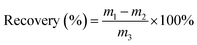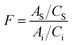Open Access Article
Open Access ArticleAn integrated strategy for quality control of the multi-origins herb medicine of Gentianae Macrophyllae Radix based on UPLC-Orbitrap-MS/MS and HPLC-DAD†
Jiangyi Luo‡
,
Hanwen Yuan‡,
Ling Liang,
Qinling Xie,
Sai Jiang,
Yangfen Fu,
Shenghuang Chen* and
Wei Wang *
*
TCM and Ethnomedicine Innovation & Development International Laboratory, Innovative Material Medical Research Institute, School of Pharmacy, Hunan University of Chinese Medicine, Changsha, China. E-mail: cshtyh@163.com; wangwei402@hotmail.com; Fax: +86-731-8845-8227; Tel: +86-136-5743-8606
First published on 16th March 2023
Abstract
Gentianae Macrophyllae Radix, the dried root of Gentiana macrophylla Pall., Gentiana crassicaulis Duthie ex Burk., Gentiana straminea Maxim., or Gentiana dahurica Fisch., is a traditional Chinese medicine with multi-origins and some adulterants. Liquid chromatography coupled to electrostatic orbitrap high-resolution mass spectrometry (LC-Orbitrap-MS) was used to search the different components of Gentianae Macrophyllae Radix of the four species. High-performance liquid chromatography (HPLC) combined with fingerprint analysis, principal components analysis (PCA), and partial least-squares discrimination analysis (PLS-DA) was also utilized to distinguish them and their adulterants based on the critical components identified by LC-MS. A single standard to determine the multi-components (SSDMC) method was established for the determination of the critical markers. A total of 93 compounds were identified from Gentianae Macrophyllae Radix, including 58 common ones. Their HPLC fingerprints show a significant difference with the adulterants. In addition, PCA and PLS-DA could make a distinction among the four species. Loganic acid, 6′-O-β-D-glucosylgentiopicroside, swertiamarine, gentiopicroside, and sweroside were identified as the critical markers and then quantified by the SSDMC method. The developed strategy is powerful for the quality control and authentication of Gentianae Macrophyllae Radix.
1. Introduction
Gentianae Macrophyllae Radix, the dried root of Gentiana macrophylla Pall., Gentiana crassicaulis Duthie ex Burk., Gentiana straminea Maxim., or Gentiana dahurica Fisch., has been used as a medicine since Han Dynasty (202 BC to 220 AD) to dispel wind-damp, clear damp-heat, ease pain, and eliminate deficiency-heat.1–4 They are cultivated in different geographic regions in China and are generally known as Qinjiao (QJ), CuJing Qinjiao (CJQJ), MaHua Qinjiao (MHQJ), and Xiao Qinjiao (XQJ) in Chinese due to their different appearances, respectively (Fig. 1).1,5–7 Numerous studies have proved that this herbal medicine is abundant in iridoids and secoiridoids, such as loganic acid and gentiopicroside, which have been recorded as quality markers in the Chinese Pharmacopoeia (2020 edition).3,8 Loganic acid, 6′-O-β-D-glucosylgentiopicroside, swertiamarine, gentiopicroside, and sweroside are reported to have various excellent activities.9–13 Studies have shown that the content of active components of the herb medicine will affect their pharmacological activities, which was the reason for the differences in activity among the Gentianae Macrophyllae Radix of the four species.14–17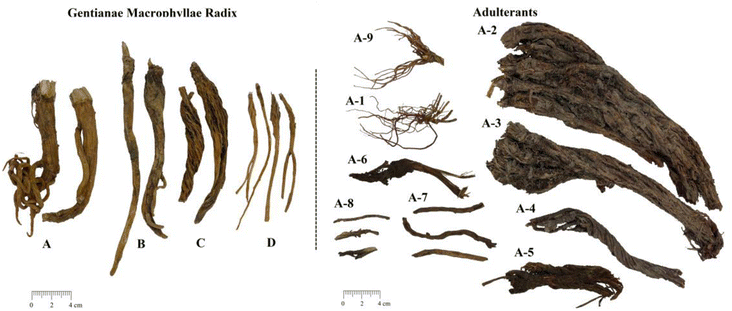 | ||
| Fig. 1 Gentianae Macrophyllae Radix and the adulterants (A: Gentiana macrophylla Pall.; B: Gentiana crassicaulis Duthie ex Burk.; C: Gentiana straminea Maxim.; D: Gentiana dahurica Fisch.). | ||
In addition, there are some adulterants used as Gentianae Macrophyllae Radix in the market (Table 1 and Fig. 1), such as Long dan (the rhizome of Gentiana scabra Bge.), Hong qin jiao (the root of Salvia Przewalskii Maxim.), and Ma bu qi (the root of Aconitum sinomontanum Nakai.). Therefore, the identification of different species and authentication are of great importance for the safety and effectiveness of Gentianae Macrophyllae Radix in clinical practice.
| Sample name | Medicine | Origin | Sample name | Medicine | Origin | Sample name | Medicine/chinese name | Origin/plant |
|---|---|---|---|---|---|---|---|---|
| QJ-1 | QJ | Yunnan | QJ-15 | CJQJ | Sichuan | QJ-29 | XQJ | Inner Mongoria |
| QJ-2 | QJ | Yunnan | QJ-16 | CJQJ | Sichuan | QJ-30 | XQJ | Qinghai |
| QJ-3 | QJ | Yunnan | QJ-17 | MHQJ | Xinjiang | QJ-31 | XQJ | Inner Mongoria |
| QJ-4 | QJ | Yunnan | QJ-18 | MHQJ | Sichuan | QJ-32 | XQJ | Inner Mongoria |
| QJ-5 | QJ | Yunnan | QJ-19 | MHQJ | Qinghai | QJ-33 | XQJ | Xinjiang |
| QJ-6 | QJ | Yunnan | QJ-20 | MHQJ | Sichuan | A-1 | Long dan | Gentiana scabra Bge. |
| QJ-7 | QJ | Yunnan | QJ-21 | MHQJ | Qinghai | A-2 | Hong qin jiao | Salvia Przewalskii Maxim. |
| QJ-8 | CJQJ | Yunnan | QJ-22 | MHQJ | Sichuan | A-3 | Hong qin jiao | Salvia Przewalskii Maxim. |
| QJ-9 | CJQJ | Yunnan | QJ-23 | MHQJ | Sichuan | A-4 | Hong qin jiao | Salvia Przewalskii Maxim. |
| QJ-10 | CJQJ | Sichuan | QJ-24 | MHQJ | Sichuan | A-5 | Hong qin jiao | Salvia Przewalskii Maxim. |
| QJ-11 | CJQJ | Sichuan | QJ-25 | MHQJ | Sichuan | A-6 | Ma bu qi | Aconitum sinomontanum Nakai. |
| QJ-12 | CJQJ | Sichuan | QJ-26 | MHQJ | Tibet | A-7 | Du yi wei | Lamiophlomis rotata (Benth.) Kudo |
| QJ-13 | CJQJ | Qinghai | QJ-27 | MHQJ | Sichuan | A-8 | Bai tou wen | Pulsatilla chienensis (Bge.) Regel |
| QJ-14 | CJQJ | Sichuan | QJ-28 | MHQJ | Qinghai | A-9 | Bai wei | Cynanchum atratum Bunge. |
Liquid chromatography coupled to electrostatic orbitrap high-resolution mass spectrometry (LC-Orbitrap-MS) has the advantages of high resolution, quality accuracy,18 and qualitative analysis of constituents by the in-house and online database. Due to its stability and controllability, high-performance liquid chromatography (HPLC) is still the classic technology for quality control of herbal medicines in pharmacopeia worldwide.
In this paper, the major constituents of Gentianae Macrophyllae Radix were analyzed by LC-Orbitrap-MS. Subsequently, variable influence on projection (VIP) score, K-means calculation, and self-organizing map (SOM) were used to obtain differentials for the compounds, which were then quantified by HPLC and a single standard for determination of multiple components (SSDMC) method.19–21 Finally, the fingerprint analysis, principal components analysis (PCA), and partial least-squares discrimination analysis (PLS-DA) were performed to distinguish QJ, CJQJ, MHQJ, and XQJ, as well as the adulterants.
2. Materials and methods
2.1. Reagents and materials
The reference standards, including loganic acid, swertiamarine, gentiopicroside, and sweroside, were purchased from NatureStandard (Shanghai, China), while 6′-O-β-D-glucosylgentiopicroside was supplied by Shanghai Yuanye Bio-Technology Co., Ltd. (Shanghai, China). HPLC-grade methanol, acetonitrile, and formic acid were purchased from Sigma-Aldrich (Sigma-Aldrich, Co., Louis, USA). All aqueous solution was prepared with purified water from C'estbon (Shenzhen, China).Thirty-three batches of Gentianae Macrophyllae Radix (including 7 batches of QJ, 9 batches of CJQJ, 12 batches of MHQJ, and 5 batches of XQJ) were collected from Xinjiang, Yunnan, Tibet, Sichuan, Inner Mongolia, and Qinghai. In addition, nine batches of its adulterants were also collected from different provinces. The details are summarized in Table 1. All the samples were authenticated by Professor Wei Wang (School of Pharmacy, Hunan University of Chinese Medicine), according to the plant morphology. Voucher specimens (QJ 1 ∼ 33, Long dan, Hong qin jiao-1 ∼ 4, Ma bu qi, Du yi wei, Bai tou wen, and Bai wei) were deposited at the TCM and Ethnomedicine Innovation & Development International Laboratory, Innovative Material Medical Research Institute, School of Pharmacy, Hunan University of Chinese Medicine, Changsha, China.
2.2. Sample preparation
The air-dried roots were pulverized and passed through a 50-mesh sieve. Then, 0.2 g of the powder was accurately weighed and ultrasonic-extracted with 20 mL methanol for 30 min. After 10 min centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and filtration with a 0.22 μm of filter membrane, the sample solution was collected. The quality control (QC) sample was prepared by mixing an equal volume of each sample solution.
000 rpm and filtration with a 0.22 μm of filter membrane, the sample solution was collected. The quality control (QC) sample was prepared by mixing an equal volume of each sample solution.
2.3. LC-MS conditions
The LC analysis was run on a Hypersil GOLDTM Aq-C18 column (20 × 2.1 mm, 1.9 μm) (Thermo Sencitific, MA, USA) with a Vanquish™ Flex UPLC system at 30 °C, using 0.01% formic acid (A) and acetonitrile (B) as the mobile phase at a flow rate of 0.3 mL min−1. The gradient elution conditions were as follows: 5–30% B (0–1 min), 30–40% B (1–5 min), 40–90% B (5–6 min), 90–95% B (6–13 min), 95% B (13–21 min), and 5% B (21–24 min). The injection volume was 4 μL. MS analysis (qualitative analysis) was performed on a Orbitrap Exploris 120 in negative ion mode with a full scan MS spectrum over the m/z range 150–1000, using ion spray voltage of 2.5 kV, sheath gas of 50 Arb, aux gas of 10 Arb, sweep gas of 1 Arb, ion transfer tube temp of 325 °C, and vaporizer temp 350 °C. The orbitrap resolution of full scan MS was 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and MS2 was 15
000 and MS2 was 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, and HCD Collision Energies (%) was kept at 30%.
000, and HCD Collision Energies (%) was kept at 30%.
2.4. HPLC analysis
In which, m1, m2, and m3 were the amount obtained, the half original amount in the sample, and the amount spiked into the sample, respectively.
where Ai and AS are the peak area of the corresponding compounds obtained from the sample solution and the peak area of gentiopicroside obtained from the standard solution. Ci and Cs are the concentration of components (mg mL−1) to be measured in the test sample and the concentration of gentiopicroside in the reference solution.
2.5. Data analysis
The MS/MS data was analyzed by Freestyle 1.8 and Compounds discover 3.3 software. The compounds were identified by mzVault, mzCloud, ChemSpider, and Mass List Search. The VIP scores, K-means Clustering, SOM, and Pearson correlations were analyzed by Metaboanalyst 5.0. The fingerprint was generated by the Similarity Evaluation System for Chromatographic Fingerprint of Traditional Chinese Medicine (Version 2012A) software. PCA, PLS-DA, and HCA were obtained by SIMCA 14.1 software.3. Results and discussion
3.1. LC-MS analysis
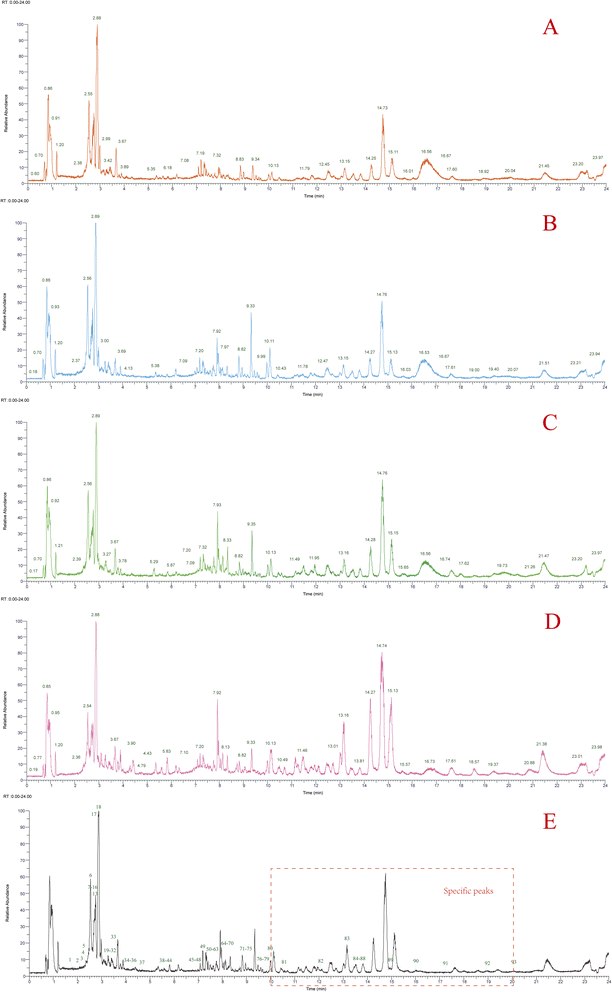
The response of Gentianae Macrophyllae Radix under the negative ion mode was better than the positive model in LC-MS analysis (Fig. S1†). The total ion chromatograms of QJ, CJQJ, MHQJ, XQJ, QC sample, and the adulterants are shown in Fig. 2 and S2.† The total ion chromatograms between Gentianae Macrophyllae Radix and its adulterants exhibit great difference in triterpenoid with retention time from 10 to 20 min (Fig. 2E). On the basis of reference standards, literature data,8,22,23 and the inhouse and online database (inluding mzVault, mzCloud, ChemSpider, and Mass List Search with scores of more than 90), a total of 93 compounds were identified from Gentianae Macrophyllae Radix, including 9 iridoids, 10 secoiridoids, 12 flavonoids, 6 lignans, 38 terpenes, and 18 other types of compounds (Table 2). There were 58 common compounds in QJ, CJQJ, MHQJ, and XQJ (Fig. 3A). Their peak areas data was uploaded to Metaboanalyst 5.0 for statistical analysis (one factor) to screen out the differential components through VIP scores (Fig. 3B). Compounds 18 (gentiopicroside), 12 (6′-O-β-D-glucosylgentiopicroside), and 13 (swertiamarine) were the critical markers due to their high VIP scores. In addition, K-means calculation and SOM specified that compounds 6 (loganic acid), 18 (gentiopicroside), 12 (6′-O-β-D-glucosylgentiopicroside), 13 (swertiamarine), and 83 (soyasapogenol B) were the most critical components (Fig. 3C and D). Therefore, the HPLC analysis focused on these compounds.| Peak no. | RT (min) | Reference ion | m/z | Diff. (ppm) | Formula | Fragment ions (m/z) | Identification | QJ | CJQJ | MHQJ | XQJ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| a The common compounds identified from Gentianae Macrophyllae Radix. | |||||||||||
| 1 | 1.886 | [M − H]− | 421.1351 | −0.11 | C17H26O12 | 375.12955, 310.12399, 255.08681, 229.04193, 213.07692, 169.05067, 125.0245 | Lamiide | √ | √ | √ | |
| 2 | 2.176 | [M − H]− | 315.0724 | 0.78 | C13H16O9 | 272.01117, 259.91162, 229.02588, 180.87544, 165.01967, 153.01947, 109.02966 | Gentisic acid-5-O-β-glucoside | √ | √ | ||
| 3a | 2.377 | [M + FA-H]− | 391.12454 | −0.35 | C15H22O9 | 246.82138, 229.03535, 211.06192, 183.06647, 167.07111, 137.06094, 121.06605 | Aucubin | √ | √ | √ | √ |
| 4 | 2.439 | [M − H]− | 375.12949 | −0.4 | C16H24O10 | 229.03514, 213.07697, 169.08714, 151.0766, 125.06094 | Mussaenosidic acid | √ | √ | √ | |
| 5 | 2.499 | [M − H]− | 188.03529 | −0.12 | C10H7NO3 | 181.76221, 159.87849, 144.04565, 116.05076, 89.20306, 81.13036 | Kynurenic acid | √ | |||
| 6a | 2.544 | [M − H]− | 375.12921 | −0.63 | C16H24O10 | 310.93152, 273.17215, 229.03088, 213.077, 169.08717, 151.07664, 125.06109 | Loganic acid | √ | √ | √ | √ |
| 7a | 2.584 | [M + FA-H]− | 725.21478 | 0.25 | C28H40O19 | 415.73318, 383.1196, 323.09967, 229.04402, 221.06702, 179.0564, 149.06079, 131.03517 | Scabrans G3 | √ | √ | √ | √ |
| 8a | 2.601 | [M − H]− | 373.11379 | −0.32 | C16H22O10 | 364.96799, 229.02231, 211.06148, 193.05083, 167.07156, 149.06096, 123.04534 | Geniposidic acid | √ | √ | √ | √ |
| 9a | 2.607 | [M − H]− | 593.15125 | 0.09 | C27H30O15 | 503.12003, 473.10944, 375.12659, 311.05649, 282.05389, 229.02704, 213.0777 | Vicenin-2 | √ | √ | √ | √ |
| 10a | 2.65 | [M − H]− | 405.13995 | −0.6 | C17H26O11 | 281.06702, 243.09027, 229.04758, 221.0452, 197.08226, 179.03546, 155.03568, 141.05588 | Shanzhiside methyl ester | √ | √ | √ | √ |
| 11 | 2.682 | [M + FA-H]− | 729.26111 | 0.17 | C32H44O16 | 383.11844, 359.1492, 329.13965, 310.11627, 139.07669, 101.02464 | (+)-Lariciresinol-4,4′-O-β-D-diglucopyranoside | √ | √ | ||
| 12a | 2.707 | [M + FA-H]− | 563.16179 | 0.09 | C22H30O14 | 374.97791, 304.93506, 229.04768, 221.06778, 193.04979, 179.05632, 149.06085 | 6′-O-β-D-glucosylgentiopicroside | √ | √ | √ | √ |
| 13a | 2.754 | [M + FA-H]− | 419.11919 | −0.65 | C16H22O10 | 361.99875, 229.04565, 169.6095, 149.06107, 141.01965 | Swertiamarin | √ | √ | √ | √ |
| 14 | 2.791 | [M − H]− | 403.12551 | 1.13 | C17H24O11 | 273.46982, 249.06258, 229.02711, 195.06598, 179.05597, 161.04498, 153.01945, 149.06076 | Gardenoside | √ | |||
| 15a | 2.798 | [M − H]− | 681.23997 | 1.3 | C32H42O16 | 519.13965, 501.16895, 381.1348, 357.13501, 339.12378, 323.07669, 309.11362, 229.02699, 203.07179, 179.05647 | Pinoresinol diglucoside | √ | √ | √ | √ |
| 16a | 2.832 | [M + FA-H]− | 639.15704 | 0.64 | C27H30O15 | 519.11658, 477.10452, 459.09302, 433.11359, 323.07831, 315.0726, 283.26459, 229.05029, 153.01952 | Saponarin | √ | √ | √ | √ |
| 17 | 2.867 | [M − H]− | 447.09268 | −1.34 | C21H20O11 | 429.08356, 357.06143, 327.05136, 297.04062, 285.0405, 269.10281, 229.03467, 161.04578 | Isoorientin | √ | √ | √ | |
| 18a | 2.881 | [M + FA-H]− | 401.10856 | −1.04 | C16H20O9 | 295.08347, 235.06143, 229.02054, 193.05118, 175.04045, 149.06096, 121.06655 | Gentiopicroside | √ | √ | √ | √ |
| 19a | 2.984 | [M − H]− | 357.11896 | 0.66 | C16H22O9 | 269.11877, 259.0976, 229.03552, 195.0313, 177.05588, 153.01932, 133.06612 | Sweroside | √ | √ | √ | √ |
| 20 | 3.026 | [M + FA-H]− | 581.1883 | 1.58 | C26H32O12 | 373.12875, 355.11829, 343.1185, 313.1084, 229.02733, 209.08191, 193.0506, 163.04053, 151.04045, 137.02448 | 8-Hydroxypinoresinol-4′-O-β-D-glucopyranoside | √ | √ | ||
| 21a | 3.062 | [M − H]− | 431.09847 | 0.24 | C21H20O10 | 413.0878, 387.07269, 341.06653, 327.05112, 311.0564, 283.05881, 255.06628, 229.04552, 205.01439, 178.99817 | Isovitexin | √ | √ | √ | √ |
| 22 | 3.083 | [M − H]− | 359.13467 | −0.15 | C16H24O9 | 271.37622, 232.08966, 229.03514, 197.08215, 153.09233, 135.08171, 109.06606, 119.03494 | 7-Deoxyloganic acid | √ | √ | √ | |
| 23a | 3.093 | [M − H]− | 521.20322 | 0.75 | C26H34O11 | 477.13144, 329.13983, 325.05759, 229.02888, 192.07962, 178.06396, 175.07663 | Lariciresinol-4-O-glucoside | √ | √ | √ | √ |
| 24a | 3.278 | [M − H]− | 417.15476 | −1.74 | C22H26O8 | 402.13223, 387.10843, 236.06927, 229.04073, 190.0636, 181.05084, 166.02734, 152.04797 | (+)-Syringaresinol | √ | √ | √ | √ |
| 25a | 3.28 | [M − H + HAc]− | 579.20863 | 0.69 | C26H32O11 | 357.13419, 342.11081, 311.12927, 229.04831, 151.04018, 135.04543 | (−)-Pinoresinol glucoside | √ | √ | √ | √ |
| 26a | 3.333 | [M − H]− | 191.03512 | 0.72 | C10H8O4 | 176.04822, 163.95222, 147.04532, 144.86652, 111.00893, 87.00882 | 6,7-Dihydroxy-4-methylcoumarin | √ | √ | √ | √ |
| 27a | 3.404 | [M − H]− | 609.18261 | 0.21 | C28H34O15 | 503.62744, 488.21753, 367.08380, 343.082, 325.07181, 301.07217, 286.04901, 257.08224, 229.04509, 179.78441, 125.02504 | Hesperidin | √ | √ | √ | √ |
| 28 | 3.452 | [M + FA-H]− | 569.15155 | 0.16 | C24H28O13 | 476.10995, 474.09473, 388.09415, 374.32324, 289.98453, 229.04309, 137.02455, 93.03461 | (+)-Seguinoside D | √ | √ | ||
| 29a | 3.473 | [M − H]− | 397.11412 | 0.34 | C18H22O10 | 356.28165, 328.25037, 326.10419, 235.0983, 229.04501, 153.01874, 149.06119 | 6′-O-Acetyl-gentiopicroside | √ | √ | √ | √ |
| 30a | 3.483 | [M + FA-H]− | 417.21305 | 0.1 | C19H32O7 | 349.17096, 252.32874, 229.04927, 161.04509, 141.23784, 123.11726 | Blumel C glucoside | √ | √ | √ | √ |
| 31 | 3.485 | [M − H]− | 697.19905 | 0.69 | C31H38O18 | 655.1897, 571.16766, 535.14624, 475.12473, 409.11395, 367.10291, 349.09293, 315.07251, 229.03539, 153.01949 | Gentistraminoside A | √ | √ | ||
| 32a | 3.571 | [M − H]− | 447.0936 | 0.82 | C21H20O11 | 357.06213, 327.05191, 285.04074, 229.03545, 177.01965, 116.92889 | Kaempferol-7-O-glucoside | √ | √ | √ | √ |
| 33 | 3.658 | [M − H]− | 521.16652 | 0.31 | C25H30O12 | 359.1142, 357.11865, 315.12424, 297.11343, 229.0264, 213.07632, 195.06635, 163.04022, 151.07657 | 2′-O-(4′′-Hydroxycinnamoyl)mussaenosidic acid | √ | √ | ||
| 34 | 3.993 | [M − H]− | 755.2045 | 0.64 | C33H40O20 | 713.19714, 613.17859, 593.15192, 571.16803, 533.12988, 451.12473, 409.11401, 391.10428, 367.10297, 349.09274, 315.07257, 229.0226, 153.01955 | Gentistraminoside B | √ | √ | ||
| 35 | 4.3 | [M − H]− | 479.15596 | 0.44 | C23H28O11 | 357.11929, 273.11514, 234.43077, 229.03462, 195.0663, 151.07655, 121.02967 | Albiflorin | √ | |||
| 36a | 4.306 | [M + FA-H]− | 493.22922 | 0.43 | C21H36O10 | 315.18182, 285.11374, 229.05032, 191.05629, 161.04556, 131.03499, 113.02454, 101.02451 | Atractyloside A | √ | √ | √ | √ |
| 37a | 4.692 | [M − H]− | 301.03555 | 0.57 | C15H10O7 | 286.04874, 257.04626, 242.0584, 233.0457, 193.01447, 164.01154, 151.00389, 125.02461 | Quercetin | √ | √ | √ | √ |
| 38 | 5.234 | [M − H]− | 301.14468 | 0.48 | C18H22O4 | 283.13483, 257.15469, 229.03175, 213.16476, 193.01418, 177.09232, 149.81148, 106.89589 | Terbucromil | √ | √ | √ | |
| 39a | 5.272 | [M − H]− | 797.21476 | 0.21 | C35H42O21 | 755.20563, 655.18994, 635.16437, 613.17889, 593.15143, 493.13611, 451.12488, 409.11401, 315.07251, 153.01958 | Rindoside | √ | √ | √ | √ |
| 40a | 5.385 | [M − H]− | 269.04572 | 0.64 | C15H10O5 | 225.0562, 200.88235, 181.91168, 159.04614, 151.00385, 117.03601 | Aloe-emodin | √ | √ | √ | √ |
| 41 | 5.55 | [M − H]− | 299.05627 | 0.54 | C16H12O6 | 284.0329, 256.03848, 229.02341, 190.84854, 169.60602, 134.90634 | Hispidulin | √ | √ | √ | |
| 42a | 5.615 | [M − H]− | 285.04067 | 0.74 | C15H10O6 | 257.04556, 241.05099, 199.04027, 193.01436, 177.0195, 151.00386, 133.02988 | Luteolin | √ | √ | √ | √ |
| 43 | 5.847 | [M − H]− | 781.21987 | 0.26 | C35H42O20 | 697.20068, 655.1911, 619.16736, 577.15662, 493.13516, 451.12427, 315.07239, 153.0195 | Trifloroside | √ | √ | ||
| 44 | 6.087 | [M − H]− | 955.49086 | 0.08 | C48H76O19 | 835.44794, 793.4389, 731.43817, 613.37518, 569.38464, 523.37915, 455.35229, 229.04059 | Gensenoside Ro | √ | |||
| 45a | 6.782 | [M − H]− | 519.33282 | 0.11 | C30H48O7 | 501.32266, 453.3027, 451.28586, 435.28998, 389.28601, 365.28641, 229.04555, 152.99568 | Cucurbitacin P | √ | √ | √ | √ |
| 46 | 6.833 | [M − H]− | 793.43851 | 0.67 | C42H66O14 | 733.41687, 673.39227, 631.38538, 613.37506, 569.38501, 455.3537, 356.71674, 317.46048, 229.02242, 175.02512, 157.01413 | Fatsiaside C | √ | |||
| 47 | 6.898 | [M − H]− | 821.39696 | 0.54 | C42H62O16 | 759.39392, 645.37128, 627.3584, 351.05676, 333.04721, 289.0556, 229.04875, 193.03555 | Glycyrrhizic acid | √ | |||
| 48 | 6.989 | [M − H]− | 319.1188 | 0.27 | C17H20O6 | 287.09247, 275.12903, 243.10214, 207.06651, 205.05106, 191.03516, 179.03526, 148.05307 | Mycophenolic acid | √ | √ | ||
| 49a | 7.2 | [M − H]− | 325.20226 | 0.62 | C18H30O5 | 307.19183, 289.18204, 263.20181, 229.03473, 195.10229, 171.1026, 151.11298, 137.09737, 125.09727, 111.08197 | 2,3-Dinor-11-β-prostaglandin F2α | √ | √ | √ | √ |
| 50a | 7.265 | [M − H]− | 485.32726 | 0.13 | C30H46O5 | 407.29648, 373.28903, 273.11261, 231.10339, 193.05061, 179.03552, 155.53018, 111.57957 | (3β,4)-3,23-Dihydroxy-1-oxoolean-12-en-28-oic acid | √ | √ | √ | √ |
| 51a | 7.311 | [M + FA-H]− | 549.34344 | 0.11 | C30H48O6 | 470.01324, 441.30182, 393.09708, 349.1084, 285.0416, 229.04987, 193.01363, 111.00887 | Arjungenin | √ | √ | √ | √ |
| 52a | 7.329 | [M − H]− | 299.05623 | 0.4 | C16H12O6 | 284.03311, 271.06165, 240.04305, 207.03024, 191.03517, 176.01167, 165.01964, 139.0403, 133.02956 | Kaempferide | √ | √ | √ | √ |
| 53 | 7.4 | [M − H]− | 373.16547 | −0.51 | C21H26O6 | 355.15488, 329.17667, 285.18646, 246.0903, 229.03528, 191.03514, 178.02711 | Ustosolate E | √ | |||
| 54a | 7.407 | [M − H]− | 487.34297 | 0.17 | C30H48O5 | 373.74661, 251.06982, 229.02534, 86.7402 | Asiatic acid | √ | √ | √ | √ |
| 55a | 7.412 | [2 M − H]− | 499.30671 | 0.47 | C15H22O3 | 229.03546, 205.16005, 189.12825, 163.00533, 141.0988, 121.06618, 116.97608 | 2-[(1S,2S,4aR,8aS)-1-Hydroxy-4a-methyl-8-methylidene-decahydronaphthalen-2-yl]prop-2-enoic acid | √ | √ | √ | √ |
| 56 | 7.536 | [M − H]− | 503.33804 | 0.85 | C30H48O6 | 490.36255, 301.03766, 247.0618, 229.04099, 193.01413, 152.99648, 116.92847 | Sericic acid | √ | |||
| 57a | 7.656 | [M − H]− | 269.04576 | 0.8 | C15H10O5 | 251.20149, 229.0273, 211.13416, 197.15456, 185.11882, 150.9537, 130.08713, 119.43418 | Apigenin | √ | √ | √ | √ |
| 58a | 7.693 | [M − H]− | 499.30665 | 0.38 | C30H44O6 | 455.31589, 423.28726, 409.31454, 247.75471, 229.04976, 139.07742, 100.93333 | 11-Deoxocucurbitacin I | √ | √ | √ | √ |
| 59 | 7.731 | [M − H]− | 403.1185 | −0.53 | C24H20O6 | 388.09537, 357.11511, 319.18762, 298.02972, 229.04922, 217.0874, 201.05643, 151.96779 | Tribenzoin | √ | √ | ||
| 60 | 7.777 | [M − H]− | 343.22738 | −1.42 | C22H32O3 | 315.25467, 297.24362, 287.22321, 269.21329, 229.02066, 201.11327, 187.0975, 139.11281 | Medroxyprogesterone | √ | √ | ||
| 61 | 7.81 | [M − H]− | 265.12357 | 0.62 | C18H18O2 | 247.1111, 229.04784, 117.73475, 111.88721 | Magnolol | √ | |||
| 62 | 7.86 | [M − H]− | 285.04067 | 0.72 | C15H10O6 | 267.19684, 257.0451, 241.21785, 229.04376, 223.20557, 174.75847, 112.1844 | Kaempferol | √ | √ | ||
| 63 | 7.89 | [M − H]− | 235.09763 | 0.54 | C13H16O4 | 229.02434, 199.85121, 189.85136, 176.08412, 163.64058, 134.89507 | 4-(3-Hydroxy-1-buten-1-yl)-3-methoxy-5-methylbenzoic acid | √ | √ | ||
| 64 | 7.956 | [M − H]− | 299.20142 | −0.77 | C20H28O2 | 281.21219, 255.23141, 237.22255, 229.03529, 197.14368, 173.10722, 157.88564 | Tretinoin | √ | |||
| 65a | 7.991 | [M − H]− | 471.34794 | −0.05 | C30H48O4 | 318.6073, 229.02156, 195.29953, 152.99611, 144.25693 | Colosolic acid | √ | √ | √ | √ |
| 66 | 8.071 | [M − H]− | 503.33821 | 0.78 | C30H48O6 | 473.32901, 459.35007, 441.33878, 425.30679, 341.26569, 279.23495, 237.15018, 152.18785 | 2,3,19,23-Tetrahydroxyolean-12-en-28-oic acid | √ | √ | √ | |
| 67 | 8.241 | [M − H]− | 315.19681 | 0.77 | C20H28O3 | 296.23151, 271.20706, 243.1758, 229.05154, 120.11904 | 15d-PGA2 | √ | |||
| 68a | 8.249 | [M − H]− | 469.33264 | 0.66 | C30H46O4 | 451.32596, 425.34454, 383.35666, 280.06595, 229.03511 | Enoxolone | √ | √ | √ | √ |
| 69a | 8.341 | [M − H]− | 455.35293 | 0 | C30H48O3 | 393.27533, 375.2677, 327.8494, 279.23322, 257.23996, 229.04012, 175.06078, 114.02003 | β-Boswellic acid | √ | √ | √ | √ |
| 70 | 8.471 | [M − H]− | 301.18104 | 0.4 | C19H26O3 | 272.23172, 254.2231, 229.03221, 218.09511, 204.11674, 189.09203 | 2-Methoxyestradiol | √ | |||
| 71a | 8.702 | [M − H]− | 487.3429 | 0.08 | C30H48O5 | 469.33502, 443.34897, 425.34457, 369.31638, 353.28592, 229.0455 | Arjunic acid | √ | √ | √ | √ |
| 72a | 8.734 | [M − H]− | 455.35301 | −0.05 | C30H48O3 | 437.3428, 379.05026, 365.32156, 297.94916, 285.42944, 229.03523, 160.8452 | Oleanolic acid | √ | √ | √ | √ |
| 73a | 8.762 | [M − H]− | 323.25943 | 0.75 | C20H36O3 | 295.26431, 265.25485, 238.83621, 229.02774, 165.12883, 125.39846 | Labdanolic acid | √ | √ | √ | √ |
| 74a | 8.857 | [M − H]− | 455.35294 | −0.24 | C30H48O3 | 437.34277, 411.33011, 229.03543, 214.91963, 144.2729, 128.21217 | Ursolic acid | √ | √ | √ | √ |
| 75 | 9.112 | [M − H]− | 485.32726 | 0.04 | C30H46O5 | 441.33749, 423.3273, 407.29764, 381.31573, 365.28815, 229.02361, 177.2758 | Melaleucic acid (6CI) | √ | |||
| 76a | 9.457 | [M − H]− | 391.28268 | −6.88 | C24H40O4 | 363.28711, 355.32327, 343.26233, 229.02185, 191.03249, 172.81613, 142.60358 | Deoxycholic acid | √ | √ | √ | √ |
| 77a | 9.53 | [M − H]− | 453.33751 | 0.38 | C30H46O3 | 435.32776, 391.28375, 355.77863, 298.2478, 229.0423, 171.10327 | Pinicolic acid | √ | √ | √ | √ |
| 78a | 9.602 | [M − H]− | 469.33236 | 0.17 | C30H46O4 | 425.3428, 411.29059, 397.31042, 367.30142, 339.26877, 229.04851 | 18-β-Glycyrrhetinic acid | √ | √ | √ | √ |
| 79a | 9.664 | [M − H]− | 453.33746 | 0.11 | C30H46O3 | 435.32736, 393.31696, 336.5705, 247.89471, 229.03549, 165.65364, 157.5334 | Glycyrrhetaldehyde | √ | √ | √ | √ |
| 80a | 10.104 | [M − H]− | 471.34791 | −0.03 | C30H48O4 | 441.33719, 427.35895, 413.30646, 397.35464, 341.28409, 251.1653, 229.02982, 191.1433, 152.99596 | Bourjotinolone A (7CI) | √ | √ | √ | √ |
| 81a | 10.432 | [M − H]− | 783.4906 | 0.74 | C42H72O13 | 737.48523, 600.46472, 575.43262, 484.41187, 323.10037, 221.06688, 179.05632, 161.04568 | Ginsenoside F2 | √ | √ | √ | √ |
| 82a | 12.115 | [M − H]− | 455.353 | 0.02 | C30H48O3 | 437.34271, 408.33716, 383.33078, 312.17545, 229.02316, 175.14978 | 3-Hydroxyurs-12-en-23-oic acid | √ | √ | √ | √ |
| 83a | 13.162 | [M − H]− | 457.36858 | −0.21 | C30H50O3 | 439.36069, 399.32846, 333.66791, 293.06851, 229.04471, 153.84746, 120.77197 | Soyasapogenol B | √ | √ | √ | √ |
| 84a | 13.44 | [M − H]− | 439.35802 | −0.25 | C30H48O2 | 313.57648, 263.74875, 229.02229, 194.77271, 163.58023, 137.89886, 120.79447 | Roburic acid | √ | √ | √ | √ |
| 85 | 13.618 | [M − H]− | 437.34251 | 0.02 | C30H46O2 | 419.33331, 365.32208, 361.2926, 345.65881, 229.03558, 152.99625, 127.24102 | 2,2′-Ethylidene-bis(4,6-di-tert-butylphenol) | √ | |||
| 86a | 13.873 | [M − H]− | 415.32155 | −0.51 | C27H44O3 | 380.88861, 326.64908, 229.02951, 216.85457, 163.04048, 145.02974, 118.04257 | Calcitriol | √ | √ | √ | √ |
| 87a | 13.951 | [M − H]− | 441.33739 | −0.07 | C29H46O3 | 402.7934, 383.35178, 355.32266, 260.00064, 243.20294, 229.04424, 193.1041, 163.04025, 145.02969 | 4-α-Methylzymosterol-4-carboxylate | √ | √ | √ | √ |
| 88a | 14.016 | [M − H]− | 425.34254 | 0.16 | C29H46O2 | 407.33124, 379.39474, 363.36276, 349.29794, 238.83777, 229.03508, 152.10498, 134.22285 | 4-β-Methylzymosterol-4-carbaldehyde | √ | √ | √ | √ |
| 89a | 14.923 | [M − H]− | 427.35806 | −0.2 | C29H48O2 | 367.33734, 288.5162, 229.05119, 174.3268, 116.92864 | (3β,24R,24′R)-fucosterol epoxide | √ | √ | √ | √ |
| 90a | 16.017 | [M − H]− | 443.35303 | −0.06 | C29H48O3 | 305.3432, 300.46295, 252.77142, 229.05147, 201.34343, 163.04047, 145.03, 139.7027, 118.04246 | 3-β-Hydroxy-4β-methyl-5α-cholest-7-en-4α-oic acid | √ | √ | √ | √ |
| 91a | 17.338 | [M − H]− | 433.36878 | 0.19 | C28H50O3 | 397.36902, 389.37885, 322.08398, 258.96381, 229.03616, 180.1319, 152.99579, 146.96379 | 6-Deoxoteasterone | √ | √ | √ | √ |
| 92a | 18.95 | [M − H]− | 471.38424 | −0.25 | C31H52O3 | 417.68497, 300.66922, 229.03661, 163.04039, 145.02951, 118.04212 | (22S,24R)-24-Methyllanosta-8-en-22,28-epoxy-3β,28α-diol | √ | √ | √ | √ |
| 93 | 20.902 | [M − H]− | 485.39998 | −0.07 | C32H54O3 | 440.36252, 397.52771, 344.73618, 302.97443, 229.05005, 145.02914, 116.92834 | 6-Deoxy-16-β-O-acetyl-leucotylin | √ | √ | ||
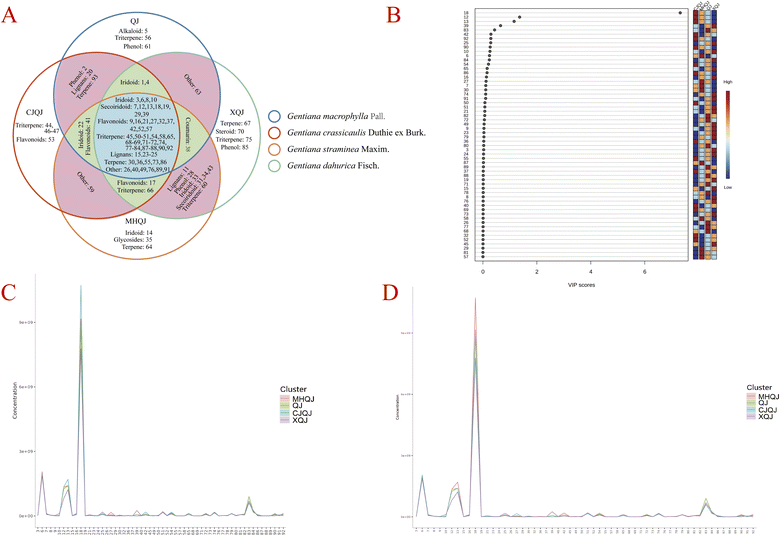 | ||
| Fig. 3 The analysis of LC-MS data (A: the common peaks in Gentianae Macrophyllae Radix; B: the VIP score of 58 common peaks; C: K-means clustering; D: SOM). | ||
3.2. HPLC analysis
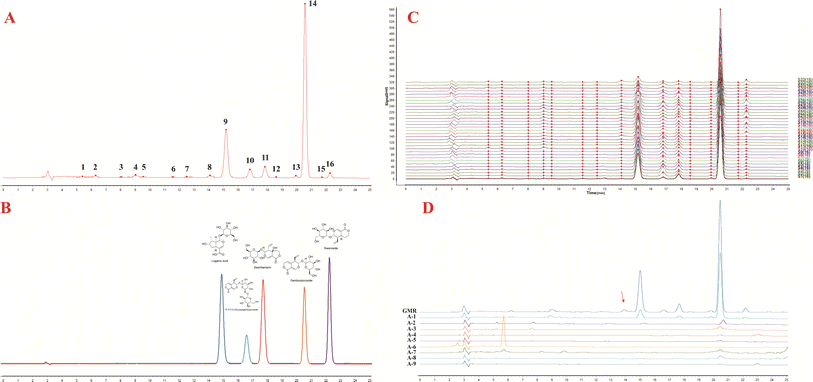
After optimization, methanol-0.04% formic acid water was finally selected as the elution system (Fig. S3†) for HPLC analysis. Five of the peaks were identified to belong to loganic acid, 6′-O-β-D-glucosylgentiopicroside, swertiamarine, gentiopicroside, and sweroside by comparison with the reference standards (Fig. 4A and B). The precision, stability, and repeatability results are shown in Tables 3 and S1.† The durability result is shown in Table S2.† All the RSD values of the five compounds were less than 3.0%, which indicated this developed method was sensitive, precise, and robust.| Analyst | Precision (n = 6) | Repeatability (n = 6) | Stability (n = 8) | |||
|---|---|---|---|---|---|---|
| RRT | RRA | RRT | RRA | RRT | RRA | |
| RSD (%) | RSD (%) | RSD (%) | RSD (%) | RSD (%) | RSD (%) | |
| Loganic acid | 0.10 | 0.09 | 0.28 | 1.82 | 0.75 | 1.63 |
| 6′-O-β-D-Glucosylgentiopicroside | 0.10 | 0.31 | 0.30 | 2.23 | 0.66 | 1.68 |
| Swertiamarine | 0.07 | 0.30 | 0.26 | 2.12 | 0.54 | 1.59 |
| Gentiopicroside | 0.04 | 0.11 | 0.18 | 2.12 | 0.31 | 1.73 |
| Sweroside | 0.03 | 0.30 | 0.14 | 2.19 | 0.23 | 1.71 |
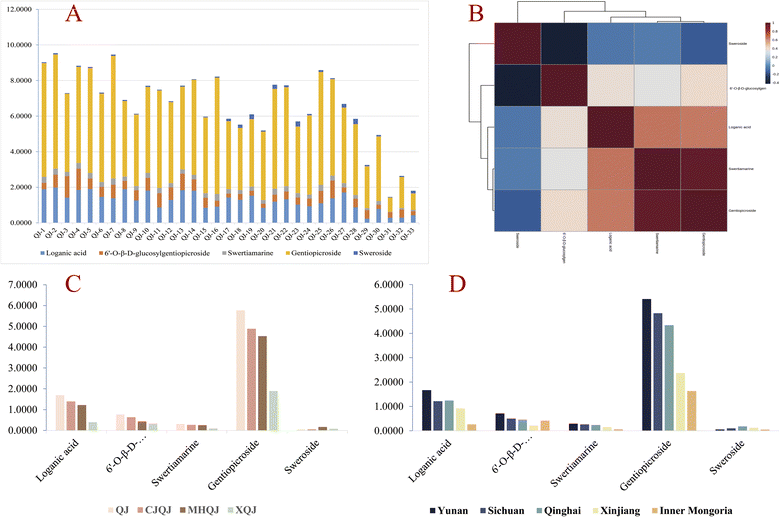
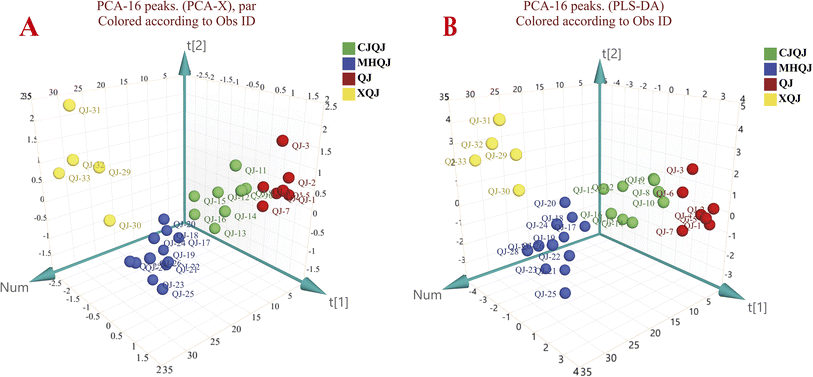
All the collected samples were analyzed according to the HPLC method. Thereafter, the data was used to establish the fingerprints. As a result, 16 common peaks were observed in QJ, CJQJ, MHQJ, and XQJ (Fig. 4C). Loganic acid, 6′-O-β-D-glucosylgentiopicroside, swertiamarine, gentiopicroside, and sweroside were relatively abundant in Gentianae Macrophyllae Radix compared with it adulterates, which are the key ingredients for the authentication (Fig. 4D). The content of active compounds is the linchpin for distinguishing Gentianae Macrophyllae Radix of the four species. Gentiopicroside had a significant relationship with swertiamarine (p < 0.01), and sweroside had a significant negative relationship with 6′-O-β-D-glucosylgentiopicroside (p < 0.05) (Table S3† and Fig. 5B). The average content of each component in XQJ is far lower than QJ, CJQJ, and MHQJ, and the content of sweroside in MHQJ is the highest (Fig. 5C). In addition, the five active components of Gentianae Macrophyllae Radix in Yunnan, Sichuan, and Qinghai all show high content, and the content of sweroside in Gentianae Macrophyllae Radix of Qinghai is the highest (Fig. 5D). In order to further confirm the findings, the peak area data of 16 common compounds was used for PCA and PLS-DA, which could also distinguish QJ, CJQJ, MHQJ, and XQJ (Fig. 6).
SSDMC method based on the optimized HPLC was developed for their simultaneous detection of the compounds. The calibration curves, linear ranges, LOD, and LOQ of the analytes are shown in Table 4. The average relative response factors (F) for loganic acid, 6′-O-β-D-glucosylgentiopicroside, swertiamarine, and sweroside were 0.78, 1.97, 0.67, and 0.81, with an RSD of 1.31%, 0.89%, 0.08%, and 1.76%, respectively (Table S4†). Additionally, the recovery of loganic acid, 6′-O-β-D-glucosylgentiopicroside, swertiamarine, gentiopicroside, and sweroside was 102.96–104.52%, 100.74–103.44%, 100.26–105.93%, 98.07–101.45%, and 99.62–102.83%, respectively (Table 5). Combined with the results from method validation in the HPLC fingerprint study, the described SSDMC approach proved to be robust, sensitive, precise, and accurate. As shown in Table 6, the results calculated by the SSDMC method showed no significant difference from the calibration curve method.
| Analyst | RT (min) | Calibration curve | R2 | Linear range (mg mL−1) | LOD (mg mL−1) | LOQ (mg mL−1) |
|---|---|---|---|---|---|---|
| Loganic acid | 14.91 | y = 7472.7x + 1.9765 | 0.9996 | 0.0065625–0.21 | 1.09 × 10−5 | 3.62 × 10−5 |
| 6′-O-β-D-Glucosylgentiopicroside | 16.51 | y = 3007.1x − 0.5279 | 0.9995 | 0.004688–0.15 | 2.81 × 10−5 | 9.36 × 10−5 |
| Swertiamarine | 17.56 | y = 8048.6x + 5.1576 | 0.9994 | 0.001894–0.0606 | 1.00 × 10−5 | 3.34 × 10−5 |
| Gentiopicroside | 20.34 | y = 5858.9x + 3.1785 | 0.9996 | 0.025313–0.81 | 1.14 × 10−5 | 3.79 × 10−5 |
| Sweroside | 22.06 | y = 7385.1x − 1.0209 | 0.9994 | 0.001656–0.053 | 8.35 × 10−6 | 2.78 × 10−5 |
| Analytes | Level | Original (mg) | Spiked (mg) | Found (mg) | Average (%) | RSD (%) |
|---|---|---|---|---|---|---|
| Loganic acid | High | 2.8498 | 2.1 | 3.684 | 104.52 | 1.157 |
| Medium | 1.575 | 3.104 | 103.47 | 1.846 | ||
| Low | 1.05 | 2.548 | 102.96 | 1.211 | ||
| 6′-O-β-D-Glucosylgentiopicroside | High | 0.4512 | 0.46 | 0.695 | 101.44 | 1.368 |
| Medium | 0.23 | 0.471 | 103.44 | 1.926 | ||
| Low | 0.15 | 0.378 | 100.74 | 1.395 | ||
| Swertiamarine | High | 0.4905 | 0.505 | 0.752 | 100.26 | 1.199 |
| Medium | 0.2525 | 0.508 | 102.09 | 1.464 | ||
| Low | 0.101 | 0.367 | 105.93 | 1.564 | ||
| Gentiopicroside | High | 7.6598 | 4.536 | 8.487 | 101.45 | 1.169 |
| Medium | 3.8475 | 7.754 | 101.00 | 1.822 | ||
| Low | 1.9278 | 5.647 | 98.07 | 1.280 | ||
| Sweroside | High | 0.2691 | 0.212 | 0.356 | 102.83 | 1.191 |
| Medium | 0.1378 | 0.271 | 99.62 | 1.853 | ||
| Low | 0.0636 | 0.203 | 102.63 | 1.616 |
| No. | Loganic acid (mg mL−1) | 6′-O-β-D-Glucosylgentiopicroside (mg mL−1) | Swertiamarine (mg mL−1) | Gentiopicroside (mg mL−1) | Sweroside (mg mL−1) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Calibration curve | SSDMC | Calibration curve | SSDMC | Calibration curve | SSDMC | Calibration curve | Calibration curve | SSDMC | |
| QJ-1 | 0.1893 | 0.1872 | 0.0355 | 0.0354 | 0.035 | 0.032 | 0.6393 | 0.0045 | 0.0044 |
| QJ-2 | 0.1985 | 0.1963 | 0.0721 | 0.0722 | 0.034 | 0.032 | 0.6421 | 0.0068 | 0.0067 |
| QJ-3 | 0.1421 | 0.1406 | 0.1208 | 0.1210 | 0.023 | 0.022 | 0.4400 | 0.0030 | 0.0029 |
| QJ-4 | 0.1846 | 0.1826 | 0.1196 | 0.1198 | 0.032 | 0.030 | 0.5406 | 0.0061 | 0.0060 |
| QJ-5 | 0.1901 | 0.1880 | 0.0589 | 0.0590 | 0.031 | 0.029 | 0.5897 | 0.0064 | 0.0063 |
| QJ-6 | 0.1459 | 0.1444 | 0.0565 | 0.0565 | 0.027 | 0.025 | 0.4982 | 0.0052 | 0.0051 |
| QJ-7 | 0.1385 | 0.1370 | 0.0759 | 0.0760 | 0.035 | 0.032 | 0.6899 | 0.0073 | 0.0073 |
| QJ-8 | 0.1891 | 0.1870 | 0.0476 | 0.0476 | 0.024 | 0.023 | 0.4250 | 0.0063 | 0.0062 |
| QJ-9 | 0.1259 | 0.1246 | 0.0578 | 0.0579 | 0.023 | 0.022 | 0.4030 | 0.0040 | 0.0039 |
| QJ-10 | 0.1814 | 0.1794 | 0.0718 | 0.0719 | 0.028 | 0.026 | 0.4835 | 0.0065 | 0.0065 |
| QJ-11 | 0.0870 | 0.0861 | 0.0800 | 0.0801 | 0.030 | 0.028 | 0.5489 | 0.0038 | 0.0037 |
| QJ-12 | 0.1284 | 0.1271 | 0.0711 | 0.0712 | 0.023 | 0.022 | 0.4566 | 0.0046 | 0.0045 |
| QJ-13 | 0.1840 | 0.1819 | 0.0928 | 0.0930 | 0.024 | 0.023 | 0.4654 | 0.0056 | 0.0055 |
| QJ-14 | 0.1810 | 0.1790 | 0.0638 | 0.0638 | 0.025 | 0.024 | 0.5344 | 0.0035 | 0.0034 |
| QJ-15 | 0.0849 | 0.0841 | 0.0558 | 0.0559 | 0.025 | 0.024 | 0.4274 | 0.0042 | 0.0041 |
| QJ-16 | 0.0911 | 0.0902 | 0.0330 | 0.0329 | 0.038 | 0.035 | 0.6545 | 0.0058 | 0.0057 |
| QJ-17 | 0.1425 | 0.1410 | 0.0226 | 0.0225 | 0.025 | 0.023 | 0.3830 | 0.0135 | 0.0135 |
| QJ-18 | 0.1297 | 0.1284 | 0.0332 | 0.0331 | 0.021 | 0.020 | 0.3502 | 0.0179 | 0.0180 |
| QJ-19 | 0.1531 | 0.1514 | 0.0275 | 0.0275 | 0.024 | 0.022 | 0.3795 | 0.0267 | 0.0269 |
| QJ-20 | 0.0835 | 0.0827 | 0.0242 | 0.0241 | 0.020 | 0.019 | 0.3851 | 0.0063 | 0.0063 |
| QJ-21 | 0.1192 | 0.1180 | 0.0396 | 0.0396 | 0.033 | 0.031 | 0.5625 | 0.0231 | 0.0233 |
| QJ-22 | 0.1325 | 0.1312 | 0.0441 | 0.0441 | 0.029 | 0.027 | 0.5574 | 0.0111 | 0.0111 |
| QJ-23 | 0.1026 | 0.1016 | 0.0397 | 0.0397 | 0.024 | 0.022 | 0.3759 | 0.0287 | 0.0290 |
| QJ-24 | 0.0933 | 0.0924 | 0.0430 | 0.0430 | 0.023 | 0.021 | 0.4458 | 0.0071 | 0.0071 |
| QJ-25 | 0.1103 | 0.1091 | 0.0694 | 0.0695 | 0.034 | 0.032 | 0.6359 | 0.0104 | 0.0104 |
| QJ-26 | 0.1383 | 0.1368 | 0.0999 | 0.1001 | 0.028 | 0.026 | 0.5438 | 0.0054 | 0.0053 |
| QJ-27 | 0.1717 | 0.1698 | 0.0278 | 0.0277 | 0.024 | 0.022 | 0.4260 | 0.0200 | 0.0201 |
| QJ-28 | 0.0875 | 0.0867 | 0.0484 | 0.0484 | 0.020 | 0.019 | 0.3991 | 0.0295 | 0.0298 |
| QJ-29 | 0.0230 | 0.0230 | 0.0480 | 0.0480 | 0.011 | 0.011 | 0.2360 | 0.0073 | 0.0073 |
| QJ-30 | 0.0783 | 0.0775 | 0.0240 | 0.0239 | 0.020 | 0.019 | 0.3645 | 0.0081 | 0.0080 |
| QJ-31 | 0.0282 | 0.0281 | 0.0320 | 0.0320 | 0.001 | 0.002 | 0.0810 | 0.0035 | 0.0034 |
| QJ-32 | 0.0288 | 0.0287 | 0.0463 | 0.0463 | 0.008 | 0.008 | 0.1744 | 0.0061 | 0.0060 |
| QJ-33 | 0.0427 | 0.0424 | 0.0232 | 0.0232 | 0.007 | 0.007 | 0.0926 | 0.0142 | 0.0143 |
4. Conclusion
In this paper, a LC-Orbitrap-MS method was established to analyze the common or characteristic components of Gentianae Macrophyllae Radix originated from four species, which led to the identification of 93 components, including 58 common ones in the four species. It also proved that Gentianae Macrophyllae Radix mainly contains terpenes (iridoids and triterpenes), flavonoids, alkaloids, lignans, and sterols. The terpenes (with retention time between 10 to 20 min) were the characteristic compounds to identify Gentianae Macrophyllae Radix and its adulterants. The established HPLC fingerprint could also distinguish this medicine and its adulterants depended on the five critical compounds of loganic acid, 6′-O-β-D-glucosylgentiopicroside, swertiamarine, gentiopicroside, and sweroside. Another compound (peak 8) is also one of the specific components of Gentianae Macrophyllae Radix, but it has not been identified (Fig. 4D). In addition, HPLC combined with PCA and PLS-DA could identify QJ, CJQJ, MHQJ, and XQJ based on the content of 16 common peaks. The SSDMC method is also powerful for the determination of five main compounds. It is very important to select the authentic and high-quality medicinal materials because the level of the compounds is directly related to the clinical efficacy.6,7,14,15,24,25In conclusion, the developed LC-Orbitrap-MS and HPLC strategy is of great importance for quality control and authentication of Gentianae Macrophyllae Radix. The further study is needed for the comparison of pharmacological effects of Gentianae Macrophyllae Radix of the four species and the impact of geographical and ecological environment on its chemicals.
Conflicts of interest
All the authors have declared no conflict of interest.Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFC1707903), Provincial Natural Science Foundation of Hunan (2022JJ40318), Scientific Research Fund of Hunan University of Chinese Medicine (2021XJJJ006), Changsha Municipal Science and Technology Bureau (kq1901095), and Innovation and Entrepreneurship Training Plan for College Students of Hunan University of Chinese Medicine.References
- Z. Li, Y. Du, Y. Yuan, X. Zhang, Z. Wang and X. Tian, Chin. Med., 2020, 15, 16 CrossRef CAS PubMed.
- X. Y. Cao and Z. Z. Wang, Phytochem. Anal., 2010, 21, 348–354 CrossRef CAS PubMed.
- C. P. Committee, Pharmacopoeia of the People's Republic of China, China Medical Science Press, Beijing, 2020 Search PubMed.
- S. B. Hou, X. Wang, R. Huang, H. Liu, H. M. Hu, W. Y. Hu, S. T. Lv, H. Zhao and G. Chen, Fitoterapia, 2020, 141, 104476 CrossRef CAS PubMed.
- J. Liang, Y. Ito, X. Zhang, J. He and W. Sun, J. Sep. Sci., 2013, 36, 3934–3940 CrossRef CAS PubMed.
- D. Zhou, D. Lv, H. Zhang, T. Cheng, H. Wang, P. Lin, S. Shi, S. Chen and J. Shen, J. Ethnopharmacol., 2021, 280, 114068 CrossRef CAS PubMed.
- M. Zhang, D. Jiang, M. Yang, T. Ma, F. Ding, M. Hao, Y. Chen, C. Zhang, X. Zhang and M. Li, Front. Plant Sci., 2021, 12, 706822 CrossRef PubMed.
- X. Zhang, G. Zhan, M. Jin, H. Zhang, J. Dang, Y. Zhang, Z. Guo and Y. Ito, Phytomedicine, 2018, 46, 142–163 CrossRef CAS PubMed.
- N. Jia, H. Ma, T. Zhang, L. Wang, J. Cui, Y. Zha, Y. Ding and J. Wang, Int. Immunopharmacol., 2022, 108, 108854 CrossRef CAS PubMed.
- E. Park, C. G. Lee, E. Lim, S. Hwang, S. H. Yun, J. Kim, H. Jeong, Y. Yong, S. H. Yun, C. W. Choi, H. S. Jin and S. Y. Jeong, Int. J. Mol. Sci., 2020, 22(1), 233 CrossRef PubMed.
- M. Kalinowska, J. Zimowski, M. Bucholc and Z. A. Wojciechowski, Phytochemistry, 1989, 28, 2931–2935 CrossRef CAS.
- Y. Xiang, W. Haixia, L. Zenggen and T. Yanduo, Nat. Prod. Res., 2019, 33, 598–601 CrossRef PubMed.
- Q. C. Wu, X. Y. Tang, Z. Q. Dai, Y. Dai, H. H. Xiao and X. S. Yao, Phytomedicine, 2020, 68, 153146 CrossRef CAS PubMed.
- R. Zeng, H. Hu, G. Ren, H. Liu, Y. Qu, W. Hua and Z. Wang, J. Chromatogr. Sci., 2015, 53, 1274–1279 CAS.
- N. Jia, Y. Li, Y. Wu, M. Xi, G. Hur, X. Zhang, J. Cui, W. Sun and A. Wen, J. Ethnopharmacol., 2012, 144, 638–645 CrossRef CAS PubMed.
- Y. M. Wang, M. Xu, D. Wang, C. R. Yang, Y. Zeng and Y. J. Zhang, J. Ethnopharmacol., 2013, 147, 341–348 CrossRef CAS PubMed.
- Q. Z. Hou, D. W. Chen, Y. P. Wang, N. Ehmet, J. Ma and K. Sun, BMC Microbiol., 2022, 22, 90 CrossRef CAS PubMed.
- S. Saito-Shida, T. Hamasaka, S. Nemoto and H. Akiyama, Food Chem., 2018, 256, 140–148 CrossRef CAS PubMed.
- J. J. Hou, W. Y. Wu, J. Da, S. Yao, H. L. Long, Z. Yang, L. Y. Cai, M. Yang, X. Liu, B. H. Jiang and D. A. Guo, J. Chromatogr. A, 2011, 1218, 5618–5627 CrossRef CAS PubMed.
- M. Lyu, Y. Liu, Y. Qiu, S. Yang, H. Yuan and W. Wang, J. AOAC Int., 2021, 104, 1652–1660 CrossRef PubMed.
- C. Wu, Q. Guan, S. Wang and Y. Rong, Pharmacogn. Mag., 2017, 13, S84–S89 CrossRef PubMed.
- J. Wu, Z. Zhao, L. Wu and Z. Wang, Biomed. Chromatogr., 2016, 30, 2061–2066 CrossRef CAS PubMed.
- H. Liu, H. Zhao, R. Huang, A. S. Ali, X. Wang, S. Meng and G. Chen, Biomed. Chromatogr., 2021, 35, e5046 CAS.
- Y. Y. Lu, Y. M. Yang, X. H. Ma, X. B. Zhang, S. D. Zhu and L. Jin, China J. Chin. Mater. Med., 2016, 41, 3176–3180 Search PubMed.
- Y. Y. Lu, X. B. Zhang, Y. M. Yang, M. X. Hui, T. T. Zhu, X. H. Yu and L. Jin, China J. Chin. Mater. Med., 2016, 41, 3132–3138 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra07591a |
| ‡ Two authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |