Open Access Article
Open Access ArticleDe novo creation of fluorescent molecules via adversarial generative modeling
Zheng Tana,
Yan Lib,
Xin Wuc,
Ziying Zhangd,
Weimei Shi *b,
Shiqing Yangb and
Wanli Zhanga
*b,
Shiqing Yangb and
Wanli Zhanga
aState Key Laboratory of Electronic Thin Films and Integrated Devices, University of Electronic Science and Technology of China, Chengdu, 610054, P. R. China
bChengdu Polytechnic, 83 Tianyi Street, Chengdu, 610000, P. R. China. E-mail: shiweimei@cdp.edu.cn
cXiyuan Quantitative Technology, 388 Yizhou Road, Chengdu, 610000, P. R. China
dGuangzhou Yinfo Information Technology, 2 Ruyi Road, Panyu District, Guangzhou, 511431, P. R. China
First published on 4th January 2023
Abstract
The development of AI for fluorescent materials design is technologically demanding due to the issue of accurately forecasting fluorescent properties. Besides the huge efforts made in predicting the photoluminescent properties of organic dyes in terms of machine learning techniques, this article aims to introduce an adversarial generation paradigm for the rational design of fluorescent molecules. Molecular SMILES is employed as the input of a GRU based autoencoder, where the encoding and decoding of the string information are processed. A generative adversarial network is applied on the latent space with a generator to generate samples to mimic the latent space, and a discriminator to distinguish samples from the latent space. It is found that the excited state property distributions of generated molecules fully match those of the original samples, with the molecular synthesizability being accessible as well. Further screening of the generated samples delivers a remarkable luminescence efficiency of molecules epitomized by the significant oscillator strength and charge transfer characteristics, demonstrating the great potential of the adversarial model in enriching the fluorescent library.
Introduction
Organic fluorescent materials, especially small-molecule organic dyes, have been extensively used across several research fields, including in sensors,1–4 organic light-emitting diodes (OLED)5–8 and bioimaging.9–11 It is thus significant to have a rational design framework so that the molecules can possess different functionality to meet various requirements. For instance, a significant luminescence quantum yield with controllable absorption/emission wavelength is generally needed for OLED molecules,12 while a large Stokes shift and high photo-stability of fluorescent dyes are a prerequisite for reliable bioimaging.13 Up to now, most of the fluorescent design work is based on experimental experience in a trial-and-error manner, which is unlikely to give rise to the development of transformative new molecules due to the complexity of chemical space. On the other hand, owing to the complicated transfer mechanism happening in the excited state hypersurface during the photon absorption/emission process, the fluorescent efficiency can be hardly correlated with the molecular structure,14 posing challenges for the structural design work.The advent of machine learning brings about a variety of opportunities for the exploration of unknown chemical space and predictions of molecular properties. In particular, numerous efforts have been made to investigate the excited state properties of organic semiconductors,15–21 and relevant de novo design frameworks14,22 have also been proposed to develop new molecules with desired functionalities. For fluorescent materials, machine learning has given successful predictions for the experimental quantum yields and emission energies,21,23 by using empirically designed chemical descriptors. The forecasting of oscillator strength and the corresponding fluorescent rate (specifically at the theoretical level) is however less satisfactory,19 where the machine learning techniques have to be used with caution. It is interesting to see an inverse design case for fluorescent molecules development by employing a reinforcement-like paradigm coupled with ‘in situ’ quantum chemical calculations.14 The generated novel compounds possess the anticipated emission wavelengths and significant Stokes shifts which are subsequently verified experimentally.
Adversarial generation, known as a powerful generative model in machine learning, has attracted great attentions in physical and chemical fields for the innovative design of drug molecules and optoelectronic materials.24 The family of adversarial models basically includes adversarial autoencoder (AAE), generative adversarial network (GAN), reinforcement learning augmented GAN, etc., where the model framework generally consists of a neural network generator and discriminator for the sample generation and discrimination respectively.
In order to introduce an alternative inverse design paradigm for fluorescent molecules, this article employs an autoencoder based generative adversarial network (AGAN) for the generation of molecular SMILES25 (simplified molecular input line entry system). The architecture is inspired by the LatentGAN,26 in which a heteroencoder is first pre-trained before the training of the GAN. AGAN performs the training in a cohesive manner by optimizing the encoder/decoder and GAN simultaneously. The SMILES strings are fed into the encoder to be transformed to a low-dimensional latent space, which is subsequently decoded back to the character sequences with the reconstruction loss being minimized. A generator is trained to produce a virtual latent space from a Gaussian prior distribution to mimic the real latent samples, while a discriminator is employed to distinguish the real and virtual latent. High throughput quantum chemical calculations are carried out to label the excited state properties of generated molecules. Further fine screening of the generated samples gives rise to molecules with significant luminescence efficiency epitomized by the large oscillator strength and remarkable charge transfer characteristics, indicating the effectiveness of the AGAN model in exploring the fluorescent search space.
The paper is organized as follows. In the Methodology section, the dataset, details of quantum chemical calculations and the model architecture are described. It is then followed by the Results and discussion section which presents the main findings for our fluorescent molecules generation. Finally in section, a Conclusion is drawn.
Methodology
Fluorescent dyes database
The original fluorescent samples are extracted from the database used in Ju et al.,23 where experimental emission wavelength and quantum yield are fitted with machine learning models. 2923 distinct SMILES strings are obtained and the singlet excited state properties are calculated for the labels of molecules. Note that the excited state properties for the 2923 original molecules are acquired in terms of high throughput calculations at the theoretical level, with no experimental data being involved here except for the experimentally synthesized molecular entries (represented as SMILES). All original SMILES are input into the AGAN model for the generative training.Quantum chemical calculations
High throughput time-dependent density functional theory (TDDFT) calculations are implemented for both the original and generated molecules to obtain the excited state properties. The fluorescent compounds are first processed with the initial geometry optimization using the MMFF94 force field27 in RDKit. The ground state geometries are then optimized using the PBE0 hybrid functional (which is shown to be the most realistic functional in predicting the emissive properties for fluorescent dyes23) with the def2SVP basis set. TDDFT calculations are then implemented at S0 geometries at the PBE0/def2SVP level, where the first three singlet transition energies and the corresponding oscillator strengths are exported. All quantum chemical calculations are performed in gas phase using Gaussian 16,28 with no solvent effect being considered for the sake of simplicity.During the fine screening stage, the selected fluorescent candidates are optimized at the S1 state with TDDFT/PBE0/def2TZVP, with the transition energies and oscillator strengths being extracted for the evaluation of emissive properties.
Autoencoder based generative adversarial network
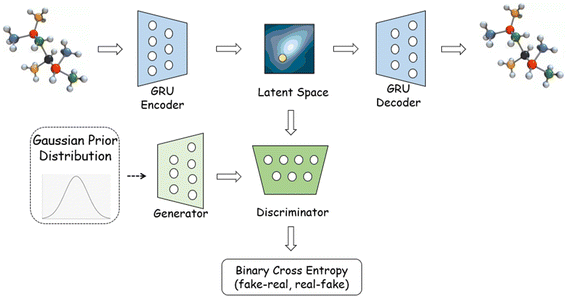
Conventional GAN cannot be used to treat SMILES or other molecular representations due to the nondifferentiability of the input data. Several solutions have been proposed to tackle with this challenge. SeqGAN29 is designed by treating the generator to create actions for sequence generations, while the discriminator is used to give feedbacks. ORGAN30 is built on SeqGAN by incorporating an RNN-like generator and a CNN-like discriminator to generate SMILES strings with a pre-set objective. MolGAN31 is developed to directly process the topological graph, with the model architecture consisting of a generator, discriminator and reward network to generate molecular topology with nearly 100% validity. The current sequence-based and graph-based GAN frameworks, however, face the issue of generation quality where the produced compounds have to satisfy the criteria of validity, uniqueness and novelty. It is known that the ORGAN can have a significant non-validity ratio during the generation,32 while MolGAN can suffer from a very low molecular uniqueness. To the best of our knowledge, no model can yet deliver an ‘ideal’ generation.In this research, we adopt a SMILES-based GAN architecture for the fluorescent molecules' generation. The SMILES strings are first transformed into a vector-based latent space via a GRU encoder (a 1-layer bidirectional Gated Recurrent Unit with 256 hidden dimensions is employed here), which is then transformed back (by using a 3-layer GRU decoder of 512 hidden dimensions) to character sequences for the reconstruction of SMILES. A one-hot embedding layer is applied before the encoder, and the teacher forcing33 is employed in the decoder to lower the reconstruction loss. As exhibited in Fig. 1, the latent vectors are viewed as the real input for the discriminator. The generator serves to produce virtual latent samples from the Gaussian noise to replicate the real latent, while the discriminator tries to distinguish the two. The training process oscillates between the autoencoder and GAN until the virtual and real latent spaces are indistinguishable. The latent space is of dimension 128. The discriminator is a two-layer neural network with hidden dimensions of 512 and 256, while the generator is a two-layer network as well with hidden dimensions of 512 and 512.
To be in details for the SMILES transformation, the character string is first converted to a one-hot layer with a length set to be the maximal number of characters within the SMILES training data. The 0/1 digital layer is then linearly transformed into an embedded matrix with a dimension of 64, which is subsequently fed into a bidirectional GRU machine. Both forward and backward encodings in GRU are performed to account for the foregoing and following information during the character vectorization. The final hidden state is eventually converted to the latent space via a linear transformation. For the decoding process, we perform an almost reverse procedure as conducted in the encoder, to transform the latent vector back into a multiclass probability matrix. The reconstruction is treated as a classification issue with cross entropy being used as the loss function.
We employ the binary cross entropy (BCE) to classify the real (latent space) and fake (virtual latent space) data in AGAN. Unlike the conventional GAN, we follow a relativistic approach34 to differentiate samples by defining the loss function as,
| G_loss = −log[sigmoid(C(xf) − C(xr))] | (1) |
| D_loss = (discr_real_loss + discr_fake_loss)/2 | (2) |
| Discr_real_loss = −log[sigmoid(C(xr) − C(xf))] | (3) |
| Discr_fake_loss = −log[1 − sigmoid(C(xf) − C(xr))] | (4) |
The training is conducted by optimizing the reconstruction loss, generator loss and discriminator loss simultaneously after a threshold epoch (set as 20 here), while before that only the autoencoder is trained. 150 training epochs are set, with the learning rate being 0.001 and batch size being 64. For the generation process, the items are randomly sampled from the Gaussian noise and then fed into the generator network, from which the output goes through the GRU decoder to produce the SMILES.
Results and discussion
Generation profiles
Fig. 2 exhibits the loss function convergences of AGAN. It is observed that recon_loss can smoothly converge to 0, demonstrating that an accurate enough mapping between the character string and the latent space is established. The generator and discriminator losses start to oscillate after the pre-set threshold epoch and converge to a level of 5 and 0.2 respectively. The finding is slightly different from the convergence profile in our previous adversarial training24 where the classification loss can approach the theoretical value of BCE for indistinguishable samples. The phenomenon can be attributed to the relativistic approach applied on the discriminator which can possibly lead to a state-of-the-art during the convergence.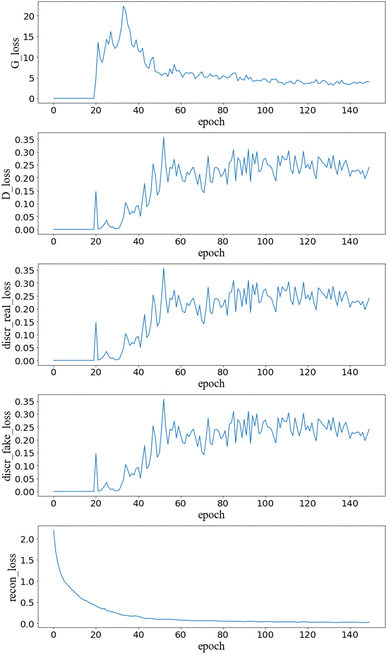 | ||
| Fig. 2 Convergences of loss functions during the training of AGAN. G_loss, D_loss, discr_real_loss, discr_fake_loss are defined in eqn (1)–(4), recon_loss denotes the cross entropy reconstruction loss for the autoencoder. | ||
The performance of generations is measured with the molecular validity, novelty, uniqueness and overall efficiency, as defined in our previous work.24 To briefly summarize, validity represents the fraction of molecules within the generated samples that can pass the RDKit's sanitization check so that the atomic valency and the consistency of bonds in aromatic rings can be maintained. Novelty is the fraction of the generated molecules that are not present in the training set. Uniqueness gives rise to the percentage of the generated samples that are unique. We compute the overall generation efficiency for molecules that simultaneously fulfill the above three criteria, given different generation sizes.
As shown in Table 1, given the small training set (which is 2923 in this case), generation sizes of 1 K to 20 K are considered. The uniqueness is found to be comparable with the profiles in Polykovskiy et al.37 (approaching 1.0 for LatentGAN at 1 K generation size) and Tan et al.24 (0.99 for AAE at 1 K generation size). For the novelty ratio, the current generation result is even better than that in Tan et al.24 (where around 0.65 novelty is reported), possibly owing to the addition of the generator network in AGAN which can potentially enhance the probability of creating new molecules not appearing in the original dataset. It is observed that, however, the validity is relatively low. The underlying reason can be tentatively ascribed to the small size of the training data, where there are not enough samples to inform the autoencoder for how to reconstruct a valid SMILES string. The overall generation efficiency is located between 13% and 14%, with 2610 molecules being efficiently produced in case of the generation size of 20 K. We would state that the current generation efficiency is fully acceptable based on the comparability of the efficiently generated size and the input size.
| Generation size | Validity | Novelty | Uniqueness | Overall efficiency |
|---|---|---|---|---|
| 1 K | 0.2870 | 0.8600 | 0.9730 | 0.1470 |
| 5 K | 0.2840 | 0.8632 | 0.9178 | 0.1326 |
| 10 K | 0.2943 | 0.8618 | 0.8928 | 0.1371 |
| 20 K | 0.2922 | 0.8677 | 0.8678 | 0.1305 |
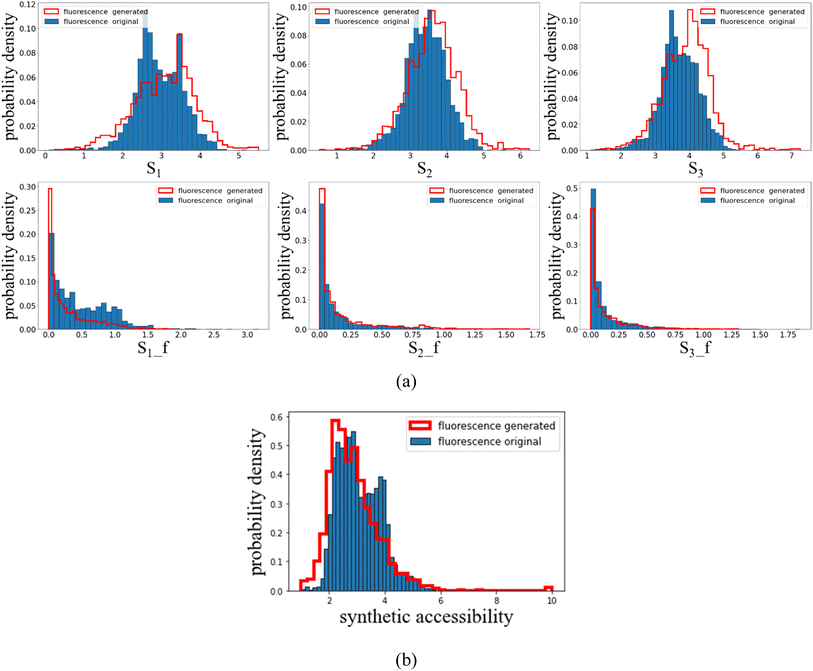
High throughput TDDFT calculations are performed to obtain the singlet transition energies and oscillator strengths for both the generated and original fluorescent dyes, the distributions of which are displayed in Fig. 3(a). It is found that the property distributions of the generation space almost reproduce those of the original space, indicating the effectiveness of the AGAN model. The first excitation energy is in the range of 1.5–4.5 eV (with the generated distribution being slightly broader), covering the full visible spectrum for the light absorption. Both the generated and original oscillator strengths tend to have a distribution near 0, implying the difficulty to find a molecule with large luminescence efficiency.
It is interesting to note that, the fine structures of S1 distributions differ between the original and generated samples, while the distributions of S2 and S3 for the generated dyes seem to have a plausible ‘blue shift’ towards higher energies. The observation is in contrast to our previous work24 where perfect matches of distributions are found when thermally activated delayed fluorescence molecules are generated. Two possible reasons might be announced to address the inconsistencies. One is for the insufficient data within the histogram which can lead to the irregularity of distribution curves. In addition, the diversity in the training set (where samples are known to be experimentally synthesized and collected from different literature), can be another cause for the distribution mismatch, while in Tan et al.24 both original and generated sets are of the same molecular type. More details regarding the diversity can be seen in the following analyses.
The synthetic accessibility (SA)38 is also examined which is measured in terms of the fragment contributions and the molecular complexity. The SA score is simply estimated as the difference between the fragment score and the complexity penalty, where the fragment score can be understood as the frequency of the current molecular fragments appearing in the empirical chemical fragment database, and the complexity penalty characterizes the presence of complex structural features that may lead to overcounts of molecular substructures. Note that the SA score spans from 1 to 10 with higher values denoting enhanced difficulties to synthesize a molecule. A major peak of SA around 2–3 is found, indicating that these fluorescent dyes are easily synthesizable. Since the original molecules are recorded to be synthesized experimentally,23 the generated samples are supposed to have a wide experimental accessibility.
Regarding the diversity of the original and generation sets, internal diversities (IntDiv1 and IntDiv2, as defined to be one minus the average Tanimoto similarity within a molecular set,24 with higher values denoting higher diversity) are computed. IntDiv1 and IntDiv2 (the indices represent the order of the diversity) for the original samples are 0.8845 and 0.8618, while those for the generated samples are 0.8854 and 0.8696. The analysis points towards an enhanced diversity compared to the values in Tan et al.,24 where a level around 0.82 is reported. The discrepancy makes sense as the molecular entries in the current training set are collected from different experimental literature, while only a focused library of donor–acceptor molecules is used in our previous work.
Fig. 4 gives rise to a two-dimensional principal components analysis (PCA) for the latent space, to further investigate the diversity of the original and generation sets. The original and generated SMILES are separately fed into the autoencoder where the models are trained with the reconstruction loss being minimized respectively. The converged autoencoder (after 150 epochs) can therefore deliver the latent space as robust numerical representations for character strings. A dimension reduction via PCA is then performed to extract the main features of the chemical space. As shown in Fig. 4, the reduced original space exhibits a certain degree of aggregation which may correspond to the similar topologies in the experimental synthesis. A broader coverage of the reduced generation space is found compared to the original one, implying a possible higher diversity of the generated samples. It is interesting that the generated space can fully capture the major characteristics of the original space including the structural aggregation, demonstrating the reliability of the adversarial algorithm in producing mirrored spaces. The detected augmentation of diversity in the generation set might partially explain the energy distribution mismatch issue mentioned above.
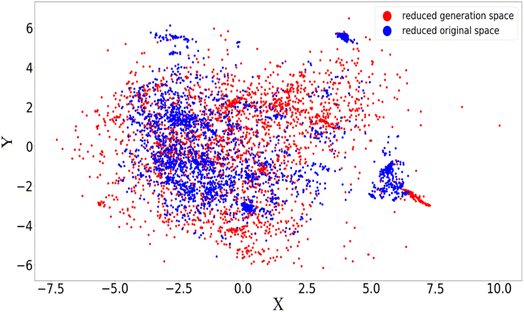 | ||
| Fig. 4 Two-dimensional PCA analysis of latent space for the autoencoder used in AGAN. The X and Y axes are the principal components selected from the PCA analysis. | ||
Fine screening
In order to explore the emissive properties of the generated molecules, we select 10 dyes with the largest absorptive oscillator strengths from the generation space to perform the excited state optimization. The S1 geometry optimization is conducted at the TDDFT/PBE0/def2TZVP level (where the PBE0 is justified to be the most accurate functional in describing the fluorescent properties according to Ju et al.23), and the emissive energies and oscillator strengths are displayed in Table 2. Significant emissive oscillator strengths are observed which is comparable with the absorptive ones, implying a remarkable fluorescent rate for these new molecules. The emission energies lie in the visible region and being slightly smaller than the absorption energies, exhibiting moderate Stokes shifts as calculated from 5 nm to 33 nm. The molecular structures of the fine screened dyes are shown in Fig. 5, where π-conjugated, polycyclic, benzonitrile-like systems are observed, as usually present in the fluorescent compounds.39| Id | Smiles | Emission | Absorption | ||
|---|---|---|---|---|---|
| E_S1 (eV) | E_S1_f | A_S1 (eV) | A_S1_f | ||
| mol_1 | CCN1/C(C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C3 C(C3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(N(C4 C(N(C4![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C4)C5 C4)C5![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C5)C C5)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3)S2) C3)S2)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NC6 NC6![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1C7 C1C7![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C8 C(C8![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C6C C6C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N8)N N8)N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C7 C7 |
2.969 | 1.4451 | 3.0344 | 1.4718 |
| mol_2 | FC(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1) C1)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1C C1C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC2 CC2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(NC(C3 C(NC(C3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3C) C3C)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)C O)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2 C2 |
3.5919 | 1.4546 | 3.6645 | 1.4568 |
| mol_3 | F[B−]1(F)[N+]![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) 2C( 2C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Cc3n1c(cc3)\C Cc3n1c(cc3)\C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C\c1oc(cc1)–c1ccc([N+]( C\c1oc(cc1)–c1ccc([N+](![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)[O−])cc1)C O)[O−])cc1)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) 2 2 |
2.3355 | 1.6269 | 2.3926 | 1.612 |
| mol_4 | OC1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC(O2) CC(O2)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C C(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1)C C1)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2/C C2/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/C3 C/C3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(N4CCCCC4)C C(N4CCCCC4)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3 C3 |
3.2705 | 1.6294 | 3.3375 | 1.6566 |
| mol_5 | OC1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C#CC2 C(C#CC2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC(C#N) CC(C#N)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C#CC3 C(C#CC3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC(C#N) CC(C#N)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3)C C3)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2)C C2)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1 C1 |
3.2075 | 1.5565 | 3.2493 | 1.5583 |
| mol_6 | F[B−]1(F)[N+]![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) 2C( 2C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Cc3n1c(cc3C)\C Cc3n1c(cc3C)\C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C\C C\C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C\c1oc(cc1)–c1oc(cc1)–c1oc(cc1)–c1ccc([N+]( C\c1oc(cc1)–c1oc(cc1)–c1oc(cc1)–c1ccc([N+](![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)[O−])cc1)C O)[O−])cc1)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) 2 2 |
1.9596 | 1.5718 | 2.0132 | 1.5316 |
| mol_7 | N#CC1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C#CC2 C(C#CC2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C#CC3 C(C#CC3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3C#N)C C3C#N)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2C#N)C C2C#N)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1 C1 |
3.2363 | 1.8274 | 3.2864 | 1.8329 |
| mol_8 | F[B−]([N+]1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2C C2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C3 C(C3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3)OC2 C3)OC2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC1 CC1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C4)(F)N5C4 C4)(F)N5C4![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC(O6) CC(O6)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C5C C5C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C6C7 C6C7![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C7C(C)C C7C(C)C |
2.2242 | 1.8164 | 2.364 | 1.7839 |
| mol_9 | FC1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C2 C(C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(C#CC3 C(C#CC3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3)C C3)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2)C C2)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1 C1 |
3.8249 | 1.4566 | 3.8856 | 1.4618 |
| mol_10 | C1(N(C2![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C2)C3 C2)C3![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C3) C3)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(/C C(/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/C4 C/C4![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(N(C5 C(N(C5![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C5)C6 C5)C6![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C6)C C6)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C4)C C4)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1 C1 |
2.945 | 1.5698 | 3.0107 | 1.6149 |
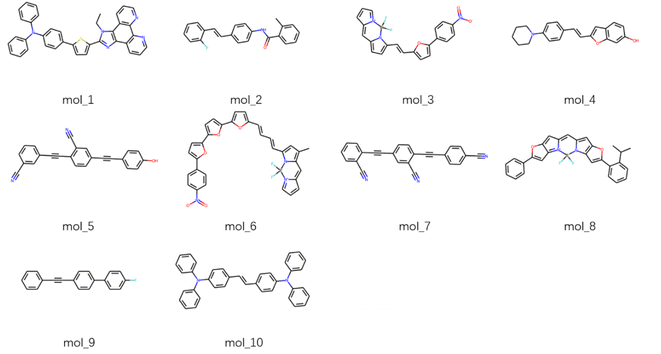 | ||
| Fig. 5 Topological structures of fluorescent molecules from the generation space with the largest 10 absorptive oscillator strengths. | ||
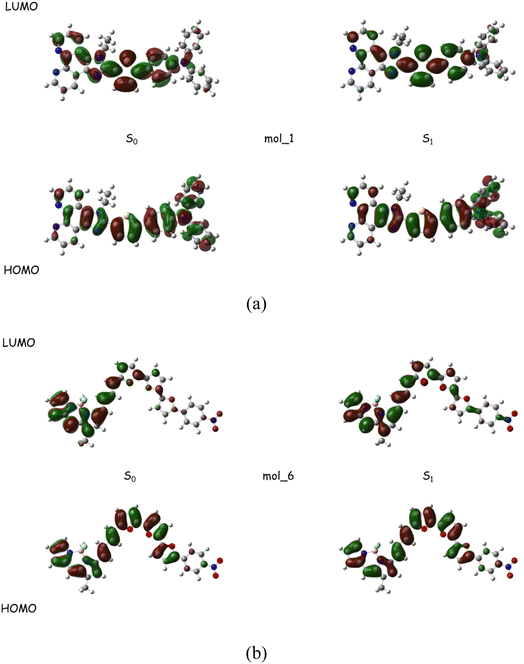
The optimized S0 and S1 geometries for the two representative molecules from Table 2 are exhibited in Fig. 6. No substantial structural displacement is found between the ground state and the excited state, which is consistent with the small Stokes shift where relaxations on the hypersurface can readily happen. The highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO) are both highly delocalized with a notable charge transfer characteristics being detected. The electronic structure of the generated samples can imitate most of the known fluorescent dyes,40,41 further validating the effectiveness of the adversarial model in creating molecules with reliable photophysical properties.
Conclusions
For a short conclusion, the article has proposed an efficient framework by employing the autoencoder based generative adversarial network to generate novel fluorescent compounds. The GRU encoder and decoder are utilized to map the string-based data onto the low-dimensional latent space. A GAN architecture is then applied to create a virtual latent space from the generator so that it can fully mimic the real latent based on a relativistic loss mechanism. The new samples are produced from the Gaussian noise and eventually decoded to molecular SMILES with the validity, novelty, uniqueness being examined. Given the current generation efficiency (around 13%), the model can deliver more than 2000 new molecules with satisfactory luminescence properties. Considering the small training input size (2923 fluorescent molecules that are experimentally synthesized), it is equivalent to doubling the fluorescent library in a very low computational cost (where the model cost-effectiveness is stated in contrast to the empirical trial and error method used in conventional molecular design which may take decades to develop thousands of fluorophores). High throughput TDDFT calculations are performed for nearly 5500 samples to obtain the excited state properties. It is found that the distributions of singlet transition energies and oscillator strengths for the generated and original molecules are highly comparable. The SA of the generation space is also similar with the original one, further validating the experimental accessibility of the novel compounds. By implementing the excited state optimization for the molecules with the largest absorptive strengths, the generated dyes are proved to possess a remarkable fluorescent rate along with a moderate Stokes shift. The charge transfer characteristics are detected in the frontier orbitals analysis, demonstrating the efficiency of AGAN in producing anticipated electronic structures.The current adversarial methodology has displayed a great potential in designing molecules with the desired fluorescent properties. It can be easily extended to the design of a large family of luminescent materials with different functionalities, with one of the most attractive design cases possibly being to develop molecules with large Stokes shifts particularly for bioscience applications. Additional improvements of the model are expected including the enhancement of the generation validity given the small input size, or by involving a predictor-augmented reinforcement algorithm42 to further elevate the generation efficiency, and the works are subject to the future research.
Data availability
All of the codes associated with the research work are made available at https://github.com/Xiyuan-Quantitative-Technology/relativistic-GAN. The data that support the findings of this study are obtainable from the corresponding author upon reasonable request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work is supported by the Sichuan Science and Technology Project (No. 2019YJ0646); Chengdu Science and Technology Project (No. 2019-YF05-00224-SN); Research Platform Foundation of Chengdu Polytechnic (No. 19KYPT01, No. 20KYTD07).References
- D. Cao, Z. Liu, P. Verwilst, S. Koo, P. Jangili, J. S. Kim and W. Lin, Coumarin-based small molecule fluorescent chemosensors, Chem. Rev., 2019, 119, 10403–10519 CrossRef CAS PubMed.
- H. Izawa, S. Nishino, M. Sumita, M. Akamatsu, K. Morihashi, S. Ifuku, M. Morimoto and H. Saimoto, A novel 1,8-naphthalimide derivative with an open space for an anion: Unique fluorescence behaviour depending on the binding anion's electrophilic Properties, Chem. Commun., 2015, 51, 8596–8599 RSC.
- M. C. L. Yeung and V. W. W. Yam, Luminescent cation sensors: from host–guest chemistry, supramolecular chemistry to reaction-based mechanisms, Chem. Soc. Rev., 2015, 44, 4192–4202 RSC.
- P. A. Gale and C. Caltagirone, Anion sensing by small molecules and molecular ensembles, Chem. Soc. Rev., 2015, 44, 4212–4227 RSC.
- Y. Qin, G. Li, T. Qi and H. Huang, Aromatic imide/amide-based organic small-molecule emitters for organic light-emitting diodes, Mater, Chem. Front., 2020, 4, 1554–1568 CAS.
- M. Mamada, K. Inada, T. Komino, W. J. Potscavage Jr, H. Nakanotani and C. Adachi, Highly efficient thermally activated delayed fluorescence from an excited-state intramolecular proton transfer system, ACS Cent. Sci., 2017, 3, 769–777 CrossRef CAS PubMed.
- H. Uoyama, K. Goushi, K. Shizu, H. Nomura and C. Adachi, Highly efficient organic light-emitting diodes from delayed fluorescence, Nature, 2012, 492, 234–238 CrossRef CAS PubMed.
- H. Kaji, H. Suzuki, T. Fukushima, K. Shizu, K. Suzuki, S. Kubo, T. Komino, H. Oiwa, F. Suzuki, A. Wakamiya, Y. Murata and C. Adachi, Purely organic electroluminescent material realizing 100% conversion from electricity to light, Nat. Commun., 2015, 6, 8476 CrossRef CAS PubMed.
- S. Banerjee, E. B. Veale, C. M. Phelan, S. A. Murphy, G. M. Tocci, L. J. Gillespie, D. O. Frimannsson, J. M. Kelly and T. Gunnlaugsson, Recent advances in the development of 1,8-naphthalimide based DNA targeting binders, anticancer and fluorescent cellular imaging agents, Chem. Soc. Rev., 2013, 42, 1601–1618 RSC.
- J. A. Carr, D. Franke, J. R. Caram, C. F. Perkinson, M. Saif, V. Askoxylakis, M. Datta, D. Fukumura, R. K. Jain, M. G. Bawendi and O. T. Bruns, Shortwave infrared fluorescence imaging with the clinically approved near-infrared dye indocyanine green, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 4465–4470 CrossRef CAS PubMed.
- R. Chouket, A. Pellissier-Tanon, A. Lemarchand, A. Espagne, T. Saux and L. Jullien, Dynamic contrast with reversibly photoswitchable fluorescent labels for imaging living cells, Chem. Sci., 2020, 11, 2882–2887 RSC.
- E. Kim, M. Koh, B. J. Lim and S. B. Park, Emission Wavelength Prediction of a Full-Color-Tunable Fluorescent Core Skeleton, 9-Aryl-1,2-dihydropyrrolo[3,4-b]indolizin-3-one, J. Am. Chem. Soc., 2011, 133(17), 6642 CrossRef CAS PubMed.
- Z. Gao, Y. C. Hao, M. L. Zheng and Y. Chen, A fluorescent dye with large Stokes shift and high stability: synthesis and application to live cell imaging, RSC Adv., 2017, 7, 7604 RSC.
- M. Sumita, K. Terayama, N. Suzuki, S. Ishihara, R. Tamura, M. K. Chahal, D. T. Payne, K. Yoshizoe and K. Tsuda, De novo creation of a naked eye–detectable fluorescent molecule based on quantum chemical computation and machine learning, Sci. Adv., 2022, 8(10), 3906 CrossRef PubMed.
- P. O. Dral and M. Barbatti, Molecular excited states through a machine learning lens, Nat. Rev. Chem., 2021, 5, 388–405 CrossRef CAS.
- B. Kang, C. Seok and J. Lee, A benchmark study of machine learning methods for molecular electronic transition: tree-based ensemble learning versus graph neural network, Bull. Korean Chem. Soc., 2022, 43, 3 CrossRef.
- B. Kang, C. Seok and J. Lee, Prediction of Molecular Electronic Transitions Using Random Forests, J. Chem. Inf. Model., 2020, 60(12), 5984–5994 CrossRef CAS PubMed.
- S. L. Luo, T. S. Li, X. J. Wang, M. Faizan and L. J. Zhang, High-throughput computational materials screening and discovery of optoelectronic semiconductors, Wiley Interdiscip. Rev. Comput. Mol. Sci., 2020, 11, 7690 Search PubMed.
- C. Q. Lu, Q. Liu, Q. M. Sun, C. Y. Hsieh, S. Y. Zhang, L. Shi and C. K. Lee, Deep Learning for Optoelectronic Properties of Organic Semiconductors, J. Phys. Chem. C, 2020, 124(13), 7048–7060 CrossRef CAS.
- S. Q. Ye, J. C. Liang and X. Zhu, The Catalyst Deep Neural Networks (Cat-DNNs) in Singlet Fission Property Prediction, Phys. Chem. Chem. Phys., 2021, 23, 20835 RSC.
- Z. R. Ye, I. S. Huang, Y. T. Chana, Z. J. Lia, C. C. Liao, H. R. Tsai, M. C. Hsieh, C. C. Changa and M. K. Tsai, Predicting the emission wavelength of organic molecules using a combinatorial QSAR and machine learning approach, RSC Adv., 2020, 10, 23834–23841 RSC.
- A. Subramanian, U. Saha, T. Sharma, N. K. Tailor and S. Satapathi, Inverse Design of Potential Singlet Fission Molecules using a Transfer Learning Based Approach, arXiv, 2020, preprint, arXiv:2003.07666, DOI:10.48550/arXiv.2003.07666.
- C. W. Ju, H. Z. Bai, B. Li and R. Z. Liu, Machine Learning Enables Highly Accurate Predictions of Photophysical Properties of Organic Fluorescent Materials: Emission Wavelengths and Quantum Yields, J. Chem. Inf. Model., 2021, 61(3), 1053–1065 CrossRef CAS PubMed.
- Z. Tan, Y. Li, Z. Y. Zhang, X. Wu, T. Penfold, W. M. Shi and S. Q. Yang, Efficient Adversarial Generation of Thermally Activated Delayed Fluorescence Molecules, ACS Omega, 2022, 7(21), 18179–18188 CrossRef CAS PubMed.
- D. Weininger, SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules, J. Chem. Inf. Comput. Sci., 1998, 28, 31–36 CrossRef.
- O. Prykhodko, S. V. Johansson, P. C. Kotsias, J. Arús-Pous, E. J. Bjerrum, O. Engkvist and H. M. Chen, A de novo molecular generation method using latent vector based generative adversarial network, J. Cheminformatics, 2019, 11, 74 CrossRef PubMed.
- P. Tosco, N. Stiefl and G. Landrum, Bringing the MMFF force field to the RDKit: implementation and validation, J. Cheminformatics, 2014, 6, 37 CrossRef.
- M. J. Frisch, et al., Gaussian 16, Revision A.03, 2016 Search PubMed.
- L. T. Yu, W. N. Zhang, J. Wang and Y. Yu, Seqgan: sequence generative adversarial nets with policy gradient, arXiv, 2017, preprint, arXiv:1609.05473, DOI:10.1609/aaai.v31i1.10804.
- G. L. Guimaraes, B. Sanchez-Lengeling, C. Outeiral, P. L. C. Farias and A. Aspuru-Guzik, Objective-Reinforced Generative Adversarial Networks (ORGAN) for Sequence Generation Models, arXiv, 2018, preprint, arXiv:1705.10843, DOI:10.48550/arXiv.1705.10843.
- N. D. Cao and T. Kipf, MolGAN: An implicit generative model for small molecular graphs, arXiv, 2018, preprint, arXiv:1805.11973, DOI:10.48550/arXiv.1805.11973.
- B. Sanchez-Lengeling, C. Outeiral, G. L. Guimaraes and A. Aspuru-Guzik, Optimizing Distributions Over Molecular Space. An Objective-Reinforced Generative Adversarial Network for Inverse-design Chemistry (ORGANIC), ChemRxiv, 2017 Search PubMed , preprint, https://chemrxiv.org/engage/chemrxiv/article-details/60c73d91702a9beea7189bc2.
- R. J. Williams and D. Zipser, A Learning Algorithm for Continually Running Fully Recurrent Neural Networks, Neural Comput., 1989, 1, 270–280 CrossRef.
- A. Jolicoeur-Martineau, The relativistic discriminator: a key element missing from standard GAN, arXiv, 2018, preprint, arXiv:1807.00734, https://doi.org/10.48550/arXiv.1807.00734 Search PubMed.
- B. Kim, S. W. Lee and J. H. Kim, Inverse design of porous materials using artificial neural networks, Sci. Adv., 2020, 6, 9324 CrossRef PubMed.
- Y. B. Dan, Y. Zhao, X. Li, S. B. Li, M. Hu and J. H. Hu, Generative adversarial networks (GAN) based efficient sampling of chemical composition space for inverse design of inorganic materials, npj Comput. Mater., 2020, 6, 84 CrossRef CAS.
- D. Polykovskiy, A. Zhebrak, B. S. Lengeling, S. Golovanov, O. Tatanov, S. Belyaev, R. Kurbanov, A. Artamonov, V. Aladinskiy, M. Veselov, A. Kadurin, S. Johansson, H. M. Chen, S. I. Nikolenko, A. Aspuru-Guzik and A. Zhavoronkov, Molecular Sets (MOSES): A Benchmarking Platform for Molecular Generation Models, Front. Pharmacol., 2020, 11, 1931 Search PubMed.
- P. Ertl and A. Schuffenhauer, Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions, J. Cheminformatics, 2009, 1, 8 CrossRef PubMed.
- S. A. Ahmad, J. Eng and T. J. Penfold, Rapid predictions of the colour purity of luminescent organic molecules, Recent advances of cyclotriphosphazene derivatives as fluorescent dyes, J. Mater. Chem. C, 2022, 10, 4785–4794 RSC.
- B. Czaplińska, K. Malarz, A. Mrozek-Wilczkiewicz, A. Slodek, M. Korzec and R. Musiol, Theoretical and Experimental Investigations of Large Stokes Shift Fluorophores Based on a Quinoline Scaffold, Molecules, 2020, 25(11), 2488 CrossRef PubMed.
- T. B. Ren, W. Xu, W. Zhang, X. X. Zhang, Z. Y. Wang, Z. Xiang, L. Yuan and X. B. Zhang, A General Method to Increase Stokes Shift by Introducing Alternating Vibronic Structures, J. Am. Chem. Soc., 2018, 140(24), 7716–7722 CrossRef CAS PubMed.
- Z. P. Zhou, S. Kearnes, L. Li, R. N. Zare and P. Riley, Optimization of Molecules via Deep Reinforcement Learning, Sci. Rep., 2019, 9, 10752 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2023 |