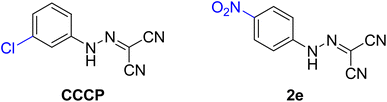Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Efflux pump inhibitor combined with ofloxacin decreases MRSA biofilm formation by regulating the gene expression of NorA and quorum sensing
Xueer Lu†
a,
Guifeng Wang†b,
Yunfeng Xieb,
Wenjian Tang c,
Biyong Liu*b and
Jing Zhang
c,
Biyong Liu*b and
Jing Zhang *b
*b
aDepartment of Clinical Laboratory, The Third People's Hospital of Hefei, Hefei, 230022, China
bAnhui Prevention and Treatment Center for Occupational Disease, Anhui No. 2 Provincial People's Hospital, Hefei, 230041, China. E-mail: lby770114@163.com; hfzj2552@163.com
cSchool of Pharmacy, Anhui Medical University, Hefei 230032, China
First published on 19th January 2023
Abstract
Carbonyl cyanide p-nitrophenylhydrazone (2e) displayed a lone or synergistic efficacy against MRSA (RSC Adv., 2020, 10, 17854). In this work, the synergistic mechanism of 2e with ofloxacin was studied. MRSA2858 had potential for biofilm formation, and the value of MBEC of 2e alone was 0.78–1.56 μM, while that of 2e + ofloxacin was 0.39–0.78 μM. 2e combined with ofloxacin showed a synergistic anti-biofilm effect against MRSA. Efflux pump inhibitor 2e can better bind to NorA protein. After MRSA2858 was treated with 2e of 1/2MIC (0.78 μM) and ofloxacin of 1/8MIC (0.097 μM), the transcript levels of efflux genes (norA) and quorum-sensing (QS) regulatory genes (agrA, sarA, icaA, hla) were substantially down-regulated, and alpha-hemolysin (Hla) was inhibited by 99.15%. 2e combined with ofloxacin was more effective than 2e alone in reducing bacterial load in vivo. All in all, efflux pump inhibitor 2e enhanced the bactericidal activities of antibiotics through regulating the gene expression of NorA and QS system.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) has become a major global health threat.1 Biofilm-associated MRSA are difficult to eradicate, which can lead to acute to chronic infections, including diabetic foot infections, cystic fibrosis, and so on.2–5 Therefore, it is urgently needed to find unique therapeutic strategies or new chemical scaffolds against MRSA to counter this health challenge.Carbonyl cyanide chlorophenylhydrazone (CCCP), an efflux pump inhibitor, is combined with antibiotics to enhance antimicrobial activity. CCCP reduces ATP production and increases membrane permeability in bacteria6–9 by interfering with the transmembrane electrochemical gradient and proton motive force.6,10 CCCP blocks the efflux of antibiotics outside the bacterial cells to decrease the biofilm-forming capacity of bacteria.11 However, it is toxic and has poor antibacterial activity when used alone.12 The efflux pumps also play a role in biofilm formation.13 The inhibition of efflux pumps helps to enhance the therapeutic efficacy of traditional antibiotics by affecting QS genes and virulence factors to control biofilm production.11
Recently, CCCP analog 2e was found to display alone or synergistic efficacy against MRSA.13 2e could inhibit biofilm formation, effectively eradicated preformed biofilm, and showed low toxicity for human hepatic L02 cells.14 Studying the anti-biofilm activity of 2e and 2e combined with ofloxacin will help to understand the role and relationship of efflux pump and biofilm formation in MRSA.
Quorum sensing (QS) is one of the signaling mechanisms which directly aids in biofilm formation, and inhibiting QS can increase susceptibility to antibiotics. Formation and maintenance of biofilms are controlled by complex regulatory circuits such as QS and SaeRS two-component system (TCS).15,16 TCS is at the core of the QS system, which play an important role in bacterial survival, adaptation and pathogenesis, biofilm formation and virulence factor production.17,18 It is interesting how efflux pump inhibitors affect biofilm formation by QS and TCS.
When 2e was combined with ofloxacin, a synergistic anti-MRSA activity was observed (FICI = 0.28). In this work, we would further explore this synergistic antibacterial mechanism of efflux pump inhibitors on the biofilm formation by down-regulating QS system. The multi-drug resistant MRSA2858 strain was isolated and identified from clinical MRSA strains, which showed the potential of biofilm formation. 2e combined with ofloxacin inhibited biofilm formation of MRSA2858 by down-regulating the transcript levels of efflux gene (norA) and QS regulatory genes (agrA, sarA, icaA, hla), and decreasing alpha-hemolysin (Hla). This study provided a novel strategy for MRSA treatment.
Materials and methods
Reagents
All chemicals, reagents and solvents were purchased from commercial sources and used without further purification. Cation-adjusted Mueller–Hinton broth (CAMHB), crystal violet, trypsin–EDTA, tryptic soy broth (TSB), tryptic soy agar (TSA), phosphate-buffered saline (PBS), Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), and 96-well plates were all purchased from commercial vendors (BD, Cat.). CCCP and compound 2e was synthesized and their structures were shown in Fig. 1.13Strains and growth media
In this work, 16 clinical multi-resistant isolates of MRSA strains capable of establishing biofilms were collected. All of MRSA strains were resistant to three or more antimicrobial agents. S. aureus reference strain was obtained from American Type Culture Collection (ATCC 6538). The ability of the 16 strains to establish biofilms was tested using crystal violet (CV) assay (Table 2). MRSA2858 was the strongest biofilm-forming strain isolated from the patient's pus. Inoculums for the antimicrobial testing and the biofilm assays were prepared overnight in Mueller–Hinton (MH) broth and incubated at 37 °C.| Primer | Sequence | Reference |
|---|---|---|
| nor A-F | GAGTGCTGGTATGGTAATGCC | Sivaranjani et al., 2019 |
| nor A-R | CCCTGGTCCTAAAATGAATCC | Sivaranjani et al., 2019 |
| agr A-F | TGCGAAGACGATCCAAAACA | Sivaranjani et al., 2019 |
| agr A-R | GGGCAATTTCCATAGGCTTTTC | Sivaranjani et al., 2019 |
| sar A-F | TGGTTCTCATCTCCCTTTGCTT | Sivaranjani et al., 2019 |
| sar A-R | GCGATGCTAATCTTCCTGGTG | Sivaranjani et al., 2019 |
| ica A-F | GGCTGGACTCATATTTGTAAGTTGG | Sivaranjani et al., 2019 |
| ica A-R | GTATTCCCTCTGTCTGGGCTTG | Sivaranjani et al., 2019 |
| hla-F | TTGGTGCAAATGTTTC | Sivaranjani et al., 2019 |
| hla-R | TCACTTTCCAGCCTACT | Sivaranjani et al., 2019 |
| gyrB-F | GGTGCTGGGCAAATACAAGT | Valliammai et al., 2019 |
| gyrB-R | TCCCACACTAAATGGTGCAA | Valliammai et al., 2019 |
| Spec. | MIC of clinical used antibioticsb (μg ml−1) | MIC of 2e (μM) | Biofilm formation (OD595)c | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SAM | AMP | AMC | CRO | CLI | CIP | ERY | GEN | LVX | LNZ | OXA | PEN | RIF | QD | SXT | TCY | VAN | ||||
| a MRSA: methicillin-resistant Staphylococcus aureus.b Antibiotics abbreviations: SAM: ampicillin/sulbactam, AMP: ampicillin, AMC: amoxicillin/clavulanic, CRO: ceftriaxone, CLI: clindamycin, CIP: ciprofloxacin, ERY: erythromycin, GEN: gentamicin, LVX: levofloxacin, LNZ: linezolid, OXA: oxacillin, PEN: penicillin, RIF: rifampin, QD: quinupristin/dalfopristin, SXT: trimethoprim/sulfa, TCY: tetracycline, VAN: vancomycin.c Each data point represents a mean value of three independent experiment. | ||||||||||||||||||||
| MRSA0102 | Sputum | ≤8/4 | >8 | ≤4/2 | ≤8 | ≤0.5 | ≤1 | >4 | ≤4 | ≤1 | ≤4 | >2 | >8 | ≤1 | ≤0.5 | ≤0.5/9.5 | ≤4 | 2 | 1.562 | 0.599 |
| MRSA0101 | Sputum | ≤8/4 | >8 | ≤4/2 | ≤8 | >4 | ≤1 | >4 | ≤4 | ≤1 | ≤4 | >2 | >8 | ≤1 | ≤0.5 | ≤0.5/9.5 | ≤4 | 2 | 1.562 | 0.972 |
| MRSA2858 | Pus | ≤8/4 | >8 | ≤4/2 | 32 | >4 | ≤1 | >4 | >8 | ≤1 | 4 | >2 | >8 | ≤1 | >2 | ≤0.5/9.5 | ≤4 | 2 | 1.562 | 1.511 |
| MRSA0612 | Sputum | 16/8 | >8 | >4/2 | >32 | ≤0.5 | >2 | >4 | >8 | >4 | ≤1 | >2 | >8 | >2 | ≤0.5 | ≤0.5/9.5 | >8 | ≤0.25 | 3.125 | 1.461 |
| MRSA10105 | Blood | 16/8 | >8 | ≤4/2 | ≤8 | ≤0.5 | ≤1 | ≤0.5 | ≤4 | ≤1 | 2 | >2 | >8 | 2 | ≤0.5 | ≤0.5/9.5 | 8 | 1 | 1.562 | 0.366 |
| MRSA05107 | Blood | 16/8 | >8 | >4/2 | >32 | >4 | >2 | >4 | ≤4 | >4 | 2 | >2 | >8 | ≤1 | ≤0.5 | ≤0.5/9.5 | ≤4 | 2 | 3.125 | 0.445 |
| MRSA1905 | Sputum | ≤8/4 | >8 | ≤4/2 | 32 | >4 | ≤1 | >4 | ≤4 | ≤1 | 2 | >2 | >8 | ≤1 | ≤0.5 | ≤0.5/9.5 | ≤4 | 1 | 1.562 | 0.530 |
| MRSA0352 | Wound secretion | ≤8/4 | >8 | ≤4/2 | ≤8 | ≤0.5 | ≤1 | >4 | ≤4 | ≤1 | 4 | >2 | >8 | ≤1 | ≤0.5 | ≤0.5/9.5 | ≤4 | 1 | 1.562 | 0.615 |
| MRSA1257 | Wound secretion | ≤8/4 | ≤2 | ≤4/2 | ≤8 | ≤0.5 | ≤1 | ≤0.5 | ≤4 | ≤1 | 2 | >2 | 0.12 | ≤1 | ≤0.5 | ≤0.5/9.5 | ≤4 | 1 | 1.562 | 0.404 |
| MRSA1252 | Wound secretion | 16/8 | >8 | >4/2 | >32 | ≤0.5 | >2 | ≤0.5 | ≤4 | ≤1 | 2 | >2 | >8 | ≤1 | ≤0.5 | ≤0.5/9.5 | ≤4 | 2 | 3.125 | 1.313 |
| MRSA0458 | Wound secretion | ≤8/4 | >8 | ≤4/2 | ≤8 | ≤0.5 | ≤1 | ≤0.5 | ≤4 | ≤1 | 2 | >2 | >8 | ≤1 | ≤0.5 | ≤0.5/9.5 | ≤4 | 1 | 1.562 | 0.699 |
| MRSA0961 | Wound secretion | 16/8 | >8 | >4/2 | ≤8 | ≤0.5 | ≤1 | ≤0.5 | ≤4 | ≤1 | 4 | >2 | >8 | ≤1 | ≤0.5 | ≤0.5/9.5 | ≤4 | 1 | 1.562 | 0.605 |
| MRSA100902 | Sputum | ≤8/4 | >8 | ≤4/2 | ≤8 | ≤0.5 | ≤1 | ≤0.5 | ≤4 | ≤1 | ≤1 | >2 | >8 | ≤1 | ≤0.5 | ≤0.5/9.5 | ≤4 | 1 | 1.562 | 0.468 |
| MRSA101262 | Wound secretion | ≤8/4 | >8 | ≤4/2 | ≤8 | >4 | ≤1 | >4 | ≤4 | ≤1 | 2 | >2 | >8 | ≤1 | ≤0.5 | ≤0.5/9.5 | >8 | 2 | 3.125 | 1.031 |
| MRSA2854 | Pus | ≤8/4 | ≤2 | ≤4/2 | ≤8 | >4 | ≤1 | >4 | ≤4 | ≤1 | 4 | >2 | 2 | ≤1 | ≤0.5 | ≤0.5/9.5 | ≤4 | 2 | 1.562 | 0.416 |
| MRSA1554 | Pus | ≤8/4 | >8 | ≤4/2 | ≤8 | >4 | ≤1 | >4 | ≤4 | ≤1 | 2 | >2 | >8 | ≤1 | ≤0.5 | ≤0.5/9.5 | ≤4 | 1 | 1.562 | 0.203 |
MIC assay
The minimum inhibitory concentration (MIC) of tested compounds was determined using MH broth micro-dilution assay established by the Clinical Laboratory Standards Institute (CLSI) in 96-well microtest plates. Bacterial strains were grown overnight in MH broth at a concentration of 108 CFU ml−1 in microtiter plates. Then, 100 μl of culture medium containing different concentrations of test compounds and reference antibiotics were added to the plates. The final bacterial concentration in the plate was 105 CFU ml−1. After incubating for 20 h at 37 °C, the MIC was determined by observing the lowest concentration of tested compound which inhibited bacterial growth. All assays were performed in quadruplicate.Minimal biofilm eradication concentration (MBEC) determination
The antibiofilm activity of 2e was assessed in vitro against a mature biofilm formed in a flat-bottomed plate by an MRSA strain. For the assay, wells were filled with 200 μl of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution of a bacterial suspension cultured overnight in MH broth. After 24 h of incubation at 37 °C in static conditions, the medium was discarded, and planktonic cells were removed by thorough washing of the biofilm with sterile PBS. Then, 200 μl of MH broth supplemented with 2-fold dilutions of the compound (in the range 100–0.045 μM) were added in each well, and the plate was incubated further for 24 h at 37 °C.19 As an antibiotic control, dilutions of vancomycin, ofloxacin, rifampin and erythromicin in the range 100–0.045 μM were included. Biofilm formation was determined by crystal violet staining. To dissolve the crystal violet, 150 μl methanol per well was added to allow for minimal biofilm eradication concentration determination (MBEC) analysis.20 The optical density (OD) of each well was measured at 630 nm using an automated spectrophotometer (Bio-Tek Epoch-2, USA). Biofilm growth inhibition (%) = 100 − (ODassay/ODpositive) × 100. Each experiment was repeated three times, with at least eight wells of each sample.
100 dilution of a bacterial suspension cultured overnight in MH broth. After 24 h of incubation at 37 °C in static conditions, the medium was discarded, and planktonic cells were removed by thorough washing of the biofilm with sterile PBS. Then, 200 μl of MH broth supplemented with 2-fold dilutions of the compound (in the range 100–0.045 μM) were added in each well, and the plate was incubated further for 24 h at 37 °C.19 As an antibiotic control, dilutions of vancomycin, ofloxacin, rifampin and erythromicin in the range 100–0.045 μM were included. Biofilm formation was determined by crystal violet staining. To dissolve the crystal violet, 150 μl methanol per well was added to allow for minimal biofilm eradication concentration determination (MBEC) analysis.20 The optical density (OD) of each well was measured at 630 nm using an automated spectrophotometer (Bio-Tek Epoch-2, USA). Biofilm growth inhibition (%) = 100 − (ODassay/ODpositive) × 100. Each experiment was repeated three times, with at least eight wells of each sample.
Molecular docking and toxicity prediction
A structure based in silico procedure was applied to discover the binding modes of the active compounds to NorA protein active site. The CDOCKER of Discovery Studio Client v18.1.0 (DS) was conducted to explain SAR of series compounds and further guide the design of more effective and concrete NorA protein inhibitors. The ligand binding to the crystal structure of NorA protein with PDB ID: 3WDO was selected as template. The target enzyme was prepared with Prepare Protein of DS to ensure the integrity of target. The ligands (CCCP and 2e) were processed by full minimization of the small molecular in DS. Then title compounds were docked into the active site of protein using CDOCKER. The view results of molecular docking were extracted after the program running end, each docking result was analyzed for interaction and their different pose. Those poses with the lowest –CDOCKER_INTERACTION_ENERGY values were regarded as the most stable and picked to analysis binding interactions with target enzyme visualized.Molecular dynamics simulations
The molecular dynamics (MD) simulations were performed in Yinfo Cloud Computing Platform using AmberTools 20 package with AMBER ff19SB and GAFF force fields for NorA and compound 2e, respectively.21,22 The system was solvated by a truncated octahedron (or cubic) water box using OPC water model with a margin of 10 Å, the net charge was neutralized by (0.15 M of NaCl) sodium ions. Periodic boundary condition (PBC) was used and the net charge neutralized by Na+ (or Cl−) ions (or 0.15 M of NaCl). Nonbonded van der Waals interactions were calculated using the Lennard-Jones 12–6 potentials with a 10 Å cutoff, while long-range electrostatics were treated using the Particle Mesh Ewald (PME) algorithm. The SHAKE algorithm was applied to constrain bonds involving hydrogen atoms. To removed improper atom contacts, the system was first minimized by (1) the 5000 steps of steepest descent and the 5000 steps of the conjugate gradient, under a harmonic constraint of 10.0 kcal (mol Å2)−1 on heavy atoms; (2) relaxing the entire system by steepest descent and conjugate gradient each in 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps. And then the system was gradually heated up to 300 K by a 20 ps NVT simulation. The equilibration phase was carried out in two steps: (1) a 200 ps NPT simulation with constraints on heavy atoms followed by (2) a 500 ps NVT simulation without restraint. The temperature and pressure were maintained at 300 K and 1 atm using the Berendsen thermostat and Monte Carlo barostat, with coupling constant and relaxation time of 1 ps. Finally, the system was subjected to a 20 ns NPT simulation with a time step of 1 fs. The root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and hydrogen bonds were analyzed by the Cpptraj module.
000 steps. And then the system was gradually heated up to 300 K by a 20 ps NVT simulation. The equilibration phase was carried out in two steps: (1) a 200 ps NPT simulation with constraints on heavy atoms followed by (2) a 500 ps NVT simulation without restraint. The temperature and pressure were maintained at 300 K and 1 atm using the Berendsen thermostat and Monte Carlo barostat, with coupling constant and relaxation time of 1 ps. Finally, the system was subjected to a 20 ns NPT simulation with a time step of 1 fs. The root-mean-square deviation (RMSD), root-mean-square fluctuation (RMSF), and hydrogen bonds were analyzed by the Cpptraj module.
MM/PB(GB)SA calculations. The binding free energies were calculated using the Molecular Mechanics Poisson–Boltzmann Surface Area (MM/PBSA) method implemented in AmberTools 20 for 200 snapshots from the MD trajectory. For each snapshot, the free energy was calculated for protein NorA, compound 2e, and the complex NorA–2e using a single trajectory approach. The total binding free energy was calculated according to the following equation:
| ΔGbind = ΔGcomplex − (ΔGreceptor − ΔGligand) |
| ΔGbind = ΔEMM + ΔGsolv − TΔS |
| ΔEMM = ΔEvdw + ΔEele |
| ΔGsolv = ΔGPB + ΔGSA |
Effect of 2e on gene expression of MRSA strains
Total RNA from MRSA cultures treated with 2e alone or in combination with ofloxacin for 24 h was extracted using the TRIzol method for RNA extraction and converted to cDNA using SuperScript™ III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen, USA). The PCRs were performed in a 20 μl volume and contained Roche SYBR Green PCR Master Mix. The fold change in gene expression was calculated by the 2−ΔΔct method with gyrB as the housekeeping gene. Specific primers are listed in Table 1. Amplification was performed in a gradient thermal cycler (Bio-Rad, Hercules, USA). All samples were analyzed in triplicate.Hemolytic activity
Hemolytic activity can be assessed by a rabbit red blood cell lysis technique, which consists of bringing the red blood cells into contact with the MRSA2858 or 2e, and quantifying the hemolytic activity of alpha-hemolysin (Hla) by spectrophotometric determination. Samples (100 μl) of MRSA2858 culture (1 × 109 CFU ml−1) were filtered and were added to 900 μl hemolysin buffer (0.145 mol per l NaCl, 0.02 mol per l CaCl2) and 25 μl of defibrinated rabbit blood. Rabbit blood was washed three times with 0.9% NaCl and resuspended to a concentration of 0.5 × 108 cells per ml, as determined by manual cell count. The solution was incubated for 15 min at 37 °C. Unlysed blood cells were pelleted by centrifugation (5500g, 1 min). The hemolytic activity of the supernatant was determined by measuring the optical density at OD541. Sterile culture medium served as the standard for 0% hemolysis, and a bacterial culture supernatant devoid of any inhibitor (control) was designated as the standard for 100% hemolysis. The percentage of hemolysis inhibition was calculated by comparison with the control culture. All assays were performed in triplicate.In vivo anti-bacterial activity
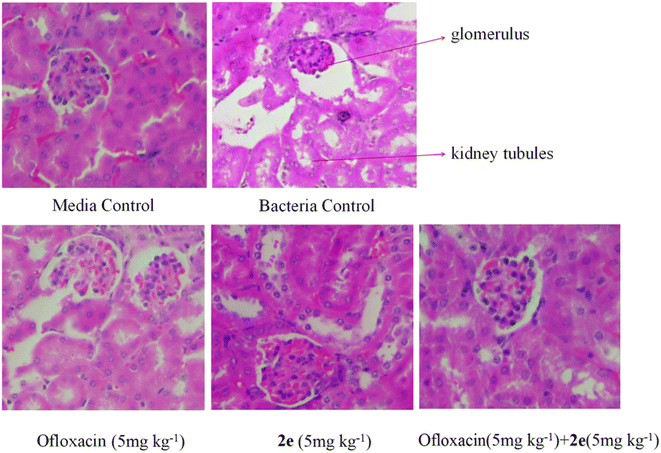
SPF female BALB/c mice were selected to avoid the re-infection induced by the fighting and barbering of males. SPF female BALB/c mice (aged 6–8 weeks; 18–21 g) were purchased from Changzhou Cavens Experimental Animal Co., Ltd, Animal certificate number: SCXK (SU) 2016-0010; mice were maintained with SPF food and water for 1 week, 6 mice per group. The test strain was refreshed on MH broth at 37 °C for 24 h. From the overnight culture preparation, a single colony was diluted into 5 ml of MH broth and incubated overnight at 37 °C. A log-phase subculture was obtained by adding 150 μl of overnight subculture in 10 ml MH broth and incubated for a further 2–3 h. A full dilution of the bacterial cell suspension in saline was achieved by washing (3220 g, 10 min) and the OD600 in saline determined. The suspension of 1.5 × 109 CFU ml−1 solution was diluted in MH broth into the final bacterial concentration of 105 CFU ml−1. Mice were infected by intraperitoneal administration of MRSA2858 in 0.2 ml of MH broth.After bacterial challenge for 2 h, mice were randomized to receive subcutaneous injection of saline as a control, ofloxacin (5 mg kg−1), 2e (5 mg kg−1), or 2e combined with ofloxacin group (5 mg kg−1 + 5 mg kg−1). To assess bacterial clearance, six mice in each group were euthanized, and bacterial counts were determined in the kidney and blood from each animal after bacterial challenge for 24 h. In the MRSA infection model, parts of kidney tissues were fixed in 10% neutral buffered formalin for 24 h, and then their morphologies were observed using H&E staining.
Statistic analysis
Statistical analysis was performed using SPSS 13.0 software package, and one-way ANOVA followed was used to assess the significance. A p-value ≤ 0.05 was set as statistically significant. Data were analyzed by Microsoft Excel 2003 spreadsheet software, and expressed as the mean ± standard deviation.Results
Antibacterial activity of 2e
The results showed that 2e performed the best against S. aureus with MIC value of 1.56 μM, and against MRSA with MIC value of 1.56 μM, which was superior to the front-line antibiotics including cefoxitin and linazolid.14 As shown in Table 2, 16 clinical MRSA were isolated from different specimens, and all were resistant to at least three antimicrobial agents of furantoin, penicillin, rifampicin, tetracycline, tegacyclin, ciprofloxacin, erythromycin, gentamicin, linezolid, benzacillin or clindamycin. Among them, MRSA2858 was a clinical isolate with multi-drug resistance and the strongest biofilm formation ability. Although certain antibiotics have MIC values even more favorable than compound 2e in Table 2, levofloxacin and ciprofloxacin are alternative or complementary drugs and are not clinical first-line anti-MRSA agents according to CLSI guidelines.Studying the synergistic mechanism of 2e with ofloxacin will help to enhance the therapeutic efficacy of traditional antibiotics against resistant bacteria. Data indicated that 2e strongly inhibited the bacterial growth of all clinical strains with the potency of MIC values less than 1.56 μM and the results had no statistically significant differences for all strains, which validated 2e as a promising anti-MRSA agent.
Anti-biofilm activity of 2e
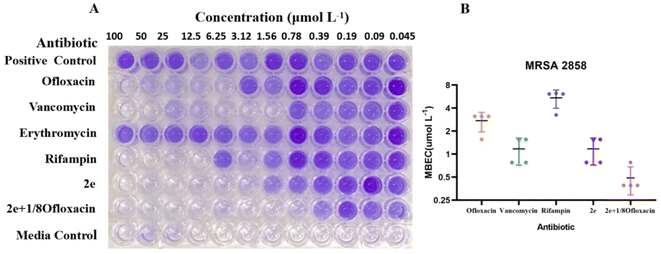
To assess the anti-biofilm potential of 2e, the MRSA clinical isolates were assayed in vitro in a preliminary set of experiments for their ability to form adherent biofilm layers under static conditions.19 Clinical strain MRSA2858 demonstrated high biofilm capacity and planktonic cells that were susceptible to 2e (Table 2). MRSA2858 biofilms were initially grown for 24 h and then treated with dilutions of 2e and vancomycin, ofloxacin, rifampin, erythromicin, as antibiotic control, for a further 24 h. Biofilm production and eradication were quantitatively evaluated using crystal violet staining.As shown in Fig. 2 and Table 3, 2e alone and combination with 1/8MIC ofloxacin was active against MRSA biofilms, the value of MBEC was 0.78–1.56 μM and 0.39–0.78 μM, similar to it MIC, superior to those of vancomycin, ofloxacin, rifampin and erythromycin (Fig. 2A). Although low level of β-lactam antibiotic was reported to induce biofilm formation of S. aureus,25,26 2e combined with subinhibitory concentration of ofloxacin (1/8MIC) showed a synergistic anti-biofilm effect against MRSA (Fig. 2B).
| Compound | Ofloxacin | Vancomycin | Rifampin | 2e | 2e + 1/8ofloxacin |
|---|---|---|---|---|---|
| MBEC (μM) | 2.73 ± 0.78 | 1.17 ± 0.45 | 5.43 ± 1.46 | 1.17 ± 0.45 | 0.48 ± 0.20 |
Molecular docking and toxicity prediction
In the molecular docking study, two ligands (CCCP and 2e) were docked into NorA protein, and they were located in the same activity site. 4-NO2 of 2e formed hydrogen bond interaction with Gln 65 and Ser 344, while CCCP cannot interact with these two amino acid residues. Therefore, 2e can better bind to NorA protein. Structure–activity relationship (SAR) analysis showed that 4-NO2 made 2e adopt a different binding method into the NorA protein (Fig. 3), which implied that 2e may be a new NorA protein inhibitor.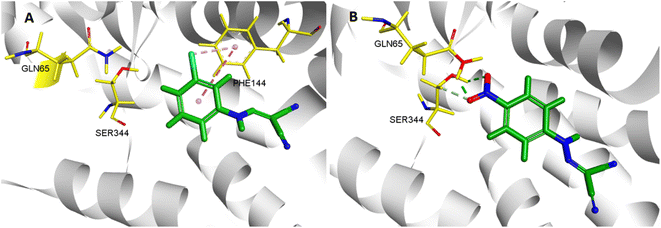 | ||
| Fig. 3 3D interaction patterns CCCP (A) and 2e (B) with NorA (PDB: 3WDO) using DS client v19.1.0 analysis. | ||
In order to assess the feasibility of 2e in vivo application, we performed an in silico analysis of physicochemical, pharmacokinetic and toxicological properties using Discovery Studio2018 software. Although 2e was predicted to be carcinogenic risk and mutagenic risk in humans, this cannot be considered a major problem in a drug discovery process, as many widely used drugs in the market are exist these risks. Outstandingly, the chronic oral lowest toxic dose of 2e (90.2 mg kg−1) is much higher than the actual dose (5 mg kg−1).
Molecular dynamics simulation
MD simulation mainly relies on Newtonian mechanics to simulate the motion of a molecular system. This approach can not only obtain the trajectory of atoms but also enables the observation of various microscopic details during atomic motion. Compound 2e was docked into the NorA protein pocket and then subjected to MD simulation to examine the structural stability of simulation system. MD simulation of complexes was performed for 20 ns period. In this study, the kinetic energy of 2e started to increase by temperature and time until a steady state was reached. As shown in Fig. 4A, before 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ps, the entire system of the complex NorA–2e was in a stable state. After 10
000 ps, the entire system of the complex NorA–2e was in a stable state. After 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ps, an obvious upward trend was emerged, especially at 12
000 ps, an obvious upward trend was emerged, especially at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ps, after which the overall system was relatively stable, and the wave range of RMSD was within 1 Å. Subsequently, calculated by the MM-PB(GB)SA method (Fig. 4B), the total binding free energy was −43.76 kcal mol−1 for the complex of NorA–2e, and the van der Waals energy was −30.01 kcal mol−1. Fig. 4C was the contributions of hot residues in the binding pocket of NorA protein. Under normal circumstances, a residue with lower interaction energy than −1 kcal mol−1 was considered to be essential for ligand recognition and combination. As disclosed in docking studies, hydrogen bonding of 2e with Gln65 (−2.44 kcal mol−1) was significant for binding to NorA protein.
000 ps, after which the overall system was relatively stable, and the wave range of RMSD was within 1 Å. Subsequently, calculated by the MM-PB(GB)SA method (Fig. 4B), the total binding free energy was −43.76 kcal mol−1 for the complex of NorA–2e, and the van der Waals energy was −30.01 kcal mol−1. Fig. 4C was the contributions of hot residues in the binding pocket of NorA protein. Under normal circumstances, a residue with lower interaction energy than −1 kcal mol−1 was considered to be essential for ligand recognition and combination. As disclosed in docking studies, hydrogen bonding of 2e with Gln65 (−2.44 kcal mol−1) was significant for binding to NorA protein.
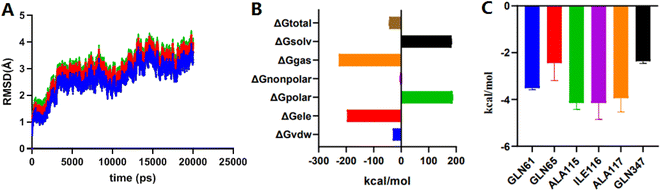 | ||
| Fig. 4 The RMSD plots of complex NorA–2e vs. time (A); total binding free energy and its component (B); residue contribution for receptor–ligand combination (C). | ||
Effect of 2e on norA gene expression
2e was used to examine the effect on the expression of norA gene in MRSA2858 using qRT-PCR (Fig. 5). 2e alone significantly down-regulated the gene expression of norA by 41.8%. Combination of 2e with 1/8MIC ofloxacin reduced the expression of norA by 65.9%. The results showed that 2e combined with low concentration of ofloxacin showed a synergistic inhibitory effect against MRSA by regulating NorA efflux pump.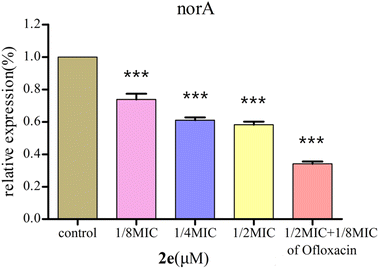 | ||
| Fig. 5 Effect of 2e and 1/8MIC ofloxacin (0.097 μM) on norA genes expression in MRSA. The control was the untreated biofilm control group (***p < 0.01). | ||
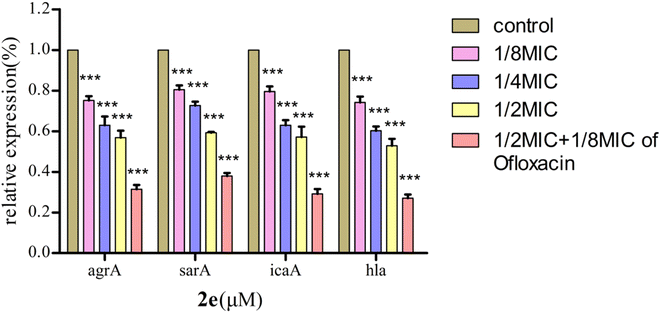
Effect on expression of QS regulatory genes
The QS system plays a role in the precise regulation of genes controlling virulence factors and biofilm formation in MRSA. 2e exerted dose-dependent inhibitory effects on virulence phenotypes (hla) regulated by QS in MRSA using qRT-PCR (Fig. 6). Moreover, the expression levels of QS regulatory genes, including agrA, sarA and icaA, were repressed after 2e combined with 1/8MIC ofloxacin treatment in a dose-dependent manner, and genes hla, agrA, sarA and icaA were down-regulated by 73.0%, 68.7%, 62.1% and 71.0%, respectively. These results confirmed the expression of QS regulatory genes (agrA, sarA, icaA, hla) were substantially down-regulated in treated with 1/2MIC 2e at 0.78 μM and 1/8MIC ofloxacin at 0.097 μM.Effect on MRSA2858 virulence
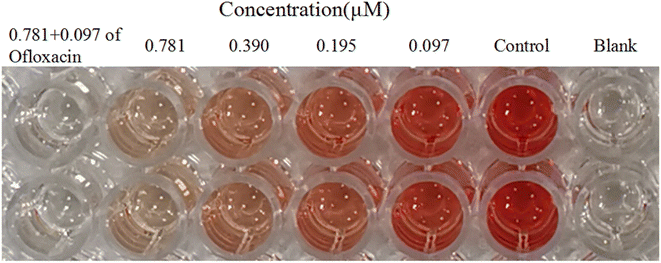
Alpha-hemolysin (Hla) was a major virulence factor in the pathogenesis of S. aureus infection, being active against a wide range of host cells. Hla created holes in the membranes of various host cells, such as immune system cells and erythrocytes. The amount of hemoglobin released from lysed red blood cells was taken as a measure of Hla production. 2e significantly decreased the level of hemolysis in a dose-dependent manner at 0.097–0.781 μM. When 2e was combined with 1/8MIC ofloxacin, the inhibition rate of α-hemolysin was 99.15%, while the inhibition rate of α-hemolysin for 2e alone was 93.75% (Table 4, Fig. 7). These results suggested that the synergistic antibacterial effect came along with the virulence inhibition of MRSA.| Compound (μM) | % inhibition of hemolysis of rabbit blood at | ||||
|---|---|---|---|---|---|
| 0.781 + 0.097 of ofloxacin | 0.781 | 0.390 | 0.195 | 0.097 | |
| 2e | 99.15 ± 0.64 | 93.75 ± 1.50 | 83.85 ± 2.24 | 68.88 ± 0.74 | 43.73 ± 0.91 |
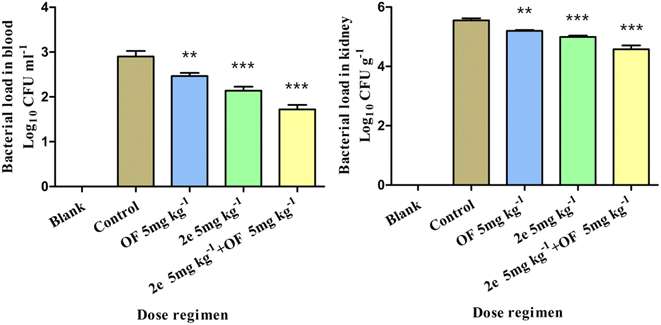
In vivo antibacterial activity
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 CFU ml−1, respectively (Fig. 8). The efficacy of 2e combined with ofloxacin was superior to that of 2e or ofloxacin, which suggested that 2e combined with ofloxacin could effectively reduce the MRSA abdominal infection.
log10 CFU ml−1, respectively (Fig. 8). The efficacy of 2e combined with ofloxacin was superior to that of 2e or ofloxacin, which suggested that 2e combined with ofloxacin could effectively reduce the MRSA abdominal infection.
| Blood parameter | Groups | ||||
|---|---|---|---|---|---|
| Negative control | Positive control | 2e (5 mg kg−1) | OF (5 mg kg−1) | 2e (5 mg kg−1) + OF (5 mg kg−1) | |
| a p < 0.05.b p < 0.01, compared with negative group. Data are presented in term of mean ± SD. | |||||
| WBC (109 l−1) | 10.48 ± 0.83 | 4.64 ± 1.12b | 5.94 ± 0.51b | 4.75 ± 1.11b | 8.52 ± 0.44a |
| NE (109 l−1) | 1.98 ± 0.39 | 0.98 ± 0.33a | 1.25 ± 0.21a | 1.15 ± 0.29a | 1.60 ± 0.51 |
| MONO (109 l−1) | 8.38 ± 0.46 | 3.00 ± 0.91b | 3.66 ± 0.39b | 4.21 ± 1.26b | 5.71 ± 1.16a |
| RBC (1012 l−1) | 11.26 ± 1.35 | 8.98 ± 0.42a | 9.70 ± 0.24 | 9.59 ± 0.23 | 9.81 ± 1.19 |
| HGB (g l−1) | 168.67 ± 18.01 | 135.33 ± 6.51a | 145.67 ± 2.08 | 143.00 ± 1.00 | 152.00 ± 14.73 |
Results and discussion
MRSA infection is a serious threat to human health. The ability of biofilm formation in MRSA can lead to resistance to most currently used antibiotics.27–29 Novel antibacterial agents can be developed to inhibit their function, to help reduce the virulence of bacteria and avoid drug resistance. There's different methods for evaluating biofilm formation, and the most common ones are CV staining for biomass and XTT (tetrazolium salt reduction) assay for viability.30 Because CV staining is often used for quantification of S. aureus biofilm formation,31 the CV method is more suitable to evaluate the inhibition of 2e for biofilm formation and biomass production.This study showed that 2e could inhibit the biofilm formation of MRSA in vitro. The transcription profiling demonstrated that the NorA efflux pathway was obviously inhibited by 2e treatment. As one of the most important efflux pump genes of S. aureus, norA gene was over expressed in 43% of strains.32,33 NorA was capable of extruding multiple structurally dissimilar substrates such as hydrophilic fluoroquinolones.34 The use of efflux pump inhibitors (EPIs) could decrease the MIC of antibacterial agents, increase susceptibility restoration in clinical resistant strains, and inhibit biofilm formation. NorA EPIs were capable of restoring drug susceptibility in resistant strains have been found, such as reserpine, verapamil, omeprazole, chlorpromazine, etc.35 The efflux pump can expel metabolites, toxic substances and antibacterial drugs out of the membrane of bacteria, which helps to maintain the vitality and virulence of starving bacteria in the biofilm. Therefore, the expression level of efflux pump was positively correlated with the formation of biofilm.34 We found that 2e markedly inhibited NorA pump activity.
The norA pump is regulated by the QS system. QS system has been known to be essential for robust biofilm production and development in numerous bacteria, including clinical multi-drug resistant isolates.36 Consistent with these observations, 2e treatment could down-regulate the expression of QS regulatory genes (agrA, sarA, icaA, hla) in a dose-dependent manner, decrease the virulence by blocking the NorA efflux pathway, and contribute to the biofilm inhibitory activity. Although biofilm development in MRSA is ica independent,37 we observed that the regulatory gene of MRSA2858 was ica dependent.
One of the key features of S. aureus infection was the production of a series of virulence factors, including secreted enzymes and toxins.38 In this study, the pore-forming toxin alpha-hemolysin (Hla) was a S. aureus secreted factor which participated in the activation of the NorA efflux pathway. This study showed that the anti-virulence activity of compound 2e combined with ofloxacin was superior to that of 2e alone against MRSA.
QS-controlled biofilm formation and virulence factor secretion by MRSA in clinical settings has remained controversial due to emerging drug resistance. Therefore, it is important to find diverse molecules for anti-biofilm or anti-QS activities.
Conclusion
It was urgent for the discovery of new anti-resistant agents with anti-biofilm and anti-efflux pump activity, especially therapeutic alternatives for multi-drug resistant MRSA. Carbonyl cyanide p-nitrophenylhydrazone 2e alone or combined with ofloxacin was found to have potent anti-MRSA activity. 2e combined with ofloxacin was exhibited anti-resistant activity through inhibiting NorA efflux pump, as well as decreasing biofilm formation, decreasing Hla and regulating QS systems of MRSA. Therefore, our results are significant and relevant to find novel therapeutic methods against MRSA to reduce the use of antibiotics.Ethical statement
In this study, MRSA strains were isolated and identified from diverse sources including blood, peritoneal fluid, and purulent leukorrhea of clinical infected patients, and the samples were collected by technically trained persons. These samples were evaluated and permitted by the Institutional Ethical Committee (IEC), Anhui No. 2 Provincial People's Hospital, China (IEC No. 2019814). The procedures for maintenance and treatment of laboratory animals were approved by the Animal Care and Use Committee of the Anhui No. 2 Provincial People's Hospital. All experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.Author contributions
Xueer Lu, Guifeng Wang and Yunfeng Xie contributed to the conception of the study and performed the experiment; Xueer Lu and Guifeng Wang contributed to analysis and manuscript preparation; Xueer Lu and Wenjian Tang helped perform the analysis with constructive discussions. Biyong Liu and Jing Zhang performed the data analyses and wrote the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.Conflicts of interest
The authors confirm that this article content has no conflict of interest.Acknowledgements
Financial support was provided by 2021 Scientific research project of Anhui Provincial Health Commission (AHWJ2021b054), and Anhui Provincial Natural Science Foundation (2008085MH261, 2208085QH282).References
- K. Ishii, F. Tabuchi, M. Matsuo, K. Tatsuno, T. Sato, M. Okazaki, H. Hamamoto, Y. Matsumoto, C. Kaito, T. Aoyagi, K. Hiramatsu, M. Kaku, K. Moriya and K. Sekimizu, Sci. Rep., 2015, 5, 17092 CrossRef CAS PubMed.
- T. Oyama, M. Miyazaki, M. Yoshimura, T. Takata, H. Ohjimi and S. Jimi, Toxins, 2016, 8, 198 CrossRef PubMed.
- F. Lamret, M. Colin, C. Mongaret, S. C. Gangloff and F. Reffuveille, Antibiotics, 2020, 9, 547 CrossRef CAS PubMed.
- T. Gonzalez, M. L. Stevens, A. Baatyrbek Kyzy, R. Alarcon, H. He, J. W. Kroner, D. Spagna, B. Grashel, E. Sidler, L. J. Martin, J. M. Biagini Myers, G. K. Khurana Hershey and A. B. Herr, Allergy, 2021, 76, 302–313 CrossRef CAS PubMed.
- D. Kaizerman-Kane, M. Hadar, R. Joseph, D. Logviniuk, Y. Zafrani, M. Fridman and Y. Cohen, ACS Infect. Dis., 2021, 7, 579–585 CrossRef CAS PubMed.
- E. C. Spindler, J. D. F. Hale, T. H. Giddings, R. E. W. Hancock and R. T. Gill, Antimicrob. Agents Chemother., 2011, 55, 1706–1716 CrossRef CAS PubMed.
- Y. K. Park and K. S. Ko, J. Microbiol., 2015, 53, 53–59 CrossRef CAS PubMed.
- Y. F. Mohamed, H. M. Abou-Shleib, A. M. Khalil, N. M. El-Guink and M. A. El-Nakeeb, Braz. J. Microbiol., 2016, 47, 381–388 CrossRef CAS PubMed.
- W. Ni, Y. Li, J. Guan, J. Zhao, J. Cui, R. Wang and Y. Liu, Antimicrob. Agents Chemother., 2016, 60, 3215–3218 CrossRef CAS PubMed.
- Z. Yu, Y. Cai, W. Qin, J. Lin and J. Qiu, PLoS One, 2015, 10, e0135198 CrossRef PubMed.
- D. Gupta, A. Singh and A. U. Khan, Nanoscale Res. Lett., 2017, 12, 454 CrossRef PubMed.
- P. J. F. Henderson, C. Maher, L. D. H. Elbourne, B. A. Eijkelkamp, I. T. Paulsen and K. A. Hassan, Chem. Rev., 2021, 121, 5417–5478 CrossRef CAS PubMed.
- X. Lu, Z. Zhang, Y. Xu, J. Lu, W. Tang and J. Zhang, RSC Adv., 2020, 10, 17854–17861 RSC.
- I. Alav, J. M. Sutton and K. M. Rahman, J. Antimicrob. Chemother., 2018, 8, 2003–2020 CrossRef PubMed.
- J. Reichling, Planta Med., 2020, 86, 520–537 CrossRef CAS PubMed.
- O. Simonetti, G. Rizzetto, G. Radi, E. Molinelli, O. Cirioni, A. Giacometti and A. Offidani, Antibiotics, 2021, 10, 1377 CrossRef CAS PubMed.
- X. P. Guo and Y. C. Sun, Front. Microbiol., 2017, 8, 2014 CrossRef PubMed.
- P. Piewngam, Y. Zheng, T. H. Nguyen, S. W. Dickey, H. S. Joo, A. E. Villaruz, K. A. Glose, E. L. Fisher, R. L. Hunt, B. Li, J. Chiou, S. Pharkjaksu, S. Khongthong, G. Y. C. Cheung, P. Kiratisin and M. Otto, Nature, 2018, 562, 532–537 CrossRef CAS PubMed.
- S. Stepanović, D. Vuković, V. Hola, G. Di Bonaventura, S. Djukić, I. Cirković and F. Ruzicka, APMIS, 2007, 115, 891–899 CrossRef PubMed.
- K. Xu, S. He, S. Chen, G. Qiu, J. Shi, X. Liu, X. Wu, J. Zhang and W. Tang, Eur. J. Med. Chem., 2018, 154, 144–154 CrossRef CAS PubMed.
- A friendly and versatile cloud computing platform for biomedicinal, material, and statistical researches, https://cloud.yinfotek.com/ Search PubMed.
- X. Jiang, Z. Zhang, J. Zuo, C. Wu, L. Zha, Y. Xu, S. Wang, J. Shi, X. H. Liu, J. Zhang and W. Tang, Eur. J. Med. Chem., 2021, 223, 113735 CrossRef CAS PubMed.
- M. Sivaranjani, K. Leskinen, C. Aravindraja, P. Saavalainen, S. K. Pandian, M. Skurnik and A. V. Ravi, Front. Microbiol., 2019, 10, 150–166 CrossRef PubMed.
- A. Valliammai, S. Sethupathy, A. Priya, A. Selvaraj, J. P. Bhaskar, V. Krishnan and S. K. Pandian, Sci. Rep., 2019, 9, 13744 CrossRef PubMed.
- J. B. Kaplan, E. A. Izano, P. Gopal, M. T. Karwacki, S. Kim, J. L. Bose, K. W. Bayles and A. R. Horswill, mBio, 2012, 3, e00198–12 CrossRef CAS PubMed.
- M. R. M. Ranieri, C. B. Whitchurch and L. L. Burrows, Curr. Opin. Microbiol., 2018, 45, 164–169 CrossRef CAS PubMed.
- D. Sharma, L. Misba and A. U. Khan, Antimicrob. Resist. Infect. Control, 2019, 8, 76 CrossRef PubMed.
- A. Warrier, K. Satyamoorthy and T. S. Murali, Future Microbiol., 2021, 16, 1003–1021 CrossRef CAS PubMed.
- J. Zhang, H. Huang, X. Zhou, Y. Xu, B. Chen, W. Tang and K. Xu, ACS Med. Chem. Lett., 2019, 10, 329–333 CrossRef CAS PubMed.
- B. Alonso, J. Lucio, M. J. Pérez-Granda, R. Cruces, C. Sánchez-Carrillo, E. Bouza and M. Guembe, J. Microbiol. Methods, 2016, 131, 110–112 CrossRef CAS PubMed.
- B. Alonso, R. Cruces, A. Perez, A. Fernandez-Cruz and M. Guembe, Front. Biosci., Scholar Ed., 2018, 10, 300–308 Search PubMed.
- P. S. Pereira, M. D. C. A. Lima, P. P. M. Neto, C. D. M. Oliveira-Tintino, S. R. Tintino, I. R. A. Menezes, J. F. de Oliveira, P. Marchand, H. D. M. Coutinho, M. D. D. Rodrigues and T. G. da Silva, Bioorg. Med. Chem., 2019, 27, 3797–3804 CrossRef CAS PubMed.
- K. L. C. Monteiro, T. M. de Aquino and F. J. B. Mendonça Junior, Curr. Top. Med. Chem., 2020, 20, 2168–2185 CrossRef PubMed.
- A. A. Neyfakh, C. M. Borsch and G. W. Kaatz, Antimicrob. Agents Chemother., 1993, 37, 128–129 CrossRef CAS PubMed.
- Y. Wang, H. Venter and S. Ma, Curr. Drug Targets, 2016, 17, 702–719 CrossRef CAS PubMed.
- M. M. Brown, J. M. Kwiecinski, L. M. Cruz, A. Shahbandi, D. A. Todd, N. B. Cech and A. R. Horswill, Antimicrob. Agents Chemother., 2020, 64, e00172–20 CrossRef PubMed.
- E. O'Neill, C. Pozzi, P. Houston, D. Smyth, H. Humphreys, D. A. Robinson and J. P. O'Gara, J. Clin. Microbiol., 2007, 45, 1379–1388 CrossRef PubMed.
- Y. Zheng, X. Lu, B. Liu, B. Li, C. Yang, W. Tang and J. Zhang, Microb. Pathog., 2022, 163, 105391 CrossRef CAS PubMed.
Footnote |
| † Xueer Lu and Guifeng Wang contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |