Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Open-source tool for real-time and automated analysis of droplet-based microfluidic†
Joana P.
Neto
 *a,
Ana
Mota
a,
Gonçalo
Lopes
b,
Beatriz J.
Coelho
*a,
Ana
Mota
a,
Gonçalo
Lopes
b,
Beatriz J.
Coelho
 ac,
João
Frazão
d,
André T.
Moura
ac,
João
Frazão
d,
André T.
Moura
 a,
Beatriz
Oliveira
c,
Bárbara
Sieira
a,
Beatriz
Oliveira
c,
Bárbara
Sieira
 a,
José
Fernandes
a,
Elvira
Fortunato
a,
José
Fernandes
a,
Elvira
Fortunato
 a,
Rodrigo
Martins
a,
Rui
Igreja
a,
Rodrigo
Martins
a,
Rui
Igreja
 a,
Pedro V.
Baptista
a,
Pedro V.
Baptista
 c and
Hugo
Águas
c and
Hugo
Águas
 *a
*a
aCENIMAT|i3N, Department of Materials Science, NOVA School of Science and Technology, NOVA University of Lisbon and CEMOP/UNINOVA, Campus de Caparica, Caparica, Portugal. E-mail: joana.neto@campus.fct.unl.pt; hma@fct.unl.pt
bNeuroGEARS Ltd., London, UK
cUCIBIO, I4HB, Department of Life Sciences, NOVA School of Science and Technology, NOVA University of Lisbon, Campus de Caparica, Caparica, Portugal
dChampalimaud Centre for the Unknown, Champalimaud Foundation, Lisbon, Portugal
First published on 14th June 2023
Abstract
Droplet-based microfluidic technology is a powerful tool for generating large numbers of monodispersed nanoliter-sized droplets for ultra-high throughput screening of molecules or single cells. Yet further progress in the development of methods for the real-time detection and measurement of passing droplets is needed for achieving fully automated systems and ultimately scalability. Existing droplet monitoring technologies are either difficult to implement by non-experts or require complex experimentation setups. Moreover, commercially available monitoring equipment is expensive and therefore limited to a few laboratories worldwide. In this work, we validated for the first time an easy-to-use, open-source Bonsai visual programming language to accurately measure in real-time droplets generated in a microfluidic device. With this method, droplets are found and characterized from bright-field images with high processing speed. We used off-the-shelf components to achieve an optical system that allows sensitive image-based, label-free, and cost-effective monitoring. As a test of its use we present the results, in terms of droplet radius, circulation speed and production frequency, of our method and compared its performance with that of the widely-used ImageJ software. Moreover, we show that similar results are obtained regardless of the degree of expertise. Finally, our goal is to provide a robust, simple to integrate, and user-friendly tool for monitoring droplets, capable of helping researchers to get started in the laboratory immediately, even without programming experience, enabling analysis and reporting of droplet data in real-time and closed-loop experiments.
Introduction
Droplet microfluidics has been globally adopted for chemistry and biology research and in a wide variety of applications, such as nucleic acid amplification, single-cell experimentation, or chemical synthesis1 – to mention a few. Moreover, microfluidics technology enables faster and decentralized analysis (e.g., point-of-care platforms) for a sustainable healthcare system. Nevertheless, a major challenge in droplet-based microfluidic platforms lies in monitoring thousands, even millions, of passing droplets. Accurate control of droplet and production parameters is critical since changes in droplet features, such as volume, for instance, lead to errors in analysis results (e.g., estimation of product concentration).2 Several monitoring methodologies have been developed to perform automated droplet detection and measurements. Real-time droplet detection methods include fluorescence,3,4 electrical measurements5–8 and other optical methods.9–15 However, fluorescence labelling is undesirable for some applications and despite the remarkable progress in the field, label-free monitoring methods are often complex and therefore require substantial effort for integration in the experimentation setup, or if commercially available they are prohibitively expensive.13 Moreover, label-free methods based on optical monitoring, which use bright-field imaging for real-time visual inspection of droplets, do not allow fast imaging processing and analysis.9–12To address this, we present a method based on bright-field imaging, which allows automatic, fast and high-throughput quantification in real-time of flowing water-in-oil droplets. The software used, Bonsai, is freely available and is capable of real-time object tracking and measuring, and is supported by a growing community of users.16,17 One of the first use cases driving the development of Bonsai was the automated real-time tracking of animal behavior using video, which involves image segmentation and binary region analysis to allow the extraction of the spatial location of an animal over time.18 Critically, Bonsai is a visual programming language, meaning that users with little or no previous coding experience can quickly use and adapt workflows for imaging analysis. Nowadays, Bonsai is widely used in neuroscience experimentation and it is capable of interfacing with most types of external hardware, as well as fast visualization, analysis and interaction with the experiments, enabling closed-loop experimental designs.19 In other words, Bonsai enables automated adjustment of experimental parameters through feedback, almost instantaneously due to its high processing speed.
The same method can easily be extended to track droplets, with adequate illumination contrast by exploiting the dark boundary surrounding the droplet, which occurs due to the refractive index difference between the two phases, and the appropriate choice of image processing steps for droplet segmentation. In this work we have translated a software widely used for recording precise measurements of behavior in animals to passing nanoliter-sized droplets in microfluidics devices. Bonsai excelled in tracking and measuring passing droplet parameters such as radius, speed or production frequency, regardless of the experience of the user or experimental conditions. Briefly, our system relies on open-source software and on conventional brightfield videos acquired through an overhanging camera. It is easily accessible due to its lower costs and can be implemented by non-trained users. It presents great flexibility and adaptability in terms of the measurements to be performed and microfluidic devices to be tested. Therefore, our method has the potential to become a widely used tool in droplet-based microfluidic field, helping to improve automation and scalability of high-throughput screening technology.
Experimental
Microfluidic device fabrication and water-in-oil droplet generation
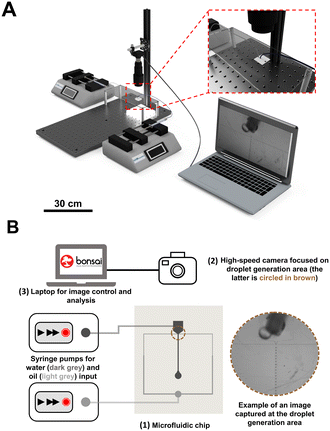
The microfluidic devices were fabricated using standard photo- and soft-lithography protocols developed earlier by the group.20–22 Water-in-oil droplets were generated at a flow-focusing junction of a microfluidic device (Fig. S1 of ESI†). Regarding the continuous phase, mineral oil with 0.5 wt% SPAN 80 (Sigma) was prepared, whereas ultra-pure water was used for the dispersed phase. Two syringes (Hamilton® GASTIGHT® syringe, 1700 series, PTFE Luer lock, 1750TLL, volume 0.5 mL) were filled with the continuous and dispersed phases, respectively. Continuous and dispersed phases were injected by syringe pumps (Legato210P, KDScientific) into the inlets of the device. The syringes were connected to the inlets via microfluidic tubes and blunt-end Luer lock syringe needles (LVF-KTU-13 and AE-23G-100x, Darwin microfluidics). The water-in-oil droplet generation began by filling the device with the continuous phase. A second syringe previously filled with ultra-pure water was then connected to the inlet. A drainage tube was connected to the outlet, to accommodate liquid overflow. Different droplet sizes, speeds and production rates were tested by adjusting the flow rates of continuous oil and dispersed aqueous phases, from 1.25 to 5 μl min−1, and 0.25 to 2 μl min−1, respectively.Optical experimental setup
The optical experimental setup consists of a high rate and sensitive CMOS camera (CM3-U3-13Y3M-CS, Chameleon 3, USB3, 1.3 MP, 149 FPS, PYTHON 1300, MONO). In high-definition mode (1280 × 1024), the camera is capable of recording up to 150 frames per second (fps), with an upper limit of around 600 fps in low-resolution mode (60 × 120). A magnification lens was also installed into the optical path (Computar, MLM3X-MP), considering the small size of the passing droplets in the microfluidic device. A home-made illumination set (LED light) was positioned under the device to facilitate image acquisition. We used a standard laptop with the following specifications: Dell XPS 15 9550 with an Intel Core i7 2.6 GHz CPU, 32 GB RAM, and Windows 10 64-bit.Bonsai workflow for droplet analysis
We developed a powerful and easy-to-use graphical user interface within the Bonsai programming language, to obtain several measurements from passing droplets. Users can find documentation, video tutorials, online support, and other materials in its accompanying website (https://bonsai-rx.org/). To quickly acquaint themselves with the basics of Bonsai, users can access example workflows online (https://bonsai-rx.org/docs/tutorials/acquisition.html). Additionally, video tutorials on how to implement workflows for data processing and storage can also be found here: https://bonsai-rx.org/learn/. Once a Bonsai project is opened, three distinct panels will appear from left to right: ‘Toolbox panel’, ‘Workflow panel’ and ‘Properties panel’ (Fig. 1). Inside the ‘Workflow panel’, there are three group nodes, which represent different tasks within the droplet pipeline analysis: i) ‘Image Acquisition’, ii) ‘Feature Extraction’ and iii) ‘Droplet Analysis’.Briefly, with these nodes, it is possible to set the type of video that is being analyzed (real-time recording or previously recorded video), to define parameters in real-time for optimizing analysis, to extract direct measurements related to the droplets, and to visualize and save droplet data. The workflow and the nodes are explained in detail in ESI† (Fig. S2–S4).
ImageJ for method validation
To validate the performance of our method, we analyzed the recorded videos using a conventional offline software, ImageJ.23 To this end, the video was imported and all frames converted into grayscale. The ‘Set Scale…’ was calibrated by introducing values of channel width in microns (200 μm) and pixels (25 pixels). The ROI was adjusted using ‘Crop’ command in such a way to include droplets and to avoid channel wall. The droplet radius and generation rate were manually determined for each experiment (i.e., flow condition) including 20 and 10 droplets per video, respectively. Regarding speed analysis, all frames were converted into binary images using ‘Make Binary’ command, where threshold is calculated for each image by default method, and the wrMTrck plugin24 was used to obtain the average droplet speed for each experiment (Fig. S5 of ESI†).Results and discussion
Setup
Fig. 2 illustrates an easy-to-implement and cost-effective setup used for monitoring droplets in microfluidic device. This setup does not require complicated or expensive hardware, as cameras are now available at an affordable cost and can be placed anywhere, provided that an adequate view of the microfluidic device is achieved (please see Fig. S6 of ESI† for more detail about the setup).Our approach does not require fluorescent tracers; instead, it operates on conventional brightfield digital videos by exploiting the dark boundary surrounding the droplet,25 which makes it compatible with a wide variety of microfluidic systems. Bonsai takes advantage of bright-field imaging which enables real-time visual inspection of passing droplets as shown in Fig. 2B. It is important to achieve excellent edge contrast while recording videos, which is why a custom-made LED lighting platform was placed below the support frame of the microfluidic device. Since the Bonsai software relies on droplet boundary to identify droplets, simple steps to maximize the video quality should be considered during experiments. Firstly, the video should have sufficiently illumination so that the droplet boundaries have high contrast. Secondly, the focal plane should be at the midplane of the droplet rather than the upper or lower plane of the channel (e.g., focus the boundary of the droplet). Finally, the microfluidic channel should be positioned in parallel to one of the video frames, x- or y-axis. It is extremely important to meet these requirements for the following imaging processing pipeline and droplet analysis. Another critical step in the analysis pipeline, that is user-dependent, is the selection of the crop and threshold values. Fig. 3 shows a screenshot of Bonsai data visualization of an experiment, where it is possible to observe an example of optimized data, such as droplet image, crop and threshold value for analysis. The user should use these visualizers to assist debugging and inspection of data, including droplet imaging and detection. For droplet detection, the threshold value sets the sensitivity of the edge detector. Lower values will be able to detect low-contrast boundaries, but at the expense of more noise (artifacts), and vice versa. A suitable value should be empirically chosen. A dataset with three videos (experiment 2, 10 and 12), as well as the workflow and a tutorial, is available online https://github.com/JoanaPNeto/Droplets. New users may resort to this information for practicing droplet analysis with Bonsai. Moreover, it is a good practice to set the crop at [0, 0, 0, 0] and threshold value at 255 (highest value) when starting analysis, and after that, adjusting them to obtain visualization outputs as the ones from Fig. 3 before saving the results. In conclusion, Bonsai operates on conventional brightfield videos by exploiting the dark boundary surrounding the droplet. Droplets are identified through a series of image processing steps and users are advised to optimize lightening conditions and the focus and zoom to capture a sharp contrast between droplets and background.
Method validation
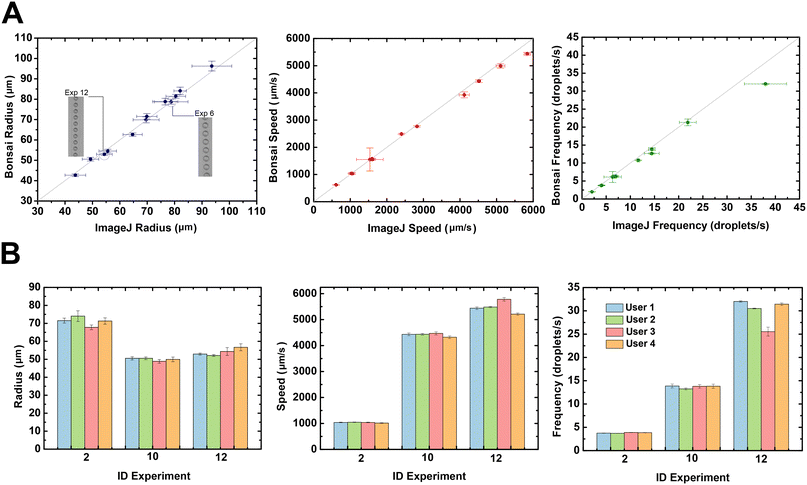
Our Bonsai workflow has been used to perform droplet measurement on a typical microfluidic droplet generator using different flow conditions. We validated our method by analyzing droplets radius, speed and production rate with the workflow described above and comparing the results obtained using ImageJ, a widely used software for offline analysis (Fig. 4). As can be seen on Fig. 4, all parameters measured by the Bonsai software (radius, speed and production frequency) are close to the results obtained with the offline imaging analysis software, ImageJ. The insets in Fig. 4A further show the ROI of passing droplets differing in size. The increased flow rates lead to smaller droplets with shorter spacing and therefore higher droplet production rates. For example, at the highest flow rate (experiment 12) the average droplet radius measured with Bonsai was 52.9 ± 0.5 μm compared to 54.4 ± 2.9 μm measured manually with ImageJ. In the case of speed, the obtained results were 5438.8 ± 50.8 μm s−1versus 5836.8 ± 82.6 μm s−1 for the Bonsai versus ImageJ measurements, respectively. For the production rate, Bonsai measured a droplet production rate of 32.0 ± 0.2 Hz, compared to the manually determined rate of 38.0 ± 4.3 Hz. For detailed information, see Table S1 in the ESI,† which summarizes all measurements. The differences between the values and associated standard deviation can be attributed to the number of droplets analyzed by each method (i.e., number of droplets averaged). The radius, speed and droplet generation rate against time show ripples and variations during the video recording. These variations are caused by instabilities of the microfluidic system, including syringe pumps and mechanical vibrations. It is likely that offline quantification is more affected by a ‘ripple region’ due to the small sampling. Bonsai values are averaged over the whole measurement time including ≈576 droplets, while for manual analysis only 10 and 20 droplets were considered for production rate and radius measurements, respectively. The measurements for radius, speed and frequency are found to be within a Bonsai and ImageJ ratio (0.97–1.03), (0.93–1.03) and (0.84–0.97), respectively. Here, it is reasonable to conclude that Bonsai is accurate in the above measurements. Moreover, we evaluated the reproducibility of our proposed analysis method by providing the videos to 4 users (with distinct levels of experience in using Bonsai), who resorted to the developed workflows to analyze the droplets.User 1 is an experienced user, whereas users 2 to 4 are new users. Experiences 2, 10 and 12 were selected due to the difference on the droplet generation rate. As illustrated in Fig. 4B (see also Fig. S7 from ESI†), all users achieve similar results in all parameters, despite their experience level with the Bonsai software. A critical step in the analysis pipeline, that is user-dependent, is the selection of the crop and threshold values in order to convert video into binary images. This step is done manually by inputting and selecting the parameters, which are evaluated by observing the data output. The differences between users can be attributed to differences in the selection of user-dependent parameters such as, ROI (i.e., crop size (width and height) and position (x and y)) and threshold value (see Fig. S8 from ESI†). The ROI defines the number of droplets analyzed per frame. As the selection is done visually, the values are different from one user to other. We have studied the fluctuation of these values and their implication to droplets measurement on the same video. As illustrated in Fig. S8A,† the radius is highly affected by the threshold value. Moreover, the standard deviation of the radius decreases for a higher number of droplets, since more droplets are averaged. Nevertheless, there is a drawback in increasing the number of droplets due to the increased probability of averaging artifacts (i.e., inclusion of unqualified droplets), as shown in Fig. S9 of the ESI.† For instance, for experiment 2, the highest number of droplets was 12 without the ‘BinaryRegionAnalysis’ node detecting artifacts, while for experiment 12, this number was higher, 21 droplets. Therefore, there is a compromise between a high droplet count and the probability of appearance of artifacts. However, the speed and production rate (i.e., frequency) are relatively constant for a wide range of droplet counts and threshold values. Nonetheless, there is also a maximum value for both number of droplets and threshold, above which the detection of droplets will be affected by artifacts and noise. Above a certain value, the speed and frequency decrease because the ‘BinaryRegionAnalysis’ node starts to extract artifacts, such as microfluidic walls (Fig. S9 of the ESI†). Thus, the user must determine the optimal balance between the quantity of droplets and the ideal threshold value to avoid distortion of the droplet detection process. In the workflow pipeline, droplets are detected by continuous contours from the binary image. The reliability of droplet detection and the accuracy of the droplet measurements relies in the threshold value that is essential to find droplet boundaries. For instance, if the value is set too large, it can distort the droplet shape, or form a bridge between two adjacent droplets. In our experiments, the experienced user (user 1) has selected empirically a ROI with 12 droplets and a threshold value of 136, which are within a range of optimal values found in Fig. S9 of the ESI.† Additionally, even the inexperienced users have obtained similar results. Hence, these results attest to the repeatability and robustness of the workflow developed in Bonsai, and provide a good indication of their simplicity and ease-of-use by inexperienced researchers. However, even the experienced user may encounter more challenging experiments where droplets are closely packed or have small radius and high generation rate (Fig. S10 of the ESI†). In summary, Bonsai enables a reliable extraction of features from the video, thus empowering a wide range of applications for the system presented herein, such as chemical synthesis, nucleic acid amplification and cell sorting and screening, among others. For each droplet found, Bonsai is capable of providing several parameters, including major and minor axis, centroid position, orientation and area. In this work we have monitoring in real-time a droplet generator performance by measuring radius, speed and generation rate of passing droplets. But with these measures is possible to further control droplet production parameters, such volume, to estimate the correct product concentration. Additionally, a plot of droplet speed against droplet centroid position may reveals subtle periodic flow perturbations due to the pulsatile motion of stepper motors from syringe pumps for droplets in serpentine channel. Moreover, the continuous quality monitoring of microfluidic systems enabled by our system is not possible with other image analysis techniques within reasonable cost, time and effort. Overall, we validated a new method for real-time analysis of droplet-based microfluidic processes.
Conclusions
One of the major challenges in droplet-based microfluidic systems is the quality control of the thousands of droplets circulating within microfluidic channels. Existing methodologies are either prohibitively expensive, or require extensive training before use, or require complex experimentation setups. Here, we report an alternative method based on Bonsai, an open-source visual programming tool, associated with an experimental optical setup that is relatively inexpensive, simple to assemble and it is able to accommodate different microfluidic devices. The software, Bonsai, is a visual programming language, meaning that users with litter or no previous experience can quickly control and develop workflows for image analysis. Briefly, Bonsai workflows are constructed by connecting functions, or ‘operators’ that come in the form of nodes, together.26 In this way, Bonsai unlocks complex image processing schemes to untrained users.In this work, a custom Bonsai workflow was used to track and evaluate droplets flowing within a microfluidic device, in terms of radius, speed and droplet production frequency. We confirmed the ease-of-use of our droplet analysis methodology though the comparison of the results obtained from an experienced user with those of new users, for the same set of videos. The outputs achieved by the experienced Bonsai operator were identical to the outputs achieved by new users. Moreover, we evaluated the functionality and sensitivity of Bonsai by comparing the output results with a standard image analysis software, ImageJ. Briefly, droplet production rate, radius, and speed measured with Bonsai matched the results obtained with ImageJ, for several tested experimental conditions.
Due to easy and fast operation and integration of the Bonsai software (Table S2†), we envision that Bonsai will be a powerful tool for real-time droplet monitoring, enabling automated analysis of passing droplets and facilitating the commercialization of droplet systems. Bonsai also allows interfacing with syringe pumps and pressure systems, thus enabling closed-loop experiments. The basic functionalities demonstrated herein set the path for various new experimental setups and research procedures, such as evaluating droplet splitting and merging, cell encapsulation, cell sorting, droplet sorting, among others.
Author contributions
JPN, AM conceptualization, methodology, software, investigation, formal analysis, writing – review & editing. GL and JF conceptualization, methodology, software. BJC, ATM, BO, BS, JF formal analysis, writing – review & editing, visualization. RM and EF funding acquisition, resources. RI, PVB and HA supervision, funding acquisition, resources, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank the support provided by NeuroGears, CENIMAT|I3N, UCIBIO and i4HB. This work was financed by national funds from FCT – Fundação para a Ciência e a Tecnologia, I. P., in the scope of the projects LA/P/0037/2020, UIDP/50025/2020 and UIDB/50025/2020 of the Associate Laboratory Institute of Nanostructures, Nanomodelling and Nanofabrication – i3N, and also under project dPCR4FreeDNA of the same research unit – PTDC/BTM-SAL/31201/2017. Furthermore, the work received funding from FCT in the scope of projects UIDP/04378/2020 and UIDB/04378/2020 of the Research Unit on Applied Molecular Biosciences – UCIBIO and the project LA/P/0140/2020 of the Associate Laboratory Institute for Health and Bioeconomy – i4HB.References
- P. Garstecki, S. T. S. Kaminski, P. Garstecki and T. S. Kaminski, Controlled droplet microfluidic systems for multistep chemical and biological assays, Chem. Soc. Rev., 2017, 46(20), 6210–6226 RSC , Available from: https://pubs.rsc.org/en/content/articlehtml/2017/cs/c5cs00717h.
- S. Sohrabi, N. Kassir and M. Keshavarz Moraveji, Droplet microfluidics: fundamentals and its advanced applications, RSC Adv., 2020, 10(46), 27560–27574 RSC , Available at: https://xlink.rsc.org/?DOI=D0RA04566G.
- E. Zamir, C. Frey, M. Weiss, S. Antona, J. P. Frohnmayer and J.-W. Janiesch, et al., Reconceptualizing Fluorescence Correlation Spectroscopy for Monitoring and Analyzing Periodically Passing Objects, Anal. Chem., 2017, 89(21), 11672–11678 CrossRef CAS PubMed , Available at: https://pubs.acs.org/doi/10.1021/acs.analchem.7b03108.
- J. Fattaccioli, J. Baudry, J.-D. Émerard, E. Bertrand, C. Goubault and N. Henry, et al., Size and fluorescence measurements of individual droplets by flow cytometry, Soft Matter, 2009, 5(11), 2232 RSC , Available from: https://pubs.rsc.org/en/content/articlehtml/2009/sm/b814954b.
- C. Elbuken, T. Glawdel, D. Chan and C. L. Ren, Detection of microdroplet size and speed using capacitive sensors, Sens. Actuators, A, 2011, 171(2), 55–62, DOI:10.1016/j.sna.2011.07.007.
- E. W. M. Kemna, L. I. Segerink, F. Wolbers, I. Vermes and A. Van Den Berg, Label-free, high-throughput, electrical detection of cells in droplets, Analyst, 2013, 138(16), 4585–4592 RSC , Available from: https://pubs.rsc.org/en/content/articlehtml/2013/an/c3an00569k.
- A. Saateh, A. Kalantarifard, O. T. Celik, M. Asghari, M. Serhatlioglu and C. Elbuken, Real-time impedimetric droplet measurement (iDM), Lab Chip, 2019, 19(22), 3815–3824 RSC , Available from: https://pubs.rsc.org/en/content/articlehtml/2019/lc/c9lc00641a.
- G. Yesiloz, M. S. Boybay and C. L. Ren, Label-free high-throughput detection and content sensing of individual droplets in microfluidic systems, Lab Chip, 2015, 15(20), 4008–4019 RSC , Available from: https://pubs.rsc.org/en/content/articlehtml/2015/lc/c5lc00314h.
- Z. Z. Chong, S. B. Tor, A. M. Gañán-Calvo, Z. J. Chong, N. H. Loh and N. T. Nguyen, et al., Automated droplet measurement (ADM): an enhanced video processing software for rapid droplet measurements, Microfluid. Nanofluid., 2016, 20(4), 66 CrossRef , Available from: https://link.springer.com/article/10.1007/s10404-016-1722-5.
- V. Anagnostidis, B. Sherlock, J. Metz, P. Mair, F. Hollfelder and F. Gielen, Deep learning guided image-based droplet sorting for on-demand selection and analysis of single cells and 3D cell cultures, Lab Chip, 2020, 20(5), 889–900 RSC , Available from: https://pubs.rsc.org/en/content/articlehtml/2020/lc/d0lc00055h.
- D. F. Crawford, C. A. Smith and G. Whyte, Image-based closed-loop feedback for highly mono-dispersed microdroplet production, Sci. Rep., 2017, 7(1), 10545 CrossRef CAS PubMed , Available at: https://www.nature.com/articles/s41598-017-11254-5.
- E. Zang, S. Brandes, M. Tovar, K. Martin, F. Mech and P. Horbert, et al., Real-time image processing for label-free enrichment of Actinobacteria cultivated in picolitre droplets, Lab Chip, 2013, 13(18), 3707–3713, 10.1039/C3LC50572C.
- C. Frey, J. Pfeil, T. Neckernuss, D. Geiger, K. Weishaupt and I. Platzman, et al., Label-free monitoring and manipulation of microfluidic water-in-oil droplets, View, 2020, 1(4), 20200101 CrossRef , Available from: https://onlinelibrary.wiley.com/doi/full/10.1002/VIW.20200101.
- R. M. Maceiczyk, D. Hess, F. W. Y. Chiu, S. Stavrakis and A. J. DeMello, Differential detection photothermal spectroscopy: towards ultra-fast and sensitive label-free detection in picoliter & femtoliter droplets, Lab Chip, 2017, 17(21), 3654–3663 RSC , Available from: https://pubs.rsc.org/en/content/articlehtml/2017/lc/c7lc00946a.
- C. Song, T. Jin, R. Yan, W. Qi, T. Huang and H. Ding, et al., Opto-acousto-fluidic microscopy for three-dimensional label-free detection of droplets and cells in microchannels, Lab Chip, 2018, 18(9), 1292–1297 RSC , Available from: https://pubs.rsc.org/en/content/articlehtml/2018/lc/c8lc00106e.
- G. Lopes, N. Bonacchi, J. Frazão, J. P. Neto, B. V. Atallah and S. Soares, et al., Bonsai: An event-based framework for processing and controlling data streams, Front. Neuroinform., 2015, 9, 7 Search PubMed , Available from: https://journal.frontiersin.org/article/10.3389/fninf.2015.00007/abstract.
- G. Lopes, Bonsai [Internet], [cited 2023 Jan 13], Available from: https://bonsai-rx.org/ Search PubMed.
- G. Lopes and P. Monteiro, New Open-Source Tools: Using Bonsai for Behavioral Tracking and Closed-Loop Experiments, Front. Behav. Neurosci., 2021, 15, 53 Search PubMed , Available from: https://www.frontiersin.org/articles/10.3389/fnbeh.2021.647640/full.
- G. Lopes, K. Farrell, E. A. Horrocks, C.-Y. Lee, M. M. Morimoto and T. Muzzu, et al., Creating and controlling visual environments using BonVision, eLife, 2021, 10, e65541 CrossRef CAS PubMed , Available from: https://elifesciences.org/articles/65541.
- P. U. Alves, R. Vinhas, A. R. Fernandes, S. Z. Birol, L. Trabzon and I. Bernacka-Wojcik, et al., Multifunctional microfluidic chip for optical nanoprobe based RNA detection – application to Chronic Myeloid Leukemia, Sci. Rep., 2018, 8(1), 381 CrossRef PubMed , Available from: https://www.nature.com/articles/s41598-017-18725-9.
- I. Bernacka-Wojcik, P. Lopes, A. Catarina Vaz, B. Veigas, P. Jerzy Wojcik and P. Simões, et al., Bio-microfluidic platform for gold nanoprobe based DNA detection—application to Mycobacterium tuberculosis, Biosens. Bioelectron., 2013, 48, 87–93 CrossRef CAS PubMed , Available from: https://linkinghub.elsevier.com/retrieve/pii/S0956566313002492.
- F. Pereira, I. Bernacka-Wojcik, R. Ribeiro, M. Lobato, E. Fortunato and R. Martins, et al., Hybrid Microfluidic Platform for Multifactorial Analysis Based on Electrical Impedance, Refractometry, Optical Absorption and Fluorescence, Micromachines, 2016, 7(10), 181 CrossRef PubMed , Available from: https://www.mdpi.com/2072-<?pdb_no 666X?>666X<?pdb END?>/7/10/181.
- ImageJ [Internet], [cited 2022 Dec 21], Available from: https://imagej.net/ij/index.html Search PubMed.
- J. S. Pedersen, wrMTrck multiple object tracker [Internet], 2011 [cited 2022 Dec 22], Available from: https://www.phage.dk/plugins/wrmtrck.html Search PubMed.
- A. S. Basu, Droplet morphometry and velocimetry (DMV): a video processing software for time-resolved, label-free tracking of droplet parameters, Lab Chip, 2013, 13(10), 1892 RSC , Available from: https://pubs.rsc.org/en/content/articlehtml/2013/lc/c3lc50074h.
- V. Ajuwon, B. F. Cruz, P. Carriço, A. Kacelnik and T. Monteiro, GoFish: A low-cost, open-source platform for closed-loop behavioural experiments on fish, Behavior Research Methods, 2023, 1, 3 Search PubMed , Available from: https://link.springer.com/10.3758/s13428-022-02049-2.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc00327b |
| This journal is © The Royal Society of Chemistry 2023 |