Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Natural multi-osmolyte cocktails form deep eutectic systems of unprecedented complexity: discovery, affordances and perspectives
Marina
Cvjetko Bubalo
 *a,
Thanos
Andreou
*a,
Thanos
Andreou
 *b,
Manuela
Panić
a,
Mia
Radović
*b,
Manuela
Panić
a,
Mia
Radović
 a,
Kristina
Radošević
a and
Ivana
Radojčić Redovniković
a
a,
Kristina
Radošević
a and
Ivana
Radojčić Redovniković
a
aUniversity of Zagreb, Faculty of Food Technology and Biotechnology, Croatia. E-mail: mcvjetko@pbf.hr
bVIO Chemicals AG, Thessaloniki, Greece. E-mail: thanos.andreou@viochem.com
First published on 19th April 2023
Abstract
While exaptation is most impressive when links between very dissimilar contexts are established, it can be even more pervasive when the previously unestablished connection seems surprisingly obvious in retrospect. We herein established such a connection between two major research fields previously advancing in parallel: osmolytes and deep eutectic solvents. Osmolytes are small molecules produced in cells as a response to external stimuli. Based on their individual interaction with macromolecules, single osmolytes are currently categorized as “kosmotropes” (stabilizing proteins) and “chaotropes” (destabilizing them). However, with two or more osmolytes, synergistic effects were also observed on top of cumulative ones. All current attempts to explain these synergistic effects have been studying osmolyte–osmolyte interactions in aqueous solutions, but none has been generally applicable so far, indicating that a new model “beyond kosmotropes and chaotropes” is needed to understand the function of osmolytes. We have gathered enough evidence to formulate a hypothesis of such a new model. First, inspired by patterns frequently observed in nature, five major stabilizing osmolytes (kosmotropes) prominent across kingdoms (trimethylamine N-oxide, sarcosine, glycerophosphorylcholine, dimethylsulfoniopropionate and ectoine) were for the first time employed to form novel two- and three-component DESs with all known natural perturbants/chaotropes (urea, guanidine hydrochloride and arginine). Going beyond the current three-component barrier, we mimicked the exact composition of multi-osmolyte cocktails widely observed in nature and we here report the rapid and consistent formation of deep eutectic systems of unprecedented complexity and the tunable potential of these new systems to stabilize a template protein. Based on these observations, we postulate that, in vivo, osmolytes form deep eutectic systems featuring new, emergent and synergistic properties which govern their interaction with macromolecules. We believe that such bio-inspired, osmolyte-based DESs can be a remarkable new tool to study complex natural systems in higher granularity and to engineer their microenvironment towards efficient and sustainable processes at scale.
Introduction
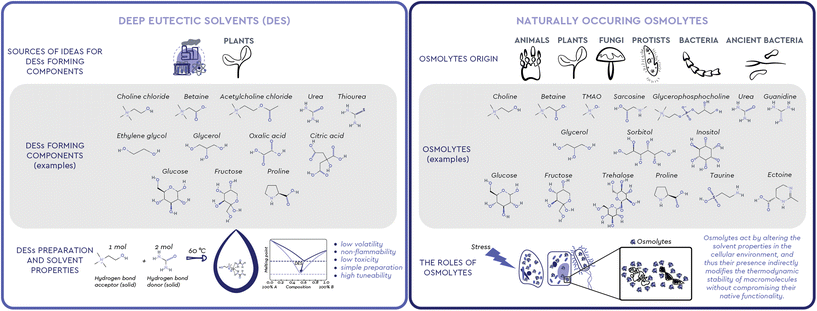
Observing nature has been one of the most successful ways to find inspiration. Humans have long used this inspiration, among other ends, to find new ways to solve complex problems through engineering, chemistry and biotechnology. Looking at the way living creatures cope with changes in the environment and understanding the functions observed in organisms and processes in nature can help us shape and create new products, processes, and systems.1 We can learn from biological systems frequently exposed to harsh environments, from extremophilic bacteria, marine organisms, sporulating microorganisms and plants, to living assemblages functioning in complex microenvironments, such as the mammal kidney. All these living systems share a similar mechanism of coping with the stressors of a harsh environment: the accumulation of small molecules commonly called osmolytes (Fig. 1). These versatile organic compounds have been attributed with several biological functions, among which the most pronounced one is the increase in the thermodynamic stability of macromolecules without compromising their native functionality. For this reason, osmolyte-induced stability of biomacromolecules has attracted considerable attention in various industrial fields. Interestingly, the exact mechanisms and interactions involved in this stabilizing effect have been a subject of debate and still remain largely unresolved.2Meanwhile, neoteric systems that effectively mimic the natural environment for various biomolecules, the so-called Deep Eutectic Solvents (DESs), have been intensively studied as nontoxic and highly tunable solvents in food, agrochemicals, cosmetics, and pharmaceuticals production (Fig. 1).3 Unlike conventional molecular solvents, a DES is a mixture of two or more, usually solid compounds combined in a suitable molar ratio to form a liquid at ambient temperature.4 A particular subgroup of these systems, Natural DESs (NADESs), only consist of compounds that occur in nature.5 In most cases, these compounds are strikingly similar, if not identical, to naturally occurring osmolytes, i.e. sugars and their derivatives, polyols, amino acids, and quaternary ammonium compounds.
Based on previous hypotheses that DES may be formed in vivo and may be responsible for the solubilisation and storage of biomolecules,6 in this paper we experimentally prove that, across kingdoms and environments, context-based combinations of osmolytes form multicomponent eutectic systems which help maintain the native conformation and functionality of proteins and other biomolecules under adverse conditions. These new findings provide an excellent opportunity to engineer new, osmolyte-based solvents and systems which are directly inspired by the natural microenvironment of biomacromolecules and can therefore mimic it effectively.
Naturally occurring osmolytes
To ensure their survival, persistence and growth, various organisms, such as animals, plants, and microorganisms, must respond in a timely manner to a myriad of stressful conditions and nutrient limitations they face in their natural environments. In particular, the loss of internal water due to drought, extreme temperatures or diseases that cause osmotic imbalance, is a common threat because it results in high concentrations of salts and organic solutes.2 To maintain osmotic balance with their environment and prevent perturbations that can cause structural changes in cellular proteins, most organisms use osmolytes, small, electrically neutral and nontoxic organic molecules.2,7While molecules acting as osmolytes vary across kingdoms, all known osmolytes can be grouped into a few main chemical categories: (i) polyols and sugar polyols (e.g. glycerol, sorbitol, xylitol) found in all kingdoms (ii) sugars and their derivatives (e.g. glucose, sucrose, trehalose) accumulated mainly in plants, insects, and polar fish; (iii) amino acids and their derivatives (e.g. glycine, proline, ectoine, taurine) found mainly in prokaryotic cells and plants; (iv) methylamines (e.g. trimethylamine N-oxide – TMAO, sarcosine, betaine) found mainly in marine fishes and plants; (v) methylsulfonium compounds (e.g. dimethylsulfoniopropionate-DMSP) found in marine organisms; (vi) Y-conjugated compounds (e.g. ureas and guanidines) used by mammals and marine life.7–9 The diversity of osmolytes, their multiple biological functions and their nontoxicity over a wide range of concentrations have led to their wide application in biotechnology, agriculture, and medicine, primarily as protein stabilizers and cell protectants (Table 1).10
| Osmolytes | Origin | Function | Examples of osmolytes applications | |
|---|---|---|---|---|
| Polyols and sugar polyols | Glycerol, mannitol, sorbitol, trehalose, inositol, o-methyl-inositol, glucosylglycerol, mannosylglycerol, arabitol, erythritol, sulfotrehalose, threitol, pinitol, D-ononitol, L-querbrachitol | Generally present in all kingdoms | • Freezing tolerance | • Mannitol, glycerol – cryoprotection |
| • Cell water retention while remaining compatible with macromolecular function | • Glycerol, sorbitol – treatment of neurodegenerative diseases | |||
| • Stabilization of proteins | • Mannitol – excipient for pharmaceutical formulations, stabilization and activation of therapeutic proteins | |||
| • Osmotic adjustment | • Sorbitol – vaccines production and stabilization | |||
| • Signaling molecules | • Erythritol – treatment of dry eye syndrome | |||
| Sugars and their derivatives | Glucose, sucrose, fructose, trahelose, maltose, mannose, raffinose, rhamnose, xylose, α-glucosylglycerate, α-mannosylglycerate | • Stabilization of membranes (sugars and their derivatives) | • Sucrose – excipient for pharmaceutical formulations | |
| • Glucose, sucrose, trehalose – cryoprotection | ||||
| • Sucrose, trehalose – vaccine production, stabilization and formulation | ||||
| • Trehalose – hypothermic storage of human organs, neuroprotection | ||||
| Amino acids and their derivatives | Glycine, alanine, proline, valine, serine, isoleucine, histidine, taurine, glutamine, hypotaurine, aspartic acid, thiotaruine, ectoine, hydroxyectoine, octopine, citrulline, poly-γ-glutamic acid, Nγ-acetyldiaminobutyrate, Nε-acetyl-L-ysine | Found principally in prokaryotic cells and vascular plants and some mammalian organs | • Precursors for most of the osmolytes | • Histidine, glycine, arginine, proline, L-glutamate – excipients for pharmaceutical formulations |
| • Alleviation of cytoplasmic acidosis | • Ectoine – skin protection, anti-inflammatory treatment, inhibitory effects in neurodegenerative diseases | |||
| • Preventing membrane damage and ion toxicity | • Taurine – congestive heart failure, anemia, neuroprotection | |||
| • strongly perturbing effects on enzyme catalytic velocity and protein structural stability | • Proline – vaccine production, flocculation and stabilization, skin healing | |||
| Methyl -ammonium compounds | Betaine, choline, trimethylamine oxide (TMAO), N-methyltaurine, sarcosine, glycerophosphorylcholine (GPC), choline sulfate, homoserine betaine | Found in every kingdom of life | • Protection against damage of membrane | • Betaine – treatment of homocystinuria, neuroprotection, skin care ingredient |
| • Stabilisation and activation of proteins and enzymes | • TMAO – vaccine stabilisation | |||
| • Regulation of ROS detoxification | ||||
| Methyl-sulfonium compounds | Dimethylsulfonium propionate (DMSP) | Found in phytoplankton and some halophytic vascular plants in large quantities | • osmoprotection, thermoprotection, and antioxidative activity | • DMSP – one of Earth's most abundant organosulfur molecules and marine osmolyte, but no uses reported so far |
| • detoxification of excess sulphur | ||||
| • precursor of oceanic DMS in Lovelock's “Gaia hypothesis” | ||||
| Y-conjugated compounds | Urea, guanidine, arginine | Used only by relatively few types of animals (e.g. cartilaginous fish) and organs (mammalian kidney) (urea) | • Deleterious effect on protein structure and function | • Urea – skin care ingredient |
| • Guanidine – treatment of muscle weakness caused by Eaton-Lambert syndrome | ||||
| • Arginine – health supplement against chest pain, high blood pressure, erectile dysfunction and peripheral arterial disease, monoclonal anitbody stabilization and formulation |
Apart from their chemical structure, osmolytes have been categorized in two major groups, according to their impact on macrobiomolecules, particularly proteins.11 Urea, guanidine hydrochloride (guanidine HCl) and, although somewhat controversial,12 arginine (Arg) are denaturing or perturbing osmolytes, commonly referred to as chaotropes, because they have been observed to disrupt the structure and function of macromolecules. On the other hand, methylamines and methylsulfonium compounds, carbohydrates, polyols, amino acids and their derivatives are referred to as kosmotropes and compatible solutes because they push the equilibrium of protein folding towards the native form in various stressful situations, often counteracting the destabilizing effect of chaotropes.12 Although generally non-toxic and compatible with cytoplasmic proteins over wide concentration ranges, many kosmotropes may result harmful at high concentrations in the absence of a chaotrope. For example, at high concentrations TMAO inhibits some enzymes and enhances formation of non-functional protein aggregates in vitro.13,14
Yancey et al.15 have shown that adding urea (a chaotrope) or glycine betaine (a kosmotrope) alone to the medium greatly reduced mammalian renal cells growth, while adding both types of osmolytes partly or fully restored normal growth. Also, when high protein diets dictate changes in the concentration of urea, the mammalian renal medulla appears to regulate one of its methylamine osmolytes, glycerophosphorylcholine (GPC), to maintain a constant GPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea concentration ratio.12 In a totally different environment, the TMAO kosmotrope content is reported to be high in deep sea organisms only when there is an obvious chaotrope present, mostly notably urea. Furthermore, cartilaginous fish and coelacanths accumulate high concentrations of methylamine compounds (mainly TMAO, with lesser amounts of betaine and sarcosine) and certain free amino acids (mainly α-alanine and taurine) in concentrations averaging 0.2 M, which is about half the concentration of urea found in the same organism.16
urea concentration ratio.12 In a totally different environment, the TMAO kosmotrope content is reported to be high in deep sea organisms only when there is an obvious chaotrope present, mostly notably urea. Furthermore, cartilaginous fish and coelacanths accumulate high concentrations of methylamine compounds (mainly TMAO, with lesser amounts of betaine and sarcosine) and certain free amino acids (mainly α-alanine and taurine) in concentrations averaging 0.2 M, which is about half the concentration of urea found in the same organism.16
The presence of osmolytes in certain combinations and proportions in nature is primarily related to the fact that, at specific molar ratios, kosmotropes, such as methylamines and amino acids, are able to counterbalance urea's deleterious effects on proteins and other macromolecules.12 Yancey et al.16 were the first to conduct a series of in vitro experiments to demonstrate that kosmotropes (TMAO, betaine, sarcosine, β-alanine and taurine) are effective stabilizers of protein structure (i.e. bovine ribonuclease, rabbit and shark lactate dehydrogenases, and bovine glutamate dehydrogenase) and that these compounds largely or completely offset the denaturing effect of urea at a molar concentration ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. Ahnad and coworkers17 performed a series of studies to measure the thermal denaturation equilibrium of α-lactalbumin in the presence of urea and methylamines (TMAO and sarcosine) and found that at a molar ratio of 1
2. Ahnad and coworkers17 performed a series of studies to measure the thermal denaturation equilibrium of α-lactalbumin in the presence of urea and methylamines (TMAO and sarcosine) and found that at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (methylamine
2 (methylamine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea), the denaturing effect of urea on the protein was perfectly compensated by TMAO and sarcosine. In addition, Khan et al.18 observed that myo-inositol provides a perfect counteraction for three proteins (RNase-A, lysozyme, and α-lactalbumin) at a ratio of 1
urea), the denaturing effect of urea on the protein was perfectly compensated by TMAO and sarcosine. In addition, Khan et al.18 observed that myo-inositol provides a perfect counteraction for three proteins (RNase-A, lysozyme, and α-lactalbumin) at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 myo-inositol to urea, whereas taurine regulates perfect counteraction in a protein-specific manner: 1.5
2 myo-inositol to urea, whereas taurine regulates perfect counteraction in a protein-specific manner: 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (RNase-A), 1.2
2 (RNase-A), 1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (lysozyme), and 1
2 (lysozyme), and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (α-lactalbumin) taurine–urea ratio. This study has also shown that the counteraction of kosmotropes on urea is not limited to the 1
2 (α-lactalbumin) taurine–urea ratio. This study has also shown that the counteraction of kosmotropes on urea is not limited to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio but depends on the protein structure and its origin.
2 molar ratio but depends on the protein structure and its origin.
Osmolyte-protein interaction models and the over-stabilizing paradox
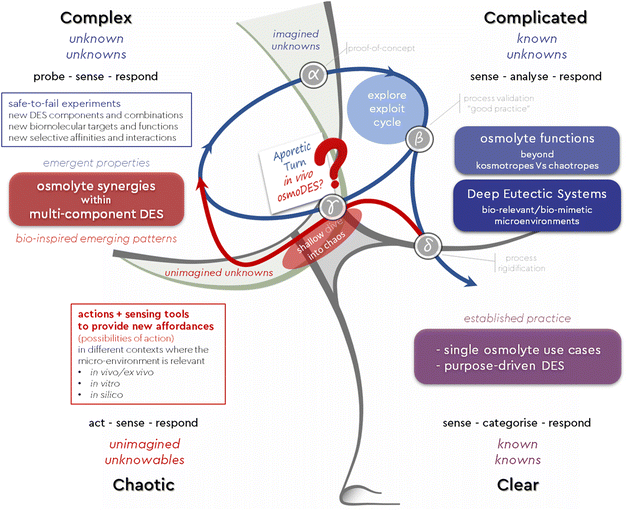
Over the past decade, many studies using both experimental and theoretical approaches have attempted to elucidate the molecular mechanism by which kosmotropes, especially TMAO, stabilize proteins in the presence of the chaotrope urea. The “indirect mechanism” states that osmolytes affect the folding behavior of proteins by changing the structure of the medium through interaction with the surrounding water molecules and subsequently modulating (weakening or strengthening) the water H-bond network and its thermodynamic properties. In contrast, the “direct mechanism” approach proposes that the osmolyte interacts directly with the peptides or amino acid side chains of the protein backbone to stabilize the native folding of the protein.19 So far, studies that consider the effects of osmolyte combinations as cumulative cannot adequately account for their complex interaction with macromolecules. The culmination of this shortcoming is the over-stabilizing paradox: it has been shown that a kosmotrope (TMAO) stabilizes a protein more in the presence of a chaotrope (urea) than alone.20,21 A similar trend to synergy has been found with mixtures of other methylamines, i.e. betaine and sarcosine, with urea.22,23 On these grounds, it has recently been suggested that it is important to consider osmolyte–protein interactions beyond the simple notions of individual kosmotropes and chaotropes affecting the folding mechanism.Rather, the effects of the combination of kosmotropes and chaotropes on the folding equilibrium of proteins need to be considered in a way that acknowledges not only cumulative but also synergistic effects between the system components.19,22 The above observations are consistent with the concept of a complex adaptive system,24 which exhibits properties that extend beyond those of the sum of its parts.25 These properties are often referred to as “emergent properties” and have been pivotal to a major field of research during the last two decades: the field of deep eutectic solvents.
Deep eutectic solvents: green solvents for a myriad of applications
Eutectic is a term used to define a homogeneous mixture of substances that melts at a single temperature which is lower than the melting point of any of its constituents. These mixtures have been known for more than a century.26 However, when Abbott et al.27 studied mixtures of quaternary ammonium salts (e.g. choline chloride, m.p. 302 °C) and urea (m.p. 133 °C) at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio in 2004, they observed a substantial decrease in the solid–liquid phase transition temperature compared to the melting point of the individual components. The observed decrease was so substantially “deeper” than the one expected for an ideal mixture that these compositions remained liquid even at room temperature. These remarkable findings prompted Abbott's group to claim a novel class of such mixtures and to coin a suitable new term to describe them: Deep Eutectic Solvents (DESs). As initially suggested, such significant depression of the melting point is attributed to the interaction between the mixture components through intermolecular hydrogen bonds in a certain range of component ratios. In practice, a DES is commonly prepared with 100% atom economy by mixing two or more, usually solid compounds, a hydrogen bond donor (HBD, e.g. amides, polyols, sugars, organic acids) and an acceptor (HBA, e.g. quaternary ammonium compounds choline chloride and betaine), in a certain molar ratio, which upon heating form a liquid at the operating temperature. While several studies on the nature of the DES component interactions have appeared,28–30 the most recent ones focus on how these interactions define the macroscopic properties of the resulting DES as a single, coherent entity,31,32 thanks to, and not despite of, the “microscopic heterogeneity” observed.33 Along the same notion, recently Martins et al.34 proposed a more pragmatic definition of a DES as “a mixture of two or more pure compounds for which the eutectic point temperature is below that of an ideal liquid mixture, presenting significant negative deviations from ideality”, significant enough so that “the mixture is liquid at the operating temperature for a certain composition range”. Based on these attributes, these neoteric solvents offer flexible physical properties such as low volatility, non-flammability, low toxicity, simple and solvent-free preparation from widely available natural raw materials and are therefore considered an excellent green alternative to conventional organic solvents and a promising tool for shaping numerous processes into being more efficient and sustainable.4 Additionally, the wide range of possible structural combinations encompassed by DES (estimated to be approximately 106), their sustainability, their unique physiochemical characteristics, as well as the possibility of fine tuning their solvent properties (e.g. pH value, polarity, hydrophilicity/hydrophobicity, viscosity) for certain purposes, make them ideal green candidates for (bio)chemical, electrochemical, and material applications, as well as for the extraction of various compounds, both inorganic (i.e. metals and CO2) and organic (i.e. plant metabolites, DNA, proteins).35 These advantages have been reflected on the interest of the academic community in these neoteric solvents, with more than 7500 scientific papers published in the last 20 years.
2 molar ratio in 2004, they observed a substantial decrease in the solid–liquid phase transition temperature compared to the melting point of the individual components. The observed decrease was so substantially “deeper” than the one expected for an ideal mixture that these compositions remained liquid even at room temperature. These remarkable findings prompted Abbott's group to claim a novel class of such mixtures and to coin a suitable new term to describe them: Deep Eutectic Solvents (DESs). As initially suggested, such significant depression of the melting point is attributed to the interaction between the mixture components through intermolecular hydrogen bonds in a certain range of component ratios. In practice, a DES is commonly prepared with 100% atom economy by mixing two or more, usually solid compounds, a hydrogen bond donor (HBD, e.g. amides, polyols, sugars, organic acids) and an acceptor (HBA, e.g. quaternary ammonium compounds choline chloride and betaine), in a certain molar ratio, which upon heating form a liquid at the operating temperature. While several studies on the nature of the DES component interactions have appeared,28–30 the most recent ones focus on how these interactions define the macroscopic properties of the resulting DES as a single, coherent entity,31,32 thanks to, and not despite of, the “microscopic heterogeneity” observed.33 Along the same notion, recently Martins et al.34 proposed a more pragmatic definition of a DES as “a mixture of two or more pure compounds for which the eutectic point temperature is below that of an ideal liquid mixture, presenting significant negative deviations from ideality”, significant enough so that “the mixture is liquid at the operating temperature for a certain composition range”. Based on these attributes, these neoteric solvents offer flexible physical properties such as low volatility, non-flammability, low toxicity, simple and solvent-free preparation from widely available natural raw materials and are therefore considered an excellent green alternative to conventional organic solvents and a promising tool for shaping numerous processes into being more efficient and sustainable.4 Additionally, the wide range of possible structural combinations encompassed by DES (estimated to be approximately 106), their sustainability, their unique physiochemical characteristics, as well as the possibility of fine tuning their solvent properties (e.g. pH value, polarity, hydrophilicity/hydrophobicity, viscosity) for certain purposes, make them ideal green candidates for (bio)chemical, electrochemical, and material applications, as well as for the extraction of various compounds, both inorganic (i.e. metals and CO2) and organic (i.e. plant metabolites, DNA, proteins).35 These advantages have been reflected on the interest of the academic community in these neoteric solvents, with more than 7500 scientific papers published in the last 20 years.
Natural Deep Eutectic Solvents (NADESs), a subgroup of DESs, were introduced and defined by Choi et al.36 as mixtures exclusively composed of two or three compounds naturally occurring in plants, often primary metabolites, such as nontoxic quaternary ammonium salts, amines, sugars, alcohols, polyols and organic acids. Precisely because of their natural origin, these solvents are expected to provide a natural, cytosol-like environment for various biomolecules, allowing them to show properties and functionality profiles that resemble the ones observed in their natural environment. Namely, in anhydrous form or with water as an additional component, these systems not only allow for excellent solubility of various biomolecules per se, but can also stabilise a wide range of commercially important molecules of natural origin (DNA, biologically active compounds, drugs, proteins, and enzymes) by providing a network of hydrogen bonds that favours stable structural conformations.37
Given the above mentioned extractive and solubilising properties, together with their capability to stabilise diverse (bio)molecules, DESs stand out as solvents with almost endless possibilities of applications. However, some key concerns on DESs application at the industrial level are their relatively high viscosity, possible corrosive activity, and in some cases, their high price. Thus, the choice of a DES for a particular use should be made carefully, having the production process and the final product in mind. Besides that, as DESs have a very low vapor pressure, isolation of the target compounds is considered to be one of the major challenges for their industrial application. Cautiously chosen DES can be safe for direct use in food, pharmaceutical, cosmetic and agrochemical products, and may thus widen the options during end product formulation.38
Osmolytes and deep eutectic systems: what happens when the two worlds collide?
