Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Controlled droplet microfluidic systems for multistep chemical and biological assays
T. S.
Kaminski
 * and
P.
Garstecki
* and
P.
Garstecki
 *
*
Institute of Physical Chemistry, Polish Academy of Sciences, Kasprzaka 44/52, 01-224 Warsaw, Poland. E-mail: tkaminski@ichf.edu.pl; garst@ichf.edu.pl
First published on 31st August 2017
Abstract
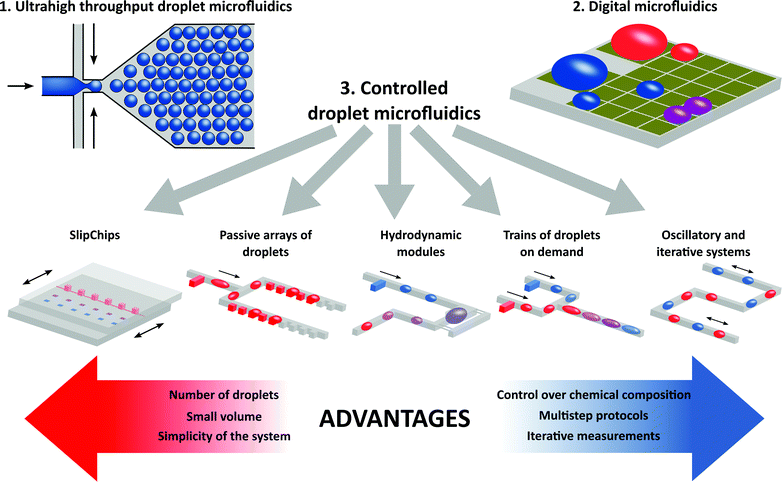
Droplet microfluidics is a relatively new and rapidly evolving field of science focused on studying the hydrodynamics and properties of biphasic flows at the microscale, and on the development of systems for practical applications in chemistry, biology and materials science. Microdroplets present several unique characteristics of interest to a broader research community. The main distinguishing features include (i) large numbers of isolated compartments of tiny volumes that are ideal for single cell or single molecule assays, (ii) rapid mixing and negligible thermal inertia that all provide excellent control over reaction conditions, and (iii) the presence of two immiscible liquids and the interface between them that enables new or exotic processes (the synthesis of new functional materials and structures that are otherwise difficult to obtain, studies of the functions and properties of lipid and polymer membranes and execution of reactions at liquid–liquid interfaces). The most frequent application of droplet microfluidics relies on the generation of large numbers of compartments either for ultrahigh throughput screens or for the synthesis of functional materials composed of millions of droplets or particles. Droplet microfluidics has already evolved into a complex field. In this review we focus on ‘controlled droplet microfluidics’ – a portfolio of techniques that provide convenient platforms for multistep complex reaction protocols and that take advantage of automated and passive methods of fluid handling on a chip. ‘Controlled droplet microfluidics’ can be regarded as a group of methods capable of addressing and manipulating droplets in series. The functionality and complexity of controlled droplet microfluidic systems can be positioned between digital microfluidics (DMF) addressing each droplet individually using 2D arrays of electrodes and ultrahigh throughput droplet microfluidics focused on the generation of hundreds of thousands or even millions of picoliter droplets that cannot be individually addressed by their location on a chip.
1. Introduction
This tutorial review will present the potential of microfluidic techniques for controlled manipulation of microdroplet (bio)chemical reactors – a unique platform for the execution of complex experimental protocols. In the first part of the review, we briefly introduce the development of general droplet microfluidics and the most important physical and chemical aspects of the formation and dynamics of microdroplets. We highlight the major advantages of this technology and present two general approaches for the manipulation of microvolumes in biphasic systems such as Electro-Wetting On Dielectric (EWOD) digital microfluidic (DMF) systems and ultrahigh throughput droplet microfluidics. Next, we present ‘controlled droplet microfluidics’ as a group of methods synergistically combining the advantages of DMF systems and ultrahigh throughput droplet microfluidics. We also present the important and outstanding applications of controlled droplet microfluidics in analytical, materials and biological chemistry. Finally, the most important and most fascinating challenges are briefly described for the future directions of development, including the putative ‘holy grail’ opportunities for this discipline.The miniaturization of progressively more and more integrated laboratory protocols, i.e. the development of Lab on a Chip devices or Micro Total Analysis Systems, began almost 30 years ago. Currently, this technology plays an increasingly important role in biomedical and chemical research, diagnostics and development of new pharmaceuticals. Microfluidics is the field of research intersecting engineering, chemistry, physics, materials science, nanotechnology, biology and medicine. The main interests of microfluidics are in studying the properties of fluids at the microscale and in developing practical and high-throughput applications, opening up new research opportunities.
Droplet microfluidic systems enable the formation and analysis of a large number of discrete microvolumes of fluids submerged in an immiscible continuous phase. A microdroplet reactor is a tiny volume of liquid that may comprise samples, reagents and components for an analytical assay, a chemical synthesis or a biological process. The continuous or matrix fluid is usually – as most of the applications target the biocompatible aqueous environments of the reactors – an oil (either simple hydrocarbon or fluorinated liquid). Sometimes air is used, particularly in EWOD-based digital droplet systems1 or in droplet platforms for chemical synthesis.2,3 The technology of microdroplets is sometimes also called bi- or multiphase microfluidics – an alternative approach to continuous-flow microfluidics.
Droplet microreactors are fundamentally different from a typical continuous flow reactor surrounded by solid walls, since they are isolated from the surrounding immiscible fluid by the liquid–liquid or liquid–air interface. The interface is usually stabilized by amphiphilic surfactants and exhibits unique physical and chemical properties. While these characteristics often present substantial experimental challenges even in properly designed systems, they also provide unique experimental opportunities, making this platform attractive in selected applications in bio-chemical sciences. The main advantages of droplet technologies include the following: (i) the simple generation and manipulation of large numbers (up to millions) of reactors, (ii) the confinement of the reaction into tiny volumes, (iii) high throughput control that is usually realized through selected operations such as droplet formation, merging or sorting, and (iv) the presence of a liquid–liquid interface that enables specific applications, e.g. studies of lipid membranes or the formation of multiple emulsions as new materials or as multicompartment reactors.
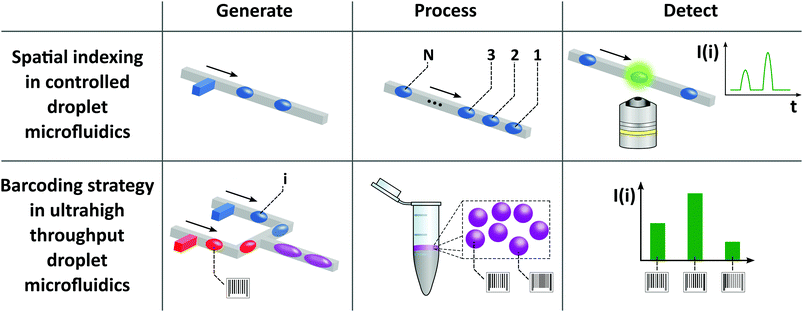
For the sake of this review, we classify the droplet microfluidic technologies as shown in Fig. 1. On the one side of the spectrum of droplet microfluidic methods we can find ultrahigh throughput microfluidic systems that generate very large numbers (thousands to millions) of monodisperse droplet reactors. On the other end of the spectrum there is digital microfluidics (DMF). DMF is a quite mature technology in which each droplet is addressed individually and can be manipulated within multistep and complex liquid handling protocols. We also introduce a new definition of controlled droplet microfluidics as a set of technologies in which the series of drops can be indexed and addressed by the spatial position of each droplet on a chip. In contrast to ultrahigh throughput systems, the number of droplets is smaller; however, complex operations can be executed on-chip during multistep protocols without the need for human intervention, which is required in classical microdroplet systems, e.g. for transfer of droplets between the chips and tubes.
2. Advantages of droplet microreactors
Microdroplets exhibit several properties that justify the use of these systems in research over classical macroscale reactors or miniaturized continuous flow microfluidic systems (Fig. 2).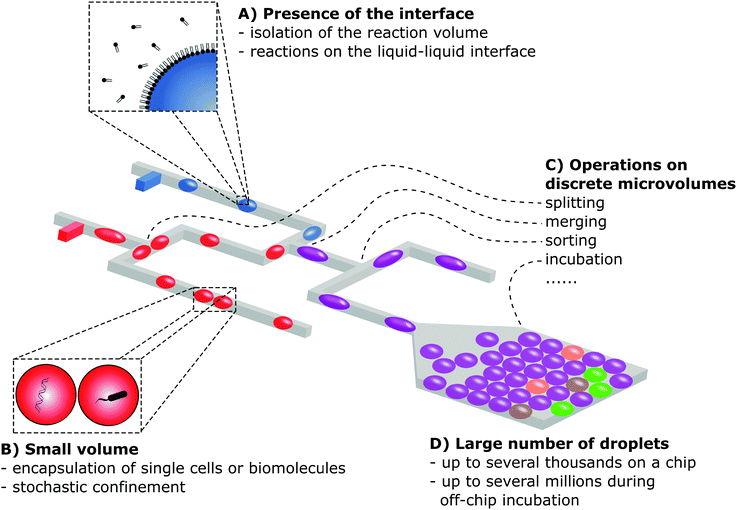 | ||
| Fig. 2 Schematic illustration presenting the four main properties of droplet microfluidic technologies. | ||
The unique properties of the fluid–fluid interface
The liquid–liquid (sometimes liquid–gas) interface serves as a boundary of the compartment, excluding or minimizing the risk of fouling, e.g. (i) formation of biofilms during the cultivation of microbes or (ii) adsorption of chemical compounds on the surface during synthesis. The interface also reduces the physical contact with a surface of a solid chip material that may be toxic for cells confined to droplets. The adsorption of amphiphilic surfactants to the interface allows for the generation of stable emulsions, and multiple droplets and vesicular structures. Droplet microfluidics is also a robust approach for the generation of a large number of monodisperse particles for multiple applications in materials science. Additionally, multiphase microfluidics was used in smart approaches for the formation of micro-assemblies in which the interface itself plays the most important role, such as liposomes, polymersomes and multisomes – structures that were used in biochemical studies of lipid bilayers and membrane proteins, and also as drug delivery systems.4 The stabilization of the interface with surfactants brings a unique opportunity to form multi-compartment chemical networks and bioelectrical circuits5,6 and to study the properties of lipid bilayers and membrane proteins.7,8Small volume
The small volumes of droplets generated in microfluidic systems ranging from pico- to microliters provide new research capabilities. First of all, microdroplets enable efficient encapsulation of single biomolecules and cells that can subsequently be detected and precisely quantified by digital assays, even in the presence of a much higher concentration of background molecules that would hinder a classical analogue assay (e.g. detection of cancer-associated mutations in the high background of wild-type DNA).9 Isolation of single cells is also required in studies on population heterogeneity,10 and the sequencing of single cell genomes11 or transcriptomes12,13 and for ultrahigh throughput screening in directed evolution campaigns.14 The small volume of droplets allows for faster accumulation of macromolecules secreted by a cell or products of biochemical reactions, as compared to standard bulk cultures. Thus, a detectable level of the signal is achieved faster – a feature useful, e.g., in the rapid detection of pathogens or in screening during directed evolution protocols.14Control over the processing of droplets
Microdroplet technology offers a high degree of control over the conditions within which bioprocesses or chemical reactions occur. Some of the aspects of improved control result from the small scale itself – which is typical also for continuous flow microfluidic systems. High surface area-to-volume ratios enable faster and more uniform heat transfer, which is important for the efficient execution of, e.g., PCR15 or in chemical syntheses.2,3,16 Isolation of the reaction volume to individual droplets brings additional benefits. First of all, in contrast to single phase microfluidics characterized by the parabolic profile of flow and the Taylor–Aris dispersion of reagents, droplet systems ensure an equal time of residence for each reaction vessel. This enables precise control over the execution of multistep protocols without the need for sophisticated automation as was recently demonstrated in the elegant example of the synthesis of quantum dots.16 Another distinctive characteristic of two-phase systems versus single phase microfluidics is rapid and thorough mixing provided by chaotic advection inside the droplets. Last but not least, compartmentalization within droplet reactors opens up a wide spectrum of possibilities for fluid handling. Droplets can be generated, transported and manipulated in the network of channels at high-throughput with up to kilohertz frequencies using mechanical actuation, electric fields or acoustic waves. There is a broader range of physical mechanisms that can be used for slower manipulation of droplets, e.g., EWOD, thermocapillary effects, magnetic forces, surface phenomena (e.g. Marangoni effect) or spontaneous translocation due to the surface tension.17,18Large numbers of droplets
One of the most significant advantages of multiphase microfluidic systems is the feasibility of generation of large numbers, from hundreds to millions, of small droplet compartments. In continuous flow systems the volume is defined by the geometry of the channels and it is difficult to reduce the volume of the reaction vessels below the nanoliter range, leaving aside the technical problem of isolating the large number of wells from each other after they are filled with the sample. In contrast, in droplet microfluidic systems it is relatively straightforward to tune the volume of microdroplets from microliters down to even the femtoliter (10−15 L) scale.19 The rapid and facile formation of large quantities of reaction vessels is beyond the capabilities of other systems and this feature opens new research opportunities – e.g. ultrahigh throughput screening of large libraries of microorganisms or molecules14 or massively parallel analysis of single cells.11–133. General description of droplet microfluidic systems
Ultrahigh throughput microfluidics
In the ultrahigh throughput approach small femto-, pico- or nanoliter droplets are generated at very high frequencies, in a passive and continuous manner, and usually using flow-focusing junctions.20 A substantial fraction of applications of these systems are single-step processes that comprise droplet formation and then usually off-chip incubation followed by re-injection of monodisperse droplet emulsion onto another chip for additional operations or for endpoint optical read-out, also conducted at high (typically kHz) rates. The transfer of droplets to and from external containers can be challenging due to the issues with emulsion stability and loss of a fraction of droplets on the bottom of the wells, tubing connections, inlets of a chip, etc. In spite of these shortcomings, this mode of operation avoids some of the problems associated with the integration of the processing and detection on a single chip, such as unwanted bubble formation during on-chip heating or challenging fabrication of large chambers for the storage of droplets. Within the off-chip approach complex incubation schemes can be easily applied using standard laboratory equipment – e.g. thermocycling in standard PCR devices. In some applications the readout is not needed – e.g. in preparatory applications, emulsion is broken after incubation and the product of the reaction can be further analyzed in the bulk. In more complex protocols, fluorescence detection can be coupled with the sorting of droplets.14 In the case of multistep reactions, successive portions of reagents are delivered via droplet fusion or by direct injection of picoliter volumes of liquids, yet such operations require a high degree of expertise and experience and are limited to a small number of laboratories.Digital microfluidics (DMF)
On the other end of the spectrum, with completely different features, is the technique of digital microfluidics – which uses the electrowetting effect. This phenomenon relies on the modulation of the interfacial energy and decreasing the contact angle between a solid surface and polar liquid, with the use of an externally applied electric field. The droplets rest on a solid surface of an electrode, usually isolated with a thin hydrophobic dielectric film – this technology is called Electrowetting-on-Dielectric (EWOD). Normally, the dielectric film repels aqueous solution, but when an electric field is applied, water due to its higher polarizability than the surrounding oil (or air) is attracted towards the regions of a higher magnitude of the electric field. Modern microfluidic EWOD systems, also referred to as Digital Microfluidics (DMF), comprise 2D arrays of multiple individually controlled electrodes.1 DMF systems, in contrast to high-throughput droplet microfluidics, typically operate on a small number (several up to tens) of droplets, yet they allow performing all fundamental operations on any individual droplet (generation, motion, splitting, or merging), and combining these unit operations into arbitrarily complex protocols. However, Hadwen et al.22 developed a system in which the conventional EWOD module was replaced with a thin film transistor (TFT) array. This demonstration proved that DMF devices can be scaled up to comprise thousands of individually addressable electrodes that theoretically can enable operations on hundreds or even thousands of droplets.The DMF technology is unique in its ability to individually and simultaneously control many droplet compartments. The freely open 2D space that is available for droplet movement allows easy implementation of even complicated procedures, including iterative protocols. Furthermore, all operations performed on droplets are encoded mostly in the software, and thus EWOD chips are reconfigurable. All these features make DMF platforms very versatile and universal.
The EWOD technology presents also limitations: DMF devices are characterised by a very low, as compared to channel microfluidics, rate of operations – the simplest operation of moving a droplet across the electrodes typically requires a significant fraction of a second; thus operations on droplets can be executed at rates typically well below single Hz. Also the fabrication of EWOD chips is much more complicated than soft lithography or micromachining commonly used to fabricate conventional channel microfluidic chips. DMF chips are also more complicated to scale up to production technologies, which is fairly straightforward for channel microfluidics via rapid industrial production of thermoplastic chips. However, recent advances in scalable fabrication make EWOD platforms more attractive and affordable, also for point of care (POC) diagnostic applications. Dixon et al.23 presented an approach for the mass production of DMF devices using simple processes of inkjet printing and roll-coating with material cost below one dollar per chip. Another important factor is the volume of the droplets. In DMF the volume of the droplets is relatively large and ranges from about single microliters down to a hundred nanoliters, while channel microfluidic systems span the scale from microliters down to femtoliters. One important challenge in the operation of EWOD chips is fouling of the dielectric surface by hydrophobic molecules. In particular, the adsorption of proteins is still challenging, despite several strategies developed to overcome this obstacle – e.g. introduction of water immiscible oils, control of the pH of the solution inside the droplets, removable films, superhydrophobic surfaces and use of surfactants and other solutions.1 Despite these challenges, the DMF technology is a very attractive platform for sample preparation, a judgement backed up by the interest of well-known biotech companies such as Illumina or Oxford Nanopore Technologies that invested in implementing this technology for the automated construction of libraries of DNA fragments for their next generation sequencing platforms.
4. Controlled droplet microfluidics
Controlled droplet microfluidics is a group of techniques based on the automation and passive control of flow that address droplets in series. In this respect, controlled droplet microfluidics positions between digital microfluidics addressing each of the droplets independently and ultrahigh throughput microfluidics that does not index the droplets by their positions and is incapable of addressing them individually apart from reading the barcodes that they may contain.Controlled droplet microfluidics ranges from almost entirely passive systems such as the SlipChip24,25 for relatively simple protocols executed at high-throughput on several hundreds of compartments, through systems requiring active, yet not necessarily precise, flow for passive formation of matrices of trapped droplets to automated droplet microfluidics that uses sophisticated control of flow to perform multistep protocols with tens or hundreds of droplet microreactors (Table 1).
| Type | Maximum number of compartments | Complexity of operation and flow control | Capability of generation of arbitrarily set chemical compositions | Compatibility with complex protocols |
|---|---|---|---|---|
| SlipChip | Up to several thousands29 | Straightforward operation of the system, and simple sources of flow can be integrated on a chip | Usually for stochastic encapsulation of cells or molecules; however, it is possible to generate gradients based on diffusion under static conditions25 | Protocols comprising up to a few steps can be carried out, but each step requires separate operation of slipping29 |
| Passive self-digitization systems with arrays of droplets | Tens of thousands30 | Easy operation of the system that requires simple and not necessarily precise sources of flow | Bioreactors usually have the same chemical compositions; single phase gradients can be introduced after gellification of droplets and exchange of oil to aqueous medium31 | Usually single step reaction; however, the delivery of nutrients is possible after exchange of oil to aqueous medium31 |
| Systems with hydrodynamic traps | Several thousands32 | Flow rate does not have to be precisely set; however, it should be controlled in time | Convenient method for the generation of various gradients and serial dilutions33,34 | Multi-step assay can be performed provided that a more precise source of flow is used33 |
| Trains of droplets | Tens of hundreds16 | Control of the system has moderate complexity and it requires more sophisticated sources of flow such as valves or precise syringe pumps | Efficient generation of chemical gradients; however, they have a limited dynamic range of concentrations35 | Multi-step operation can be executed in relatively high throughput; however, it requires a larger footprint of the devices16 and/or can be performed only for a smaller number of droplets |
| Oscillatory and iterative systems | Up to around one thousand10 | Complex control of the system requiring precise sources of flow with short response times; devices are also equipped with sensors providing information about the positions of the droplets in the system and the feedback loop to control the operation of the flow sources27 | Chemical composition of each droplet can be arbitrarily set and dynamically changed in the course of the assay | Oscillatory transport and on-demand splitting of droplets provide the capability of execution of multistep operation regardless of the length of the network of channels27 |
The location of a series of droplets on a chip can be determined either statically by dedicated microstructures (wells, hydrodynamic traps and rails) or dynamically by the flow of a continuous phase controlled by external, usually mechanical, forces. The positions of the droplet compartments can be either fixed during the whole experiment24,26 or very dynamic as in automated droplet oscillators.27,28 Usually, static methods with a higher degree of confinement and irreversible immobilization of droplets can handle more droplets in a single experiment, at the expense of a lower complexity of the possible protocols. There are also numerous demonstrations of mixed approaches that take advantage of both – the confinement of droplets and the controlled flow of the continuous phase – e.g. systems with automated hydrodynamic traps.7
Controlled droplet microfluidics usually operates with larger volumes than ultrahigh throughput microfluidics but still in the nanoliter range that enables, e.g., single-cell analysis and provides an opportunity to generate up to several thousands of compartments – a much higher throughput than is available in DMF systems.
Predominately passive systems – with their operation defined predominantly by micro-confinement
The Ismagilov group developed the SlipChip24 – an almost entirely passive device composed of two glass plates that are separated by a continuous phase of oil. Both layers comprise wells and ducts that are initially positioned to form a network of channels. This network is filled at the beginning of the protocol with a to-be-dispersed phase – e.g. samples, reagents. Filling the SlipChip with liquids is the only step of the protocol that requires the active control of the flow. Next, one of the layers is slipped and translated with respect to the other one, the wells are separated from the ducts, and the stream of the sample or reagent liquid is broken by the sliding walls to form an array of droplets. Additional slipping steps can be performed to introduce further operations such as droplet merging, splitting or removal from the chip. So far, the SlipChip has found interesting applications as a tool for performing protein crystallization,25 digital PCR, immunoassays and microbial studies such as the identification and isolation of new strains of bacteria.29 This technique is very robust and promising due to its ingenious simplicity. The limitations of the SlipChip technology include slow, principally diffusive, mixing of liquids. The fabrication of SlipChip systems is challenging as it must ensure tightly flat and smooth surfaces of both layers. The main technique used so far is wet chemical etching of glass, which is laborious and not compatible with mass production. Nevertheless, the commercialization process of this technology that should provide more accessible and cheaper devices made of thermoplastics is currently in progress.Passive arrays of droplets
2D arrays of droplets can be formed without complete definition of the traps by solid walls – as is done in the SlipChip system. Usually arrays of traps are used to capture nanoliter droplet microbioreactors. The two most interesting and most robust exemplary systems are microfluidic bottom-well arrays for self-digitization samples30 and the generic platform with arrays of anchored droplets.31 Both of them use the same principle – an aqueous plug is initially squeezed between the walls of the channels or between the floor and the ceiling of the channel. The confinement does not allow the droplet to assume the spherical shape that represents the minimum of interfacial energy. Then the droplet flows into a section of the channel or chamber that comprises deeper wells (also called anchors). Flowing over the anchor, the droplet relaxes its shape locally and becomes trapped by the capillary forces. Initially, the whole device is filled with oil which is displaced by the aqueous sample in the course of the filling of the chip. Then, the oil is injected again and the aqueous sample is removed from the whole device except from the wells or anchors to form a predesigned array of droplet reactors. This process of self-digitization can be very efficient for facile and relatively quick formation of high-density arrays of thousands of droplets without the need for a sophisticated control of the flow.Arrays of anchored droplets were used for the cultivation of colonies derived from single bacterial cells in aqueous and hydrogel droplets.31 In the case of gelled agarose droplets, the oil can be once again replaced by an aqueous continuous phase in order to provide stimuli, e.g., antibiotics for bacterial susceptibility testing. Finally, the hydrogel droplet can be melted using an infrared laser and extracted from the array for further off-chip analysis.
Hydrodynamic traps and modules for complex operations
Hydrodynamic traps can be used not only to immobilize droplets but also for other, more complex operations on droplets such as metering, merging, diluting, controlled releasing and assembling of the droplets into functional chemical networks (Fig. 4). In one of the most interesting examples, Niu et al.34 developed a geometry for the generation of serial dilution by first locking a larger ‘mother’ droplet with a sample in a dedicated hydrodynamic chamber with pillar structures. Next the trapped droplet is sequentially ‘washed’ with a series of smaller buffer droplets – each small droplet merges with the trapped one and the same excessive amount of liquid is ejected from the trap. This process generates an output series of droplets that present a gradation of the concentration of the sample.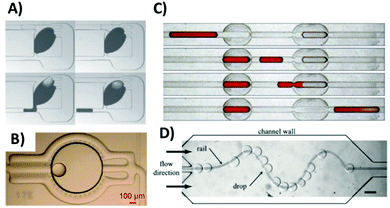 | ||
| Fig. 4 Examples of hydrodynamic modules used for droplet handling on a chip. (A) Microdilutor comprising a large “mother droplet” trapped within a microfluidic chamber. The series of smaller buffer droplets can be merged, mixed and re-split with the large droplet to generate an output sequence of droplets with decreasing concentration of a reagent.34 Reprinted by permission from Macmillan Publishers, Ltd: Nature Chemistry, ref. 34, copyright 2011. (B) Storage chamber containing a nanoliter droplet formed via merging of sequences of picoliter droplets containing reagents and microbial cells.36 Reproduced from ref. 36 with permission. Copyright 2012 National Academy of Sciences. (C) A set of two hydrodynamic traps for precise metering (trap on the left) and merging (trap on the right) of nanoliter droplets.33 Reproduced from ref. 33 with permission from The Royal Society of Chemistry. (D) Droplets in the wide channel are guided by micro-fabricated “rails” – grooves that are used to localize droplets due to the reduction of their surface energy.37 Reproduced from ref. 37 with permission from The Royal Society of Chemistry. | ||
We also proposed a set of hydrodynamic capillary traps that allow for a broader array of operations. An individual module – a ‘trap’ – comprises a barrier – a constriction in the main channel that stops the flowing droplet, and bypasses – shallower auxiliary side channels that facilitate the flow of the oil around the locked droplet and prevent the build-up of pressure that would push the droplet through the constriction out of the trap. The appropriate and careful design of various geometries of barriers and bypasses allowed for the development of several versions of traps to perform operations such as (i) dilution,34 (ii) metering and diluting,33 (iii) merging and releasing,33 (iv) locking and shifting,33 (v) derailing and storing,18 (vi) metering and storing17 and (vii) splitting large droplets into a library of subnanoliter droplets.32 These functions are realized by a set of hard-wired geometric modules that can be assembled into networks – systems that perform multistep complex liquid handling protocols.
Importantly, in contrast to SlipChips and self-digitization systems, these systems allow complex and sequential operations for, e.g., serial dilution that spans a wide range of concentrations of the samples or reagents. The rate or the timing of switching the flow direction does not need to be controlled precisely and the chip can be operated even by a very simple flow generator, e.g., standard automatic pipette.17
Automated droplet microfluidics for the generation of trains of droplets on demand
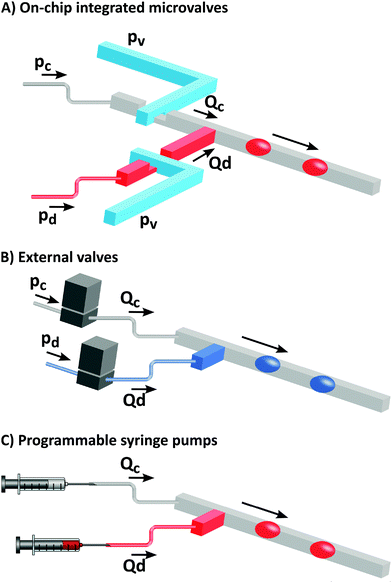
The parallel approach to control the operations on droplets is to precisely control the velocity of the flow of the continuous phase using pumps, valves and other external forces (Fig. 5). We can further distinguish the systems that use integrated on-chip valves36 or off-chip actuation.10,27,28,35,38,39 Both approaches have their strengths and weaknesses. On-chip valves made of elastomer have very small dead-volumes and thus can handle minute subnanoliter volumes36 and their capabilities for the execution of multiplex assays can be further improved by efficient multiplexing via microfluidics large-scale integration. However, their fabrication through multi-layer soft lithography is much more challenging than the fabrication of simple systems with a single fluidic layer with channels. Additionally, the PDMS elastomer is not compatible with the majority of liquids and even the use of aqueous solution requires some cautionary steps to prevent problems with evaporation. Off-chip pressure-driven actuation based on external valves is not as precise as in the case of integrated valves; however, the introduction of flow resistors between the valve and the chip mitigates the influence of the valve's microliter dead volume and enables operation in the nanoliter range of volumes on a chip.27,28,35 Microfluidic devices with displacement-driven actuation, such as programmed syringe pumps, can work properly only if the resistance of the chip itself is relatively low – otherwise the systems can experience significant time-lags and unwanted fluctuations of the flow that can significantly compromise the precision of the execution of the protocol. Off-chip actuation and the external control of the flow have a significant advantage in that the chip itself can be very simple and disposable, reduced to just networks of channels that can be produced with the simplest and least expensive techniques.Automated droplet microfluidic systems can be built with a range of levels of integration and complexity. In the simplest case actuation is active only in the first stage of the protocol to provide control over the volume and frequency of the formation of droplets.35,38 Control over droplet formation could be used to generate trains of droplets with arbitrarily set chemical compositions, e.g., gradations of concentrations35 or libraries of various chemistries drawn from multiwell plates.38
Oscillatory and iterative droplet systems
In more complex automated systems, trains of droplets can be circulated back and forth in order to perform time-course measurements and to provide long incubation times without the need for stopping the flow.2,3,39–41 The process with constantly moving droplets provides two important advantages: (i) it efficiently stirs the content of the droplet, and (ii) it preserves a non-zero thickness of the lubricating film of the continuous liquid on the wall of the channel, which reduces the risk of wetting/fouling of the surface of the channel by the content of the droplet reactor. The periodic reversal of the flow is usually achieved either (i) by placing at each end of the channel pairs of valves connected to different pressure sources, or (ii) by implementation of programmable syringe pumps that can operate in the infuse/refill mode interchangeably. More complicated systems go beyond simple cycling of the droplets back and forth – to offer the possibility of adding or titrating portions of reagents to already formed droplets28 and the most advanced systems are capable of on-demand droplet splitting and removal of a fraction of the volume of each droplet from the system.27 It is worth pointing out that the flow of droplets does not depend simply on the flow velocity of the continuous phase and the linear distances between drop microreactors might change over time and the ordered sequence of drops in long trains can be easily broken.42 For that reason, the control of flow is usually coupled with the real-time detection of the positions of droplets and the feedback loop controlling the operation of valves and/or pumps.27,28,39,405. Applications of controlled droplet microfluidics
In this section we present examples of applications of controlled droplet microfluidics in chemical and biological experimental sciences. The field of controlled droplet microfluidics has become wide and fruitful, and within the format of a tutorial review we are forced to present only a subjective selection of exemplary methods and systems.Chemical synthesis
Organic synthesis in well-controlled microsystems provides improved and more uniform chemical and thermal conditions that result in better batch-to-batch reproducibility. Most of the currently used flow microreactors are single phase systems, either capillary- or chip-based. Despite providing very good control over the conditions of the process, these systems suffer from limitations of continuous flow microfluidics, such as dispersion of reagents' time of residence due to the parabolic profile of flow, and, more importantly, the risk of fouling and deposition of reactants and products on the surface of microfluidic channels. Droplet-based flow can be an interesting alternative to single-phase microflows, since the reaction volume is confined and isolated in a droplet and the unwanted deposition of reagents is limited. Also, in controlled microfluidic systems, droplets occupy the whole cross-section of the channel and there is no velocity dispersion during their transport. Most importantly, the droplet format allows testing multiple reaction conditions (e.g. stoichiometry) in a single experiment. It is worth noting that the reaction does not always take place within droplets and there are several examples of synthesis that occur in inter-droplet slugs of the continuous phase.2,3The first example of the application of droplet systems to chemical synthesis was the work of Shestopalov et al.43 The publication presented the synthesis of nanoparticles in trains of nanoliter aqueous droplets (Fig. 6A). The microfluidic system operated at room temperature enabled execution of the two-step protocol of the synthesis of CdS core–shell nanoparticles within the reaction times on the order of milliseconds. One of the limitations of the droplet-based system is the difficulty in adding new portions of reagents due to the presence of a surfactant that could prevent the merging of droplets. Additionally, variations in droplet velocities during the transport along the channel could result in the desynchronization of the operations on a chip such as droplet merging. In a recent study Nightingale et al.16 demonstrated a robust and easy-to-implement method for the addition of subsequent portions of reagents and applied this approach in the microfluidic system for the multistep synthesis of nanocrystals. The introduction of gas as a third immiscible phase into the system in the form of plugs separating droplets enables reliable injection of subsequent portions of precursors in the multistep synthesis of CdSe nanoparticles. High gas-to-liquid surface tension ensured uniform spacing of droplets and prevented their break-up and formation of small satellites during the process of dosing of the precursor material (Fig. 6B and C).
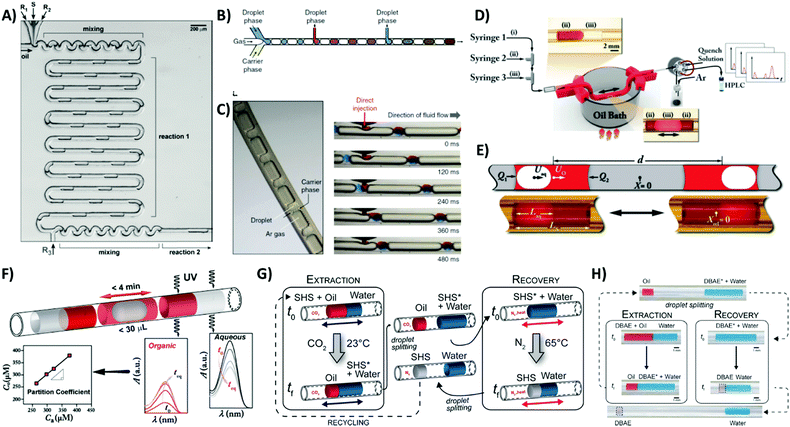 | ||
| Fig. 6 Droplet microfluidic systems offering a high degree of control over chemical synthesis. (A) Two-step nanoparticle synthesis in the train of aqueous droplets.43 Reproduced from ref. 43 with permission from The Royal Society of Chemistry. (B and C) Multistep synthesis of CdSe quantum dots in a three-phase droplet reactor.16 Reprinted by permission from Macmillan Publishers, Ltd: Nature Communications, ref. 16, copyright 2014. (D and E) Three-phase oscillatory droplet microfluidic system for studies of bi-phasic carbon–carbon (C–C) and carbon–nitrogen (C–N) cross coupling reactions.3 (D) Schematic of the microfluidic system comprising 3 syringes controlling the flow of gas (Ar), an organic phase (THF) and an aqueous liquid. (E) Schematic and two snapshots presenting various positions of the inner aqueous droplet engulfed in the oscillating organic plug. Reproduced from ref. 3 with permission from The Royal Society of Chemistry. (F) Measurement of the water–octanol partition coefficients of hydrophobic and hydrophilic molecules performed using an oscillatory microfluidic system.40 Reprinted with permission from ref. 40. Copyright 2015 American Chemical Society. (G and H) Similar oscillatory approaches used for screening of the performance of switchable hydrophobicity solvents (SHSs) for extraction of CO2.41 Reprinted with permission from ref. 41. Copyright 2017 American Chemical Society. | ||
Until recently, droplet microfluidic systems could not be applied for the execution of slower chemical reactions due to the limited length of the microfluidic channels and consequently reduced mixing efficiency and residence times of droplets on a chip. Moreover, hard-wired designs of multiphase droplet systems restrict the flexibility of the multi-step protocols since the residence time of each step cannot be adjusted independently. The elegant approach that solved these limitations is based on the use of oscillatory droplet microfluidic platforms as was recently proposed in the series of publications presented by Abolhasani et al.2,3 One of the most interesting examples is the two-phase gas–liquid oscillatory platform developed for studies and optimization of conditions for the synthesis of semiconductor nanocrystals. The oscillatory transport of the droplet in the heated tubular Teflon reactor was combined with in situ temporal spectral characterization and automated injection of the solutions with precursors. The system was used for studies of the effect of the temperature on the dynamics of the growth and nucleation of II–VI and III–V quantum dots. In another example an oscillatory approach was extended to three-phase systems.3 During the reaction, a bi-phasic slug composed of an aqueous droplet submerged in tetrahydrofuran (THF) was circulated back and forth in the tubular reactor filled with argon (Fig. 6D and E). The higher surface tension of the inner phase resulted in its facile movement and complete engulfment of the aqueous droplet in the organic slug. As a result, the increase of the available surface area provided for better mass transfer in the bi-phasic carbon–carbon (C–C) and carbon–nitrogen (C–N) cross coupling reactions. The oscillating flow reactor was next used for studies of Suzuki–Miyaura and Buchwald–Hartwig reactions. Three phase oscillating systems produced similar yields to batch reactions, but significantly higher than using a continuous flow bi-phasic reactor.
Physical chemistry
The experimental approach exploiting three-phase droplet oscillatory microfluidics has also been recently used for the rapid measurement of partition coefficients of chemical compounds between aqueous and organic liquids40 (Fig. 6F). The automated oscillatory flow strategy enabled a rapid determination of the partition coefficients of hydrophilic (caffeine) and hydrophobic (acetaminophen) drugs between water and 1-octanol within a few minutes and using just microliter volumes of solutions.This technology might bear the potential for future applications in the screening of conditions for liquid–liquid extraction in the pharmaceutical industry or in the removal of organic contaminates of water. Lestari et al.41 proposed to use a three-phase oscillatory droplet microfluidic platform in the studies of CO2 extraction towards the evaluation of the performance and recovery of switchable hydrophobicity solvents (SHSs) that are of great interest in sustainable chemical processing. The experimental assessment of the performance of an exemplary SHS, 2-(dibutylamino)ethanol (DBAE), was conducted within 1.5 hours in the entire cycle of CO2-mediated extraction and recovery (Fig. 6G and H). The droplet microfluidic oscillatory platforms can serve as an efficient method, reducing time, labor and costs needed for screening of SHS performances and in more general context for the optimization of chemical solvent properties.
Biochemistry: enzyme kinetics and protein crystallization
Controlled droplet microfluidics has also found several applications in protein biochemistry. One of the first demonstrations of the potential of droplet microfluidics was the seminal work of Ismagilov's group presenting the system for studies of millisecond enzymatic kinetics.44 Efficient mixing inside microdroplets allowed for the measurement of both fast and slow kinetics using nanoliter solutions of reagents. Droplet microfluidics has been used multiple times in recent years for measurements of the kinetics of enzymatic reactions. One of the recent interesting demonstrations presented the fully unsupervised platform for the automated generation of trains of a few hundred droplets for the determination of the Michaelis–Menten kinetics of an exemplary enzymatic reaction.38 The nanoliter droplets were withdrawn from the bottom of the sample tubes submerged in an oil-filled carousel (Fig. 7A). In the next step, the pairs of the corresponding droplets containing enzymes and substrates were merged, and the resulting train of droplets was cycled back and forth in long Teflon tubing. This simple and robust system allowed for obtaining high-quality data, which were used to calculate kcat and Km values describing the kinetics of the enzymatic hydrolysis of 4-nitrophenyl glucopyranoside by sweet almond β-glucosidase.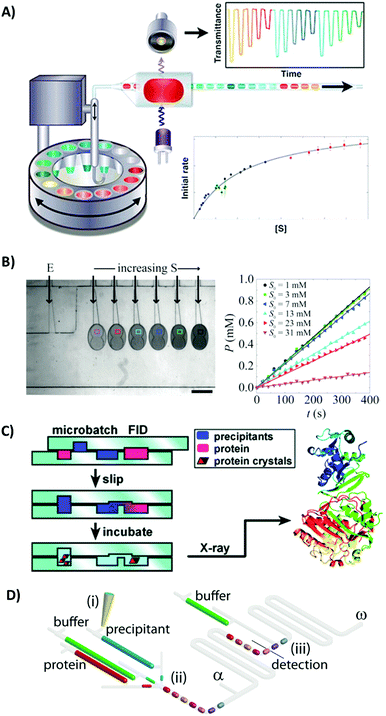 | ||
| Fig. 7 Examples of controlled droplet microfluidic systems used in protein biochemistry. (A) Fully unsupervised system for the generation and oscillation of trains of droplets for the analysis of the kinetics of an enzyme.38 Reprinted with permission from ref. 38. Copyright 2015 American Chemical Society. (B) Passive microfluidic system for the measurement of the slow and fast enzymatic reactions occurring in static arrays of nanoliter droplets.18 Reprinted with permission from ref. 18. Copyright 2015 American Chemical Society. (C) SlipChip for screening conditions of protein crystallization.25 Reprinted with permission from ref. 25. Copyright 2010 American Chemical Society. (D) Determination of protein solubility diagrams in an automated droplet system.28 Reproduced from ref. 28 with permission from The Royal Society of Chemistry. | ||
Passive droplet microfluidics can also be used for efficient and cost effective determination of parameters of enzymatic reactions. Fradet et al.18 developed a microfluidic platform comprising several pairs of stationary droplets that were next used for the analysis of both fast and slow enzymatic reactions (Fig. 7B). The aqueous compartments were generated and brought into pairs using hydrodynamic geometries providing gradients of confinement, and next the reaction was initiated by the fusion of droplets triggered by the laser beam. In case of a slow enzymatic reaction, the analysis is straightforward, since the time needed for mixing of reagents due to the convection was much shorter than the time required for the detection of reaction products. However, fast reactions occurred in much shorter times than the mixing process. The authors used reaction–diffusion theory and numerical calculations to develop a model that enabled determination of kinetic parameters of the enzymatic reaction. The results obtained for fast kinetics were semi-quantitative, probably due to the unwanted interaction of the alkaline phosphatase enzyme with the water–oil interface. This study and the results obtained offer a good example of how the unique properties of the interface can modify the results of the reaction occurring inside the droplet. It also confirms the need for additional research concerning phenomena associated with interfaces in droplet systems, and the development of new classes of better and more biocompatible surfactants.
Protein crystallization is a yet another field of biochemistry that was successfully explored using controlled bi-phasic microfluidic tools. The Ismagilov group pioneered this field developing several systems based on the generation of trains of droplets in channels and tubes dedicated for the optimization of conditions for protein crystallization. However, the SlipChip technology seems to be the most interesting, easy-to-use and robust.24 The single SlipChip device was capable of execution of both types of strategies used in protein crystallization – microbatch and free interface diffusion (FID).25 The technology was validated for screening crystallization conditions for two proteins with a library of 48 different reagents and 5 different equilibrium times for each reagent (Fig. 7C). In total, 480 experiments were performed using just three devices.
The controlled droplet microfluidic technology can also be used for the determination of the solubility diagrams of proteins. Dolega et al.28 presented a droplet oscillatory system for the determination of the phase diagrams of proteins (Fig. 7D). Sets of external valves were used to generate a train comprising 30 submicroliter droplets with various concentrations of proteins and precipitants. Next, the sequence was cycled back and forth in the channel and the content of each droplet was periodically monitored using UV-Vis spectrometry during 5 hours. In order to determine the full solubility diagram, a fresh portion of the buffer can be merged with the droplets comprising combinations of proteins and precipitants positioned in the nucleation zone. Dilution resulted in the decrease of the sample scattering and the increase of transmittance through the droplet reactor below a critical value allowed for the determination of the solubility line. The system presented by Dolega et al. was the first demonstration of the iterative titration with each droplet and dynamic control of the chemical composition of each compartment in the oscillatory droplet microfluidic system.
Microbiology
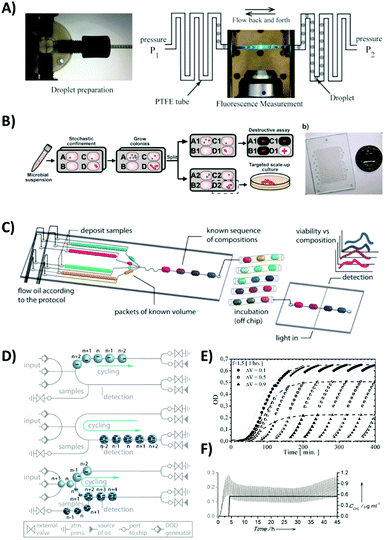 | ||
| Fig. 8 Study of microbial growth in parallel microdroplet systems. (A) Oscillatory droplet system for measuring the growth of clonal bacterial populations.10 Reproduced from ref. 10 with permission from PLoS ONE. (B) Modified version of a SlipChip for combined analysis of microbial phenotype and genotype.26 Letters A, B, C, and D are used on the schematic to index the spatial position of each compartment. The SlipChip is next split into two copies – one (indexes A1, B1,…) is used for a destructive genetic assay towards identification of bacteria, and the other one (A2, B2,…) for scaling up the microbial culture. Reproduced from ref. 26 with permission from The Royal Society of Chemistry. (C) Automated droplet microfluidic system for the determination of MICs and studying pairwise interactions of antibiotics.35 Reproduced from ref. 35 with permission from The Royal Society of Chemistry. (D) Schematics depicting consecutive operations performed on droplet bioreactors,27 and (E and F) growth curves obtained during long term cultivation in droplet chemostats.27 Reproduced with permission from ref. 27. Copyright 2013 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. | ||
Leung et al.36 presented a programmable microfluidic system for the execution of multistep protocols for the analysis of microbial cells in an array of 95 individually addressable nanoliter droplets. The device, as an example of a multilayer elastomer chip with hundreds of integrated valves, had a relatively complex design. However, it was capable of sorting bacteria, encapsulating them, cultivating them and finally subjecting them to multiparameter genetic analysis based on PCR and whole genome amplification and sequencing.
The SlipChip technology was also used for a similar purpose of isolation of monoclonal populations of microbes of interest (Fig. 8B). The selection and identification of bacteria have for the first time been based both on phenotypic and genetic analysis of selected colonies derived from single cells.26,29 In the first step, single bacterial cells are stochastically confined and cultured in nanoliter compartments to obtain clonal populations of bacteria. In contrast to oscillatory systems, a SlipChip does not support the monitoring of growth. However, the execution of the protocol is simple and each micro-compartment containing the cultured colony could be split into two droplets – each located on a separate plate of a SlipChip. Subsequently, one of the two daughter droplets from each colony could be used for the destructive genetic analysis for the identification of microbes of interest. In the next step, colonies of interest can be retrieved from the corresponding locations of droplets on the other plate and used for scaling up the culture and for obtaining a sufficient amount of biomass for further detailed off-chip characterization. In the follow-up study, the SlipChip device was used for the targeted isolation, cultivation and characterization of previously unidentified genera of bacteria from complex clinical samples.29
All the microfluidic systems, including microdroplet technology, are characterized by a small volume of the reaction vessel. This feature, despite being an advantage in most of the experimental protocols, can present a limitation in some applications. For example, the bacterial growth inside droplets can be observed for a relatively short time – spanning just a few generation of microbes due to the limited amount of nutrient sources inside the tiny droplet. However, there are many microbiological phenomena that occur at much longer timescales – e.g. adaptation of the bacterial population to the changing environment and the process of development of antibiotic resistance. Jakiela et al.27 constructed a unique microfluidic device that broke the limitation of short-term monitoring of bacterial growth in microdroplets (Fig. 8D–F). The system allowed for the cultivation of bacteria in trains of hundreds of droplets, with periodic and fully controllable exchange of a subvolume of each droplet. The system performed partial removal of saturated bacterial liquid culture and iterative infusion of fresh growth medium to each droplet. The device was used for the demonstration of long-term bacterial cultivation and the analysis of the adaptive response of the E. coli population to the gradually increased antibiotic concentration. The microfluidic system of parallel droplet chemostats can also be very useful in future studies of genotypic and phenotypic adaptation of bacteria for applications in biotechnology and sustainable chemistry.
Microdroplet systems for multi-compartment reactions and for studies of processes at liquid–liquid interfaces
Automated and passive droplet microfluidics systems can also be readily used for studying the properties of biologically-relevant interfaces – e.g. artificial lipid membranes composed of phospholipids. Lipid monolayers self-organize on the interfaces of droplets submerged in oil with dissolved lipids. When two such droplets are brought into contact, a bilayer at their interface is spontaneously formed. This technique is called Droplet Interface Bilayers (DIBs) and it provides several advantages as compared to classical methods for formation of artificial membranes. These include the (i) exceptional stability that allows for long term electrophysiological measurements, (ii) minute consumption of reagents, (iii) facile re-formation of the bilayers for sequential measurements and (iv) ability to form asymmetric bilayers composed of chemically distinct monolayers. Two recent demonstrations7,8 presented droplet microfluidic systems for the formation of droplet interface bilayers using hydrodynamic modules or traps. Electrodes integrated on the chip enabled characterization of the electrophysiological properties of the artificial membranes and execution of the efficient screening of the activity of membrane proteins and peptides incorporated in the bilayers (Fig. 9A). The automated droplet system allowed for the sequential formation of lipid bilayers at the interface between a pair of nanoliter aqueous droplets surrounded by oil. The protocol also comprised the steps of exchange of one droplet of the pair to form a new bilayer, and execution of current measurements on a level of single protein nanopores, also in the presence of inhibitor binding to the protein molecules and decreasing the transport of ions through the membrane.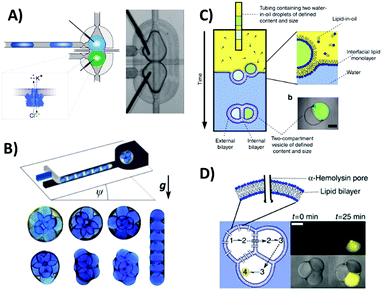 | ||
| Fig. 9 Controlled droplet microfluidic systems for the generation of tailored emulsions and for the study of interfaces. (A) Automated system for the sequential generation of DIBs and the screening of membrane protein activity.7 Schematic pictures and a snapshot presenting a microfluidic module used for locking the pair of nanoliter droplets. The transport and exchange of droplets were controlled by a set of external valves and a feedback loop based on optical sensors detecting the positions of droplets on a chip. The hydrodynamic properties of a passive microfluidic trap ensured the stable positions of the droplets during the formation of the bilayer and the measurement of the activity of the protein nanopores. Reproduced from ref. 7 with permission from The Royal Society of Chemistry. (B) System for the on-demand generation of tailored emulsions.6 Reprinted with permission from ref. 6. Copyright 2015 American Physical Society. (C and D) A droplet microfluidic approach for the formation of cell-like vesicular structures mimicking basic signalling pathways.5 Reprinted by permission from Macmillan Publishers, Ltd: Nature Communications, ref. 5, copyright 2014. | ||
Ultrahigh throughput microfluidics proved to be a very robust preparatory technique. Microfluidic systems allow high-speed generation of monodisperse emulsions and particles such as micro-gels, capsules, liposomes and polymerosomes that can be subsequently used in materials chemistry research. Controlled droplet microfluidic systems cannot compete in terms of efficiency, or throughput, with the flow-focusing planar geometry or with axisymmetric microcapillary systems for the production of emulsions. However, controlled droplet microfluidics proved to be advantageous in terms of control over the flow to generate tailored emulsions and assemblies of droplet interfaces. Guzowski et al.6 presented microfluidic systems with active control over the flow of three immiscible phases for the on-demand generation of tailored multiple droplets one-by-one. Both the number of cores (inner phase of the multiple droplet) and their volume, and the volume of the droplet shell (outer phase), could be arbitrarily set for each multiple droplet. The system allowed for the generation of tailored emulsions with individual control over the number of cores and their volume in each droplet. By varying the ratio of core to shell volumes, the desired 2D or 3D spatial arrangement of the droplet components could be achieved (Fig. 9B). The authors also analysed the thermodynamic properties of the generated metastable multiple droplets. Elani et al.5 took another experimental approach to generate multi-compartment vesicles with defined structure (Fig. 9C and D). First, the set of droplets with arbitrarily set volumes and chemical contents were formed by aspiration of nanoliter volumes to Teflon tubing. Next, the set of droplets were transferred from an oil to aqueous solution in order to engulf multiple droplets in a bilayer and therefore form multi-compartment vesicles – also called ‘multisomes’. The vesicular structure was then used to perform a multi-step enzymatic assay in such a way that single reactions from the pathway were isolated in distinct compartments. The products of each step were transported with the aid of protein nanopores to adjacent vesicles in order to trigger the subsequent reaction. The system served as a demonstration of the signalling biochemical cascade in a multi-compartment artificial tissue-like system.
Passive controlled droplet microfluidics also provided the unique capability to generate multilamellar structures imitating the double bilayer membrane. Matosevic and Paegel45 presented a system for the layer-by-layer assembly of phospholipid bilayers with defined chemical compositions and asymmetry. In the first step of the protocol, aqueous droplets were trapped in a static array of traps. Next, subsequent lipid monolayers were deposited via multiple passages of oil and aqueous phases. The system supported the insertion of functional protein nanopores into the double membrane and was allowed to demonstrate basic cell-like metabolism inside liposomes.
6. Challenges and future directions
The liquid nature of droplets presents a number of technical challenges associated with their formation, manipulation, incubation and execution of experimental protocols. In the following section the most important of these challenges are presented along with a perspective on how chemical research can contribute to the improvement of the droplet microfluidic technology.Surfactants
A major challenge in droplet microfluidics is associated with the properties of the liquid–liquid interface surrounding the aqueous droplet compartment. This interface is usually stabilized with a surfactant. One of the fundamental assumptions in droplet microfluidic protocols is the integrity and isolation of the aqueous content of a droplet. The droplets must be thus stabilized against unwanted merging (coalescence) of droplets, and against wetting of the droplet phase to the solid surface of the chip or partition of the molecules between both, dispersed and continuous, phases. One of the main challenges is to provide an optimum chemical composition of the reaction liquids and prevent unwanted interactions with the interface. Chemical precursors and products generated during a reaction inside the droplets can disturb the physico-chemical properties of the interface and consequently compromise the stability of the emulsion. And vice versa – an appropriate design of a microdroplet system requires compatibility of the reaction with the surfactant of choice – in order to avoid, e.g., protein denaturation due to the presence of the surfactant.46Another challenge is the “leakage” of the products of the reaction from droplets to the oil or to the neighboring droplets. This phenomenon decreases the performance of analytical assays by changing the concentrations of reagents and by averaging the signal between droplets – e.g. positive and negative compartments in digital assays. So far, unwanted mass transfer through the interface has been fully prevented only in selected applications – mainly in the case of compounds presenting high solubility in a droplet phase, e.g., DNA and protein molecules in aqueous droplets. There are recent elegant examples of investigation of the leakage phenomenon in droplet systems;47 however, research in this area should be further expanded, both at the level of basic science for defining and understanding the mechanism and dynamics of the transport across interface and in attempts towards the development and selection of new classes of surfactants.46 An interesting alternative approach in this respect is based on multiple screening rounds towards optimization of the composition of liquids used in droplet bi-phasic systems.48
Materials
Another challenge lies in the appropriate choice and development of new materials for fabrication of the chips. The appropriate selection of the solid material constituting a microfluidic chip should eliminate any wetting of its surface by the droplet phase. This is because any pinned residues of the aqueous phase broken off the droplets may be transferred to subsequent droplets and cause cross contamination between the droplet bioreactors. Additionally, materials for microfluidic chip fabrication should possess good optical characteristics, fair thermal conductivity and biological or chemical compatibility with both droplet liquids and surfactants. The choice between elastic (PDMS) or stiff polymers (PC, COC, PMMA, PTFE, FEP) and other materials (glass, silicone) should also be considered in terms of the method used for droplet manipulation. Unfortunately, most of the polymeric materials are characterized by transient wetting properties between hydrophobicity and hydrophilicity that do not support proper formation of droplets. For that reason, chemical modification of the surface of the channels is essential, especially in controlled droplet microfluidics where the droplets always interact mechanically with the walls of the channels, since they are typically squeezed between at least two walls. The surface chemistry of the walls must warrant a continuous wetting film of the continuous phase between the squeezed droplet and the wall. One of the recent examples of research in this area presents a smart method for coating the surface of poly(methyl methacrylate) (PMMA) and other thermoplastics with layers of silica nanoparticles (SiNPs) in order to achieve superhydrophilic and superhydrophobic properties of the channel surface.49New probes and sensors for optical detection
Another field of research in which chemists can greatly contribute to the improvement of droplet microfluidic systems is the development of new chemicals for the monitoring and detection of the reactions and processes occurring in droplet microreactors. One example is the synthesis of new hydrophilic substrates for the enzymatic reactions, which are characterized by minimized leakage from aqueous droplets to the oil.50 In another collaborative project between our microfluidic research group and analytical chemists, we developed the system for monitoring the oxygen concentration and conditions for bacterial growth inside nanoliter trains of droplets.39 The method took advantage of dedicated oxygen-sensitive indicator dyes embedded in nanoparticles. This technology offers several advantages in comparison to other techniques for measuring the oxygen concentration in microfluidic devices. Hydrophilic nanobeads do not leak from the droplets and they are excited by wavelengths in the red and near-infrared (NIR) light ranges, which helps in avoiding the background auto-fluorescence from biological samples. The measured value is the lifetime of phosphorescence which is an intrinsic property of oxygen sensor nanoparticles and does not depend on the variations of the performance of the detecting instrument, the turbidity of a sample or the influence of the ambient environment.New detection schemes
Another important research direction in the field of droplet microfluidics is the development of new detection schemes, based on optical measurement of fluorescence, absorbance or light scattering. In one of the recent demonstrations integrated nanowire based ion-sensitive field effect transistors (ISFETs) were used as nanosensors for the in-flow electrical detection and analysis of the chemical content of the droplets.51 We envisage further progress in the development of various strategies for optic-less strategies for the measurement of the result of the reaction. For example, chemical processes occurring inside droplets can trigger changes in physical parameters such as surface tension or viscosity that can be measured as a change in droplet shape or speed in the microfluidic channel.42 Also the electric and magnetic properties of the droplets can be measured in order to determine the outcome of the reactions in droplets.7. Conclusions
Controlled droplet microfluidics is an interesting area that not only intersects multiple disciplines of science – from physics and chemistry to flow, materials and systems engineering, but also is a boiling pot of multiple microfluidic techniques and approaches. The mix of methods using solid walls, geometries, capillary forces, flows, electric fields, and automation have generated a versatile range of microfluidic systems that have already found multiple applications in synthetic and materials chemistry, microbiology and biochemistry. We envisage a lot of room for further development by hybridizing other specialized microfluidic technologies, especially in combining controlled droplet microfluidics with other droplet and continuous flow microfluidic systems.For example, controlled droplet microfluidics can be combined with ultrahigh throughput microfluidics in such a way that critical operations are executed first on relatively large droplets in the nanoliter volume range. Next, these large mother droplets can be split into populations of small picoliter droplets for single cell or single molecule analysis and for parallel executions of digital assays. An interesting and recent example is the generation of libraries of picoliter droplets by step emulsification of large plugs with arbitrarily defined compositions.32 Another possible synergistic improvement might be a hybrid device composed of EWOD based modules for universal and versatile operations on individually addressed droplets combined with parts of controlled droplet channel microfluidics for the increase of the throughput and capacity of the whole system. First examples of such systems were already presented,52 but the potential of the synergistic combination of DMF and channel droplet microfluidics has not yet been fully exploited. And finally the combination of continuous flow microfluidics with droplet technology was not fully exploited. Single phase microfluidics is the first choice for the microscale and parallel cell culturing and provides a unique possibility of the formation of highly controllable and stable chemical gradients. However, the use of droplets would ensure controlled spatiotemporal delivery of discrete portions of reagents, nutrients and stimuli to continuous flow bioreactors and/or probing of the dynamics of the changes in the chemical composition of the cell culture medium. This challenge could also be described as a revival of the technology of the droplet chemistrode53 – an experimental concept proposed several years ago by the Ismagilov group and used for the analysis of conventional cell culture. However, the potential of the chemistrode is wider than so far exploited since this technology was ahead of the progress in automated microfluidics and the advancement in microfluidic cell culture.
Controlled droplet microfluidics is yet to provide a clear “killer application” that would attract many researchers from other fields, as it has happened, e.g., with droplet digital PCR9,21 and single cell sequencing11–13 using ultrahigh throughput droplet microfluidics. We anticipate that further development of techniques for controlled operations on droplets will be focused on multiple ways. There should be a stable increase of interest in the application of systems for the on-demand generation and handling of trains of hundreds of droplets in chemical synthesis and in microbiology for studies of the dynamics of bacterial populations. Another direction is the development of user-friendly and simple devices for the rapid generation of a large number of nanoliter reaction compartments – analogues and improved versions of SlipChips and self-digitization systems. And, finally the progress in automation and liquid handling should eventually lead to the development of controlled ultrahigh throughput droplet microfluidic systems in which dozens or hundreds of droplets with defined chemical compositions can be addressed by their spatial position. Recently presented technology of Printed Droplet Microfluidics is a forerunner of such research trends. In this demonstration Cole et al.54 developed a system for fluorescence-activated droplet sorting coupled with deposition of arrays of thousands of picoliter droplets on a specialized substrate in a controlled and deterministic way.
Further development of controlled droplet microfluidic methods would not only allow for the demonstration of new applications, but also widen our knowledge about the physicochemical properties and hydrodynamics of fluids at the microscale. In parallel, we expect commercialization of these technologies and much broader introduction into research fields that are not directly associated with microfluidics and microengineering.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
P. G. acknowledges support from the National Science Centre grant DEC-2014/12/W/NZ6/00454 (Symfonia). T. S. K. acknowledges the support from the Ministry of Science and Higher Education through the scholarship for outstanding young researchers (0722/E-64/STYP/10/295) and the support from the Foundation for Polish Science within the TEAM Tech/2016-2/10 program.Notes and references
- K. Choi, A. H. C. Ng, R. Fobel and A. R. Wheeler, Annu. Rev. Anal. Chem., 2012, 5, 413–440 CrossRef CAS PubMed.
- M. Abolhasani, C. W. Coley, L. Xie, O. Chen, M. G. Bawendi and K. F. Jensen, Chem. Mater., 2015, 27, 6131–6138 CrossRef CAS.
- M. Abolhasani, N. C. Bruno and K. F. Jensen, Chem. Commun., 2015, 51, 8916–8919 RSC.
- D. van Swaay and A. deMello, Lab Chip, 2013, 13, 752–767 RSC.
- Y. Elani, R. V. Law and O. Ces, Nat. Commun., 2014, 5, 5305 CrossRef CAS PubMed.
- J. Guzowski and P. Garstecki, Phys. Rev. Lett., 2015, 114, 1–5 CrossRef PubMed.
- M. A. Czekalska, T. S. Kaminski, S. Jakiela, K. Tanuj Sapra, H. Bayley and P. Garstecki, Lab Chip, 2015, 15, 541–548 RSC.
- M.-A. Nguyen, B. Srijanto, C. P. Collier, S. T. Retterer and S. A. Sarles, Lab Chip, 2016, 16, 1191–1208 Search PubMed.
- D. Pekin, Y. Skhiri, J.-C. Baret, D. Le Corre, L. Mazutis, C. Ben Salem, F. Millot, A. El Harrak, J. B. Hutchison, J. W. Larson, D. R. Link, P. Laurent-Puig, A. D. Griffiths and V. Taly, Lab Chip, 2011, 11, 2156–2166 RSC.
- D. Cottinet, F. Condamine, N. Bremond, A. D. Griffiths, P. B. Rainey, J. A. G. M. de Visser, J. Baudry and J. Bibette, PLoS One, 2016, 11, e0152395 Search PubMed.
- A. Rotem, O. Ram, N. Shoresh, R. A. Sperling, A. Goren, D. A. Weitz and B. E. Bernstein, Nat. Biotechnol., 2015, 33, 1165–1172 CrossRef CAS PubMed.
- E. Z. Macosko, A. Basu, R. Satija, J. Nemesh, K. Shekhar, M. Goldman, I. Tirosh, A. R. Bialas, N. Kamitaki, E. M. Martersteck, J. J. Trombetta, D. A. Weitz, J. R. Sanes, A. K. Shalek, A. Regev and S. A. McCarroll, Cell, 2015, 161, 1202–1214 CrossRef CAS PubMed.
- A. M. Klein, L. Mazutis, I. Akartuna, N. Tallapragada, A. Veres, V. Li, L. Peshkin, D. A. Weitz and M. W. Kirschner, Cell, 2015, 161, 1187–1201 CrossRef CAS PubMed.
- J. J. Agresti, E. Antipov, A. R. Abate, K. Ahn, A. C. Rowat, J.-C. J.-C. Baret, M. Marquez, A. M. Klibanov, A. D. Griffiths and D. a. Weitz, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4004–4009 CrossRef CAS PubMed.
- Y. Schaerli, R. C. Wootton, T. Robinson, V. Stein, C. Dunsby, M. A. A. Neil, P. M. W. French, A. J. DeMello, C. Abell and F. Hollfelder, Anal. Chem., 2009, 81, 302–306 CrossRef CAS PubMed.
- A. M. Nightingale, T. W. Phillips, J. H. Bannock and J. C. de Mello, Nat. Commun., 2014, 5, 3777 CAS.
- L. Derzsi, T. S. Kaminski and P. Garstecki, Lab Chip, 2016, 16, 893–901 RSC.
- E. Fradet, C. Bayer, F. Hollfelder and C. N. Baroud, Anal. Chem., 2015, 87, 11915–11922 CrossRef CAS PubMed.
- J. U. Shim, R. T. Ranasinghe, C. A. Smith, S. M. Ibrahim, F. Hollfelder, W. T. S. Huck, D. Klenerman and C. Abell, ACS Nano, 2013, 7, 5955–5964 CrossRef CAS PubMed.
- P. Garstecki, H. A. Stone and G. M. Whitesides, Phys. Rev. Lett., 2005, 94, 1–4 Search PubMed.
- A. Didelot, S. K. Kotsopoulos, A. Lupo, D. Pekin, X. Li, I. Atochin, P. Srinivasan, Q. Zhong, J. Olson, D. R. Link, P. Laurent-Puig, H. Blons, J. B. Hutchison and V. Taly, Clin. Chem., 2013, 59, 815–823 CAS.
- B. Hadwen, G. R. Broder, D. Morganti, A. Jacobs, C. Brown, J. R. Hector, Y. Kubota and H. Morgan, Lab Chip, 2012, 12, 3305–3313 RSC.
- C. Dixon, A. H. C. Ng, R. Fobel, M. B. Miltenburg and A. R. Wheeler, Lab Chip, 2016, 16, 4560–4568 RSC.
- W. Du, L. Li, K. P. Nichols and R. F. Ismagilov, Lab Chip, 2009, 9, 2286 RSC.
- L. Li, W. Du and R. F. Ismagilov, J. Am. Chem. Soc., 2010, 132, 112–119 CrossRef CAS PubMed.
- L. Ma, S. S. Datta, M. A. Karymov, Q. Pan, S. Begolo and R. F. Ismagilov, Integr. Biol., 2014, 6, 796–805 RSC.
- S. Jakiela, T. S. Kaminski, O. Cybulski, D. B. Weibel and P. Garstecki, Angew. Chem., Int. Ed., 2013, 52, 8908–8911 CrossRef CAS PubMed.
- M. E. Dolega, S. Jakiela, M. Razew, A. Rakszewska, O. Cybulski and P. Garstecki, Lab Chip, 2012, 12, 4022 RSC.
- L. Ma, J. Kim, R. Hatzenpichler, M. a. Karymov, N. Hubert, I. M. Hanan, E. B. Chang and R. F. Ismagilov, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 9768–9773 CrossRef CAS PubMed.
- T. Schneider, G. S. Yen, A. M. Thompson, D. R. Burnham and D. T. Chiu, Anal. Chem., 2013, 85, 10417–10423 CrossRef CAS PubMed.
- G. Amselem, C. Guermonprez, B. Drogue, S. Michelin and C. N. Baroud, Lab Chip, 2016, 16, 4200–4211 RSC.
- W. Postek, T. Kaminski and P. Garstecki, Lab Chip, 2017, 17, 1323–1331 RSC.
- P. M. P. Korczyk, L. Derzsi, S. Jakieła and P. Garstecki, Lab Chip, 2013, 13, 1–7 RSC.
- X. Niu, F. Gielen, J. B. Edel and A. J. deMello, Nat. Chem., 2011, 3, 437–442 CrossRef CAS PubMed.
- K. Churski, T. S. Kaminski, S. Jakiela, W. Kamysz, W. Baranska-Rybak, D. B. Weibel and P. Garstecki, Lab Chip, 2012, 12, 1629–1637 RSC.
- K. Leung, H. Zahn, T. Leaver, K. M. Konwar, N. W. Hanson, A. P. Page, C.-C. Lo, P. S. Chain, S. J. Hallam and C. L. Hansen, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 7665–7670 CrossRef CAS PubMed.
- P. Abbyad, R. Dangla, A. Alexandrou and C. N. Baroud, Lab Chip, 2011, 11, 813–821 RSC.
- F. Gielen, L. Van Vliet, B. T. Koprowski, S. R. A. Devenish, M. Fischlechner, J. B. Edel, X. Niu, A. J. Demello and F. Hollfelder, Anal. Chem., 2013, 85, 4761–4769 CrossRef CAS PubMed.
- M. Horka, S. Sun, A. Ruszczak, P. Garstecki and T. Mayr, Anal. Chem., 2016, 88, 12006–12012 CrossRef CAS PubMed.
- M. Abolhasani, C. W. Coley and K. F. Jensen, Anal. Chem., 2015, 87, 11130–11136 CrossRef CAS PubMed.
- G. Lestari, M. Alizadehgiashi, M. Abolhasani and E. Kumacheva, ACS Sustainable Chem. Eng., 2017, 5, 4304–4310 CrossRef CAS.
- S. Jakiela, S. Makulska, P. M. Korczyk and P. Garstecki, Lab Chip, 2011, 11, 3603–3608 RSC.
- I. Shestopalov, J. D. Tice and R. F. Ismagilov, Lab Chip, 2004, 4, 316–321 RSC.
- H. Song and R. F. Ismagilov, J. Am. Chem. Soc., 2003, 125, 14613–14619 CrossRef CAS PubMed.
- S. Matosevic and B. M. Paegel, Nat. Chem., 2013, 5, 958–963 CrossRef CAS PubMed.
- M. Pan, F. Lyu and S. K. Y. Tang, Anal. Chem., 2015, 87, 7938–7943 CrossRef CAS PubMed.
- K. S. P. Gruner, B. Riechers, B. Semin, J. Lim, A. Johnston and J. C. Baret, Nat. Commun., 2016, 7, 9 Search PubMed.
- L. A. Bawazer, C. S. McNally, C. J. Empson, W. J. Marchant, T. P. Comyn, X. Niu, S. Cho, M. J. McPherson, B. P. Binks, A. J. DeMello and F. C. Meldrum, Sci. Adv., 2016, 2, 1–12 Search PubMed.
- R. Ortiz, J. L. Chen, D. C. Stuckey and T. W. J. Steele, ACS Appl. Mater. Interfaces, 2017, 9, 13801–13811 CAS.
- J. Fenneteau, D. Chauvin, A. D. Griffiths, C. Nizak and J. Cossy, Chem. Commun., 2017, 53, 5437–5440 RSC.
- J. Schütt, B. Ibarlucea, R. Illing, F. Zörgiebel, S. Pregl, D. Nozaki, W. M. Weber, T. Mikolajick, L. Baraban and G. Cuniberti, Nano Lett., 2016, 16, 4991–5000 CrossRef PubMed.
- S. C. C. Shih, P. C. Gach, J. Sustarich, B. A. Simmons, P. D. Adams, S. Singh and A. K. Singh, Lab Chip, 2015, 15, 225–236 RSC.
- D. Chen, W. Du, Y. Liu, W. Liu, A. Kuznetsov, F. E. Mendez, L. H. Philipson and R. F. Ismagilov, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 16843–16848 CrossRef CAS PubMed.
- R. H. Cole, S.-Y. Tang, C. A. Siltanen, P. Shahi, J. Q. Zhang, S. Poust, Z. J. Gartner and A. R. Abate, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 8728–8733 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |