMed.
- S. Muniandy, K. L. Thong, J. N. Appaturi, C. W. Lai and B. F. Leo, Electrochemical aptasensor for Salmonella detection using Nafion-doped reduced graphene oxide, Sens. Diagn., 2022,(1), 1209–1217 RSC.
- E. Barbier, T. Fouchet, A. Hartmann, E. Cambau, F. Mougari, C. Dubois, M. Lubetzki and M. Rochelet, Rapid electrochemical detection of Mycobacterium tuberculosis in sputum by measuring Ag85 activity with disposable carbon sensors, Talanta, 2023, 253, 123927 CrossRef CAS PubMed.
- M. L. Mujica, A. Tamborelli, A. Castellaro, D. Barcudi, M. D. Rubianes, M. C. Rodríguez, H. A. Saka, J. L. Bocco, P. R. Dalmasso and G. A. Rivas, Impedimetric and amperometric genosensors for the highly sensitive quantification of SARS-CoV-2 nucleic acid using an avidin-functionalized multi-walled carbon nanotubes biocapture platform, Biosens. Bioelectron.: X, 2022, 12, 100222 CAS.
- C. Wan, A. Qu, L. Deng, X. Liu and C. Wu, Preparation of an electrochemical biosensor based on indium tin oxide and its performance in detecting antibiotic resistance genes, Microchem. J., 2022, 182, 107953 CrossRef CAS.
- X. Lin, Y. Mei, C. He, Y. Luo, M. Yang, Y. Kuang, X. Ma, H. Zhang and Q. Huang, Electrochemical biosensing interface based on carbon dots-Fe3O4 nanomaterial for the determination of Escherichia coli O157:H7, Front. Chem., 2021, 9, 769648 CrossRef CAS PubMed.
- E. E. Altuner, V. C. Ozalp, M. D. Yilmaz, M. Sudagidan, A. Aygun, E. E. Acar, B. B. Tasbasi and F. Sen, Development of electrochemical aptasensors detecting phosphate ions on TMB substrate with epoxy-based mesoporous silica nanoparticles, Chemosphere, 2022, 297, 134077 CrossRef CAS PubMed.
- H. R. A. Hasanjani and K. Zarei, DNA/Au-Pt bimetallic nanoparticles/graphene oxide-chitosan composites modified pencil graphite electrode used as an electrochemical biosensor for sub-picomolar detection of anti-HIV drug zidovudine, Microchem. J., 2021, 164, 106005 CrossRef.
- J. I. A. Rashid and N. A. Yusof, The strategies of DNA immobilization and hybridization detection mechanism in the construction of electrochemical DNA sensor: a review, Sensing and Bio-Sensing Research, 2017, 16, 19–31 CrossRef.
- Y. Dessie and S. Tadesse, Optimization of polyvinyl alcohol binder on PANI coated pencil graphite electrode in doubled chamber microbial fuel cell for glucose biosensor, Sensing and Bio-Sensing Research, 2022, 36, 100484 CrossRef.
- A. Afzalinia and M. Mirzaee, Ultrasensitive fluorescent miRNA biosensor based on a “sandwich” oligonucleotide hybridization and fluorescence resonance energy transfer process using an Ln(III)-MOF and Ag nanoparticles for early cancer diagnosis: application of central composite design, ACS Appl. Mater. Interfaces, 2020, 12(14), 16076–16087 CrossRef CAS PubMed.
- M. Kumari and S. K. Gupta, Response surface methodological (RSM) approach for optimizing the removal of trihalomethanes (THMs) and its precursor's by surfactant modified magnetic nanoadsorbents (sMNP)-an endeavor to diminish probable cancer risk, Sci. Rep., 2019, 9(1), 1–11 CrossRef PubMed.
- J. D. Callahan, S.-J. L. Wu, A. Dion-Schultz, B. E. Mangold, L. F. Peruski, D. M. Watts, K. R. Porter, G. R. Murphy, W. Suharyono and C.-C. King, Development and evaluation of serotype-and group-specific fluorogenic reverse transcriptase PCR (TaqMan) assays for dengue virus, J. Clin. Microbiol., 2001, 39(11), 4119–4124 CrossRef CAS PubMed.
- J. I. Abdul Rashid, N. A. Yusof, J. Abdullah, U. Hashim and R. Hajian, Surface modifications to boost sensitivities of electrochemical biosensors using gold nanoparticles/silicon nanowires and response surface methodology approach, J. Mater. Sci., 2016, 51(2), 1083–1097 CrossRef CAS.
- R. Verma, S. Sood, R. Singh, G. Sumana, M. Bala, V. K. Sharma, J. C. Samantaray, R. M. Pandey and B. D. Malhotra, Coupling electrochemical response of a DNA biosensor with PCR for Neisseria gonorrhoeae detection, Diagn. Microbiol. Infect. Dis., 2014, 78(1), 16–23 CrossRef CAS PubMed.
- I. Berktas, A. N. Ghafar, P. Fontana, A. Caputcu, Y. Menceloglu and B. S. Okan, Facile synthesis of graphene from waste tire/silica hybrid additives and optimization study for the fabrication of thermally enhanced cement grouts, Molecules, 2020, 25(4), 886 CrossRef CAS PubMed.
- R.-J. Chung and H.-T. Shih, Preparation of multifunctional Fe@Au core-shell nanoparticles with surface grafting as a potential treatment for magnetic hyperthermia, Materials, 2014, 7(2), 653–661 CrossRef PubMed.
- D. Zheng, S. K. Vashist, M. M. Dykas, S. Saha, K. Al-Rubeaan, E. Lam, J. H. Luong and F.-S. Sheu, Graphene versus multi-walled carbon nanotubes for electrochemical glucose biosensing, Materials, 2013, 6(3), 1011–1027 CrossRef CAS PubMed.
- M. K. Patel, P. R. Solanki, S. Khandelwal, V. V. Agrawal, S. Ansari and B. Malhotra, Self-assembled monolayer based electrochemical nucleic acid sensor for Vibrio cholerae detection, J. Phys.: Conf. Ser., 2012, 012009 CrossRef.
- S. K. Dash, M. Sharma, S. Khare and A. Kumar, rmpM genosensor for detection of human brain bacterial meningitis in cerebrospinal fluid, Appl. Biochem. Biotechnol., 2013, 171(1), 198–208 CrossRef CAS PubMed.
- S. M. Shet, P. Bharadwaj, M. Bisht, M. M. Pereira, S. K. Thayallath, V. Lokesh, G. Franklin, N. S. Kotrappanavar and D. Mondal, Presenting B-DNA as macromolecular crowding agent to improve efficacy of cytochrome c under various stresses, Int. J. Biol. Macromol., 2022, 215, 184–191 CrossRef CAS PubMed.
- H. U. Khan, M. E. Roberts, O. Johnson, W. Knoll and Z. Bao, The effect of ph and DNA concentration on organic thin-film transistor biosensors, Org. Electron., 2012, 13(3), 519–524 CrossRef CAS.
- Z. Xiao, X. Guo and L. Ling, Sequence-specific recognition of double-stranded DNA with molecular beacon with the aid of Ag+ under neutral pH environment, Chem. Commun., 2013, 49(34), 3573–3575 RSC.
- Q. Wang, N. Xu, J. Lei and H. Ju, Regulative peroxidase activity of DNA-linked hemin by graphene oxide for fluorescence DNA sensing, Chem. Commun., 2014, 50(51), 6714–6717 RSC.
- K.-J. Feng, Y.-H. Yang, Z.-J. Wang, J.-H. Jiang, G.-L. Shen and R.-Q. Yu, A nano-porous CeO2/chitosan composite film as the immobilization matrix for colorectal cancer DNA sequence-selective electrochemical biosensor, Talanta, 2006, 70(3), 561–565 CrossRef CAS PubMed.
- A. Ulianas, L. Y. Heng, S. A. Hanifah and T. L. Ling, An electrochemical DNA microbiosensor based on succinimide-modified acrylic microspheres, Sensors, 2012, 12(5), 5445–5460 CrossRef CAS PubMed.
- J. Zhang, H. P. Lang, G. Yoshikawa and C. Gerber, Optimization of DNA hybridization efficiency by pH-driven nanomechanical bending, Langmuir, 2012, 28(15), 6494–6501 CrossRef CAS PubMed.
- R. K. Dubey and D. Tripathi, A study of thermal denaturation/renaturation in dna using laser light scattering: a new approach, Indian J. Biochem. Biophys., 2005, 42(5), 301 CAS.
- R. Dias and B. Lindman, DNA interactions with polymers and surfactants, John Wiley & Sons, 2008 Search PubMed.
- G. Maglia, M. Henricus, R. Wyss, Q. Li, S. Cheley and H. Bayley, DNA strands from denatured duplexes are translocated through engineered protein nanopores at alkaline pH, Nano Lett., 2009, 9(11), 3831–3836 CrossRef CAS PubMed.
- G. Eidelshtein, S. Halamish, I. Lubitz, M. Anzola, C. Giannini and A. Kotlyar, Synthesis and Properties of Conjugates between Silver Nanoparticles and DNA-PNA Hybrids, J. Self-Assem. Mol. Electron., 2013, 1(1), 69–84 CAS.
- X. Wang, H. J. Lim and A. Son, Characterization of denaturation and renaturation of DNA for DNA hybridization, Environmental Health and Toxicology, 2014, 29, e2014007 CrossRef PubMed.
- X. C. Zhou, L. Q. Huang and S. F. Y. Li, Microgravimetric DNA sensor based on quartz crystal microbalance: comparison of oligonucleotide immobilization methods and the application in genetic diagnosis, Biosens. Bioelectron., 2001, 16(1), 85–95 CrossRef CAS PubMed.
- J. Fuchs, J.-B. Fiche, A. Buhot, R. Calemczuk and T. Livache, Salt concentration effects on equilibrium melting curves from DNA microarrays, Biophys. J., 2010, 99(6), 1886–1895 CrossRef CAS PubMed.
- L. Wang, S. Liu, W. Liang, D. Li, J. Yang and Y. He, Detection of DNA utilizing a fluorescent reversible change of a biosensor based on the electron transfer from quantum dots to polymyxin B sulfate, J. Colloid Interface Sci., 2015, 448, 257–268 CrossRef CAS PubMed.
- S.-J. Li, N. Xia, B.-Q. Yuan, W.-M. Du, Z.-F. Sun and B.-B. Zhou, A novel DNA sensor using a sandwich format by electrochemical measurement of marker ion fluxes across nanoporous alumina membrane, Electrochim. Acta, 2015, 159, 234–241 CrossRef CAS.
- J. Zheng, C. Chen, X. Wang, F. Zhang and P. He, A sequence-specific DNA sensor for Hepatitis B virus diagnostics based on the host–guest recognition, Sens. Actuators, B, 2014, 199, 168–174 CrossRef CAS.
- A. B. Steel, T. M. Herne and M. J. Tarlov, Electrochemical quantitation of DNA immobilized on gold, Anal. Chem., 1998, 70(22), 4670–4677 CrossRef CAS PubMed.
- C. Li, S. Liu, L.-H. Guo and D. Chen, A new chemically amplified electrochemical system for DNA detection in solution, Electrochem. Commun., 2005, 7(1), 23–28 CrossRef CAS.
- W. Wang, L. Song, Q. Gao, H. Qi and C. Zhang, Highly sensitive detection of DNA using an electrochemical DNA sensor with thionine-capped DNA/gold nanoparticle conjugates as signal tags, Electrochem. Commun., 2013, 34, 18–21 CrossRef CAS.
- Z. Mazzochette and A. Mugweru, Electrochemical DNA Hybridization Sensor Using Poly[vinylpyridine Os(bipyridine)2Cl]-co-Ethylamine Redox Polymer, Chem. Mater. Res., 2014, 6(5), 55–64 Search PubMed.
- I. Y. Wong and N. A. Melosh, An electrostatic model for DNA surface hybridization, Biophys. J., 2010, 98(12), 2954–2963 CrossRef CAS PubMed.
- A. Benvidi, N. Rajabzadeh, M. Mazloum-Ardakani and M. M. Heidari, Comparison of impedimetric detection of DNA hybridization on chemically and electrochemically functionalized multi-wall carbon nanotubes modified electrode, Sens. Actuators, B, 2015, 207, 673–682 CrossRef CAS.
- G.-U. Flechsig and T. Reske, Electrochemical detection of DNA hybridization by means of osmium tetroxide complexes and protective oligonucleotides, Anal. Chem., 2007, 79(5), 2125–2130 CrossRef CAS PubMed.
- Y. Bo, H. Yang, Y. Hu, T. Yao and S. Huang, A novel electrochemical DNA biosensor based on graphene and polyaniline nanowires, Electrochim. Acta, 2011, 56(6), 2676–2681 CrossRef CAS.
- S. W. Dutse, N. A. Yusof, H. Ahmad, M. Z. Hussein and Z. Zainal, An electrochemical DNA biosensor for ganoderma boninense pathogen of the oil palm utilizing a new ruthenium complex, [Ru(dppz)2(qtpy)]Cl2, Int. J. Electrochem. Sci., 2012, 7, 8105–8115 CAS.
- G. Rizzi, F. W. Østerberg, A. D. Henriksen, M. Dufva and M. F. Hansen, On-chip magnetic bead-based DNA melting curve analysis using a magnetoresistive sensor, J. Magn. Magn. Mater., 2014, 380, 215–220 CrossRef.
- S. W. Dutse, N. A. Yusof, H. Ahmad, M. Z. Hussein, Z. Zainal, R. Hushiarian and R. Hajian, An Electrochemical Biosensor for the Determination of Ganoderma boninense Pathogen Based on a Novel Modified Gold Nanocomposite Film Electrode, Anal. Lett., 2014, 47(5), 819–832 CrossRef CAS.
- L. Tang, G. Zeng, G. Shen, Y. Li, C. Liu, Z. Li, J. Luo, C. Fan and C. Yang, Sensitive detection of lip genes by electrochemical DNA sensor and its application in polymerase chain reaction amplicons from Phanerochaete chrysosporium, Biosens. Bioelectron., 2009, 24(5), 1474–1479 CrossRef CAS PubMed.
- C. Liu, G.-M. Zeng, L. Tang, Y. Zhang, Y.-P. Li, Y.-Y. Liu, Z. Li, M.-S. Wu and J. Luo, Electrochemical detection of Pseudomonas aeruginosa 16S rRNA using a biosensor based on immobilized stem–loop structured probe, Enzyme Microb. Technol., 2011, 49(3), 266–271 CrossRef CAS PubMed.
- L. Qicai, Y. Qiang, W. Wennan, W. Yu, L. Liqing, Z. Chengfei and L. Xinhua, DNA Electrochemical Sensor for Detection of PRSS1 Point Mutation Based on Restriction Endonuclease Technique, Prep. Biochem. Biotechnol., 2015, 45(5), 430–437 CrossRef PubMed.
- L. Zhu, R. Zhao, K. Wang, H. Xiang, Z. Shang and W. Sun, Electrochemical Behaviors of Methylene Blue on DNA Modified Electrode and Its Application to the Detection of PCR Product from NOS Sequence, Sensors, 2008, 8(9), 5649–5660 CrossRef CAS PubMed.
- M. Mix, T. Reske, H. Duwensee and G. U. Flechsig, Electrochemical detection of asymmetric PCR products by labeling with osmium tetroxide, Electroanalysis, 2009, 21(7), 826–830 CAS.
- P. D. Tam, M. A. Tuan, N. Van Hieu and N. D. Chien, Impact parameters on hybridization process in detecting influenza virus (type A) using conductimetric-based DNA sensor, Phys. E, 2009, 41(8), 1567–1571 CrossRef CAS.
- M. Jacobsen and G.-U. Flechsig, Temperature Control in Electrochemical DNA Sensing, Curr. Phys. Chem., 2011, 1(4), 292–298 CrossRef CAS.
- H. Gui, B. Wei and J. Wang, The hybridization and optimization of complementary DNA molecules on organic field-effect transistors, Mater. Sci. Semicond. Process., 2015, 30, 250–254 CrossRef CAS.
- X. Mao, J. Jiang, X. Xu, X. Chu, Y. Luo, G. Shen and R. Yu, Enzymatic amplification detection of DNA based on “molecular beacon” biosensors, Biosens. Bioelectron., 2008, 23(10), 1555–1561 CrossRef CAS PubMed.
- Z. Li, G. Zeng, L. Tang, Y. Zhang, Y. Li, Y. Pang, J. Luo and Y. Liu, Electrochemical DNA sensor for simultaneous detection of genes encoding two functional enzymes involved in lignin degradation, Biochem. Eng. J., 2011, 55(3), 185–192 CrossRef CAS.
- G. Marrazza, G. Chiti, M. Mascini and M. Anichini, Detection of human apolipoprotein E genotypes by DNA electrochemical biosensor coupled with PCR, Clin. Chem., 2000, 46(1), 31–37 CrossRef CAS.
- R. Peytavi, L.-y. Tang, F. R. Raymond, K. Boissinot, L. Bissonnette, M. Boissinot, F. J. Picard, A. Huletsky, M. Ouellette and M. G. Bergeron, Correlation between microarray DNA hybridization efficiency and the position of short capture probe on the target nucleic acid, BioTechniques, 2005, 39(1), 89 CrossRef CAS PubMed.
- Y. Zhang, K. Zhang and H. Ma, Electrochemical DNA biosensors based on gold nanoparticles/cysteamine/poly(glutamic acid) modified electrode, Am. J. Biomed. Sci., 2009, 1(2), 115–125 CrossRef CAS.
- M. Xu, J. Zhuang, X. Chen, G. Chen and D. Tang, A difunctional DNA–AuNP dendrimer coupling DNAzyme with intercalators for femtomolar detection of nucleic acids, Chem. Commun., 2013, 49(66), 7304–7306 RSC.
- L. Chen, Y. Qiu and G. Wang, An amplified electrochemical aptasensor based on hybridization chain reactions and catalysis of silver nanoclusters, Nanoscale, 2015, 7(7), 3300–3308 RSC.
- B. Meric, K. Kerman, D. Ozkan, P. Kara, S. Erensoy, U. S. Akarca, M. Mascini and M. Ozsoz, Electrochemical DNA biosensor for the detection of TT and Hepatitis B virus from PCR amplified real samples by using methylene blue, Talanta, 2002, 56(5), 837–846 CrossRef CAS PubMed.
- G. Xu, K. Jiao, J. Fan and W. Sun, Electrochemical detection of specific gene related to CaMV35S using methylene blue and ethylenediamine-modified glassy carbon electrode, Acta Chim. Slov., 2006, 53(4), 486 CAS.
- G. Mandong, L. Yanqing, G. Hongxia, W. Xiaoqin and F. Lifang, Electrochemical detection of short sequences related to the Hepatitis B virus using MB on chitosan-modified CPE, Bioelectrochemistry, 2007, 70(2), 245–249 CrossRef PubMed.
- M. Tichoniuk, M. Ligaj and M. Filipiak, Application of DNA hybridization biosensor as a screening method for the detection of genetically modified food components, Sensors, 2008, 8(4), 2118–2135 CrossRef CAS PubMed.
- R. Issa, N. Hamdan and M. Noh, Differential pulse voltammetric determination of DNA hybridization using methylene blue on screen printed carbon electrode for the detection of Mycobacterium tuberculosis, Biotechnology, 2010, 9(3), 304–311 CrossRef CAS.
- S. Taufik, N. A. Yusof, T. W. Tee and I. Ramli, Bismuth oxide nanoparticles/chitosan/modified electrode as biosensor for DNA hybridization, Int. J. Electrochem. Sci., 2011, 6, 1880–1891 CAS.
- L. H. Chen and M. Wilson, Non-Vector Transmission of Dengue and Other Mosquito-Borne Flaviviruses, Dengue Bulletin, 2005, 29, 18–31 Search PubMed.
- H. L. Lee Yu, C. M. Montesa, N. S. L. Rojas and E. P. Enriquez, Nucleic-Acid Based Lateral Flow Strip Biosensor via Competitive Binding for Possible Dengue Detection, Journal of Biosensor and Biolectronics, 2012, 3(5), 128 Search PubMed.
- L. L. Li, H. Cai, T. M. H. Lee, J. Barford and I. Hsing, Electrochemical detection of PCR amplicons using electroconductive polymer modified electrode and multiple nanoparticle labels, Electroanalysis, 2004, 16(1–2), 81–87 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2023 |