The molecules with strong H-bonding potential which have been so far considered as raw materials for the preparation of DES are mainly quaternary ammonium compounds, polyols, sugars, organic acids, and amino acids. The majority of these are in fact similar or identical to known natural osmolytes. For example, choline chloride is the most commonly used methylamine compound for the preparation of DESs. Structurally similar and naturally occurring methylamines, such as TMAO, betaine, GPC and sarcosine, are all particularly important osmolytes that stabilize macromolecules by counteracting the deleterious effects of perturbant osmolytes like urea.7 Out of these four natural methylamines, so far only betaine has been considered as a HBA for DES preparation, along with a few other osmolytes from other chemical classes, such as polyols (e.g. glycerol) and sugar polyols (e.g. glucose, sucrose, xylose, fructose, sorbitol, trehalose) and amino acids (e.g. proline, lysine, arginine and histidine) (Table 2).| DES | Components | Components molar ratio | Water content (wt%) | Ref. | |||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||||
| a DES were prepared as follows: the appropriate amount of the components was placed in a 25 mL round bottom flask and the mixture was heated at 60 °C with stirring for 2 hours until a clear homogeneous liquid was formed. Upon cooling to room temperature, the mixture was left on a bench for a week in order to observe possible solidification or precipitation. Before use, DES forming compounds were dried under vacuum. b Liquids stable at 40 °C. | |||||||
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U U |
Betaine | Urea | Water | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
17 | 56 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly Gly |
Betaine | Glycerol | Water | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
10 | 57 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sol Sol |
Betaine | Sorbitol | Water | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.2 1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 6 |
25 | 58 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Treh Treh |
Betaine | Trehalose | Water | — | 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Unk. | 5 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Xyl Xyl |
Betaine | Xylose | Water | — | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
12 | 59 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc Glc |
Betaine | Glucose | — | — | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
— | 5 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Man Man |
Betaine | Mannose | — | — | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
Unk. | 5 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Lys Lys |
Betaine | Lysine | Water | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 27–180 27–180 |
65–93 | 39 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) His His |
Betaine | Histidine | Water | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 28–190 28–190 |
65–93 | 39 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg Arg |
Betaine | Arginine | Water | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30–200 30–200 |
65–93 | 39 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Arg Arg
|
Betaine | Arginine | Water | — |
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 28 28
|
40 | |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Betaine | Guanidine HCl | Water | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 3 3
|
17 | |
TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) U U
|
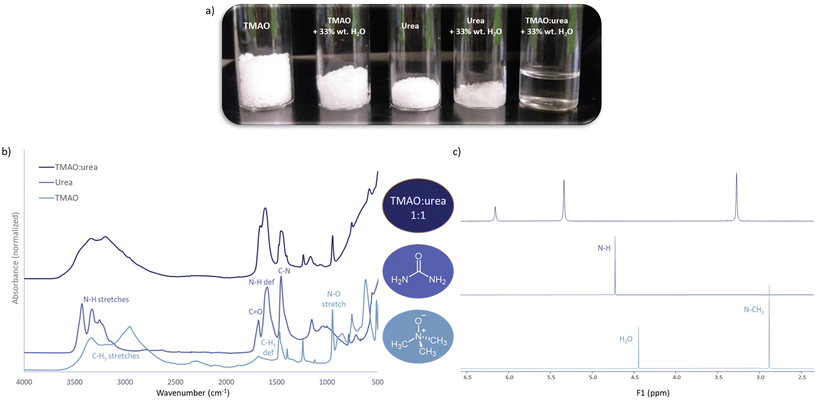
Trimethylamine N-oxide | Urea | Water | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 3.5 3.5
|
33 | |
TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Gly Gly
|
Trimethylamine N-oxide | Glycerol | Water | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2
|
17 | |
TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Sol Sol
|
Trimethylamine N-oxide | Sorbitol | Water | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2
|
12 | |
TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Glc Glc
|
Trimethylamine N-oxide | Glucose | Water | — |
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 12 12
|
23 | |
TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Treh Treh
|
Trimethylamine N-oxide | Trehalose | Water | — |
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 8 8
|
18 | |
TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Xylol Xylol
|
Trimethylamine N-oxide | Xylitol | Water | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 4 4
|
24 | |
TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Trimethylamine N-oxide | Guanidine HCl | Water | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2
|
18 | |
TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Arg Arg
|
Trimethylamine N-oxide | Arginine | Water | — |
15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 4 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 56 56
|
37 | |
Sar![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) U U
|
Sarcosine | Urea | Water | — |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 5 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 9 9
|
25 | |
Sar![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Sarcosine | Guanidine HCl | Water | — |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 5 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 11 11
|
23 | |
Sar![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Arg Arg
|
Sarcosine | Arginine | Water | — |
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 19 19
|
36 | |
Sar![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Gly Gly
|
Sarcosine | Glycerol | Water | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 4 4
|
20 | |
GPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) U U
|
Glycerophosphorylcholine | Urea | Water | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1.5 1.5
|
7 | |
GPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Glycerophosphorylcholine | Guanidine HCl | Water | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1
|
4 | |
GPC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Arg Arg
|
Glycerophosphorylcholine | Arginine | Water | — |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 4 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 80 80
|
30 | |
DMSP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) U U
|
Dimethylsulfonopropionate HCl | Urea | Water | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2
|
11 | |
DMSP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Dimethylsulfonopropionate HCl | Guanidine HCl | Water | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 3 3
|
17 | |
DMSP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Arg Arg
|
Dimethylsulfonopropionate HCl | Arginine | Water | — |
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 4 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 45 45
|
37 | |
DMSP![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Gly Gly
|
Dimethylsulfonopropionate HCl | Glycerol | Water | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 5 5
|
20 | |
Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fru Fru |
Glucose | Fructose | Water | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 8 |
30 | 60 |
Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly Gly |
Glucose | Glycerol | — | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
— | 59 |
Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Treh Treh |
Glucose | Trehalose | Water | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13 13 |
21 | 59 |
Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly:U Gly:U |
Glucose | Glycerol | Urea | — | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
— | 61 |
Fru![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Treh Treh |
Fructose | Trehalose | Water | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13 13 |
20 | 59 |
Fru![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U U |
Fructose | Glycerol | Urea | — | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
— | 61 |
Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Sol Sol
|
Glycerol | Sorbitol | — | — |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1
|
— | |
Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Treh Treh |
Glycerol | Trehalose | — | — | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
— | 59 |
Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U U |
Glycerol | Urea | — | — | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
Unk. | 62 |
Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G G |
Glycerol | Guanidine HCl | — | — | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
— | 63 |
Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Arg Arg |
Glycerol | Arginine | — | — | 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
— | 64 |
Pro![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc Glc |
Proline | Glucose | — | — | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
Unk. | 5 |
Pro![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly Gly |
Proline | Glycerol | — | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
— | 65 |
Pro![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sol Sol |
Proline | Sorbitol | — | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Unk. | 5 |
Pro![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc Suc |
Proline | Sucrose | — | — | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3 1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Unk. | 5 |
Pro![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U U |
Proline | Urea | — | — | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Unk. | 66 |
Pro![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Proline | Guanidine HCl | Water | — |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 8 8
|
30 | |
Pro![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Arg Arg
|
Proline | Arginine | Water | — |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 11 11
|
33 | |
Pro![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) GA GA |
Proline | Glutamic acid | — | — | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
Unk. | 67 |
Ser![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc Glc |
Serine | Glucose | — | — | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
Unk. | 5 |
Car![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U U |
Carnitine | Urea | — | — | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
— | 68 |
Ect![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) U U
|
Ectoine | Urea | Water | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2
|
12 | |
Ect![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Ectoine | Guanidine HCl | Water | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 3 3
|
14 | |
Ect![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Arg Arg
|
Ectoine | Arginine | Water | — |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 10 10
|
30 | |
Ect![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Gly Gly
|
Ectoine | Glycerol | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 4.5 4.5
|
20 | ||
Sol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) U U
|
Sorbitol | Urea | Water | — |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 3 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1
|
3 | |
Sol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Sorbitol | Guanidine HCl | Water | — |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 3 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1
|
3 | |
Sol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U U |
Sorbitol | Glycerol | Urea | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
— | 61 |
Sol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Gly Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Sorbitol | Glycerol | Guanidine HCl | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2
|
— | |
Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) U U
|
Sucrose | Urea | Water | — |
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 8 8
|
9 | |
Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Sucrose | Guanidine HCl | Water | — |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 12 12
|
23 | |
Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Arg Arg
|
Sucrose | Arginine | Water | — |
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 16 16
|
12 | |
Treh![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Glc Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Gly Gly
|
Trehalose | Glucose | Glycerol | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 5 5
|
— | |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sol Sol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U U |
Betaine | Sorbitol | Urea | Water | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.2 1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 7 |
25 | 58 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Sol Sol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Betaine | Sorbitol | Guanidine HCl | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1.2 1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 7 7
|
23 | |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Treh Treh![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Raf Raf |
Betaine | Trehalose | Raffinose | Water | 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 35 |
25 | 5 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pro Pro |
Betaine | Sucrose | Proline | Water | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21 21 |
20 | 59 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc Suc |
Betaine | Glycerol | Sucrose | Water | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
10 | 59 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Treh Treh |
Betaine | Glycerol | Trehalose | Water | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
13 | 59 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pro Pro |
Betaine | Sucrose | Proline | Water | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21 21 |
20 | 59 |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Tau Tau![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Gly Gly
|
Betaine | Taurine | Glycerol | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 3 3
|
— | |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Ect Ect![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Gly Gly
|
Betaine | Ectoine | Glycerol | — |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 3 3
|
— | |
B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Ect Ect![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Sor Sor
|
Betaine | Ectoine | Sorbitol | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 3 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 5 5
|
9 | |
TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) U U![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Gly Gly
|
Trimethylamine N-oxide | Urea | Glycerol | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2
|
8 | |
TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Gly Gly
|
Trimethylamine N-oxide | Guanidine HCl | Glycerol | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2
|
9 | |
TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) U U![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Sol Sol
|
Trimethylamine N-oxide | Urea | Sorbitol | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2
|
6 | |
TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Sol Sol
|
Trimethylamine N-oxide | Guanidine HCl | Sorbitol | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2
|
6 | |
TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) B B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) U U
|
Trimethylamine N-oxide | Betaine | Urea | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2
|
10 | |
TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) B B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Trimethylamine N-oxide | Betaine | Guanidine HCl | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 6 6
|
21 | |
Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fru Fru |
Sucrose | Glucose | Fructose | Water | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11 11 |
22 | 59 |
Fru![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Treh Treh |
Fructose | Glucose | Trehalose | Water | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11 11 |
21 | 59 |
Fru![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Glc Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Fructose | Glucose | Guanidine HCl | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1
|
4 | |
Fru![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Gly Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Fructose | Glycerol | Guanidine HCl | Water |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1
|
4 | |
Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Gly Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Glucose | Glycerol | Guanidine HCl | Water |
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 6 6
|
16 | |
Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Treh Treh![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) U U
|
Glucose | Trehalose | Urea | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 3 3
|
7 | |
Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Treh Treh![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) G G
|
Glucose | Trehalose | Guanidine HCl | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 3 3
|
8 | |
Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Suc Suc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sor Sor |
Glycerol | Sucrose | Sorbitol | Water | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
16 | 59 |
Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Treh Treh![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sor Sor |
Glycerol | Trehalose | Sorbitol | Water | 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
16 | 59 |
Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sor Sor |
Glycerol | Glucose | Sorbitol | Water | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
12 | 59 |
Treh![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sor Sor |
Trehalose | Glucose | Sorbitol | Water | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13 13 |
17 | 59 |
Treh![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Glc Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Sor Sor
|
Trehalose | Glucose | Sorbitol | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 4 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 13 13
|
20 | |
Treh![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Pro Pro![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Gly Gly
|
Trehalose | Proline | Glycerol | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 4 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 8 8
|
15 | |
Treh![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Ect Ect![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Gly Gly
|
Trehalose | Ectoine | Glycerol | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 4 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) 6 6
|
11 | |
Pro![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fru Fru![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly Gly |
Proline | Fructose | Glycerol | Water | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
20 | 69 |
Pro![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly Gly |
Proline | Glucose | Glycerol | Water | 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
21 | 59 |
Pro![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Gly Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/b_char_2009.gif) Sor Sor
|
Proline | Glycerol | Sorbitol | Water |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
12 | |
As a group working in the field of DESs preparation and characterization, we were not only intrigued by the structural similarity between osmolytes and common DES components, but also by the fact that osmolytes are usually present in cells and tissues in certain combinations and often in rather strict proportions, and that these combinations and proportions are often strikingly similar or identical to those used for the preparation of synthetic DESs. Based on these observations, we decided to explore the pool of naturally occurring osmolytes and the patterns of their natural distribution in order to form novel, osmolyte-based DESs.
Eutectic systems based solely on naturally occurring osmolytes have been reported, mostly consisting of simple sugars, polyols, amino acids, and the methylamine betaine (Table 2). In general, most of these systems were prepared aiming at specific industrial applications, by combining known DES components. Here, we explored various new combinations of osmolytes, including TMAO, sarcosine, GPC, ectoine, proline, DMSP, guanidine HCl, arginine, taurine, sorbitol, and trehalose, and discovered a variety of distribution patterns that form liquids stable at room temperature (Table 2). Several molar ratios of known and unknown osmolyte combinations were explored experimentally, building on the distribution patterns observed in natural systems.
All known natural methylamine osmolytes (TMAO, betaine, sarcosine and GPC), as well as DMSP, the only reported natural methylsulfonium kosmotrope, formed liquids with all known perturbant osmolytes (urea, guanidine HCl and arginine), indicating a horizontal kosmotrope![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chaotrope pattern of two-component Deep Eutectic Solvent formation (Fig. 2). Considering the melting points of the individual components (sarcosine 210 °C, GPC 143 °C, TMAO 100 °C, DMSP 123 °C, urea 133 °C, guanidine HCl 182 °C, arginine 244 °C), obtaining liquids at room temperature or even at 40 °C arguably satisfies the strict criterion for the “deep” designation. This trend, also observed for glycerol with chaotropes urea and guanidine, was extended to another chaotrope, arginine, and further corroborated for sugar polyols sucrose (m.p. 180 °C) and sorbitol (m.p. 95 °C). The potential of the amino acid osmolyte proline (m.p. 252 °C) to form DES was also confirmed for all known chaotropes (urea, guanine and arginine), together with another widely applied osmolyte and amino acid cyclic derivative, ectoine (m.p. 280 °C), which also provided stable liquid mixtures with all studied chaotropes.
chaotrope pattern of two-component Deep Eutectic Solvent formation (Fig. 2). Considering the melting points of the individual components (sarcosine 210 °C, GPC 143 °C, TMAO 100 °C, DMSP 123 °C, urea 133 °C, guanidine HCl 182 °C, arginine 244 °C), obtaining liquids at room temperature or even at 40 °C arguably satisfies the strict criterion for the “deep” designation. This trend, also observed for glycerol with chaotropes urea and guanidine, was extended to another chaotrope, arginine, and further corroborated for sugar polyols sucrose (m.p. 180 °C) and sorbitol (m.p. 95 °C). The potential of the amino acid osmolyte proline (m.p. 252 °C) to form DES was also confirmed for all known chaotropes (urea, guanine and arginine), together with another widely applied osmolyte and amino acid cyclic derivative, ectoine (m.p. 280 °C), which also provided stable liquid mixtures with all studied chaotropes.
As a representative of the methylamine osmolytes class, TMAO also formed stable liquids when combined with glycerol and major sugar polyols, following the example of betaine. The results previously published for betaine39 prompted us to further explore high water content in selected two-component DES. In particular, betaine reportedly formed liquid mixtures with three amino acids at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, but the water content of these mixtures was mentioned to be between 65 and 93 wt%, which is substantially higher than the upper limit usually considered for water-containing DES and will be further discussed in the next section. Given the abundance and application potential of betaine, we decided to probe additional molar ratios for its combination with arginine (Arg), which is also a somewhat controversial40,41 but widely employed osmolyte. Indeed, in our hands betaine did form a stable liquid with arginine at a 4
1, but the water content of these mixtures was mentioned to be between 65 and 93 wt%, which is substantially higher than the upper limit usually considered for water-containing DES and will be further discussed in the next section. Given the abundance and application potential of betaine, we decided to probe additional molar ratios for its combination with arginine (Arg), which is also a somewhat controversial40,41 but widely employed osmolyte. Indeed, in our hands betaine did form a stable liquid with arginine at a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio, while it also required a higher water content (40 wt%) compared to betaine
1 molar ratio, while it also required a higher water content (40 wt%) compared to betaine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea and betaine
urea and betaine![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) guanidine DESs, which both required 17 wt%. The same trend of very particular kosmotrope
guanidine DESs, which both required 17 wt%. The same trend of very particular kosmotrope![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) arginine ratios and relatively higher water content than usual chaotropes was observed for most of the novel Arg-based DESs. Interestingly, the arginine combinations with the other natural methylamine kosmotropes TMAO, sarcosine and GPC only remained liquid at temperatures of at least 40 °C.
arginine ratios and relatively higher water content than usual chaotropes was observed for most of the novel Arg-based DESs. Interestingly, the arginine combinations with the other natural methylamine kosmotropes TMAO, sarcosine and GPC only remained liquid at temperatures of at least 40 °C.
A hint to additional possbilities worth probing for the behaviour of two-component mixtures was revealed when we investigated the potential of the commercially promising TMAO–urea combinations. While these two-component systems were unstable at room temperature at the typical methylamine-to-urea molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 found in nature, the addition of a third osmolyte component, glycerol or sorbitol, to a final TMAO
2 found in nature, the addition of a third osmolyte component, glycerol or sorbitol, to a final TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea
urea![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polyol molar ratio of 1
polyol molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, smoothly provided a stable eutectic mixture stable at the same conditions. These two polyols were also critical as a third DES component when added to the binary mixtures betaine–ectoine (molar ratio of 1
2, smoothly provided a stable eutectic mixture stable at the same conditions. These two polyols were also critical as a third DES component when added to the binary mixtures betaine–ectoine (molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) and trehalose–glucose (molar ratio of 1
2) and trehalose–glucose (molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), once more providing three-component liquid mixtures stable at room temperature. A similar need for a third component to obtain stable liquids was observed for the binary mixture trehalose–glucose, this time with urea, affording a liquid stable at room temperature for the three-component combination Treh
1), once more providing three-component liquid mixtures stable at room temperature. A similar need for a third component to obtain stable liquids was observed for the binary mixture trehalose–glucose, this time with urea, affording a liquid stable at room temperature for the three-component combination Treh![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glu
Glu![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U (Table 2). Following this trend, several new three-component eutectic mixtures were obtained, ranging from mixed kosmotropes
U (Table 2). Following this trend, several new three-component eutectic mixtures were obtained, ranging from mixed kosmotropes![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) chaotrope systems, reminiscent of natural osmolyte distribution patterns, to industrially promising all-sugar and ectoine-based systems.
chaotrope systems, reminiscent of natural osmolyte distribution patterns, to industrially promising all-sugar and ectoine-based systems.
Some of the prepared DESs were anhydrous (i.e. Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sol, Gly
Sol, Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G, B
G, B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Tau
Tau![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly, B
Gly, B![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ect
Ect![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly, Treh
Gly, Treh![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Glc
Glc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U and Sor
U and Sor![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly
Gly![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G), while most of them required approximately 5–35 wt% of water to remain in a stable liquid form at room temperature for an extended period of time (Table 2). This was to be expected since water has been shown to play a key role in the formation of DESs by modifying the physicochemical properties of the corresponding DESs supramolecular network. For instance, it has been observed that the small amount of water present in hydrated DESs can strengthen the hydrogen bond network with water monomers confined into the DES voids.42–45 Hammond et al.43 demonstrated that water acts as a second small HBD when aqueous mixtures of ChCl
G), while most of them required approximately 5–35 wt% of water to remain in a stable liquid form at room temperature for an extended period of time (Table 2). This was to be expected since water has been shown to play a key role in the formation of DESs by modifying the physicochemical properties of the corresponding DESs supramolecular network. For instance, it has been observed that the small amount of water present in hydrated DESs can strengthen the hydrogen bond network with water monomers confined into the DES voids.42–45 Hammond et al.43 demonstrated that water acts as a second small HBD when aqueous mixtures of ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) malic acid in a 1
malic acid in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio contain 1–2 mol of water per mole of DES. This was also confirmed by López-Salas et al.45 who showed that in the “water-in-DES” system of a ternary DES composed of resorcinol, urea and choline chloride (RUChClnW, where n represents mol of water per mole of ternary DES), the tetrahedral structure of water was distorted as a consequence of its incorporation, as an additional HBD or HBA, into the hydrogen bond complexes formed among the original DES components. The same group showed that “water-in-DES” regime occurs in DES dilutions with nonaqueous hydrogen-bond-forming organic solvents (e.g. benzyl alcohol).44
1 molar ratio contain 1–2 mol of water per mole of DES. This was also confirmed by López-Salas et al.45 who showed that in the “water-in-DES” system of a ternary DES composed of resorcinol, urea and choline chloride (RUChClnW, where n represents mol of water per mole of ternary DES), the tetrahedral structure of water was distorted as a consequence of its incorporation, as an additional HBD or HBA, into the hydrogen bond complexes formed among the original DES components. The same group showed that “water-in-DES” regime occurs in DES dilutions with nonaqueous hydrogen-bond-forming organic solvents (e.g. benzyl alcohol).44
The point of transition from water-in-DES regime to DES-in-water regime (described as a simple aqueous solution of the individual components) depends on the DES composition, but it is generally considered that up to about 40–50 wt% of water (in some cases even up to 57%,46 the strong interactions between the components of the DESs slowly weaken with the supramolecular structure preserved,47 while at higher dilutions, the network can be disrupted, leading to a mixture that exhibits behavior closer to that of the individual components in an aqueous solution.48
As mentioned, none of the newly prepared mixtures required more than 40 wt% of water, which is consistent with the strict definition of these mixtures as DESs.
Overall, we here present several major advances: TMAO, sarcosine, GPC, ectoine, DMSP, arginine and guanidine HCl, the most widely encountered osmolytes and representatives of both kosmotrope and chaotrope families, are for the first time experimentally combined to form new DESs. The potential of these seven osmolytes to form eutectic systems is further established by the preparation of 54 novel two-or three-component DESs with other osmolytes. And for the first time, these new eutectic systems are directly inspired by the distribution patterns of kosmotropes and chaotropes observed in nature. In terms of DES sustainability, choline chloride, the most widely used methylamine for DESs preparation so far, is industrially manufactured from fossil-based ethylene oxide.49 The systems herein reported are based only on natural methylaimines, and thus avoid the use of choline chloride, while exhibiting novel properties and providing new affordances.
Mimicking nature: new multicomponent DESs based on osmolyte distribution patterns in biological contexts
Single osmolytes rarely accumulate alone in a given biological context, but rather within patterns comprising one chaotrope and one or more additional kosmotropes. For example, the serum of winter-acclimatized fish contains urea, TMAO and glycerol,50 while terrestrially hibernating amphibians accumulate urea, glucose and glycerol,51 an ubiquitous grass species use proline, betaine and sucrose to improve salt tolerance,52 and sharks combine urea, TMAO and betaine in their body fluids.53 We have herein shown that, among others, these exact three-component combinations (TMAO–urea–gylcerol, glucose–urea–glycerol, proline–betaine–sucrose and TMAO–betaine–urea) can form DESs (Table 2). We have further shown that in the case of the TMAO–urea–glycerol DES, the addition of the third osmolyte to the initial binary mixture was crucial to obtain a liquid at room temperature. Therefore, it would be reasonable to expect the formation of such ternary or even more complex osmolyte-based DESs inspired by additional and even more complex natural distribution patterns.Encouraged by these findings and by the fact that no DES of such complexity has been reported, we decided to break the three-component barrier and explore the patterns found in the kidney of cartilaginous fish (shark) and mammals (rabbit – inner renal medulla), as well as in the muscles of sharks and skates (Fig. 3). These organs/organisms accumulate urea to high concentrations as part of their osmoregulatory strategy and a cocktail of kosmotropes to counteract the perturbing effects of the urea chaotrope on protein structure.7,54
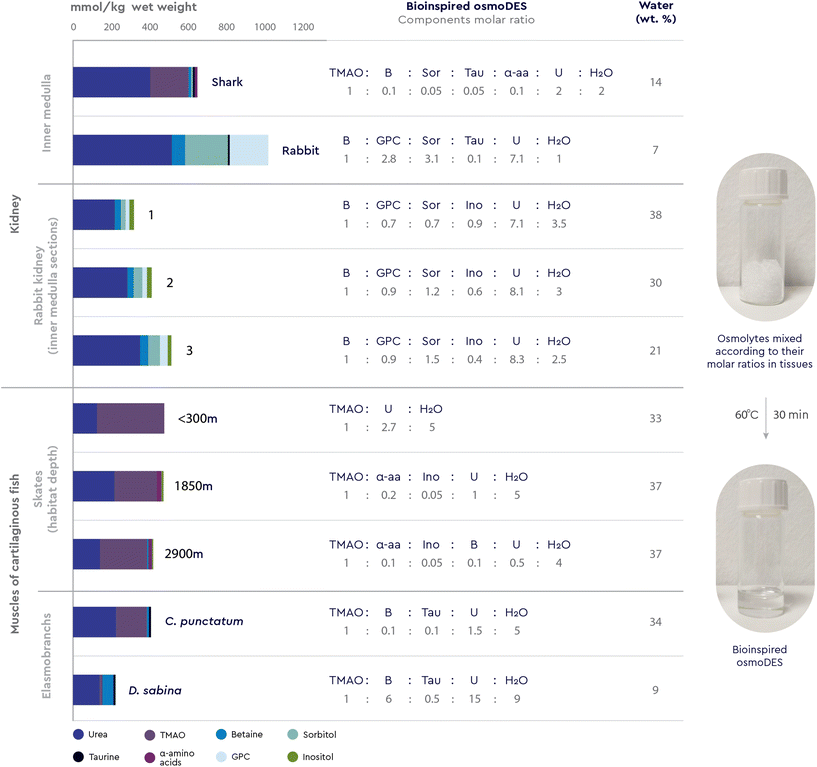 | ||
| Fig. 3 Bioinspired multicomponent osmoDES prepared by following osmolytes distribution patterns in the cartilaginous fish (shark Squalus acanthias)7 and mammal (rabbit Oryctolagus cuniculus)7 kidneys, and muscles of cartilaginous fish (sharks Chiloscyllium punctatum and Dasyatis sabina, and skates Raja hollandi).73,157 The molar ratios of DES forming components were calculated from their concentrations in the respective tissues and the mixtures were subjected to the standard DES-forming conditions. | ||
In particular, during antidiuresis (under water-deficient conditions), mammals accumulate high concentrations of compatible osmolytes in renal medullary cells, among them sorbitol, GPC, inositol, and betaine, whereas in cartilaginous fish, TMAO dominates a cocktail of urea, betaine, inositol and α-aminoacids.54 As for marine elasmobranchs, to be approximately isosmotic with seawater, these organisms accumulate high concentrations of urea in their body fluids and tissues, together with TMAO, betaine and taurine to “neutralise” its harmful effects.55 Based on these observations, we calculated the molar ratios of each osmolyte cocktail directly from their concentrations in the respective tissues and subjected the mixtures to the standard DES-forming conditions (Fig. 3).
All 10 osmolyte combinations provided liquids that were stable at room temperature with water contents less than 40 wt% (Fig. 3) and most of them were rapidly formed within minutes, compared to almost 1 hour required for usual DES formation.
This is the first time that a specific molar combination of multiple osmolytes, as encountered in a specific native environment, is experimentally proven to form a deep eutectic system of unprecedented complexity (osmoDES). It is worth noting that the tested osmolyte combinations are distinct, not only in terms of which organism they belong to, but also in terms of the topology of the studied tissue (different sections of the rabbit kidney medulla, Fig. 3) as well as the specific relative localization of a given organism (skates in different sea depths, Fig. 3). Therefore, the differences observed in the osmolyte distribution patterns may reflect the response to the respective conditions, functions and stresses related to each specific microenvironment. Similar multi-osmolyte cocktails are also found in other plant, animal and human organs and tissues, such as the tissues of other marine animals (e.g. fish, molluscs, crustaceans, shrimps, octopods, snails and worms),7,70,71 plants exposed to salt stress,72 hibernating organisms,51 and in the human brain.73 Based on this novel viewpoint, tracking osmolyte patterns in specific biological contexts could help successfully create novel, bioinspired two-, three-, and multicomponent DESs. Perhaps more significantly, such systems could be very effective in mimicking the natural microenvironment of proteins and other biomacromolecules operating within these biological contexts, by deciphering, understanding and hopefully replicating and tuning the molecular interactions between these new media and the targeted macromolecular structures.
Methylamines stabilize proteins in the presence of urea: could this phenomenon be explained by the formation of a eutectic system?
As discussed above, the molecular mechanism by which methylamines, as kosmotropes, stabilize proteins in the presence of the chaotrope urea remains controversial. At a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, the thermodynamic effects of combined methylamines and urea on protein stability and function were believed to be algebraically additive.17 However, in numerous cases methylamines in the presence of urea are more potent in stabilizing a protein than alone,20 resulting in the over-stabilizing paradox mentioned earlier.
2, the thermodynamic effects of combined methylamines and urea on protein stability and function were believed to be algebraically additive.17 However, in numerous cases methylamines in the presence of urea are more potent in stabilizing a protein than alone,20 resulting in the over-stabilizing paradox mentioned earlier.
When Abbott reported the choline chloride![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) urea DES (ChCl
urea DES (ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U, molar ratio 1
U, molar ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2),27 and when later the same DES not only did not denature74 but instead showed a strong stabilizing effect on enzymes,75 the mechanism by which methylamines stabilize proteins in the presence of the denaturant urea was not properly discussed in the context of synergies and interferences between these two in the form of a eutectic mixture. The only study that addressed this issue showed that, as the concentration of betaine–urea mixtures increased up to 75 wt% (corresponding to deep eutectic conditions), the ability of the lysozyme to refold upon cooling increased accordingly, while an increase in urea concentration alone could cause denaturation of the lysozyme.56 Zeng et al.56 were the first to show that betaine as a kosmotrope and urea as a chaotrope form a DES at molar ratios in the range of 1
2),27 and when later the same DES not only did not denature74 but instead showed a strong stabilizing effect on enzymes,75 the mechanism by which methylamines stabilize proteins in the presence of the denaturant urea was not properly discussed in the context of synergies and interferences between these two in the form of a eutectic mixture. The only study that addressed this issue showed that, as the concentration of betaine–urea mixtures increased up to 75 wt% (corresponding to deep eutectic conditions), the ability of the lysozyme to refold upon cooling increased accordingly, while an increase in urea concentration alone could cause denaturation of the lysozyme.56 Zeng et al.56 were the first to show that betaine as a kosmotrope and urea as a chaotrope form a DES at molar ratios in the range of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.56
3.56
In addition to this observation, we have here shown that the four most prominent methylamines distributed across all kingdoms of life (betaine, TMAO, sarcosine and GPC) form eutectic systems with urea, guanidine HCl and arginine (Table 2). Furthermore, we have also shown that multicomponent DESs are consistently formed when combining prominent natural kosmotropes (betaine, TMAO, GPC, DMSP, taurine, ectoine, sarcosine, inositol and α-amino acids) with natural chaotropes urea and guanidine in molar ratios found in mammal and fish kidney and muscles (Fig. 3).
Based on these findings, we wondered whether it was possible for osmolytes to coexist in the vicinity of proteins or other macromolecules in the form of a eutectic mixture, rather than as components dissolved in water, and whether this might provide additional information on the molecular mechanism of the stabilizing effect of natural methylamine kosmotropes on proteins in the presence of a natural chaotrope.
The mechanism underlying the counteraction of synthetic choline chloride to urea under eutectic conditions is well studied. Two independent experimental and molecular dynamics studies have shown that small helix-rich protein76 and lipase75 exhibit excellent conformational stability in ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U and that high concentrations of both choline chloride and urea (at a 1
U and that high concentrations of both choline chloride and urea (at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio) are critical for this stabilizing effect. At these conditions, which correspond to a deep eutectic state, the authors suggest that choline chloride acts as an efficient protein stabilizer by attracting urea and excluding it from the protein surface.76 This assumption was further corroborated by Monhemi et al.75 who showed that, at deep eutectic conditions, urea molecules have a low diffusion coefficient due to hydrogen bonding with choline and chloride ions and cannot reach protein domains. Thus, the above research on synthetic ChCl with urea may serve as a basis for the hypothesis that, within cells, strong hydrogen bonds between natural kosmotropes and chaotropes under deep eutectic state are responsible for the stabilization of proteins and other biomolecules in vivo. Similar to synthetic ChCl, the newly discovered fact that all major natural methylamines form deep eutectic mixtures with urea and guanidine HCl can give us guidelines on how to decipher the molecular mechanism behind the protein-stabilizing effects of all-natural methylamine-based kosmotropes, specifically. At the same time, the expansive interpretation of these observations raises the question whether all osmolyte-induced changes in the conformational equilibrium of macromolecules in vivo are related to the formation of osmolyte-based DESs, whose emergent properties within the wider system of the cytoplasm extend beyond those of separate osmolytes in aqueous solution.
2 molar ratio) are critical for this stabilizing effect. At these conditions, which correspond to a deep eutectic state, the authors suggest that choline chloride acts as an efficient protein stabilizer by attracting urea and excluding it from the protein surface.76 This assumption was further corroborated by Monhemi et al.75 who showed that, at deep eutectic conditions, urea molecules have a low diffusion coefficient due to hydrogen bonding with choline and chloride ions and cannot reach protein domains. Thus, the above research on synthetic ChCl with urea may serve as a basis for the hypothesis that, within cells, strong hydrogen bonds between natural kosmotropes and chaotropes under deep eutectic state are responsible for the stabilization of proteins and other biomolecules in vivo. Similar to synthetic ChCl, the newly discovered fact that all major natural methylamines form deep eutectic mixtures with urea and guanidine HCl can give us guidelines on how to decipher the molecular mechanism behind the protein-stabilizing effects of all-natural methylamine-based kosmotropes, specifically. At the same time, the expansive interpretation of these observations raises the question whether all osmolyte-induced changes in the conformational equilibrium of macromolecules in vivo are related to the formation of osmolyte-based DESs, whose emergent properties within the wider system of the cytoplasm extend beyond those of separate osmolytes in aqueous solution.
Do osmolytes form DESs in cells exposed to stress?
The cytoplasm has been traditionally viewed as a well-mixed and spatially homogeneous mixture of monomers and small complexes. However, modern molecular biology techniques are giving us unprecedented access to cell structure and dynamics at micro- and meso-length scales.77 Recent studies reveal that, at these scales, a number of interesting liquid phase transitions occur in two or three dimensions resulting in separate fluid phases within cells.78 Intermolecular hydrogen bonding between osmolytes is highly likely to occur in cells at the nanoscale, as part of dynamic (to overcome various forms of stress)79 as well as apparently static systems (e.g. in seeds for long-term survival).33 High concentrations of solutes, such as sugars, alcohols, organic acids and amino acids, has provoked scientists to wonder if these solutes contribute to the nano/mesoscale organization in certain biological systems by forming DESs.36,80 If so, DESs formation in vivo should not be seen as a simple, randomly dispersed liquid phase, but as a layer or a liquid cluster in different cellular regions or subcellular assemblies such as plastids and vesicles.80The presence of DESs in vivo was first hypothesized for plant cells by Choi et al.,36 based on the observation that plant secretions, such as sap and nectar, accumulate primary metabolites which are known DES components (choline, betaine, proline, organic acids such as malic, succinic and citric) in molar ratios typical of DES formation. The authors went on to propose that plant cells contain a third type of medium, which plays a vital role in solubilizing, storing, and transporting poorly water-soluble metabolites, adjusting the water content of plants, and protecting cells when in harsh conditions. Durand et al.80 recently raised the hypothesis that DES may also be involved in cell function regulation through interactions with membranes, by affecting their properties and therefore regulating the transport and diffusion of molecules or the circulation of fluids.
Going one step further, here we experimentally show that several natural distributions of osmolytes in several species across kingdoms and environments form new multicomponent DESs of previously unimagined complexity. We thus believe that the question is now ripe: are osmolyte-based eutectic systems formed in the cytoplasm of all species to support the stability and function of biomolecules under external stress? Furthermore, could the concept of DESs formation in the cell at the mesoscale be a key to understanding the in vivo stabilization of biomacromolecules in general?
To support this hypothesis, we identified measurable indicators of the proposed existence of DESs in vivo. It has been shown that enzymes such as laccases,5 dehydrogenases,81 lipases and other hydrolytic enzymes60,82 show little or no activity in pure synthetic DESs or in DESs at low water content. However, by adding water to DESs, the activity of the enzymes increases and reaches its maximum in highly diluted mixtures (80–90 wt%). On the other hand, low water content in DES has been shown to positively affect the stability of the same enzymes.36 We may postulate that mixtures of osmolytes form DESs at nano/microscale in vivo and in this way preserve enzymes in catalytically inactive form during cryoprotection, drought, resistance and germination. Once the stressful conditions are overcome and water enters the cell, the eutectic systems are diluted and the enzymes are activated.
Is the microenvironment a low-energy, context-aware regulator of biomacromolecule function?
Yancey et al.13 postulate that transitions between torpid and active states may not require metabolic regulation at the level of enzyme degradation or synthesis, but may rather depend on readily reversible transitions between inactive and active enzyme states in response to alterations in its microenvironment. Furthermore, this hypothesis could be relevant not only to organisms undergoing stress, but also as a general regulatory mechanism for the storage and on demand activation of enzymes. In addition, versatile enzymes (e.g. laccases,83 lipases,84 proteases85) show excellent thermal stability in synthetic DESs composed of osmolytes such as betaine, polyols and sugar alcohols. Delorme et al.83 showed that laccase is much less thermostabilized in the presence of betaine and xylitol alone than in a DES formed by those two osmolytes. This leads us to assume that formation of eutectic systems in vivo could contribute to the stabilization of proteins in organisms that must cope with high temperatures. Going beyond proteins, the nucleic acid-templated synthesis (NATS) of a dipeptide was studied as a simple model of non-enzymatic translation in DESs.86 The authors showed that, compared with aqueous buffer media, various glycholine-based DESs have a positive effect on the stability of DNA-conjugated activated esters which are necessary for non-enzymatic peptide formation. In this DES, peptide synthesis was prevented due to a significant decrease in the reactivity of amines. When the DNA-conjugated activated esters were transferred back to aqueous buffers, peptide synthesis was once again observed. This suggests that the hypothesis of DES-based regulation could be relevant to the storage and “on-demand” activation of additional biomacromolecules beyond proteins.When thinking of osmolytes forming DESs in the cytoplasm, it is worthwhile to consider the way cells react when osmolytes reach potentially toxic concentrations of up to 400 mM.87 Jagannathan et al.87 proposed that mixtures of different osmolytes in specific molar ratios may help to reduce toxicity of accumulated osmolytes. Depletion in osmolytes toxicity could be a consequence of them forming DESs. Namely, it has been shown on several occasions that phosphonium-,88 ammonium-,89 choline-90,91 and betaine-based91 DESs show lower toxicity toward immortalized animal cell lines than the aqueous solutions of their individual components.
The accumulation of osmolytes in cells is a dynamic and reversible process. Thus, to plausibly consider that osmolytes in a cytosol form eutectic systems and thus stabilize biomacromolecules, the formation of these systems must also be reversible. As previously noted, DESs have the potential to bind up to 40–50% water and their components still interact with each other,48 whereas further dilution behaves closer to a solution of its individual components. By performing hygroscopicity measurements of the glucose![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) choline chloride
choline chloride![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water DES (molar ratio 2
water DES (molar ratio 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) Dai et al.6 proved that this process is indeed reversible, indicating that, within cells, DES may easily arise and disappear at micro-scale level. More importantly, this inherent ability may enable fine-tuning of the cytoplasm water level as a means to control physicochemical properties (e.g. pH, osmolality, polarity and viscosity) which are crucial for maintaining cell homeostasis under various environmental conditions.
5) Dai et al.6 proved that this process is indeed reversible, indicating that, within cells, DES may easily arise and disappear at micro-scale level. More importantly, this inherent ability may enable fine-tuning of the cytoplasm water level as a means to control physicochemical properties (e.g. pH, osmolality, polarity and viscosity) which are crucial for maintaining cell homeostasis under various environmental conditions.
The way cells regulate the stabilization of macromolecules at low water content conditions (freezing and drought) could also be explained by the reversibility of DESs formation and dilution. In psychrophilic bactera,92 diapausing insects93,94 (e.g. grasshoppers and spiders), amphibians51 (e.g. frogs) and reptiles95,96 (e.g. snakes and turtles) the accumulation of compatible solutes such as betaine, glucose, sucrose, trehalose, sorbitol, mannitol, glycerol, glycine, taurine, and urea results in the lowering of the cytoplasmic freezing point, thereby providing protection against frost. Analogous to the mechanism proposed for plants,6 freezing may result in the formation of a separate liquid phase (DES) that keeps both water and (macro)molecules in a stable liquid form, as discussed earlier. Indeed, the interactions between the DES components and water are strong enough to maintain water liquid in the form of a glassy DES solution even at extremely low temperatures.97 Dilution with water, once an organism is back to ambient temperature, restores the cellular structures resulting in a living cell or organism. An analogous course of action could be applicable for organisms in dormant state. Namely, in certain anhydrobiotic organisms, ranging from bacteria and yeast (in cyst or spore stages) to nematodes and to certain arthropods that have entered a dormant state, water content may drop well below 10% and such anhydrobiotic states exhibit no detectable metabolism.98 These organisms can remain viable in this form for years, even millennia in the case of some prokaryotes.99 The initially obscure underlying mechanisms have been only partially clarified in the recent decades. While the effect of trehalose in anhydrobiotic states was initially touted as conclusive, recent studies indicate a more complex phenomenon,99 which might benefit by acknowledging synergistic effects between trehalose and other osmolytes accumulated in the cytoplasm, such as glycerol, proline, ectoine, betaine and TMAO,100 possibly in the form of eutectic systems. The loss of water from the cytosol at anhydrobiotic conditions could result in the formation of a DES that keeps biomolecules in an inactive form until needed. Knowing that DES easily absorb and desorb water from the environment, the existence of DESs as a third and possibly transient medium inside a cell appears as a simple and non-demanding way to tolerate water-related changes in the environment.6
Case study: (thermo)stabilisation of a model protein in novel osmolyte-based DESs
To showcase some of the novel osmolyte-based DESs as a liquid medium for storage of biomolecules, particularly proteins, we monitored the stability of a model protein, lysozyme, in these solvents (Fig. 5). The enzyme was incubated at 25 and 45 °C for 7 days in several DESs comprising natural methylamines (betaine, TMAO and sarcosine), methyl sulfone DMSP and cyclic amino acid ectoine as HBA, and glycerol as HBD, as well as in two bioinspired osmoDESs (all containing 20 wt% of water). It has been previously shown that compared to buffered solutions, lysozyme activity was greatly reduced in ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly having low water content.101,102 However, the enzyme activity was completely recovered upon dilution. Herein, we have also observed the same trend: in all tested osmolyte-based DESs the loss in enzyme activities compared to the reference buffer was observed (>90%, data not shown), while upon dilution in sodium phosphate buffer the enzyme activity was restored completely. Thus, for the purpose of measuring lysozyme stability in osmolyte-based DESs, the enzyme was incubated in DESs and residual activity upon its dilution in the buffer was measured. As can be seen form Fig. 4, DESs comprising sarcosine or DMSP with glycerol, at both incubation temperatures tested, enabled higher residual lysozyme activities after 7 days of incubation (in the range from 95 to 103%) in comparison to both controls, ChCl
Gly having low water content.101,102 However, the enzyme activity was completely recovered upon dilution. Herein, we have also observed the same trend: in all tested osmolyte-based DESs the loss in enzyme activities compared to the reference buffer was observed (>90%, data not shown), while upon dilution in sodium phosphate buffer the enzyme activity was restored completely. Thus, for the purpose of measuring lysozyme stability in osmolyte-based DESs, the enzyme was incubated in DESs and residual activity upon its dilution in the buffer was measured. As can be seen form Fig. 4, DESs comprising sarcosine or DMSP with glycerol, at both incubation temperatures tested, enabled higher residual lysozyme activities after 7 days of incubation (in the range from 95 to 103%) in comparison to both controls, ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly and the reference buffer101 with residual activities <82%. In contrast, TMAO as HBA, when paired with glycerol, did not show stabilizing effect on the tested enzyme, while the presence of ectoine was beneficial for lysozyme stabilization at 25 °C.
Gly and the reference buffer101 with residual activities <82%. In contrast, TMAO as HBA, when paired with glycerol, did not show stabilizing effect on the tested enzyme, while the presence of ectoine was beneficial for lysozyme stabilization at 25 °C.
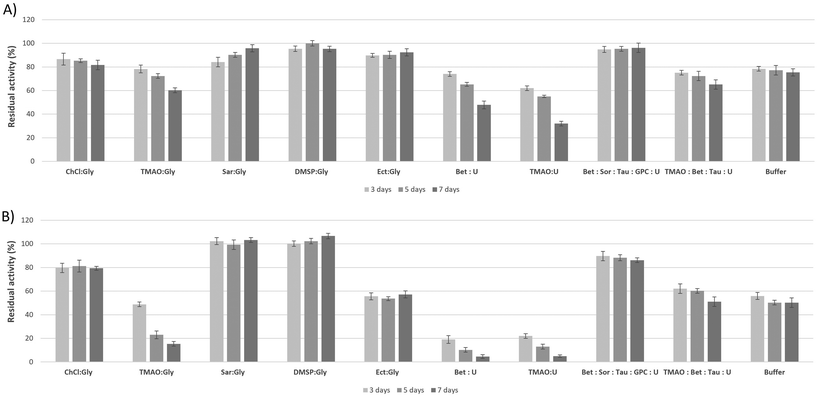 | ||
| Fig. 4 Residual lysozyme activity after incubation in osmolyte-based DES (containing 20 wt% of water) and sodium phosphate buffer solution at 25 °C (A) and 45 °C (B). Lysozyme activity was determined according to the method of Shugar et al.158 Lysozyme solutions at a concentration of 0.1 mg ml−1 were prepared in different DESs and in 10 mM sodium phosphate buffer solution (pH 7) and incubated for 7 days at 25 and 45 °C. To measure residual enzyme activity, to 525 μl of 10 mM sodium phosphate buffer solution (pH 7) in a plastic disposable cuvette 30 μl of Micrococcus lysodeikticus bacteria suspension in sterile PBS buffer (7 mg ml−1) and 30 μl of the lysozyme solution were added. Immediately after mixing, the cuvette was placed in a UV/VIS spectrophotometer and the absorbance was measured at a wavelength of 450 nm over a period of linear turbidity decline. The relative activity (%) was calculated from the initial reaction rate obtained by the enzyme after incubation, compared to the one obtained without previous exposure. | ||
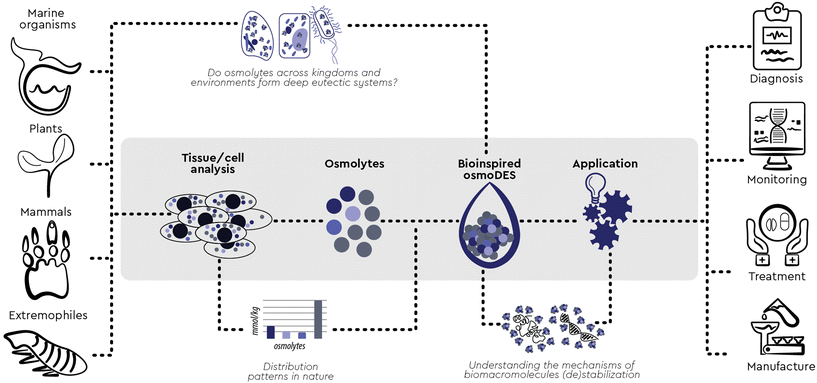 | ||
| Fig. 5 Mimicking nature: design, preparation and potential applications of bioinspired osmolyte-based DESs. | ||
Two-component DESs of betaine and TMAO with urea as HBD showed similar destabilizing effects. However, the bioinspired multicomponent osmo-DESs containing the same osmolytes, betaine and urea, exhibited excellent protein stabilisation ability, especially at 25 °C (residual activity of 96.2%), indicating that the presence of additional osmolytes in the cocktail (sorbitol, GPC, and taurine) in the ratios replicated from a natural context drastically enhanced DES ability to stabilise lysozyme. The same was observed for TMAO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) U and its multicomponent osmoDES cocktail counterpart, which besides TMAO and urea also contained betaine and taurine.
U and its multicomponent osmoDES cocktail counterpart, which besides TMAO and urea also contained betaine and taurine.
These results suggest that osmolyte-based DESs in all their variability have a potential to stabilize a template protein stored at ambient, as well as at elevated temperatures, in a significantly better and longer-lasting manner than the currently known DES stabilizing medium (ChCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Gly) and the standard buffer for lysozyme storage. Replicating osmolyte patterns inspired by specific biological contexts can, therefore, help design novel multicomponent DESs with exquisite stabilising abilities, providing new options for the preservation of proteins, ideally without the requirement of additional excipients.
Gly) and the standard buffer for lysozyme storage. Replicating osmolyte patterns inspired by specific biological contexts can, therefore, help design novel multicomponent DESs with exquisite stabilising abilities, providing new options for the preservation of proteins, ideally without the requirement of additional excipients.
Conclusions and future perspectives
It has been argued that adaptation accounts for most incremental evolutionary changes in nature, while exaptation has been linked to the most disruptive and arguably the most fascinating ones, such as the wing or the lung.103,104 While exaptation is most impressive when links between very dissimilar contexts are established as a result, it can be even more pervasive when the previously unestablished connection seems surprisingly obvious in retrospect. Most importantly, when the newly formed link can act as an enabling constraint,105 then a new way of thinking is revealed, based on which additional interdependencies can be envisaged and new opportunities of action can emerge.Here we have established such a connection between the previously discrete fields of osmolytes and DESs. We first showed that osmolytes prominent across all kingdoms, TMAO, sarcosine, ectoine, GPC, DMSP, arginine and guanidine HCl, may be used to prepare new DESs. Going one step further, we followed the inspiration from several natural distributions of osmolytes in several species and environments to reveal new multicomponent DESs of unprecedented complexity and briefly showcased their tunable ability to stabilize a template protein.
These new biorelevant DESs are context-aware and context-specific snapshots of a micronarrative which reflects the nuances of the cellular microenvironment through space, time and function. This feature can provide additional and potentially orthogonal angles when looking at biological systems, whether one is trying to understand and describe them,9,106–114 monitor or regulate them in vitro and in vivo,115–120 replicate and mimic them ex vivo or “on-a-chip”,121–125 or model them in silico.126–129
On top of that, these novel DESs are also bioinspired engineered media which may serve as new tools and open up new opportunities for action in diagnosis, monitoring, treatment and manufacture across several research fields and market sectors,9,130,131–138,139–143 starting from applications and current use cases where single osmolytes or aqueous mixtures have shown promise144 (Fig. 5). Most importantly, this new concept can serve as a constructor145 and adequate scaffolding146 can be used to devise industry-relevant substrates, probe their attributes and engineer scalable processes ased on legitimate input/output states. To explore this newly revealed space, we are aided and inspired by the Cynefin® Framework.147,148 Cynefin® is a “multi-ontology sense-making framework” which can help make sense of the systems we find ourselves in so that we can act within them.149 Perhaps the most crucial confusion elucidated by this framework is the distinction between the complex and the complicated domains. Complicated is a system where we know what information we are missing (known unknowns) and we must employ the adequate expertise to obtain it. In the complex domain of “unknown unknowns”, we do not know which questions we need to answer, and we must conduct safe-to-fail probes to discover important information. Based on these experiments, the next plausible question and the next step forward is defined and a path towards a potential solution emerges.
Being aware of the properties and interdependencies of each type of system can help us respond in meaningful ways. We have used such responses to trigger deliberate movement within or across the Cynefin domains encompassed by the osmoDES concept, creating dynamics, as we recently described elsewhere150 in detail (Fig. 6). As an example, our “shallow dive into Chaos” can trigger the emergence of a set of patterns (“osmolyte cocktails form multiDESs”) as an enabling constraint. Using this constraint as a springboard can help one explore “unimagined unknowns” about, for example, empirically observed natural substances of largely unbeknownst complexity of functions, components and distributions, such as saps. Actively exploring this space may help move beyond the postulation phase about eutectic mixtures and reveal actionable experiments to understand which questions are worth asking (“unknown unknowns”- e.g. is there an overarching preferred ratio between total methylamine kosmotropes and total perturbants that forms a DES?). Experimenting with such exploratory questions can eventually lead to exploitable practices by resolving specific problems through expertise (“known unknowns”- e.g. which osmolyte distribution and ratio best stabilise a particular recombinant protein contained in that sap in order to avoid product loss due to aggregation?). To the best of our knowledge, no prior art precedent has been reported for such a complexity-based approach, neither in the field of osmolytes nor in that of Deep Eutectic Systems. To further exploit the osmoDES concept as a constructor, we first need to acknowledge both known unknowns and unknown unknowns within the current state of the system we are in. For the first, Wardley mapping151 can be a useful tool to assess what kind of knowledge and expertise would be most relevant for each targeted use case and which current capacities require additional resources (Fig. 7).
Very often, when we dedicate resources to linked components of a Wardley map we become more aware of the space between disciplines and the amount of potential it holds. In this liminal space, we can look for alternative answers to existing questions (known unknowns), but also find additional plausible questions worth asking (unknown unknowns). A suitable approach to explore this space is the Entangled Trios method, which involves a triad of people “from radically different backgrounds with no prior interaction but with a common purpose. This entangling leads to a more extensive and diverse exploration of the issue at hand which may lead to new ideas that can be explored. The three are kept together by a shared purpose and by well-crafted sets of actions which ritualize the exchange of knowledge and lead to new possibilities”.152 Potential questions within such triads which could instigate a shared purpose would be: do single osmolytes always function as DESs with additional components we do not know yet? Do other osmolyte roles exist on top of kosmo- and chaotropic? Do other DES component roles exist on top of H-bond donor and acceptor? Are current synthetic DESs simplified versions of more nuanced natural eutectic systems? What is the role of water as a DES component? Could the “three states” postulated for hydrogels153 be relevant in DES? And how does water regulate the interaction of the deep eutectic system with biomolecules?
For additional unknown unknowns, we must first act on an attribute we can affect and have the analytical tools to sense how the system responds to the perturbation,147 to become aware of the affordances, the possibilities for further action, provided within the area of each experiment. Some of these actions could be, for example:
• to replicate more natural osmolyte cocktails and monitor their ability to form eutectic systems;
• to impose specific eutectic microenvironments on known enzymes and monitor for potential dormancy/wake-up triggers;
• to force cell protein production in specific eutectic microenvironments and monitor their effect on protein aggregation;
• to induce cell proliferation in DES-induced “meta-stress” conditions and monitor fitness landscapes and gene evolution for protein engineering;
• to subject different varieties of nucleic acid substrates to relevant DES and monitor their ability to interact and the topologies of their interaction;
• to generate new computational modules and tools based on relevant eutectic microenvironments and monitor their applicability and functionality limits.
Questions and actions will continue to arise the more we question and act, and the more diverse minds question and act the more probable it is for these initial thoughts and experiments to become a path. We believe that the inherent beauty of this path is the potential to change the way we understand information flows in nature154 by introducing the following assemblage:
Through their constant interaction with biomacromolecules, osmolyte-based microenvironments regulate a myriad of cellular properties, enable intra- and extracellular information flows155 and facilitate the “enactive” functions of the components of biological systems.156 Could osmolyte-based deep eutectic media be employed to better monitor these flows in complex natural systems and allow for the emergence of new properties and affordances we can explore and exploit? And could this new knowledge be leveraged to enable information flows within engineered systems?
Author contributions
The concept for the article was jointly conceived and developed by M.C.B. and T.A. who also designed the experiments, wrote the first draft, compiled figures and tables, and provided direction. I.R.R. and K.R. helped refine the concept. T.A. was responsible for the conceptual and contextual implementation of the Cynefin® framework and the complexity-related methods. M.C.B., M.P. and M.R. performed the experiments and prepared the DESs. The paper was reviewed and edited by all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
A sincere thank you to Marko Rogošić for his diligent proofreading of the paper. Vilko Smrečki is gratefully acknowledged for recording the NMR spectra. Thanks to Anna Panagiotou of Cynefin Co for the sense-making guidance and to Eleni Margoudi (@elenimargou) and Athanasia Lantouri (@_lant) for the artwork.References
- G. Zhang, Organogenesis, 2012, 8, 101–102 CrossRef PubMed.
- L. Czech, L. Hermann, N. Stöveken, A. A. Richter, A. Höppner, S. H. J. Smits, J. Heider and E. Bremer, Genes, 2018, 9, 1–58 CrossRef PubMed.
- H. Vanda, R. Verpoorte, P. G. L. Klinkhamer and Y. H. Choi, in Deep Eutectic Solvents: Synthesis, Properties, and Applications, 2019, pp. 61–81 Search PubMed.
- B. B. Hansen, S. Spittle, B. Chen, D. Poe, Y. Zhang, J. M. Klein, A. Horton, L. Adhikari, T. Zelovich, B. W. Doherty, B. Gurkan, E. J. Maginn, A. Ragauskas, M. Dadmun, T. A. Zawodzinski, G. A. Baker, M. E. Tuckerman, R. F. Savinell and J. R. Sangoro, Chem. Rev., 2021, 121, 1232–1285 CrossRef CAS PubMed.
- Y. Dai, J. Van Spronsen and G. Witkamp, Anal. Chim. Acta, 2013, 766, 61–68 CrossRef CAS PubMed.
- Y. Dai, E. M. Varypataki, E. A. Golovina, W. Jiskoot, G. J. Witkamp, Y. H. Choi and R. Verpoorte, Natural deep eutectic solvents in plants and plant cells: In vitro evidence for their possible functions, Elsevier Ltd, 1st edn, 2021, vol. 97 Search PubMed.
- P. H. Yancey, J. Exp. Biol., 2005, 208, 2819–2830 CrossRef CAS PubMed.
- T. Hasan, K. Kumari, S. C. Devi, J. Handa, T. Rehman, N. A. Ansari and L. R. Singh, Hum. Vaccines Immunother., 2019, 15, 514–525 CrossRef PubMed.
- N. Kushwah, V. Jain and D. Yadav, Biomolecules, 2020, 10, 132 CrossRef CAS PubMed.
- M. B. Burg and J. D. Ferraris, J. Biol. Chem., 2008, 283, 7309–7313 CrossRef CAS PubMed.
- T. O. Street, D. W. Bolen and G. D. Rose, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 13997–14002 CrossRef CAS PubMed.
- P. H. Yancey, Sci. Prog., 2004, 87, 1–24 CrossRef CAS PubMed.
- P. H. Yancey, M. E. Clark, S. C. Hand, R. D. Bowlus and G. N. Somero, Science, 1982, 217(4566), 1214–1222 CrossRef CAS PubMed.
- G. L. Devlin, H. Parfrey, D. J. Tew, D. A. Lomas and S. P. Bottomley, Am. J. Respir. Cell Mol. Biol., 2001, 24, 727–732 CrossRef CAS PubMed.
- P. H. Yancey and M. B. Burg, Am. J. Physiol.: Regul., Integr. Comp. Physiol., 1990, 258 DOI:10.1152/ajpregu.1990.258.1.r198.
- P. H. Yancey and G. N. Somero, Biochem. J., 1979, 183, 317–323 CrossRef CAS PubMed.
- L. R. Singh, T. Ali Dar, I. Haque, F. Anjum, A. A. Moosavi-Movahedi and F. Ahmad, Biochim. Biophys. Acta, Proteins Proteomics, 2007, 1774, 1555–1562 CrossRef CAS PubMed.
- S. Khan, Z. Bano, L. R. Singh, M. I. Hassan, A. Islam and F. Ahmad, PLoS One, 2013, 8, e72533 CrossRef CAS PubMed.
- M. Mukherjee and J. Mondal, J. Phys. Chem. B, 2020, 124, 11316–11323 CrossRef CAS PubMed.
- J. Rösgen and R. Jackson-Atogi, J. Am. Chem. Soc., 2012, 134, 3590–3597 CrossRef PubMed.
- J. Rösgen, J. Phys. Chem. B, 2015, 119, 150–157 CrossRef PubMed.
- L. M. F. Holthauzen, M. Auton, M. Sinev and J. Rösgen, Protein stability in the presence of cosolutes, 2011, vol. 492 Search PubMed.
- N. Kumar and N. Kishore, J. Chem. Phys., 2013, 139, 115104 CrossRef PubMed.
- J. H. Holland, J. Syst. Sci. Complexity, 2006, 19, 1–8 CrossRef.
- S. Cachel, The Growth of Biological Thought Revisited, 1986, vol. 88 Search PubMed.
- F. Guthrie, London, Edinburgh, Dublin Philos. Mag. J. Sci., 2010, 17, 462–482 CrossRef.
- A. P. Abbott, G. Capper, D. L. Davies, R. K. Rasheed and V. Tambyrajah, Chem. Commun., 2003, 70–71 RSC.
- C. Florindo, A. J. S. McIntosh, T. Welton, L. C. Branco and I. M. Marrucho, Phys. Chem. Chem. Phys., 2017, 20, 206–213 RSC.
- H. Wang, S. Liu, Y. Zhao, J. Wang and Z. Yu, ACS Sustainable Chem. Eng., 2019, 7, 7760–7767 CrossRef CAS.
- M. T. Zafarani-Moattar, H. Shekaari and F. Ghaffari, J. Mol. Liq., 2020, 311, 113347 CrossRef CAS.
- K. Töpfer, A. Pasti, A. Das, S. M. Salehi, L. I. Vazquez-Salazar, D. Rohrbach, T. Feurer, P. Hamm and M. Meuwly, J. Am. Chem. Soc., 2022, 144, 14170–14180 CrossRef PubMed.
- L. Percevault, A. Jani, T. Sohier, L. Noirez, L. Paquin, F. Gauffre and D. Morineau, J. Phys. Chem. B, 2020, 124, 9126–9135 CrossRef CAS PubMed.
- S. Spittle, D. Poe, B. Doherty, C. Kolodziej, L. Heroux, M. A. Haque, H. Squire, T. Cosby, Y. Zhang, C. Fraenza, S. Bhattacharyya, M. Tyagi, J. Peng, R. A. Elgammal, T. Zawodzinski, M. Tuckerman, S. Greenbaum, B. Gurkan, C. Burda, M. Dadmun, E. J. Maginn and J. Sangoro, Nat. Commun., 2022, 13, 219 CrossRef CAS PubMed.
- M. A. R. Martins, S. P. Pinho and J. A. P. Coutinho, J. Solution Chem., 2019, 48, 962–982 CrossRef CAS.
- Y. P. Mbous, M. Hayyan, A. Hayyan, W. F. Wong, M. A. Hashim and C. Y. Looi, Biotechnol. Adv., 2017, 35, 105–134 CrossRef CAS PubMed.
- Y. H. Choi, J. van Spronsen, Y. Dai, M. Verberne, F. Hollmann, I. W. C. E. Arends, G.-J. Witkamp and R. Verpoorte, Plant Physiol., 2011, 156, 1701–1705 CrossRef CAS PubMed.
- F. Legrand, C. Nguyen, L. Augis, S. Fourmentin, G. Barratt, F.-X. Legrand, F. Legrand, C. Nguyen, L. Augis, S. Fourmentin and G. Barratt, Deep Eutectic Solvents for Innovative Pharmaceutical Formulations, 2021 Search PubMed.
- D. Rente, M. C. Bubalo, M. Panić, A. Paiva, B. Caprin, I. R. Redovniković and A. R. C. Duarte, J. Cleaner Prod., 2022, 380, 135147 CrossRef CAS.
- Y. Liang, W. Duan, X. An, Y. Qiao, Y. Tian and H. Zhou, Bioresour. Technol., 2020, 310, 123389 CrossRef CAS PubMed.
- M. Ishibashi, K. Tsumoto, M. Tokunaga, D. Ejima, Y. Kita and T. Arakawa, Protein Expression Purif., 2005, 42, 1–6 CrossRef CAS PubMed.
- S. Sharma, S. Sarkar, S. S. Paul, S. Roy and K. Chattopadhyay, Sci. Rep., 2013, 3, 1–9 Search PubMed.
- S. Rozas, C. Benito, R. Alcalde, M. Atilhan and S. Aparicio, J. Mol. Liq., 2021, 344, 117717 CrossRef CAS.
- O. S. Hammond, D. T. Bowron, A. J. Jackson, T. Arnold, A. Sanchez-Fernandez, N. Tsapatsaris, V. G. Sakai and K. J. Edler, J. Phys. Chem. B, 2017, 121, 7473–7483 CrossRef CAS PubMed.
- N. López-Salas, J. M. Vicent-Luna, E. Posada, S. Imberti, R. M. Madero-Castro, S. Calero, C. O. Ania, R. J. Jiménez-Riobóo, M. C. Gutiérrez, M. L. Ferrer and F. del Monte, ACS Sustainable Chem. Eng., 2020, 8, 12120–12131 CrossRef.
- N. López-Salas, J. M. Vicent-Luna, S. Imberti, E. Posada, M. J. Roldán, J. A. Anta, S. R. G. Balestra, R. M. M. Castro, S. Calero, R. J. Jiménez-Riobóo, M. C. Gutiérrez, M. L. Ferrer and F. del Monte, ACS Sustainable Chem. Eng., 2019, 7, 17565–17573 CrossRef.
- M. C. Gutiérrez, M. L. Ferrer, C. R. Mateo and F. Del Monte, Langmuir, 2009, 25, 5509–5515 CrossRef PubMed.
- A. Gutiérrez, M. Atilhan and S. Aparicio, J. Mol. Liq., 2021, 339, 116758 CrossRef.
- L. Cicco, G. Dilauro, F. M. Perna, P. Vitale and V. Capriati, Org. Biomol. Chem., 2021, 19, 2558–2577 RSC.
- Choline chloride process, https://matthey.com/products-and-markets/chemicals/process-licensing/choline-chloride-process, (accessed 1 March 2023).
- J. A. Raymond, Fish Physiol. Biochem., 1994, 13, 13–22 CrossRef CAS PubMed.
- J. P. Costanzo and R. E. Lee, J. Exp. Biol., 2005, 208, 4079–4089 CrossRef PubMed.
- G. Pittaro, L. Cáceres, C. Bruno, A. Tomás, D. Bustos, M. Monteoliva, L. Ortega and E. Taleisnik, Grass Forage Sci., 2016, 71, 683–698 CrossRef CAS.
- P. C. Withers, G. Morrison and M. Guppy, Physiol. Zool., 1994, 67, 693–705 CrossRef CAS.
- P. H. Yancey and M. B. Burg, Am. J. Physiol.: Renal, Fluid Electrolyte Physiol., 1989, 257, 602–607 Search PubMed.
- P. C. Withers, G. Morrison and M. Guppy, Physiol. Zool., 1994, 67, 693–705 CrossRef CAS.
- C. X. Zeng, S. J. Qi, R. P. Xin, B. Yang and Y. H. Wang, J. Mol. Liq., 2016, 219, 74–78 CrossRef CAS.
- M. Benlebna, M. Ruesgas-Ramón, B. Bonafos, G. Fouret, F. Casas, C. Coudray, E. Durand, M. C. Figueroa-Espinoza and C. Feillet-Coudray, J. Agric. Food Chem., 2018, 66, 6205–6212 CrossRef CAS PubMed.
- A. M. Syakfanaya, F. C. Saputri and A. Mun'im, Pharmacogn. J., 2019, 11, 267–271 CrossRef CAS.
- A. R. Jesus, L. Meneses, A. R. C. Duarte and A. Paiva, Cryobiology, 2021, 101, 95–104 CrossRef CAS PubMed.
- M. Panić, M. Andlar, M. Tišma, T. Rezić, D. Šibalić, M. C. Bubalo and I. R. Redovniković, Waste Manage., 2021, 120, 340–350 CrossRef PubMed.
- M. Zdanowicz, Int. J. Biol. Macromol., 2021, 176, 387–393 CrossRef CAS PubMed.
- S. P. Simeonov and C. A. M. Afonso, RSC Adv., 2016, 6, 5485–5490 RSC.
- L. Zhong, C. Wang, G. Yang, J. Chen, F. Xu, C. G. Yoo and G. Lyu, Bioresour. Technol., 2022, 343, 126022 CrossRef CAS PubMed.
- S. N. Pedro, M. S. M. Mendes, B. M. Neves, I. F. Almeida, P. Costa, I. Correia-Sá, C. Vilela, M. G. Freire, A. J. D. Silvestre and C. S. R. Freire, Pharmaceutics, 2022, 14, 827 CrossRef CAS PubMed.
- M. A. Karadendrou, I. Kostopoulou, V. Kakokefalou, A. Tzani and A. Detsi, Catalysts, 2022, 12, 1–17 CrossRef.
- S. Sut, M. Faggian, V. Baldan, G. Poloniato, I. Castagliuolo, I. Grabnar, B. Perissutti, P. Brun, F. Maggi, D. Voinovich, G. Peron and S. Dall'Acqua, Molecules, 2017, 22, 1–11 Search PubMed.
- M. Faggian, S. Sut, B. Perissutti, V. Baldan, I. Grabnar and S. Dall'Acqua, Molecules, 2016, 21, 1–11 CrossRef PubMed.
- F. Ilgen and B. König, Green Chem., 2009, 11, 848–885 RSC.
- T. Wang, J. Jiao, Q.-Y. Gai, P. Wang, N. Guo, L.-L. Niu and Y.-J. Fu, J. Pharm. Biomed. Anal., 2017, 145, 339–345 CrossRef CAS PubMed.
- R. H. Kelly and P. H. Yancey, Biol. Bull., 1999, 196, 18–25 CrossRef CAS PubMed.
- W. E. S. Carr, J. C. Netherton, R. A. Gleeson and C. D. Derby, The Biological bulletin, 1996, 190, 149–160, DOI:10.2307/1542535.
- A. K. Parida and A. B. Das, Ecotoxicol. Environ. Saf., 2005, 60, 324–349 CrossRef CAS PubMed.
- P. H. Yancey, W. R. Blake and J. Conley, Comp. Biochem. Physiol., Part A: Mol. Integr. Physiol., 2002, 133, 667–676 CrossRef PubMed.
- J. T. Gorke, F. Srienc and R. J. Kazlauskas, Chem. Commun., 2008, 1235–1237 RSC.
- H. Monhemi, M. R. Housaindokht, A. A. Moosavi-Movahedi and M. R. Bozorgmehr, Phys. Chem. Chem. Phys., 2014, 16, 14882–14893 RSC.
- S. Sarkar, S. Ghosh and R. Chakrabarti, RSC Adv., 2017, 7, 52888–52906 RSC.
- R. P. Sear, I. Pagonabarraga and A. Flaus, BMC Biophys., 2015, 8, 4–9 CrossRef PubMed.
- R. P. Sear, Faraday Discuss., 2008, 139, 21–34 RSC.
- N. H. O'Donnell, B. L. Møller, A. D. Neale, J. D. Hamill, C. K. Blomstedt and R. M. Gleadow, Plant Physiol. Biochem., 2013, 73, 83–92 CrossRef PubMed.
- E. Durand, P. Villeneuve, C. Bourlieu-lacanal and F. Carrière, Adv. Bot. Res., 2021, 97, 133–158 Search PubMed.
- J. P. Bittner, N. Zhang, L. Huang, P. Domínguez De María, S. Jakobtorweihen and S. Kara, Green Chem., 2022, 24, 1120–1131 RSC.
- M. Panić, M. Radović, I. Maros, A. J. Tušek, M. C. Bubalo and I. R. Redovniković, Process Biochem., 2021, 102, 1–9 CrossRef.
- A. E. Delorme, J. M. Andanson and V. Verney, Int. J. Biol. Macromol., 2020, 163, 919–926 CrossRef CAS PubMed.
- S. L. Cao, X. Deng, P. Xu, Z. X. Huang, J. Zhou, X. H. Li, M. H. Zong and W. Y. Lou, J. Agric. Food Chem., 2017, 65, 2084–2088 CrossRef CAS PubMed.
- K. P. Lin, G. J. Feng, F. L. Pu, X. D. Hou and S. L. Cao, Front. Bioeng. Biotechnol., 2020, 8, 1–8 CrossRef PubMed.
- S. Núñez-Pertíñez and T. R. Wilks, Front. Chem., 2020, 8, 1–11 CrossRef PubMed.
- R. Jagannathan, P. M. Abraham and P. Poddar, J. Phys. Chem. B, 2012, 116(50), 14533–14540, DOI:10.1021/jp3050516.
- M. Hayyan, M. A. Hashim, A. Hayyan, M. A. Al-Saadi, I. M. AlNashef, M. E. S. Mirghani and O. K. Saheed, Chemosphere, 2013, 90, 2193–2195 CrossRef CAS PubMed.
- M. Hayyan, C. Y. Looi, A. Hayyan, W. F. Wong and M. A. Hashim, PLoS One, 2015, 10, 1–18 CrossRef CAS PubMed.
- K. Radošević, M. Cvjetko Bubalo, V. Gaurina Srček, D. Grgas, T. Landeka Dragičević and I. Radojčić Redovniković, Ecotoxicol. Environ. Saf., 2015, 112, 46–53 CrossRef PubMed.
- S. J. Bryant, M. N. Awad, A. Elbourne, A. J. Christofferson, A. V. Martin, N. Meftahi, C. J. Drummond, T. L. Greaves and G. Bryant, J. Mater. Chem. B, 2022, 10, 4546–4560 RSC.
- P. De Maayer, D. Anderson, C. Cary and D. A. Cowan, EMBO Rep., 2014, 15, 508–517 CrossRef CAS PubMed.
- Y. Song, W. Huang, Y. Zhou, Z. Li, R. Ji and X. Ye, Arch. Insect Biochem. Physiol., 2021, 108, e21846 CrossRef CAS PubMed.
- K. Tanaka, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1995, 110, 539–545 CrossRef.
- K. B. Storey, J. M. Storey, S. P. J. Brooks, T. A. Churchill and R. J. Brooks, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 8350–8354 CrossRef CAS PubMed.
- T. A. Churchill and K. B. Storey, Can. J. Zool., 1992, 70, 99–105 CrossRef.
- A. Jani, T. Sohier and D. Morineau, J. Mol. Liq., 2020, 304, 112701 CrossRef CAS.
- L. Sherwood, H. Klandorf and P. H. Yancey, Animal Physiology: From Genes to Organisms, 2013.
- J. H. Crowe, Subcell. Biochem., 2015, 71, 263–280 CAS.
- M. Manzanera, Environ. Microbiol., 2021, 23, 3351–3359 CrossRef PubMed.
- A. Sanchez-Fernandez, S. Prevost and M. Wahlgren, Green Chem., 2022, 24(11), 4437–4442 RSC.
- R. Esquembre, J. M. Sanz, J. G. Wall, F. Del Monte, C. R. Mateo and M. L. Ferrer, Phys. Chem. Chem. Phys., 2013, 15, 11248–11256 RSC.
- T. Pievani and E. Serrelli, J. Anthropol. Sci., 2011, 89, 9–23 Search PubMed.
- M. Frenkel-Pinter, A. S. Petrov, K. Matange, M. Travisano, J. B. Glass and L. D. Williams, J. Mol. Evol., 2022, 90, 166–175 CrossRef CAS PubMed.
- E. Manning and B. Massumi, Thought in the Act: Passages in the Ecology of Experience, University of Minnesota Press, 2014 Search PubMed.
- W. Y. Go, X. Liu, M. A. Roti, F. Liu and S. N. Ho, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 10673–10678 CrossRef CAS PubMed.
- A. Thiemicke and G. Neuert, Sci. Adv., 2021, 7, 1–13 Search PubMed.
- T. Kumar, M. Yadav and L. R. Singh, Curr. Pharm. Des., 2016, 22, 3050–3057 CrossRef CAS PubMed.
- Y. Chen, S. Yang, J. Tavormina, D. Tampe, M. Zeisberg, H. Wang, K. K. Mahadevan, C.-J. Wu, H. Sugimoto, C.-C. Chang, R. R. Jenq, K. M. McAndrews and R. Kalluri, Cancer Cell, 2022, 40, 818–834 CrossRef CAS PubMed.
- M. Warepam, K. Ahmad, S. Rahman, H. Rahaman, K. Kumari and L. R. Singh, Biomolecules, 2020, 10, 286 CrossRef CAS PubMed.
- B. De Paepe, C. Merckx, J. Jarošová, M. Cannizzaro and J. L. De Bleecker, Biomolecules, 2020, 10, 521 CrossRef CAS PubMed.
- M. A. Bhat, K. Ahmad, M. S. A. Khan, M. A. Bhat, A. Almatroudi, S. Rahman and A. T. Jan, Biomolecules, 2020, 10, 863 CrossRef CAS PubMed.
- M. Gao and R. Winter, J. Diabetes Res., 2015 DOI:10.1155/2015/849017.
- D. C. Mikles, V. Bhat, B. J. Schuchardt, C. B. McDonald and A. Farooq, Biopolymers, 2015, 103, 74 CrossRef CAS PubMed.
- M. Mcdermott, C. Kemper, W. Barone, G. Jost and J. Endrikat, Br. J. Radiol., 2020, 93 Search PubMed.
- G. N. Gomes and Z. A. Levine, J. Phys. Chem. B, 2021, 125, 1974–1996 CrossRef CAS PubMed.
- H. Marshall, M. Venkat, N. S. Hti Lar Seng, J. Cahn and D. H. Juers, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2012, 68, 69–81 CrossRef CAS PubMed.
- D. Le Bihan, Phys. Med. Biol., 2007, 57–90 CrossRef PubMed.
- H. M. Taïeb, L. Bertinetti, T. Robinson and A. Cipitria, PLoS One, 2022, 17, e0268297 CrossRef PubMed.
- S. Aravindan, S. Chen, H. Choudhry, C. Molfetta, K. Y. Chen and A. Y. C. Liu, Sci. Rep., 2020, 10, 1–12 CrossRef PubMed.
- R. Morizane, A. Q. Lam, B. S. Freedman, S. Kishi, M. T. Valerius and J. V. Bonventre, Nat. Biotechnol., 2015, 33, 1193–1200 CrossRef CAS PubMed.
- M. Takasato, P. X. Er, H. S. Chiu, B. Maier, G. J. Baillie, C. Ferguson, R. G. Parton, E. J. Wolvetang, M. S. Roost, S. M. C. De Sousa Lopes and M. H. Little, Nature, 2015, 526, 564–568 CrossRef CAS PubMed.
- A. Ganguli, A. Mostafa, C. Saavedra, Y. Kim, P. Le, V. Faramarzi, R. W. Feathers, J. Berger, K. P. Ramos-Cruz, O. Adeniba, G. J. P. Diaz, J. Drnevich, C. L. Wright, A. G. Hernandez, W. Lin, A. M. Smith, F. Kosari, G. Vasmatzis, P. Z. Anastasiadis and R. Bashir, Sci. Adv., 2021, 7 DOI:10.1126/SCIADV.ABC1323.
- P. Mai, J. Hampl, M. Baca, D. Brauer, S. Singh, F. Weise, J. Borowiec, A. Schmidt, J. M. Küstner, M. Klett, M. Gebinoga, I. S. Schroeder, U. R. Markert, F. Glahn, B. Schumann, D. Eckstein and A. Schober, Bioengineering, 2022, 9, 220 CrossRef CAS PubMed.
- B. Dorgau, M. Georgiou, A. Chaudhary, M. Moya-Molina, J. Collin, R. Queen, G. Hilgen, T. Davey, P. Hewitt, M. Schmitt, S. Kustermann, F. Pognan, D. H. Steel, E. Sernagor, L. Armstrong and M. Lako, Stem Cells Transl. Med., 2022, 11, 159–177 CrossRef PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Nature, 2021, 596, 583–589 CrossRef CAS PubMed.
- E. T. Powers and L. M. Gierasch, J. Mol. Biol., 2021, 433(20) DOI:10.1016/J.JMB.2021.167197.
- G. D. Rose, Biochemistry, 2021, 60, 3753–3761 CrossRef CAS PubMed.
- J. P. Bittner, L. Huang, N. Zhang, S. Kara and S. Jakobtorweihen, J. Chem. Theory Comput., 2021, 17, 5322–5341 CrossRef CAS PubMed.
- N. B. Pham and W. S. Meng, Int. J. Pharm., 2020, 585, 119523 CrossRef CAS PubMed.
- J. R. Christopher, Curr. Opin. Biotechnol., 2014, 30, 211–217 CrossRef PubMed.
- A. Singh, V. Upadhyay, A. K. Upadhyay, S. M. Singh and A. K. Panda, Microb. Cell Fact., 2015, 14, 1–10 CrossRef CAS PubMed.
- D. Andrijevic, Z. Vrselja, T. Lysyy, S. Zhang, M. Skarica, A. Spajic, D. Dellal, S. L. Thorn, R. B. Duckrow, S. Ma, P. Q. Duy, A. U. Isiktas, D. Liang, M. Li, S. K. Kim, S. G. Daniele, K. Banu, S. Perincheri, M. C. Menon, A. Huttner, K. N. Sheth, K. T. Gobeske, G. T. Tietjen, H. P. Zaveri, S. R. Latham, A. J. Sinusas and N. Sestan, Nature, 2022, 608, 405–412 CrossRef CAS PubMed.
- K. Athirathinam, S. Nandakumar and R. Kandasamy, Macromol. Res., 2022, 30, 599–608 CrossRef CAS PubMed.
- C. H. Pi, G. Yu, A. Petersen and A. Hubel, Sci. Rep., 2018, 8, 1–13 CAS.
- A. M. R. Kabir, T. Munmun, T. Hayashi, S. Yasuda, A. P. Kimura, M. Kinoshita, T. Murata, K. Sada and A. Kakugo, ACS Omega, 2022, 7, 3796–3803 CrossRef CAS PubMed.
- A. Argiolas, G. L. Puleo, E. Sinibaldi and B. Mazzolai, Sci. Rep., 2016, 6, 1–8 CrossRef PubMed.
- M. Zheng, Z. Wang, H. Chang, L. Wang, S. W. T. Chew, D. C. S. Lio, M. Cui, L. Liu, B. C. K. Tee and C. Xu, Adv. Healthcare Mater., 2020, 9, 1901683 CrossRef CAS.
- X. Wang and I. Rivière, Mol. Ther. – Oncolytics, 2016, 3, 16015 CrossRef CAS PubMed.
- D. B. Kell, G. J. Laubscher and E. Pretorius, Biochem. J., 2022, 479, 537–559 CrossRef CAS PubMed.
- M. Vendruscolo and M. Fuxreiter, Nat. Commun., 2022, 13, 1–11 Search PubMed.
- M. C. Gutirrez, M. L. Ferrer, L. Yuste, F. Rojo and F. del Monte, Angew. Chem. Int. Ed, 2010, 49, 2158–2162 CrossRef PubMed.
- S. Nardecchia, M. C. Gutiérrez, M. L. Ferrer, M. Alonso, I. M. López, J. C. Rodríguez-Cabello and F. del Monte, Biomacromolecules, 2012, 13, 2029–2036 CrossRef CAS PubMed.
- S. R. Wlodarczyk, D. Custódio, A. Pessoa and G. Monteiro, Eur. J. Pharm. Biopharm., 2018, 131, 92–98 CrossRef CAS PubMed.
- D. Deutsch, Synthese, 2013, 190, 4331–4359 CrossRef.
- J. Turner, D. Snowden and N. Thurlow, Systems, 2022, 10(7) DOI:10.3390/systems10010007.
- D. Snowden, J. Knowl. Manage., 2002, 6, 100–111 CrossRef.
- D. Snowden, Z. Goh and S. Borchardt, Cynefin – Weaving Sense-Making into the Fabric of Our World, Wilmington, DE, USA, 2021 Search PubMed.
- E. Commission, J. R. Centre, A. Rancati and D. Snowden, Managing complexity (and chaos) in times of crisis : a field guide for decision makers inspired by the Cynefin framework, Publications Office, 2021 Search PubMed.
- T. Andreou, in Natural Resources, Green Technology And Sustainable Development, 2022, pp. 111–116 Search PubMed.
- S. Wardley, “A Wardley Map”, https://blog.gardeviance.org/2014/02/a-wardley-map.html, 2017.
- Entangled trios – Cynefin.io, https://cynefin.io/wiki/Entangled_trios, (accessed 9 September 2022).
- B. L. Dargaville and D. W. Hutmacher, Nat. Commun, 2022, 13, 1–10 Search PubMed.
- W. S. Imari and P. C. W. Davies, J. R. Soc., Interface, 2013, 10, 1–9 Search PubMed.
- H. M. Taïeb, D. S. Garske, J. Contzen, M. Gossen, L. Bertinetti, T. Robinson and A. Cipitria, Sci. Rep., 2021, 11, 13455 CrossRef PubMed.
- T. Froese and E. A. Di Paolo, Pragmat. Cognit., 2011, 19, 1–36 CrossRef.
- J. R. Treberg, B. Speers-Roesch, P. M. Piermarini, Y. K. Ip, J. S. Ballantyne and W. R. Driedzic, J. Exp. Biol., 2006, 209, 860–870 CrossRef CAS PubMed.
- D. Shugar, Biochim. Biophys. Acta, 1952, 8, 302–309 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |