Open Access Article
Open Access ArticleA comprehensive review on CRISPR and artificial intelligence based emerging food packaging technology to ensure “safe food”
Anamika
Nayak
 a and
Debjani
Dutta
a and
Debjani
Dutta
 *b
*b
aDepartment of Biotechnology, National Institute of Technology Durgapur, Mahatma Gandhi Avenue, Durgapur 713209, West Bengal, India. E-mail: nayak.megha14@gmail.com; Tel: +91 9679493812
bDepartment of Biotechnology, National Institute of Technology Durgapur, Mahatma Gandhi Avenue, Durgapur 713209, West Bengal, India. E-mail: debjani.dutta@bt.nitdgp.ac.in; Tel: +91 9434788067
First published on 30th June 2023
Abstract
In the food industry, food quality and safety are vital, and in this case, appropriate packaging technology can significantly ensure the quality of food for consumers. Therefore, innovation in food packaging technology is believed to cater to the rising global demand for food quality and safety. The application of CRISPR technology for the ultra-sensitive detection of pathogens and food contaminants in the headspace of packaged food can be an emerging strategy to protect the health of consumers. Consequently, CRISPR-based intelligent packaging technology is likely to pave the way for the food industry to realize the real-time surveillance of food contaminants. Moreover, exploring the technological aspect of how deep learning empowers food quality inspection may facilitate global food safety and lead to a healthy secure environment. Therefore, in this review, we illustrate technological advancement with regards to traceability, computing automation, and big-data analysis, which can be an alternative approach to circumvent the limitations of existing packaging analytics. Additionally, the combination of CRISPR technology with interconnected disciplines may prompt new interest in the next-generation packaging sector.
Sustainability spotlightIn the food industry, food quality and safety are of prime concern and proper packaging technology can significantly ensure the quality for final consumers. Therefore, innovation in the food packaging technology is believed to cater to the rising global demand for food quality and safety. The application of CRISPR technology for the ultra-sensitive diagnosis of pathogens and food contaminants present in the headspace of packaged food can be an emerging strategy to protect consumers' health. The CRISPR-based intelligent packaging technology is thus likely to pave the route for food industries as the real-time surveillance system of food contaminants. Moreover, exploring a brief technological aspect of how deep learning empowered food quality inspection may provide global food safety and lead to a healthy secured environment. This review illustrates the technological advancement with traceability, computing automation, and big-data analysis that can be an alternative approach to circumvent the paucity of existing packaging analytics. Additionally, the infusion of CRISPR technology with interconnected disciplines may ignite a novel ray of hope in the next-generation packaging sector. |
1. Introduction
Food safety is one of the most significant challenges faced by both developed and developing countries worldwide.1–3 Food safety has been acknowledged by the World Health Organization (WHO) and its member nations as a crucial aspect of public health4 due to the increased mortality, morbidity, and economic burden caused by food contamination. Furthermore, public health and societal wellbeing are significantly dependent on food safety. Foodborne infections range from mild gastroenteritis to life-threatening neurologic, hepatic, and renal syndromes, which can be caused either by the toxin produced by the “disease-causing” bacterium or the body's response to the microbe itself.5 Although the majority of foodborne illnesses are mild and self-limiting, severe cases might occur in high-risk individuals such as immunocompromised persons, young children, infants, and the elderly, resulting in high mortality and morbidity. According to the WHO, an estimated 600 million individuals or about 1 in 10 people each year become ill due to consuming contaminated food, which causes 420![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths and the loss of 33 million healthy life years (DALYs). The globalization of the food industry, climate change, and shifting patterns of human consumption, such as current preferences for fresh, minimally processed food, have created new obstacles in the fight against foodborne infections6 (FAO, 2017). Foodborne illnesses negatively impact public health and affect the local economy.
000 deaths and the loss of 33 million healthy life years (DALYs). The globalization of the food industry, climate change, and shifting patterns of human consumption, such as current preferences for fresh, minimally processed food, have created new obstacles in the fight against foodborne infections6 (FAO, 2017). Foodborne illnesses negatively impact public health and affect the local economy.
Food safety management systems are comprised of production, processing, packaging, storage, and transportation. The most commonly applied internationally approved strategies to cope with food safety issues are HACCP (Hazard Analysis Critical Control Point), ISO 22000 (International Organization for Standardization), and PAS 220 (Publicly Available Specification). The five starting steps and seven principles of HACCP have been implemented to protect and control the potential risk that threatens food safety. The basic principle of ISO 22000 is to control the entire food chain, including infrastructure, equipment, and staff, to protect consumers from food-borne diseases. Good agricultural practices are another strategy to cope with food safety. The FAO (Food and Agriculture Organization) defines it as “practices that address environmental, economic, and social sustainability for on-farm processes and result in safe and quality food and non-food agricultural products”. Food packaging also plays a significant role in food safety. According to a literature survey, the COVID-19 pandemic extended the packaging market due to increased online purchases and a shift in eating habits.7 This pandemic scenario raised the public awareness of the relevant role of packaging for food protection and preservation, which has also intensified the potential hygienic-sanitary benefits of these packages.8 Increased takeaway food deliveries are closely associated with food packaging requirements. Therefore, food packaging should be functional, i.e., effectively protecting food against infection or transmittance of all pathogens, while simultaneously maintaining resistance against the loss of nutritional and aesthetic values. During the COVID-19 pandemic, the size of the global packaging market increased from USD 909.2 billion in 2019 to USD 1012.6 billion in 2021 at a Compound Annual Growth Rate (CAGR) of 5.5%, with the best-case scenario reflecting a 9.2% growth and the worst-case scenario 2.2%.9 Packaging saves food by providing mechanical protection and good barrier characteristics, maintaining the quality, reducing food losses, prolonging the shelf life of food, and consequently securing the entire food system. To reduce food loss or waste, it is essential to focus on the packaging design. The development of advanced packaging technology should focus on two main domains, i.e., packaging material and food safety.
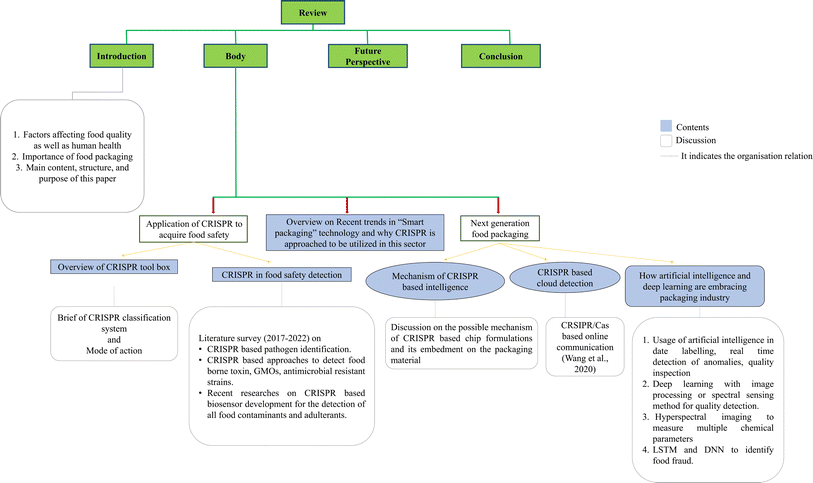
The application of next-generation information technologies, e.g., RFID (radio frequency identification) systems, global positioning systems (GPS), wireless sensor networks, and the Internet of Things (IoT), in the food chain ensures food safety according to adequate standards. The use of smart tags and embedded chips on food packaging materials and their connectivity to cloud computing infrastructures are compliant with food safety and traceability standards. However, the development of advanced food packaging systems that target specific biological markers or microbial contaminants in food still remains a key issue. The choice of a target marker is based on prior knowledge of the relevant microbiological agents, their occurrence under various conditions in various types of food products, and the release of chemicals created during the spoilage process. The “best before” or “use by” dates have become the standard in the food sector, but they do not provide information on the condition of the food inside the package, and thus “dynamic shelf-life systems” should be implemented for easy interpretation. In this regard, the environmental performance of packaging materials is a problem. Hence, research on mapping the packaging performance in the environment is necessary to provide engineers with direction on packaging design. Thus, research on CRISPR-based food safety detection is booming and surging as a new wave, representing new trends in this area.10 Besides a few recently published research works in this regard, it is encouraging that the NMPA (National Medical Products Administration) of China and the US FDA (Food and Drug Administration) have both approved CRISPR/Cas-based detection assays for the detection of SARS-CoV-2, which is considered a biohazard in contaminated cold-chain foods and their packaging.11 In this aspect, Fig. 1 shows the organisation of this review. Briefly, in this review, we focus on the utilization of CRISPR biology to target pathogen identification based on research conducted from 2017 to 2022. We aim to outline the current scientific research and emerging technological advancements that present new prospects for developing next-generation intelligent packaging systems. Considering this trending requisition, pioneering CRISPR technology is an immature research field together with the entire supply chain from farm to fork. Subsequently, special focus is given to the emergence of the IoT, implementing a modular approach to integrate deep learning, making great strides in remote serving approaches in the packaging industry.
In recent decades, IoT integrated with artificial intelligence (AI) technology has gained momentum in the food packaging sector.12,13 In this review, we present the current approaches, challenges and future trends in food packaging by highlighting the use of CRISPR and the integration of IoT and AI technologies.
2. Food packaging technology
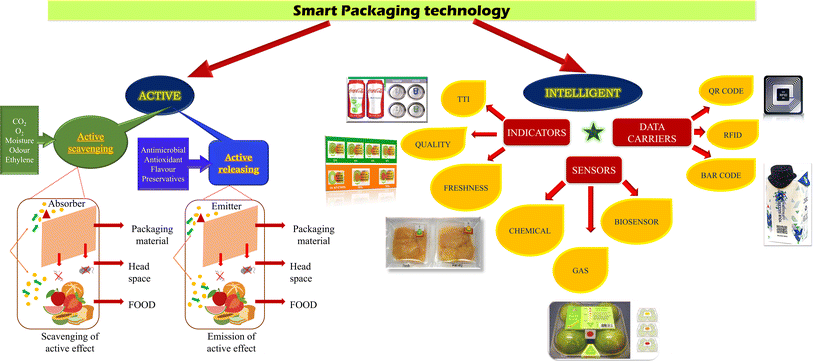
Containment, protection, communication, and convenience are the four key determinants that sustain the extrinsic and intrinsic characteristics of packaging.14 Packaging plays a vital role in protecting the content from damage associated with shipment and protects it from oxygen, dust, water vapor, and various other contaminants.15 Besides retaining the shelf life of food, packaging also provides essential information on the food quality, composition, nutritional value, date of expiration, etc. Moreover, packaging is considered an identification and communication tool, representing the usage direction, storage condition, quantity of product and quality status to help indirectly reduce waste.16 This key pillar of logistics ensures wholesomeness, aiming at the techno-commercial function of maximizing sales with optimized delivery costs. Further, it circumvents the scope of contamination at any point of the food supply chain with the assurance of zero spillage until end users open the seal. Thus, the selection of appropriate food packaging materials is an integral part of the product design and food process. In this case, metals, glass, paper, wood, and polymers make up most food packaging materials. Some packing materials are made up of two or more materials from the above-mentioned classes. These composite materials include enamelled (lacquered) metal and laminates created by binding layers of paper, polymer, and aluminium foil.Considering future consumer demands and market trends, it is necessary to alter distribution practices that traditional packaging cannot sufficiently satisfy. In recent years, significant development has been observed in the field of emerging technologies, such as smart packaging, with emphasis on educating consumers about food quality. Traditional packaging technology is limited to providing passive, inert, and inactive barriers, which keep contaminants, oxygen, and moisture out of food products, while maintaining their acceptable quality and protecting them from mechanical and chemical damage. In contrast, smart packaging incorporates novel functionality in packaging materials to detect and report food quality in real-time, thus enhancing food safety [Fig. 2]. The two main considered aspects of smart packaging are active packaging and intelligent packaging. Active packaging is redefined as “packaging in which subsidiary constituents have been deliberately included in or on either the packaging material or the package headspace to enhance the performance of the package system”.17 According to recent research, packaging can extend the shelf life of seafood products by inhibiting Gram-negative aerobic microorganisms.18 In the past two decades, vacuum packaging (VP) and modified atmosphere packaging (MAP) have gained increasing popularity in preserving seafood products.19 A study showed that MAP with a gas composition of 50% N2 and 50% CO2 was more effective in extending the shelf-life of mussels (Mytilus galloprovincialis) by 11 days compared to MAP with 100% CO2.20 In this context, Masniyom et al. (2011) concluded that the presence of oxygen in modified atmosphere packaging is required to inhibit the growth of anaerobic toxigenic bacteria.21 Studies revealed that packaging with a high-oxygen-modified atmosphere negatively impacted the quality of goose meat due to the decrease in polyunsaturated fatty acid (PUFA)/saturated fatty acids (SFA) and PUFA, and the increase in SFA. Vacuum packaging preserved goose meat for 11 days without changing its fatty acid profile due to the limited oxygen exposure.22 In the study by Marcinkowska-Lesiak et al. (2015), an increase in pH was observed in meat packed in both vacuum packages and modified atmospheres packages with extensive storage time.23 H. Ashenafi studied the significant effect of packaging on fruits and vegetables with respect to the decay percentage, color score, physiological weight loss, marketability, and overall acceptability.24 However, current research suggests that a longer shelf life with better quality is evident for functional packaging systems.25 With respect to cereal packaging, it was observed that the microbial shelf life was extended in the packaged condition where the oxygen concentration was less than 0.1%.26 A multilayer coextruded bag associated with an oxygen scavenger was examined for its impact on the shelf life of preservative-free tortillas under various storage conditions (such as accelerated storage, room temperature, and refrigerator storage).27 The outcomes of this study showed that the packages with the oxygen scavenger system had a protective effect. In contrast to the control, yeast and mold development was inhibited in the packages containing the oxygen scavenger (room temperature and accelerated storage). Furthermore, the use of active ingredients such as oregano essential oil prevented yeast and mold growth in sliced bread.28 Additionally, methylcellulose edible films incorporated with clove and oregano essential oils enhanced the shelf-life of sliced bread to 15 days at 25 °C ± 2 °C.29 Alternatively, intelligent packaging is defined as “a mode of packaging capable of carrying out intelligent functions, e.g., detecting, sensing, recording, tracking, communicating, and applying scientific logic to facilitate decision making to extend the shelf life, enhance the safety, improve the quality, provide information, and warn about possible problems”.30 In this context, in the following section, we summarize the recent development in the field of intelligent packaging. John Hopkins University developed an inexpensive and tiny molecularly imprinted polymer-based sensor with an affinity for binding to amines and changing their color, thereby helping users to understand food spoilage.31 Similarly, MIT scientists fabricated modified tags called chemically actuated resonant devices (cards), which alter their resistance with a change in the surrounding chemical environment.31 Other devices named MQ4 gas sensor and DHT 11 sensor, which were developed by an IoT project to sense methane gas and temperature fluctuation, respectively, depend on a light-dependent resistor.32 In contrast, Tufts University developed an edible sensor using silk and gold to sense the freshness status of food, bypassing the guesswork of “best before” dates.33 In 2022, Duan et al. developed a cellulose sensor to detect fish spoilage via the synthesis of the donor–π–acceptor (D–π–A) compound DPABA, which responds to amine vapour generated by fish.34
3. CRISPR approaches to ensure “safe food”
3.1. CRISPR toolbox
Over the past decade, the pioneering CRISPR-Cas has gained widespread popularity in the fields of biotechnology, agriculture, and medicine for the genesis of the next generation, resulting in a vast scale of genome editing across the globe. CRISPR spacer arrays repeat with Cas protein, encoded DNA, and RNA-mediated heritable phage resistance as integral parts of both vaccination and immunity in bacteria. According to the latest classification system, six diverse sets (Type I–VI) of CRISPR-Cas systems differ by their signature genes, precursor CRISPR RNA (pre-crRNA) processing, and DNA/RNA degradation mechanism.35,36 The mode of action of these systems is harmonized/directed through adaptation, expression, and interference. Adaptation results in the implantation of an additional spacer in the repeat spacer array, expression is the biogenesis of crRNAs, and finally interference occurs via cleavage of the offending target sequence. The adaptation step depends on the universal Cas1–Cas2 complex and integrates the acquired novel sequence into its repeat spacer array via copy-paste machinery.35,37,38 Type I, II, and V systems depend on the adjacent protospacer motif (PAM) for selectively flanking the protospacer and distinguishing between the host and foreign DNA. Once vaccinated from mobile genetic elements (MGEs), a repeat spacer array transcribes and processes into small mature crRNAs, followed by assembling into a crRNA-effector complex during the expression stage.39 Thereafter, it guides endonucleolytic Cas protein for sequence-specific recognition and degradation of complementary sequences.40 The detailed mechanism of critical CRISPR cell immunity varies with its two main classes, six main types, and 34 subtypes, and the outcome relies on idiosyncratic nucleases.41,423.2. Recent advances in CRISPR-based food safety detection
Real-time monitoring of food quality across the food supply chain is a potential method for controlling foodborne diseases. In general, the present methods for food safety detection are still unsatisfactory in certain aspects, including time consumption, inability to display desired specificity and sensitivity standards, and the comparative limitations of point-of-care and multiplexed testing.43 In this context, the major food-spoiling organisms and conventional detection methods used to date are listed in Table 1.| Food spoiling organism | Possible disease caused | Commonly used detection system | Reference | |
|---|---|---|---|---|
| Six common foodborne pathogens | ||||
| 1 | Escherichia coli | Bloody diarrhea, thrombotic thrombocytopenic purpura (TTP), hemolytic uremic syndrome (HUS), and hemorrhagic colitis | Culture-based method, blocking ELISA (enzyme-linked immunoassay), ruggedized pathogen identification modified real-time PCR (polymerase chain reaction), and FRET (fluorescence resonance energy transfer) | 44 |
| 2 | Salmonella spp. | Diarrhea, fever, vomiting, and abdominal pain | Fluorescence biosensor, DNA microarray technique, PCR-based method, and sandwich ELISA, FRET | 45 and 46 |
| 3 | Shigella spp. | Gastrointestinal infections, watery diarrhea, fever, fatigue, abdominal cramps, and malaise | Lateral flow immunoassay, MALDI-TOF, and fluorescence biosensor | 47 and 48 |
| 4 | Staphylococcus aureus | Vomiting, abdominal cramps, nausea, chills, perspiration, headache, dizziness, general weakness, and muscular cramping | LAMP (loop-mediated isothermal amplification), PCR-based method, DNA microarray technique | 49 and 50 |
| 5 | Listeria monocytogenes | Meningitis, gastroenteritis, and septicemia | LAMP, sandwich ELISA, and DNA microarray | 51 |
| 6 | Vibrio parahaemolyticus | Stomachache, diarrhea, and vomiting | LAMP, fluorescence, and PCR-based method | 52 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Others | ||||
| 7 | Clostridium spp. | Abdominal cramping and diarrhea | PCR-based method and DNA microarray technique | 53 and 54 |
| 8 | Campylobacter spp. | Diarrhea, campylobacteriosis | Glod-labeled immunosorbent assay, NASBA, and colorimetric biosensor | 55 |
| 9 | Streptococcus spp. | Epidemic pharyngitis, pneumonia, and skin infection | Amperometric biosensor | 56 |
| 10 | Leuconostoc spp. | Leukocytosis, fever, and gastroenteritis | PCR-based method, LAMP, and DNA microarray technique | 57 |
| 11 | Bacillus spp. | Diarrheal and emetic-type foodborne illness | PCR, LAMP, biosensor, etc. | 58 |
| 12 | Brucella spp. | Brucellosis | Raman spectroscopy | 59 |
| 13 | Yersinia spp. | Yersiniosis | Culture-based method, ELISA | 60 |
| 14 | Mycobacterium tuberculosis | Tuberculosis | Voltametric biosensor, and real-time PCR | 61 |
| 15 | Cronobacter sakazakii | Sepsis, necrotizing enterocolitis, and meningitis | DNA micro array, voltametric biosensor, and nucleic acid lateral flow immunoassay (NALFIA) | 62 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Foodborne Viruses | ||||
| 16 | Hepatitis | Viral hepatitis | PCR-based methods | 63 |
| 17 | Norovirus | Viral gastroenteritis | TaqMan-based RT-qPCR assays | 64 |
| 18 | Avian influenza viruses | Mild upper respiratory tract infection (fever and cough), early sputum production and rapid progression to severe pneumonia, sepsis with shock, and acute respiratory distress syndrome | PCR based method, ELISA, and cell-mimetic biosensors | 65 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Foodborne Parasites | ||||
| 1 | Giardia spp. | Giardiasis-diarrheal infection | Multiplex PCR | 66 |
| 2 | Toxoplasma gondii | Mononucleosis-type symptoms, buttran-placental infection, causing death during pregnancy. Pneumonitis, myocarditis, meningoencephalitis, hepatitis, chorioretinitis, or combinations of these illnesses can develop in immune-compromised people. AIDS patients are frequently prone to cerebral toxoplasmosis | ELISA, stem-loop-primer-assisted isothermal amplification | 67 and 68 |
| 3 | Cryptosporidium parvum | Diarrhea, fever, abdominal pain, and anorexia | PCR assay | 69 |
Recent studies have demonstrated that the ever-expanding CRISPR-Cas-based biosensing technology is contributing to this area, potentially improving or perhaps replacing existing technologies.70 Innovative scientific discovery has accelerated the CRISPR toolbox from chopping up invading viral DNA to cut–paste machinery for genome editing. It is designed to sense pathogenic nucleic acid at an attomolar concentration at breakneck speed. The traditional tedious and cumbersome protein-based assay has been replaced by nucleic acid detection in the CRISPR array, which exhibits greater accuracy and precision. The main issues in the food industry include viral contamination, which displays no organoleptic changes and threatens the life of the vulnerable population. Severe disease-causing enteric viruses such as Hepatitis A virus (HAV) and norovirus (NOV) escape harsh processing conditions and are difficult to detect. Listeriosis and other particular viral issues such as the avian influenza virus (AIV) have attracted attention to prohibit their further mutation into a more transmissible zoonotic form.71 Antimicrobial resistance coupled with the dearth of new antimicrobial drugs represents an alarming concern, and thus it is necessary to develop alternative strategies to guarantee farm-to-table food security. Due to their trans-cleavage capabilities, Cas effector proteins can provide surveillance of foodborne pathogens, especially in ready-to-eat food.
In this case, several studies have established the usability of CRISPR-based pathogen identification. For instance, F. Li et al. (2021) introduced recombinase-assisted amplification (RAA)-based CRISPR/Cas12a in an E-DNA biosensor platform for the ultra-sensitive and specific detection of L. monocytogenes.72 In this E-CRISPR technique, Cas12a cleaves the target DNA upon recognizing the PAM sequence by crRNA. The working principle relies on gold, platinum, and Ag/AgCl as the working electrode, counter electrode, and reference electrode, respectively. In the presence of target DNA, methylene blue single-stranded DNA (ssDNA) reporter cleaved from the electrode surface, producing a differentiable electrochemical signal acquired by the squarewave voltammetry, large-amplitude differential technique. This platform enabled the detection of 940 cfu g−1 of L. monocytogenes and is also applicable for other Cas systems (Cas9, Cas12b, Cas13a, and Cas13b) as a convenient method for pathogen diagnostic tool, leading to better quality of food. The specific highly sensitive enzymatic reporter unlocking (SHERLOCK) adapted RNA targeting CRISPR-associated enzyme Cas13 was used to discriminate single nucleotide differences at low concentrations.73 Further, a multiplexed platform was created for the detection of Zika virus (ZIKV) ssRNA in the HEX channel and Dengue virus (DENV) ssRNA in the FAM channel based on Cas12a (cpf1). Gradually, SHERLOCKv2 was engineered to exclude lab setup, designing lateral flow read-out based on the destruction of the FAM biotin reporter. The instrument-free detection of ZIKV or DENV ssRNA was possible with a sensitivity as low as 2 attomolar. Again, the modification of SHERLOCKv2 with lateral flow and csm6 provided an opportunity to detect viruses without power or portable readers. Improved CRISPR-based diagnostic (CRISPRdx) with portable deployment enables the sensing of nucleic acid, which is vital for application in human health. In response to the imperative determination of all putative food-related pathogens in the foodstuff, a dual foodborne pathogen detection assay was produced.74 A lateral flow strip was combined with Cas9 nickase-triggered amplification, employing specially designed sgRNA. Dual detection of Salmonella typhimurium and E. coli was the major focus of this one-pot assay. A fledgling cartridge system was also developed by He et al. (2020) using CRISPR/Cas12a and CRISPR RNA for the detection of double-stranded DNA virus with a limit of detection (LOD) of 1 pm in 2 h and 100 fM in 24 h, devoid of nucleic acid amplification.75 With respect to microfluidic lab-on-a-chip, Hajian et al. (2019) developed CRISPR-chip to broaden its prospects using on-chip electrical nucleic acid detection.76 In the presence of target DNA on the chip, nuclease deactivated Cas9 (dCas9) forms a complex with the target-specific sgRNA and emits an illuminating electrical signal read-out. This chip was comprised of a graphene field-effect transistor (gFET), where the specific sgRNA was immobilized on the surface of graphene. The invention of a reversible valve-assisted chip coupled with CRISPR/Cas12a offers new possibilities to meet public concern regarding Vibrio parahaemolyticus, the cause of seafood-associated illness.77 This simple, cost-effective chip design relies on the deployment of a silica membrane for nucleic acid capture and fluorescent read-out signal, which is distinguishable by the naked eye. Cas12-based HOLMES (a one-hour low-cost multipurpose highly efficient system) and DETECTR (DNA endonuclease targeted CRISPR trans-reporter) effectively monitored DNA and RNA viruses with attomolar sensitivity and high strain specificity.78 To identify E. coli in spring water, skimmed milk, and orange juice,79 a CRISPR-Cas9-based detection system was developed, coupling strand displacement amplification/rolling circle amplification (SDA/RCA).79 The CRISPR-Cas9 (H840A)-sgRNA complex detected and cleaved the target, which led to rolling circle and strand displacement amplification. Subsequently, the circular probe was hybridized into the products. A platform made of a metal–organic framework combined with a fluorescent signal was captured the data. Ackerman et al. developed combinatorial arrayed processes for the multiplexed assessment of nucleic acids in combination with the Cas13 (CARMEN-Cas13) detection approach, and it successfully detected multiple pathogens in 2020.80 This technique involved pairing droplets of PCR- or RPA-amplified materials with CRISPR-Cas detection mix emulsions containing Cas13 protein, certain crRNAs, and reporters. Then, as an optical identification, each experimental sample or test mixture was mapped to a solution-based fluorescent color code. The multiplexed detection of several sequences in a single reaction was also described in 2021, which was named LEOPARD (leveraging engineered tracrRNAs and on-target DNAs for parallel RNA detection).81 It was designed with a combination of gel-based readouts and tailored tracrRNAs for cross-validation and ultrasensitive detection of pathogenic bacteria.
In addition to cleaving the target DNA or RNA (known as cis-cleavage), Cas12a, Cas13a, and Cas14a have also been shown to exhibit intriguing collateral, nonspecific activities (known as trans-cleavage) on random ssDNA or ssRNA, respectively, upon target recognition regardless of their nucleic acid sequence.82,83 This emerging molecular biosensing tool is capable of detecting nucleic acid and other biomarkers with its unique properties of high sensitivity, high target-recognition specificity, room temperature reaction condition, single-base resolution, and target-induced collateral cleavage (Cas12a/Cas13a/Cas14a).84 This novel biosensing method has enormous potential for use in the food industry with the ability to detect viruses, genetically modified organisms (GMOs), toxins, and food additives in addition to bacterial pathogens.85–87 The negative perceptions by consumers regarding genetically modified organisms represent one of the biggest challenges to their worldwide adoption. In this case, CRISPR/Cas-based single-base resolution gene testing will be valuable to evaluate whether standardized regulations have been met or adhered to. The traditional qPCR method is unable to detect down to the single-nucleotide level with high specificity. In this context, M. Zhang et al. 2020, found that RPA-Cas12a enabled the detection of single-nucleotide polymorphism.88 Another severe threat to the food industry and human health is the increase in antimicrobial resistance strains due to their transmission from the food supply to humans, causing life-threatening diseases. According to the report in 2018 provided by the U.S. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS), the percentage of antimicrobial-resistant bacteria (such as Salmonella, Campylobacter, E. coli, and Enterococcus) detected in food has increased with time.89 Furthermore, the Centers for Disease Control (CDC, 2019) reported that in the United States, more than 2.8 million individuals get antibiotic-resistant infections yearly, and more than 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people die.90 This statistical data depicts the substantial need for accurate detection systems with attomolar-level sensitivity, which cannot be satisfied by existing detection systems. In this prospect, Quan et al. (2019) integrated CRISPR with next-generation sequencing to detect antimicrobial resistance.91 Cas9 was used in this system to cleave the desired target, which is later ligated to universal sequencing adaptors, and then bound to the sequencing-flow cell. Food safety-associated non-nucleic acid targets consist of analytes ranging from small molecules, ions, and proteins (exotoxins, allergens) to lipopolysaccharides (bacterial endotoxins). The fundamental principle behind employing the CRISPR-Cas system to identify non-nucleic acid targets is to dexterously translate these targets to nucleic acid signals using functional nucleic acids, which act as signal transduction elements, such as responsive DNAzymes and aptamers. Peng, Zhou, Liu, et al. (2020) produced a CRISPR-Cas12a-mediated aptamer fluorescent biosensor to trace adenosine triphosphate (ATP) with outstanding selectivity and sensitivity within 40 min.92 In this “turn-off” process, the aptamers acted as the target for CRISPR-Cas12a detection and preferentially bound to ATP when it was present. Therefore, ATP served as the switch of the CRISPR-Cas12a system, and the fluorescence intensity became an indicator for monitoring ATP concentration. Similar to food-borne pathogens, toxins also constitute a serious food safety issue. Existing aflatoxin analysis tools include immunoaffinity columns, thin-layer chromatography, ELISA, and immunochromatographic strips.93 The use of CRISPR technology not only detect toxins by identifying relevant genes but also editing these genes to alleviate their effects.94–96 Abnous et al. (2021) employed CRISPR/Cas12a and colorimetric assays to detect aflatoxin M1.85 They utilized aptamers for the specific recognition of aflatoxin and combined CRISPR/Cas12a with gold nanoparticles to enhance the sensitivity of toxin detection beyond nucleic acid-based approaches. Numerous studies have demonstrated the potential of CRISPR-based sensors to target all food contaminants. For instance, J. Li et al. (2020) developed a CRISPR-based biosensor to detect Pb2+ in the presence of other interfering cations including Ca2+, K+, Zn2+, Mn2+, Fe3+, Cd2+, Ni2+, Co2+, and Cu2+.97 In this system, DNAzyme was used to cleave the nucleic-acid sequence in the presence of Pb2+, and later recognized by CRISPR/Cas12a, which then amplified the fluorescence signal via its trans-cleavage activity. Another study reported the use of CRISPR to detect melamine in raw milk.87 An aptamer-locker was designed to bind and change the structure of the aptamer with the release of the aptamer-locker in the presence of melamine. Subsequently, it triggered the trans-cleavage activity of Cas12a and helped detect melamine in raw milk. CRISPR-Cas12a coupled with the liposome amplification strategy was applied to differentiate meat adulteration.98 A low adulteration rate could also be detected without interference from complex foods even through the naked eye. Moreover, the application of allergen-specific aptamers and their integration into point-of-care detection have undoubtedly taken allergen detection to the next level.99 The various CRISPR toolbox utilities in the food sector are listed in Table 2.
000 people die.90 This statistical data depicts the substantial need for accurate detection systems with attomolar-level sensitivity, which cannot be satisfied by existing detection systems. In this prospect, Quan et al. (2019) integrated CRISPR with next-generation sequencing to detect antimicrobial resistance.91 Cas9 was used in this system to cleave the desired target, which is later ligated to universal sequencing adaptors, and then bound to the sequencing-flow cell. Food safety-associated non-nucleic acid targets consist of analytes ranging from small molecules, ions, and proteins (exotoxins, allergens) to lipopolysaccharides (bacterial endotoxins). The fundamental principle behind employing the CRISPR-Cas system to identify non-nucleic acid targets is to dexterously translate these targets to nucleic acid signals using functional nucleic acids, which act as signal transduction elements, such as responsive DNAzymes and aptamers. Peng, Zhou, Liu, et al. (2020) produced a CRISPR-Cas12a-mediated aptamer fluorescent biosensor to trace adenosine triphosphate (ATP) with outstanding selectivity and sensitivity within 40 min.92 In this “turn-off” process, the aptamers acted as the target for CRISPR-Cas12a detection and preferentially bound to ATP when it was present. Therefore, ATP served as the switch of the CRISPR-Cas12a system, and the fluorescence intensity became an indicator for monitoring ATP concentration. Similar to food-borne pathogens, toxins also constitute a serious food safety issue. Existing aflatoxin analysis tools include immunoaffinity columns, thin-layer chromatography, ELISA, and immunochromatographic strips.93 The use of CRISPR technology not only detect toxins by identifying relevant genes but also editing these genes to alleviate their effects.94–96 Abnous et al. (2021) employed CRISPR/Cas12a and colorimetric assays to detect aflatoxin M1.85 They utilized aptamers for the specific recognition of aflatoxin and combined CRISPR/Cas12a with gold nanoparticles to enhance the sensitivity of toxin detection beyond nucleic acid-based approaches. Numerous studies have demonstrated the potential of CRISPR-based sensors to target all food contaminants. For instance, J. Li et al. (2020) developed a CRISPR-based biosensor to detect Pb2+ in the presence of other interfering cations including Ca2+, K+, Zn2+, Mn2+, Fe3+, Cd2+, Ni2+, Co2+, and Cu2+.97 In this system, DNAzyme was used to cleave the nucleic-acid sequence in the presence of Pb2+, and later recognized by CRISPR/Cas12a, which then amplified the fluorescence signal via its trans-cleavage activity. Another study reported the use of CRISPR to detect melamine in raw milk.87 An aptamer-locker was designed to bind and change the structure of the aptamer with the release of the aptamer-locker in the presence of melamine. Subsequently, it triggered the trans-cleavage activity of Cas12a and helped detect melamine in raw milk. CRISPR-Cas12a coupled with the liposome amplification strategy was applied to differentiate meat adulteration.98 A low adulteration rate could also be detected without interference from complex foods even through the naked eye. Moreover, the application of allergen-specific aptamers and their integration into point-of-care detection have undoubtedly taken allergen detection to the next level.99 The various CRISPR toolbox utilities in the food sector are listed in Table 2.
| Food borne organism | Food present | CRISPR-based antimicrobial | CRISPR-based biosensing | References | |
|---|---|---|---|---|---|
| Food pathogen | |||||
| 1 | Escherichia coli O157:H7 | Predominant in grounded meat and raw milk | Endogenous type I Cascade-Cas3 machinery | Cas9 nickase-based amplification reaction (Cas9nAR) combined with lateral flow strip | 74 |
| 2 | Salmonella spp. | Predominant in chicken eggs | Cas9 effector protein | CRISPR-Cas13a (allosteric probe-initiated catalysis C-Cas) | 100 |
| 3 | Listeria monocytogenes | Predominant in ready-to-eat (RTE) food i.e., meat, cheese, and fish products | Antibacterial drone (ABD) carrying deactivated or mutated Cas9 (dCas9) virulence-blocking module | RAA-based E-CRISPR with the trans-cleavage activity of CRISPR/Cas12a | 72 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Food spoilage organism | |||||
| 4 | Streptococcus spp. | Predominant in eggs, steamed lobster, milk, curd, pudding rice, etc. | Cas9 guided by guide RNA to the sequence-specific site, leading to the lethal cleavage of double-stranded DNA endo-nucleolytically | Cas13-based methods, automated microfluidic device or HUDSON + SHERLOCK | 101 and 102 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||
| Food poisoning organism | |||||
| 5 | Campylobacter jejuni | Predominant in raw meat and milk | Cas12a protein, which sequentially cleaves the non-targeting strand and the targeting strand to form DSBs | Cas12-based methods, hpDNA-based electrochemical biosensor or MAV-chip | 103 and 104 |
3.3. Why CRISPR biology in food packaging?
To date, the proliferation of contaminating pathogens is diagnosed by barcode-based biosensors in intelligent packaging. Commercially available pathogen indicators anticipate the formation of an unreadable dark bar on the barcode upon the interaction of specific antibodies with the target pathogens such as Salmonella spp., Escherichia coli O157:H7, and Listeria monocytogenes.105 Eventually, Ontario, Canada developed the Toxin Guard™ pathogen indicator by incorporating it in plastic packaging film. However, any foodborne pathogen can pose a significant health threat. Furthermore, especially for pathogen identification, the validation parameters such as accuracy, repeatability, precision, selectivity/specificity, linearity, and quantification limit should meet the requirements. In this case, it is essential to give a brief overview of microbes associated with packaged food products such as E. coli, Pseudomonas spp. (associated with fruits and vegetables), Rahnella aquatilis, Carnobacterium divergens, Lactobacillus sakei (associated with meat products), P. fluorescens, Providencia spp. (associated with milk), Salmonella enteritidis (associated with egg) and Clostridium botulinum (associated with seafood). Therefore, the ever-expanding global food packaging market calls for more accurate, sensitive, and reliable strategies without undue impact on the safety and cost of final food products, which traditional detectors may not provide. Clustered regularly interspaced short palindromic repeat (CRISPR) associated protein (CRISPR-Cas) molecular machines have radically changed the global agricultural era by generating improved livestock breeds, crop traits such as Candidatus Liberibacter-resistant citrus varieties, reduced gluten-containing wheat, improved rice grain yields, and many more.106 This comprehensive genome editing tool promisingly modulates the community structure of the food fermentation process to optimize and enhance the functional properties of probiotics and prebiotics, hence broadly impacting the desirable organoleptic characteristics of various foods. CRISPR-Cas antibacterial property facilitates food safety by selectively targeting the essential genes required to eliminate spoilage micro-organisms from the microbial population. CRISPR-Cas nuclease targets undesirable genotypes such as toxin-encoding genes, virulence factors, and unique antibiotic-resistance cassettes in pathogenic bacteria.107 Although the emergence of CRISPR has not yet been broadly applied to packaging systems, the application of CRISPR in diagnosis and antimicrobials demonstrates the potential use of CRISPR tools, which may be extended in the packaging industry. Thus, this review emphasizes the exploration of CRISPR in smart packaging technology. The diagnostic CRISPR system can be applied in intelligent packaging to detect the harmful pathogens present in packaged food.4. CRISPR biology: a new revolution in next-generation packaging
4.1. CRISPR search tool in intelligent packaging: mechanism of intelligence
Conventional intelligent packaging includes time–temperature indicators (TTIs), radiofrequency identification (RFID), gas indicators, and pathogen indicators as smart devices (i.e., small labels or tags) printed on or incorporated in packaging materials. Its basic working principle relies on the enzymatic, electronic, and biochemical interaction, which identifies irreversible changes through an electromagnetic wave, barcode-based sensing, electronic devices, and/or color changes. An innovative advancement in the field of intelligent packaging is automatic identification (Auto-ID). This is a grouped terminology of barcodes, QR-codes, magnetic inks, voice recognition, biometrics, etc., which functions beyond sensing and indication by automatization, counterfeit protection, and antitheft prevention.The doubling global demand for food is suspected to steer the incidence of foodborne disease outbreaks with a high degree of certainty in the next few decades.71 Accordingly, advancements in packaging techniques with tracking facilities may help mitigate these obstacles. Therefore, the CRISPR search tool is expected to meet the consumer demands for transparent and rapid traceability systems, especially when the supply chain reaches further.
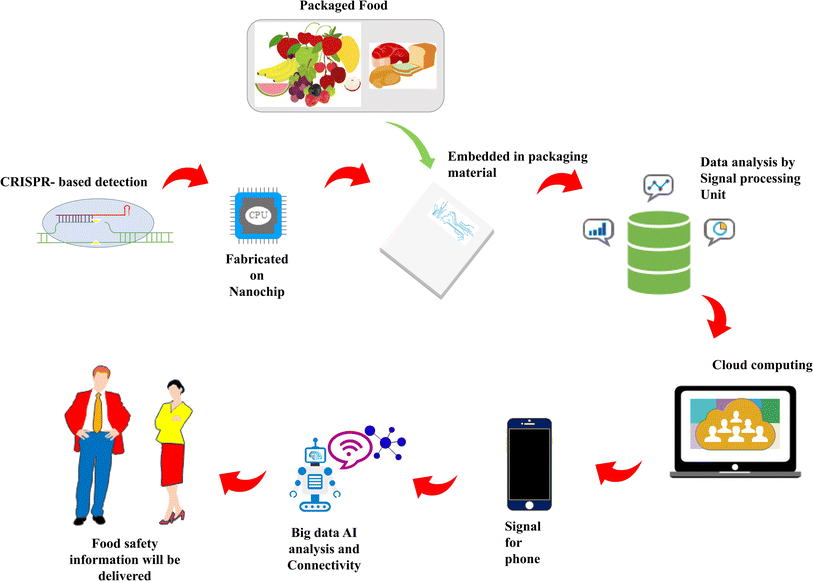
Integrated microsystems and nano-bio sensors can play a crucial role in detecting deteriorative changes in food packaging. To ensure food safety, quality, and process control, current food packaging development calls for the application of appropriate sensing technologies to identify contaminants at the parts per trillion level. Fig. 3 presents an overview of CRISPR-based next-generation intelligent packaging. Firstly, CRISPR-based biosensors can be developed employing different Cas proteins and guide RNA, which will selectively target the suspected microbe or other targets of concern in the packaged food. The principle of detection is illustrated as follows. In biosensor-based systems, the sensor responds by producing a signal when an observable event occurs. For instance, in the presence of the target, the biotinylated single-stranded DNA reporter would be trans-cut by the respective Cas protease with color development. For a better understanding, it is suggested to refer to the study conducted by.108 Another possible mechanism of intelligence is using specific genes of target food contaminants to activate the trans-cleavage activity of CRISPR-Cas, consequently triggering the single-stranded DNA cleavage activity.109 Subsequently, K+ will be added to ssDNA with guanine sequences to form a stable G-quadruplex DNAzyme. Then, DNase will catalyze the TMB-H2O2 (tetramethyl benzidine/hydrogen peroxide) reaction in the presence of hemin with a color change. Naked eyes and smartphones can easily recognize these changes. Then, the biosensor can be fabricated on a nanochip, which is embedded in a biopolymeric film. The chip can be inserted in a matrix in at least two ways, as follows: (a) face-up and (b) face-down. (a) During the chip attachment process, the backside of the chip is attached to the foil in a face-up fashion and (b) during the embedding lamination process, the front side of the chip is embedded in the dielectric layer.110 Its backside is embedded in the dielectric layer, (c) during the via drilling process, for the face-up fashion, the drilling is from the top and stops at the pads of the chip. For the face-down fashion, the drilling is performed from the bottom and stops at the pads of the chip, and (d) during the Cu plating and etching processes, both the face-up and face-down fashions electroplate Cu to fill the vias and make/etch the circuit traces. Subsequently, CRISPR will hybridize with the target present in the headspace of the packaged food and generate a readout signal. In this case, the readout signal will impart information even at trace level of contamination. Upon encountering the readout signal, it will be processed and analyzed, and all the information will be stored in the cloud computing facility, which can be further utilized by convolutional neural network (CNN) and transmitted through the IoT. With deep learning and artificial neural network (ANN), the outcome of CRISPR point of care diagnosis is believed to improvise the previous packaging methods.
The absence of an adequate surveillance mechanism to track foodborne illness channels the thinking process towards a more practical and harmonized system. Complete microbial inactivation is a prime objective in packaging and to achieve this, where generation of stress-resistant organisms has been associated with packaging trends in recent years. Accordingly, a CRISPR-based world with a lab-on-a-chip facility may be a very useful solution with regard to intelligent packaging. Given that no “magic bullet” exists, CRISPR has certain drawbacks, but with an innovative mindset, it is possible to open new frontiers by pushing the boundaries of CRISPR search tools in the packaging sector. However, consumer acceptance is crucial, and therefore educating the public with novel technologies and their possible benefits is necessary. Moreover, transparency in use and accuracy of the outcome should be consistent with food safety regulations for the sensible deployment of new technologies.
4.2. CRISPR-Cas-based cloud detection as an early alarm
The online purchase of food using 4G/5G technologies has efficiently replaced the “bricks and mortar”-based system. However, food standard should not be compromised with the globally increased mobile adoption and broadband penetration in this context. Thus, packaging needs to be audited with respect to storage conditions, transportation failures, or other breakdowns or inadequacies. The creation of big data culture is on the verge of transforming the food industry. It is of tremendous benefit to track the source of contamination, facilitating cloud computing approaches. Well-resourced technological advancement often produces a sound electronic traceability system, but the communication system is very time-consuming, creating an overarching issue in exchanging information between the links in the supply chain. The diversity and proprietary nature of the respective internal systems represent a dilemma in global interconnectedness and spreading information regarding food safety issues to consumers. Thus, opportunities exist for online communication, building an early alarm system based on artificial intelligence (AI)-combined CRISPR/Cas tools.111 This has been well reported to sense CRISPR/Cas-based read-out via android APP and the data stored in the cloud for further processing through 5G service. In the era of big data, it thus possible to realize “extra safe” food by determining the impending danger, and the proposed AI-powered machine learning of diagnostic data will more accurately reduce the risk and give suggestions for countermeasures. The synergistic use of CRISPR and cloud computing represents a promising way to develop authentic and traceable models. Although there is still much room for improvement, integrating big data analytics or neural networks with CRISPR-chip-based sensors is crucial for root cause and retrospective analysis. Contributing to the challenges, it is necessary to evaluate high-impact areas and intensify research in this field for future direction.4.3. AI overwhelms intelligent packaging, whilst others are embracing it
The importance of food packaging has led researchers to look for continuous improvements. Therefore, manual procedures are being replaced by automated production lines, resulting in the achievement of more efficient and effective processes. Quality control is one of the most important operations in the packing stage given that it inspects the packed product to ensure it meets the quality requirements before it reaches the final user. This process can be automated by computer vision and artificial intelligence tools.With the dawn of intelligent automation, the packaging sector is being equipped with the IoT to realize certain benefits. The aid of machine learning in date labeling has become the new norm in the packaging industry, ensuring greater efficiencies with fewer errors.112 AI-powered vision systems are potent in quality inspection and real-time tracking to identify anomalies. To date, AI has covered many goals such as clear traceability related to standard compliance, the concept of zero downtime, and better predictive maintenance. Indeed, with the enormous transformative experiences of the packaging industry, AI technology is leaving its mark across the entire supply chain. From this perspective, herein, we proposes to link the gap between CRISPR-based detection and wireless transmission of information using artificial intelligence.
4.4. Packaging machinery powered by deep learning-based quality assessment
It is necessary to replace time-consuming, error-prone manual packaging information to avoid financial penalties or legislation risk regarding food security. High contrast imaging and surface extraction technology are well-reputed automation defect detectors. The revolutionary concept of deep learning in the food domain outperforms conventional food packaging systems, especially in food quality inspection, which is the key theme concerning the public perception of food safety.In contrast to food science, it is essential to convey a brief description of CNN (convolutional neural network) and deep learning. CNN is the most popular machine intelligence model for image analysis, which is diversified for two- and three-dimensional data formats.113 Additionally, deep learning is used to achieve deep data representation, allowing a machine to extract features from raw data. This helps in detection and regression by utilizing a deep ANN (artificial neural network).113 Global food security encompasses significant challenges for minimally processed “natural” foods such as fruits and vegetables. The development of deep learning with image processing or spectral sensing method efficiently copes with the quality detection issues of disease or damage detection. Here, a few of the exemplary research outcomes in this context are briefly discussed. A combined approach of stacked sparse autoencoder (SSAE) and CNN was adopted to detect cucumber defects based on hyperspectral imaging.114 In the study by Tan et al., they used CNN to screen defected regions based on images in RGB channels, and thus SSAE represents and classifies the mean spectra of the defective region. Consequently, CNN-SSAE chose the whole defected cucumber as the input with 91% accuracy. Other researchers aimed to apply five-layered CNN to alert pest and diseases in apples.115 Here, images of apple skin lesions were collected via an infrared video sensor network and processed with rotational translation of four different angles. Yu, Lu et al. (2018) established a new deep learning methodology to estimate the firmness and soluble content of pears, which was achieved by extracting high-level features of raw spectra and putting them into a fully connected network block (FNN).116 Concurrently, deep learning and visible/near-infrared spectral sensing estimated these two attributes and labeled the pears with the reference value. Moreover, hyperspectral image sensing was introduced with deep learning to discriminate the damaged and normal samples with respect to mechanical damages or lesions under the skin of berries.117 W. Zhang et al. (2018) invented integrated AI technology in the design of smart refrigerators to recognize different types of fruit and indicate their firmness, nutrient content, disease/damage degree, natural maturity, etc.118 Fusion of multiple CNN architectures and discrimination by deep learning predicted parameters reflected in the RGB image or spectral information of samples for quality estimation and safety measures. In the multilayer deep CNN architecture designed by Pandey et al. (2017),119 AlexNet was used as a baseline. Thereafter, AlexNet, GoogleNet, and ResNet subnetworks were used for the development of the whole ensemble network. Rodríguez et al. (2018) chose the AlexNet architecture to discriminate plum varieties at early maturation stages.120 Furthermore, CNN coupled with fourfold cross-validation reached 97.5% classification accuracy when surface defects were detected in mangosteen.121 Similarly, AlexNet achieved 90% classification accuracy based on hyperspectral sensing and RGB imaging to screen artificially ripened bananas.
Excess water in food packaging is a major global concern given that it causes deterioration and degradation of food quality, leading to increased health risk and limiting the shelf life of produce.105 In this case, the measurement of multiple chemical parameters is necessary to determinate food safety. Yu, Tang, et al. (2018) designed a vis/near-infrared hyperspectral imaging technique with deep learning algorithm to investigate the freshness grade of shrimp during cold storage.122 Determination of the total volatile basic nitrogen (TVB-N) content in shrimp was studied by Yu et al. (2019) with the help of spectrometer-correlated hyperspectral data.123 Innovation of hyperspectral image processing in remote sensing positively influences the food domain with respect to poor food quality, deterioration region, etc. Simultaneously, Al-Sarayreh et al. (2018) provided an adulteration identification architecture for red meat.124 In this study, hyperspectral images of various meat samples were scanned to build the dataset to analyze meat quality, focusing on spectral data and spatial information applying deep CNN and FNN block. Hyperspectral images mostly contain abundant spectral-spatial information and among the other methods used, CNN has high potential of classifying those spectral features.
Recently, 1-D and 2-D CNN architectures have been applied for the quality evaluation of liquids. For instance, milk adulteration has been determined by Neto et al. (2019), exploiting deep feature extraction of CNN to extract Fourier transform infrared spectra (FTIR).125 This study deployed Random Forest (RF) and gradient boosting machine (GBM) classifiers for the component feature analysis.
Food fraud is a serious concern in food safety throughout the global supply chain. Frauds are mainly mislabeling expiration dates, improper health certificates, fraudulent import declaration, etc.126 The deep method showed better performance in this section. Mao et al. (2018) presented credit evaluation texts such as “fruit does not look very fresh” and “the quality is good,” which were converted into a simple indicator and labeled as “negative” and “positive,” respectively.127 Here, a deep learning network named long short term memory (LSTM) extracted sentence features and put the data as an input of deep neural network (DNN)-based classifier.
Future studies need to explore new algorithms for the CNN acceleration of computation speed despite prominent performances. Enormous potential modification of mobile hyperspectral imaging system by virtue of snapshot HSI and 3-D CNN model will improve the real-time monitoring of food. In these prospects, the fast acquisition of spectral data and ultra-portability of HSI are the crucial lead.128 It is hoped that understanding the features extracted by 1-D, 2-D, 3-D CNN leads to its critical application in mobile devices for the remote serving of qualitative analysis.
5. Future perspective
Food packaging seeks consistent augmentation in response to the ever-increasing demand for high-quality food, involving extended shelf life, sensory properties, etc. Intelligent packaging does more than just protect the food. It is proficient in filling the gap with decisive communication prevailing in the market application by evaluating various food. The need of the hour is technological advancement to cope with food security globally, which will lead to a new adaptation strategy. Simultaneously, the IoT efficiently eliminates the gap between consumer response and regulatory agencies for the development of fast communication and improved dissemination across the global supply chain. Big data mining and processing are an advanced approach to overcoming food challenges.Based on the current understanding, the entirety of CRISPR/Cas biosensing has just begun with SHERLOCK, HOLMES, NASBACC (nucleic acid sequence-based amplification-CRISPR cleavage), Cas EXPAR, RCH (rolling circle amplification-CRISPR-split-HRP) system, and PC (paired dCas9) reporter, which may unlock a new paradigm in next-generation packaging technology. However, they have many limitations due to their dependency on known nucleic acid sequences, off-target effects, and target amplification requirements, which may make the system inconvenient and less robust. The efficiency of CRISPR toolkits raises doubt given that these systems are unable to detect food-borne pathogens that have undergone epigenetic changes. Thus, to deliver CRISPR tools to packaging systems, the cost-effectiveness, receptor availability, and practicability of the system need to be further improved. To date, the CRISPR-Cas-based detection method is still in the laboratory stage and requires professional technicians. Thus, similar portable devices need extensive advancement and significant refinement for engineering and commercialization. Furthermore, the durability of CRISPR-based food packaging systems and the shelf life of the packaged food are also a matter of concern. Nevertheless, advocacy for the outstanding performance exhibited by the system allows its broadened application in diverse fields in the near future. With superior specificity and sensitivity in both local and global basis, the biocontrol of contaminating organisms is of utmost importance for food safety and innovation. CRISPR-driven flexible programmability with regards to targeted killing empowers food scientists to fight foodborne pathogens. However, although CRISPR-Cas technology has outstanding advantages in real-time detection, technical barriers remain in its transformation from emerging technologies to practical applicability. Economic feasibility and public acceptability are the key questions to answer to employ CRISPR technology in food packaging applications. Considering these issues, it is indispensable to consider that by 2050, the next generation of food packaging should significantly reduce waste in food and packaging materials and its adverse effect on the environment (such as resource consumption, greenhouse gas emissions, and pollution). In fact, the yearly carbon footprint of food production and non-consumption in the EU is roughly estimated to be 100 million tonnes or 495 million tonnes of CO2. Nearly 210 million hectares of land are used to produce food that is not consumed. According to modeling, food waste may reach over 200 million tonnes by 2050 if nothing is done.129,130 By promoting the market adoption of packaging innovations, it can extend and better manage food shelf-life, and therefore 50% reduction in food waste at the retail and consumer level is anticipated by 2050.131 Consequently, this can save approximately 100 million tonnes of food, which equates to an absolute decrease of 250 million tonnes of CO2-equivalent and approximately 100 million hectares of land recovered.132 Moreover, around 48 million people get sick in United State alone, and among them 3000 die due to foodborne diseases. The associated economic loss is estimated to be $15.6 billion.133 According to the World Health Organization (WHO), there were 600 million cases of foodborne infections worldwide in 2010 alone, resulting in 420![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fatalities.134 Other yearly estimates place these numbers considerably higher. Indeed, the present approach is proposed to consider the safety issues of humans with regard to disease prevention. Besides the future technical challenges, various CRISPR biology tools and applications should fall under different rules and guidelines to prevent their misuse. The application of CRISPR-based AI technology in the food packaging domain is dependent on a secure network service, where any violation in the system may lead to a potential risk. Moreover, any advance technology has risk towards environment safety and may cause exploitation of workers. One significant strategy to target this is the education or imparting knowledge to evaluate this powerful technique. Indeed, harnessing the packaging sector with CRISPR is a novel concept for enhancing safe and sustainable next-generation food. The comprehensive assessment described herein can be a guiding tool for researchers in tailoring a scientifically integrated packaging system.
000 fatalities.134 Other yearly estimates place these numbers considerably higher. Indeed, the present approach is proposed to consider the safety issues of humans with regard to disease prevention. Besides the future technical challenges, various CRISPR biology tools and applications should fall under different rules and guidelines to prevent their misuse. The application of CRISPR-based AI technology in the food packaging domain is dependent on a secure network service, where any violation in the system may lead to a potential risk. Moreover, any advance technology has risk towards environment safety and may cause exploitation of workers. One significant strategy to target this is the education or imparting knowledge to evaluate this powerful technique. Indeed, harnessing the packaging sector with CRISPR is a novel concept for enhancing safe and sustainable next-generation food. The comprehensive assessment described herein can be a guiding tool for researchers in tailoring a scientifically integrated packaging system.
Feature learning and transfer learning are enticing characteristics of deep learning. Most deep learning applications exploit pre-trained CNN models based on ImageNet as a large dataset, and fine-tuning these models on their target dataset reduces intricacies. Real-time data processing with deep learning involves a more complex model structure. Theano, Tensorflow, Caffe, PyTorch, MXNet, Keras, and MatConvNet for Matlab are some popular software designs that can cut the gordian knot to program complex neural networks.113 By contrast, a graphics processing unit (GPU) coupled with Compute Unified Device Architecture (CUDA) Toolkit and the NVIDIA CUDA Deep Neural Network library (cuDNN, a GPU-accelerated library of primitives for DNNs) produced by the NVIDIA company accelerated deep learning computation as hardware support.113 Despite its numerous shortcomings, including the acquisition of a reliable big data set, hardware restrictions, and time-consuming annotation, the real-world application of deep learning in the food domain is unavoidable. Implementing the RGB database, hyperspectral imaging, and thermal imaging in food recognition and quality inspection is essential. Miniaturized sensing equipment excludes the laboratory setup of a spectrometer and state-of-the-art performance achieved by combining portable devices with machine learning technology and the IoT for fruit quality evaluation.135 Handheld smart devices should outpace the food security issue by moving towards food identification, safety, and quality evaluation by embedding food recognition models in mobile apps.
6. Conclusion
The CRISPR-Cas toolkit plays a significant role in the food industry. In this review, we mostly focused on the application of various Cas proteins to ensure food safety. However, various technical barriers such as amplification free surveillance system, sequence limitation, cost, and large-scale implication limit its practical applicability. Besides, the implementation of AI has transformed the food industry with its ability to forecast product markets, waste reduction, round-the-clock monitoring, cost management, etc. In this context, the usage of image processing, spectral sensing, hyperspectral imaging, and deep neural network in food quality detection, chemical parameter measurement, and food fraud identification have also been overviewed. Overall, the aim of this review article is to encourage researchers to perform more experiments to overcome all the hurdles related to CRISPR-based cloud computing and deep learning method for the evolution of next-generation intelligent packaging, hence supporting food security. The integration of food-related databases acquired by modern sensors is esteemed to leverage the packaging industry.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Institute of Technology, Durgapur and DST-INSPIRE.References
- H. M. Lam, J. Remais, M. C. Fung, L. Xu and S. S. M. Sun, Food supply and food safety issues in China, Lancet, 2013, 381, 2044–2053 CrossRef PubMed
.
- P. L. Wang, L. H. Xie, E. A. Joseph, J. R. Li, X. O. Su and H. C. Zhou, Chem. Rev., 2019, 119, 10638–10690 CrossRef CAS PubMed
.
- M. Y. C. Wu, M. Y. Hsu, S. J. Chen, D. K. Hwang, T. H. Yen and C. M. Cheng, Trends Biotechnol., 2017, 35, 288–300 CrossRef CAS PubMed
.
- M. D. Kirk, S. M. Pires, R. E. Black, M. Caipo, J. A. Crump, B. Devleesschauwer, D. Döpfer, A. Fazil, C. L. Fischer-Walker, T. Hald, A. J. Hall, K. H. Keddy, R. J. Lake, C. F. Lanata, P. R. Torgerson, A. H. Havelaar and F. J. Angulo, PLoS Med., 2015, 12, e1001921 CrossRef PubMed
.
- M. E. Nyenje and R. N. Ndip, https://academicjournals.org.
-
S. Hoffmann and E. Scallan, in Foodborne Diseases, Third edn, 2017 Search PubMed
.
- A. S. Barone, J. R. V. Matheus, T. S. P. de Souza, R. F. A. Moreira and A. E. C. Fai, Green-based active packaging: opportunities beyond COVID-19, food applications, and perspectives in circular economy—a brief review, Compr. Rev. Food Sci. Food Saf., 2021, 20(5), 4881–4905 CrossRef CAS PubMed
.
- B. J. Deka, V. Bohra, W. Alam, S. Sanasam, J. Guo, L. Borana and A. K. An, Environment impact assessment of COVID-19, Integrated risk of pandemic: Covid-19 impacts, resilience and recommendations, 2020, pp. 169–195 Search PubMed.
- COVID-19 Impact on Packaging Market by Material Type, Application And Region – Global Forecast to 2021, https://www.reportlinker.com/p05892825/COVID-19-Impact-on-Packaging-Market-by-Material-Type-Application-And-Region-Global-Forecast-to.html?utm_source=PRN, accessed 17 August 2022.
- J. Shin, M. Miller and Y. C. Wang, Compr. Rev. Food Sci. Food Saf., 2022, 21, 3010–3029 CrossRef CAS PubMed
.
- National Medical Products Administration, http://english.nmpa.gov.cn/, accessed 17 August 2022.
- R. Kler, G. Elkady, K. Rane, A. Singh, M. S. Hossain, D. Malhotra, S. Ray and K. K. Bhatia, Machine learning and artificial intelligence in the food industry: a sustainable approach, J. Food Qual., 2022, 2022, 1–9 CrossRef
.
- N. R. Mavani, J. M. Ali, S. Othman, M. A. Hussain, H. Hashim and N. A. Rahman, Application of artificial intelligence in food industry—a guideline, Food Eng. Rev., 2022, 14, 134–175 CrossRef
.
-
G. L. Robertson, Food Packaging, Principle and Practices, 2013 Search PubMed
.
-
A. Gordon and R. Williams, The role and importance of packaging and labeling in assuring food safety, quality and regulatory compliance of export products II: packaging & labeling considerations, Food Safety and Quality Systems in Developing Countries, Technical and Market Considerations, 2020, vol. III, pp. 285–341 Search PubMed
.
- L. Brennan, S. Langley, K. Verghese, S. Lockrey, M. Ryder, C. Francis, N. T. Phan-Le and A. Hill, The role of packaging in fighting food waste: a systematised review of consumer perceptions of packaging, J. Cleaner Prod., 2021, 281, 125276 CrossRef
.
-
G. L. Robertson, Food Packaging: Principles and Practice, 3rd edn, 2013 Search PubMed
.
- O. A. Odeyemi, C. M. Burke, C. C. J. Bolch and R. Stanley, Seafood spoilage microbiota and associated volatile organic compounds at different storage temperatures and packaging conditions, Int. J. Food Microbiol., 2018, 280, 87–99 CrossRef CAS PubMed
.
- F. Özogul, A. Polat and Y. Özogul, The effects of modified atmosphere packaging and vacuum packaging on chemical, sensory and microbiological changes of sardines (Sardina pilchardus), Food Chem., 2004, 85(1), 49–57 CrossRef
.
- A. E. Goulas, I. Chouliara, E. Nessi, M. G. Kontominas and I. N. Savvaidis, Microbiological, biochemical and sensory assessment of mussels (Mytilus galloprovincialis) stored under modified atmosphere packaging, J. Appl. Microbiol., 2005, 98(3), 752–760 CrossRef CAS PubMed
.
- P. Masniyom, O. Benjama and J. Maneesri, Extending the shelf-life of refrigerated green mussel (Perna viridis) under modified atmosphere packaging, Songklanakarin J. Sci. Technol., 2011, 33, 2 Search PubMed
.
- A. Orkusz and M. Michalczuk, Research note: effect of packaging atmosphere on the fatty acid profile of intramuscular, subcutaneous fat, and odor of goose meat, Poult. Sci., 2020, 99(1), 647–652 CrossRef CAS PubMed
.
- M. Marcinkowska-Lesiak, Z. Zdanowska-Sąsiadek, A. Stelmasiak, K. Damaziak, M. Michalczuk, E. Poławska, J. Wyrwisz and A. Wierzbicka, Effect of packaging method and cold-storage time on chicken meat quality, CyTA–J. Food, 2016, 14(1), 41–46 CrossRef CAS
.
- H. Ashenafi and S. Tura, Shelf life and quality of tomato (Lycopersicon esculentum Mill.) fruits as affected by different packaging materials, Afr. J. Food Sci., 2018, 12(2), 21–27 CrossRef
.
- A. Bhardwaj, T. Alam and N. Talwar, Recent advances in active packaging of agri-food products: A review, J. Postharvest Technol., 2019, 7(1), 33–62 Search PubMed
.
- A. Salminen, K. Latva-Kala, K. Randell, E. Hurme, P. Linko and R. Ahvenainen, The effect of ethanol and oxygen absorption on the shelf-life of packed sliced rye bread, Packag. Technol. Sci., 1996, 9(1), 29–42 CrossRef CAS
.
- P. D. Antunez, M. Botero Omary, K. A. Rosentrater, M. Pascall and L. Winstone, J. Food Sci., 2012, 77(1), S1–S9 CrossRef CAS PubMed
.
- A. T. P. Passarinho, N. F. Dias, G. P. Camilloto, R. S. Cruz, C. G. Otoni, A. R. F. Moraes and N. D. F. F. Soares, Sliced bread preservation through oregano essential oil-containing sachet, J. Food Process Eng., 2014, 37(1), 53–62 CrossRef CAS
.
- C. G. Otoni, S. F. O. Pontes, E. A. A. Medeiros and N. D. F. F. Soares, Edible films from methylcellulose and nanoemulsions of clove bud (Syzygium aromaticum) and oregano (Origanum vulgare) essential oils as shelf life extenders for sliced bread, J. Agric. Food Chem., 2014, 62(22), 5214–5219 CrossRef CAS PubMed
.
- T. Janjarasskul and P. Suppakul, Active and intelligent packaging: the indication of quality and safety, Crit. Rev. Food Sci. Nutr., 2018, 58(5), 808–831 CrossRef PubMed
.
- Detect Food Spoilage with Sensors|Concepts & Innovations, https://www.electronicsforu.com/technology-trends/detect-food-spoilage-sensors, accessed 17 August 2022.
- IoT Based Food Monitoring System, https://iotdesignpro.com/projects/iot-based-food-monitoring-system, accessed 17 August 2022.
- H. Tao, M. A. Brenckle, M. Yang, J. Zhang, M. Liu, S. M. Siebert, R. D. Averitt, M. S. Mannoor, M. C. McAlpine, J. A. Rogers, D. L. Kaplan and F. G. Omenetto, Adv. Mater., 2012, 24, 1067–1072 CrossRef CAS PubMed
.
- Y. Duan, Y. Liu, H. Han, H. Geng, Y. Liao and T. Han, A dual-channel indicator of fish spoilage based on a D-π-A luminogen serving as a smart label for intelligent food packaging, Spectrochim. Acta, Part A, 2022, 266, 120433 CrossRef CAS PubMed
.
- K. S. Makarova, Y. I. Wolf, O. S. Alkhnbashi, F. Costa, S. A. Shah, S. J. Saunders, R. Barrangou, S. J. J. Brouns, E. Charpentier, D. H. Haft, P. Horvath, S. Moineau, F. J. M. Mojica, R. M. Terns, M. P. Terns, M. F. White, A. F. Yakunin, R. A. Garrett, J. van der Oost, R. Backofen and E. v. Koonin, An updated evolutionary classification of CRISPR-Cas systems, Nat. Rev. Microbiol., 2015, 13(11), 722–736 CrossRef CAS PubMed
.
- K. S. Makarova, D. H. Haft, R. Barrangou, S. J. J. Brouns, E. Charpentier, P. Horvath, S. Moineau, F. J. M. Mojica, Y. I. Wolf, A. F. Yakunin, J. van der Oost and E. v. Koonin, Evolution and classification of the CRISPR–Cas systems, Nat. Rev. Microbiol., 2011, 9(6), 67–77 Search PubMed
.
- R. Barrangou, C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero and P. Horvath, CRISPR provides acquired resistance against viruses in prokaryotes, Science, 2007, 315(5819), 1709–1712 CrossRef CAS PubMed
.
- H. Deveau, R. Barrangou, J. E. Garneau, J. Labonté, C. Fremaux, P. Boyaval, D. A. Romero, P. Horvath and S. Moineau, Phage response to CRISPR-encoded resistance in Streptococcus thermophilus, J. Bacteriol., 2008, 190(4), 1390–1400 CrossRef CAS PubMed
.
- S. J. J. Brouns, M. M. Jore, M. Lundgren, E. R. Westra, R. J. H. Slijkhuis, A. P. L. Snijders, M. J. Dickman, K. S. Makarova, E. V. Koonin and J. van der Oost, CRISPR provides acquired resistance against viruses in prokaryotes., Science, 2007, 315(5819), 1709–1712 CrossRef PubMed
.
- J. E. Garneau, M. È. Dupuis, M. Villion, D. A. Romero, R. Barrangou, P. Boyaval, C. Fremaux, P. Horvath, A. H. Magadán and S. Moineau, The CRiSPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA, Nature, 2010, 468, 67–71 CrossRef CAS PubMed
.
- G. J. Knott and J. A. Doudna, CRISPR-Cas guides the future of genetic engineering, Science, 2018, 361, 866–869 CrossRef CAS PubMed
.
- E. Stout, T. Klaenhammer and R. Barrangou, CRISPR-Cas technologies and applications in food bacteria, Annu. Rev. Food Sci. Technol., 2017, 8, 413–437 CrossRef CAS PubMed
.
- B. Priyanka, R. K. Patil and S. Dwarakanath, A review on detection methods used for foodborne pathogens, Indian J. Med. Res., 2016, 144, 327 CrossRef CAS PubMed
.
- A. Garrido-Maestu, S. Azinheiro, F. Roumani, J. Carvalho and M. Prado, Application of short pre-enrichment, and double chemistry real-time pcr, combining fluorescent probes and an intercalating dye, for same-day detection and confirmation of salmonella spp. and escherichia coli o157 in ground beef and chicken samples, Food Microbiol., 2020, 11, 591041 Search PubMed
.
- R. M. Renuka, N. Maroli and K. Ponmalai, Spectrochim. Acta, Part A, 2020, 243, 118662 CrossRef PubMed
.
- H. B. Yin, C. H. Chen, B. Katchman, C. Newland, M. May and J. Patel, Food Microbiol., 2022, 107, 104086 CrossRef CAS PubMed
.
- M. Demirci, A. Yigin, S. Altun, H. Uysal, S. Saribas and B. Kocazeybek,
In vitro investigation of donkey milk efficacy against standard staphylococcus aureus strains, Niger. J. Clin. Pract., 2021, 7(2), 145–149 Search PubMed
.
- N. Elahi, M. Kamali, M. H. Baghersad and B. Amini, A fluorescence Nano-biosensors immobilization on Iron (MNPs) and gold (AuNPs) nanoparticles for detection of Shigella spp, Mater. Sci. Eng. C, 2019, 105, 110113 CrossRef CAS PubMed
.
- J. Xu, Y. Hu, J. Guo, Y. Yang, J. Qiu, X. Li and Z. Xin, A loop-mediated isothermal amplification integrated G-quadruplex molecular beacon
(LAMP-GMB) method for the detection of Staphylococcus aureus in food, Food Anal. Methods, 2019, 12, 422–430 CrossRef
.
- G. Srimongkol, B. Ditmangklo, I. Choopara, J. Thaniyavarn, D. Dean, S. Kokpol, T. Vilaivan and N. Somboonna, Rapid colorimetric loop-mediated isothermal amplification for hypersensitive point-of-care Staphylococcus aureus enterotoxin A gene detection in milk and pork products, Sci. Rep., 2020, 10(1), 7768 CrossRef CAS PubMed
.
- S. Wachiralurpan, T. Sriyapai, S. Areekit, P. Sriyapai, S. Augkarawaritsawong, S. Santiwatanakul and K. Chansiri, Rapid colorimetric assay for detection of Listeria monocytogenes in food samples using LAMP formation of DNA concatemers and gold nanoparticle-DNA probe complex, Front. Chem., 2018, 6, 90 CrossRef PubMed
.
- X. Cao, L. Zhao, J. Zhang, X. Chen, L. Shi, X. Fang, H. Xie, Y. Chang and L. Wang, Food Control, 2019, 103, 145–152 CrossRef CAS
.
- R. V. Saini, P. Vaid, N. K. Saini, S. S. Siwal, V. K. Gupta, V. K. Thakur and A. K. Saini, Recent advancements in the technologies detecting food spoiling agents, J. Funct. Biomater., 2021, 12(4), 67 CrossRef CAS PubMed
.
-
P. K. Mandal and A. K. Biswas, Meat Quality Analysis: Advanced Evaluation Methods, Techniques, and Technologies, 2020, pp. 287–303 Search PubMed
.
- N. Yadav, A. K. Chhillar and J. S. Rana, Sensors International, 2020, 1, 100028 CrossRef
.
-
S. Akgönüllü, D. Çimen, M. Bakhshpour, N. Bereli, H. Yavuz and A. Denizli, Commercial Biosensors and Their Applications: Clinical, Food, and Beyond, 2020, pp. 89–106 Search PubMed
.
- M. Lee, J. H. Song, W. B. Shim and J. Y. Chang, Food Chem., 2020, 333, 127343 CrossRef CAS PubMed
.
- J. Vidic, C. Chaix, M. Manzano and M. Heyndrickx, Food sensing: detection of Bacillus cereus spores in dairy products, Biosensors, 2020, 10(3), 15 CrossRef PubMed
.
- S. Sil, R. Mukherjee, D. Kumbhar, D. Reghu, D. Shrungar, N. S. Kumar, U. K. Singh and S. Umapathy, Raman spectroscopy and artificial intelligence open up accurate detection of pathogens from DNA-based sub-species level classification, J. Raman Spectrosc., 2021, 52(12), 2648–2659 CrossRef CAS
.
- L. A. Rusak, R. de Castro Lisboa Pereira, I. G. Freitag, C. B. Hofer, E. Hofer, M. D. Asensi and D. C. Vallim, J. Microbiol. Methods, 2018, 154, 107–111 CrossRef CAS PubMed
.
- M. R. Hasan, N. Anzar, P. Sharma, S. Singh, H. Hassan, M. R. Hasan, N. Anzar, P. Sharma, S. Singh, H. Hassan, C. Rawat and J. Narang, Mycobacterium tuberculosis diagnosis from conventional to biosensor-a systematic review, Int. J. Environ. Anal. Chem., 2022, 1–16 DOI:10.1080/03067319.2022.2147427
.
- Ö. Akineden, T. Wittwer, K. Geister, M. Plötz and E. Usleber, Food Control, 2020, 109, 106952 CrossRef
.
- C. Hennechart-Collette, O. Dehan, M. Laurentie, A. Fraisse, S. Martin-Latil and S. Perelle, Int. J. Food Microbiol., 2021, 337, 108931 CrossRef CAS PubMed
.
- X. Gao, Z. Wang, Y. Wang, Z. Liu, X. Guan, Y. Ma, H. Zhou, Y. Jiang, W. Cui, L. Wang and Y. Xu, Food Microbiol., 2019, 82, 119–126 CrossRef CAS PubMed
.
- G. Park, J. W. Lim, C. Park, M. Yeom, S. Lee, K. S. Lyoo, D. Song and S. Haam, Biosens. Bioelectron., 2022, 212, 114407 CrossRef CAS PubMed
.
- P. Shankar, J. Mishra, V. Bharti, D. Parashar and S. Singh, Multiplex PCR assay for simultaneous detection and differentiation of Entamoeba histolytica, Giardia lamblia, and Salmonella spp. in the municipality-supplied drinking water, J. Lab. Physicians, 2019, 11(3), 275–280 CrossRef CAS PubMed
.
- J. Plaza, F. Dámek, I. Villena, E. A. Innes, F. Katzer and C. M. Hamilton, Food Waterborne Parasitol., 2020, 20, e00086 CrossRef PubMed
.
- S. Zhang, S. Lin, L. Zhu, Z. Du, J. Li, L. Wang and W. Xu, Sens. Actuators, B, 2022, 372, 132544 CrossRef CAS
.
- Q. Yao, X. Yang, Y. Wang, J. Wang, S. Huang, J. Song and G. Zhao, Animals, 2022, 12, 1953 CrossRef PubMed
.
- Y. Li, S. Man, S. Ye, G. Liu and L. Ma, Compr. Rev. Food Sci. Food Saf., 2022, 21, 3770–3798 CrossRef CAS PubMed
.
- T. King, M. Cole, J. M. Farber, G. Eisenbrand, D. Zabaras, E. M. Fox and J. P. Hill, Food safety for food security: Relationship between global megatrends and developments in food safety, Trends Food Sci. Technol., 2017, 68, 160–175 CrossRef CAS
.
- F. Li, Q. Ye, M. Chen, B. Zhou, J. Zhang, R. Pang, L. Xue, J. Wang, H. Zeng, S. Wu, Y. Zhang, Y. Ding and Q. Wu, An ultrasensitive CRISPR/Cas12a based electrochemical biosensor for Listeria monocytogenes detection, Biosens. Bioelectron., 2021, 179, 113073 CrossRef CAS PubMed
.
- J. S. Gootenberg, O. O. Abudayyeh, M. J. Kellner, J. Joung, J. J. Collins and F. Zhang, Joung J. Collins JJ Zhang F., Science, 2018, 360, 439–444 CrossRef CAS PubMed
.
- L. Wang, X. Shen, T. Wang, P. Chen, N. Qi, B. C. Yin and B. C. Ye, A lateral flow strip combined with Cas9 nickase-triggered amplification reaction for dual food-borne pathogen detection, Biosens. Bioelectron., 2020, 165, 112364 CrossRef CAS PubMed
.
- Q. He, D. Yu, M. Bao, G. Korensky, J. Chen, M. Shin, J. Kim, M. Park, P. Qin and K. Du, High-throughput and all-solution phase African Swine Fever Virus (ASFV) detection using CRISPR-Cas12a and fluorescence based point-of-care system, Biosens. Bioelectron., 2020, 154, 112068 CrossRef CAS PubMed
.
- R. Hajian, S. Balderston, T. Tran, T. deBoer, J. Etienne, M. Sandhu, N. A. Wauford, J. Y. Chung, J. Nokes, M. Athaiya, J. Paredes, R. Peytavi, B. Goldsmith, N. Murthy, I. M. Conboy and K. Aran, Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor, Nat. Biomed. Eng., 2019, 3, 427–437 CrossRef CAS PubMed
.
- H. Wu, Y. Chen, Q. Yang, C. Peng, X. Wang, M. Zhang, S. Qian, J. Xu and J. Wu, A reversible valve-assisted chip coupling with integrated sample treatment and CRISPR/Cas12a for visual detection of Vibrio parahaemolyticus, Biosens. Bioelectron., 2021, 188, 113352 CrossRef CAS PubMed
.
- Y. Li, S. Li, J. Wang and G. Liu, Trends Biotechnol., 2019, 37, 730–743 CrossRef CAS PubMed
.
- X. Sun, Y. Wang, L. Zhang, S. Liu, M. Zhang, J. Wang, B. Ning, Y. Peng, J. He, Y. Hu and Z. Gao, CRISPR-Cas9 triggered two-step isothermal amplification method for E. coli O157: H7 detection based on a metal–organic framework platform, Anal. Chem., 2020, 92(4), 3032–3041 CrossRef CAS PubMed
.
- C. M. Ackerman, C. Myhrvold, S. G. Thakku, C. A. Freije, H. C. Metsky, D. K. Yang, S. H. Ye, C. K. Boehm, T. S. F. Kosoko-Thoroddsen, J. Kehe, T. G. Nguyen, A. Carter, A. Kulesa, J. R. Barnes, V. G. Dugan, D. T. Hung, P. C. Blainey and P. C. Sabeti, Massively multiplexed nucleic acid detection with Cas13, Nature, 2020, 582(7811), 277–282 CrossRef CAS PubMed
.
- C. Jiao, S. Sharma, G. Dugar, N. L. Peeck, T. Bischler, F. Wimmer, Y. Yu, L. Barquist, C. Schoen, O. Kurzai, C. M. Sharma and C. L. Beisel, Noncanonical crRNAs derived from host transcripts enable multiplexable RNA detection by Cas9, Science, 2021, 372(6545), 941–948 CrossRef CAS PubMed
.
- Y. Xiong, J. Zhang, Z. Yang, Q. Mou, Y. Ma, Y. Xiong and Y. Lu, Functional DNA regulated CRISPR-Cas12a sensors for point-of-care diagnostics of non-nucleic-acid targets, J. Am. Chem. Soc., 2019, 142(1), 207–213 CrossRef PubMed
.
- S. Stella, P. Alcón and G. Montoya, Structure of the Cpf1 endonuclease R-loop complex after target DNA cleavage, Nature, 2017, 546(7659), 559–563 CrossRef CAS PubMed
.
- L. Yin, S. Man, S. Ye, G. Liu and L. Ma, CRISPR-Cas based virus detection: Recent advances and perspectives, Biosens. Bioelectron., 2021, 193, 113541 CrossRef CAS PubMed
.
- K. Abnous, N. M. Danesh, M. Ramezani, M. Alibolandi, M. A. Nameghi, T. S. Zavvar and S. M. Taghdisi, A novel colorimetric aptasensor for ultrasensitive detection of aflatoxin M1 based on the combination of CRISPR-Cas12a, rolling circle amplification and catalytic activity of gold nanoparticles, Anal. Chim. Acta, 2021, 1165, 338549 CrossRef CAS PubMed
.
- D. Huang, J. Qian, Z. Shi, J. Zhao, M. Fang and Z. Xu, CRISPR-Cas12a-assisted multicolor biosensor for semiquantitative point-of-use testing of the nopaline synthase terminator in genetically modified crops by unaided eyes, ACS Synth. Biol., 2020, 9(11), 3114–3123 CrossRef CAS PubMed
.
- B. Qiao, J. Xu, W. Yin, W. Xin, L. Ma, J. Qiao and Y. Liu, “Aptamer-locker” DNA coupling with CRISPR/Cas12a-guided biosensing for high-efficiency melamine analysis, Biosens. Bioelectron., 2021, 183, 113233 CrossRef CAS PubMed
.
- M. Zhang, C. Liu, Y. Shi, J. Wu, J. Wu and H. Chen, Selective endpoint visualized detection of Vibrio parahaemolyticus with CRISPR/Cas12a assisted PCR using thermal cycler for on-site application, Talanta, 2020, 214, 120818 CrossRef CAS PubMed
.
- 2018 NARMS Update: Integrated Report Summary Interactive Version|FDA, https://www.fda.gov/animal-veterinary/national-antimicrobial-resistance-monitoring-system/2018-narms-update-integrated-report-summary-interactive-version, accessed 17 August 2022.
- K. A. T. Verheijden, Digesting the role of specific pre-and synbiotics in the prevention of house dust mite asthma: Thinking out of the lung, Doctoral dissertation, Utrecht University, 2015 Search PubMed.
- J. Quan, C. Langelier, A. Kuchta, J. Batson, N. Teyssier, A. Lyden, S. Caldera, A. McGeever, B. Dimitrov, R. King, J. Wilheim, M. Murphy, L. P. Ares, K. A. Travisano, R. Sit, R. Amato, D. R. Mumbengegwi, J. L. Smith, A. Bennett, R. Gosling, P. M. Mourani, C. S. Calfee, N. F. Neff, E. D. Chow, P. S. Kim, B. Greenhouse, J. L. DeRisi and E. D. Crawford, FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences, Nucleic Acids Res., 2019, 47(14), e83 CrossRef CAS PubMed
.
- L. Peng, J. Zhou, G. Liu, L. Yin, S. Ren, S. Man and L. Ma, CRISPR-Cas12a based aptasensor for sensitive and selective ATP detection, Sens. Actuators, B, 2020, 320, 128164 CrossRef CAS
.
- M. Mwanza, A. Abdel-Hadi, A. M. Ali and M. Egbuta, Evaluation of analytical assays efficiency to detect aflatoxin M1 in milk from selected areas in Egypt and South Africa, J. Dairy Sci., 2015, 98(10), 6660–6667 CrossRef CAS PubMed
.
- G. Bindal, R. Krishnamurthi, A. S. N. Seshasayee and D. Rath, CRISPR-Cas-mediated gene silencing reveals RacR to be a negative regulator of YdaS and YdaT toxins in Escherichia coli K-12, mSphere, 2017, 2(6), 10–1128 CrossRef PubMed
.
- Q. Fu, S. Li, Z. Wang, W. Shan, J. Ma, Y. Cheng, H. Wang, Y. Yan and J. Sun, H-NS Mutation-Mediated CRISPR-Cas activation inhibits phage release and toxin production of Escherichia coli Stx2 phage lysogen, Food Microbiol., 2017, 8, 652 Search PubMed
.
- X. Wang, Y. Xu, J. Huang, W. Jin, Y. Yang and Y. Wu, CRISPR-mediated knockout of the ABCC2 gene in Ostrinia furnacalis confers high-level resistance to the Bacillus thuringiensis Cry1Fa toxin, Toxins, 2020, 12(4), 246 CrossRef CAS PubMed
.
- J. Li, S. Yang, C. Zuo, L. Dai, Y. Guo and G. Xie, Applying CRISPR-Cas12a as a signal amplifier to construct biosensors for non-DNA targets in ultralow concentrations, ACS Sens., 2020, 5(4), 970–977 CrossRef CAS PubMed
.
- J. Liu, J. Chen, D. Wu, M. Huang, J. Chen, R. Pan, Y. Wu and G. Li, CRISPR-/Cas12a-mediated liposome-amplified strategy for the surface-enhanced Raman scattering and naked-eye detection of nucleic acid and application to food authenticity screening, Anal. Chem., 2021, 93(29), 10167–10174 CrossRef CAS PubMed
.
- S. Stidham, V. Villareal, V. Chellappa, L. Yoder, O. Alley, W. Shreffler, J. Spergel, D. Fleischer, H. Sampson and A. Gilboa-Geffen, Aptamer based point of care diagnostic for the detection of food allergens, Sci. Rep., 2022, 12(1), 1303 CrossRef CAS PubMed
.
- J. Shen, X. Zhou, Y. Shan, H. Yue, R. Huang, J. Hu and D. Xing, Sensitive detection of a bacterial pathogen using allosteric probe-initiated catalysis and CRISPR-Cas13a amplification reaction, Nat. Commun., 2020, 11(1), 267 CrossRef CAS PubMed
.
- A. Kostyusheva, S. Brezgin, Y. Babin, I. Vasilyeva, D. Glebe, D. Kostyushev and V. Chulanov, CRISPR-Cas systems for diagnosing infectious diseases, Methods, 2022, 203, 431–446 CrossRef CAS PubMed
.
- C. Escalona-Noguero, M. López-Valls and B. Sot, CRISPR/Cas technology as a promising weapon to combat viral infections, BioEssays, 2021, 43(4), 2000315 CrossRef CAS PubMed
.
- M. Wang, R. Zhang and J. Li, CRISPR/cas systems redefine nucleic acid detection: principles and methods, Biosens. Bioelectron., 2020, 165, 112430 CrossRef CAS PubMed
.
- B. Li, S. M. Clohisey, B. S. Chia, B. Wang, A. Cui, T. Eisenhaure, L. D. Schweitzer, P. Hoover, N. J. Parkinson, A. Nachshon, N. Smith, T. Regan, D. Farr, M. U. Gutmann, S. I. Bukhari, A. Law, M. Sangesland, I. Gat-Viks, P. Digard, S. Vasudevan, D. Lingwood, D. H. Dockrell, J. G. Doench, J. K. Baillie and N. Hacohen, Genome-wide CRISPR screen identifies host dependency factors for influenza A virus infection., Nat. Commun., 2020, 11(1), 164 CrossRef CAS PubMed
.
- J. W. Han, L. Ruiz-Garcia, J. P. Qian and X. T. Yang, Food packaging: A comprehensive review and future trends, Compr. Rev. Food Sci. Food Saf., 2018, 17, 860–877 CrossRef PubMed
.
- CRISPR in Agriculture: An Era of Food Evolution, https://www.synthego.com/blog/crispr-agriculture-foods, accessed 23 July 2021.
- R. Barrangou and R. A. Notebaart, CRISPR-directed microbiome manipulation across the food supply chain, Trends Microbiol., 2019, 27, 489–496 CrossRef CAS PubMed
.
- O. Mukama, J. Wu, Z. Li, Q. Liang, Z. Yi, X. Lu, Y. Liu, Y. Liu, M. Hussain, G. G. Makafe, J. Liu, N. Xu and L. Zeng, An ultrasensitive and specific point-of-care CRISPR/Cas12 based lateral flow biosensor for the rapid detection of nucleic acids, Biosens. Bioelectron., 2020, 159, 112143 CrossRef CAS PubMed
.
- L. Yin, N. Duan, S. Chen, Y. Yao, J. Liu and L. Ma, Ultrasensitive pathogenic bacteria detection by a smartphone-read G-quadruplex-based CRISPR-Cas12a bioassay, Sens. Actuators, B, 2021, 347, 130586 CrossRef CAS
.
-
J. H. Lau, in Fan-Out Wafer-Level Packaging, 2018 Search PubMed
.
- X. Wang, X. Shang and X. Huang, Next-generation pathogen diagnosis with CRISPR/Cas-based detection methods, Emerging Microbes Infect., 2020, 9(1), 1682–1691 CrossRef CAS PubMed
.
- 4 Ways AI is changing the packaging industry|Monolith AI, https://www.monolithai.com/post/4-ways-ai-is-changing-the-packaging-industry, accessed 23 July 2021.
- L. Zhou, C. Zhang, F. Liu, Z. Qiu and Y. He, Application of deep learning in food: a review, Compr. Rev. Food Sci. Food Saf., 2019, 18, 1793–1811 CrossRef PubMed
.
- J. Zheng, L. Zou and Z. Jane Wang, Mid-level deep food part mining for food image recognition, IET Comput. Vis., 2018, 12(3), 298–304 CrossRef
.
- W. Tan, C. Zhao and H. Wu, Robotic kiwifruit harvesting using machine vision, convolutional neural networks, and robotic arms, Multimed. Tools Appl., 2019, 181, 140–156 Search PubMed
.
- X. Yu, H. Lu and D. Wu, Development of deep learning method for predicting firmness and soluble solid content of postharvest Korla fragrant pear using Vis/NIR hyperspectral reflectance imaging, Postharvest Biol. Technol., 2018, 141, 39–49 CrossRef
.
- Z. Wang, M. Hu and G. Zhai, Application of deep learning architectures for accurate and rapid detection of internal mechanical damage of blueberry using hyperspectral transmittance data, Sensors, 2018, 18(4), 1126 CrossRef PubMed
.
- W. Zhang, Y. Zhang, J. Zhai, D. Zhao, L. Xu, J. Zhou, Z. Li and S. Yang, Multi-source data fusion using deep learning for smart refrigerators, Comput. Ind., 2018, 95, 15–21 CrossRef
.
- P. Pandey, A. Deepthi, B. Mandal and N. B. Puhan, FoodNet: recognizing foods using ensemble of deep networks, IEEE Signal Process. Lett., 2017, 24(12), 1758–1762 Search PubMed
.
- F. J. Rodríguez, A. García, P. J. Pardo, F. Chávez and R. M. Luque-Baena, Study and classification of plum varieties using image analysis and deep learning techniques, Prog. Artif. Intell., 2018, 7, 119–127 CrossRef
.
-
L. M. rifatul Azizah, S. F. Umayah, S. Riyadi, C. Damarjati and N. A. Utama, in Proceedings – 7th IEEE International Conference on Control System, Computing and Engineering, ICCSCE, vol. 2017, November 2017 Search PubMed
.
- X. Yu, L. Tang, X. Wu and H. Lu, Nondestructive freshness discriminating of shrimp using visible/near-infrared hyperspectral imaging technique and deep learning algorithm, Food Anal. Methods, 2018, 11, 768–780 CrossRef
.
- X. Yu, J. Wang, S. Wen, J. Yang and F. Zhang, A deep learning based feature extraction method on hyperspectral images for nondestructive prediction of TVB-N content in Pacific white shrimp (Litopenaeus vannamei), Biosyst. Eng., 2019, 178, 244–255 CrossRef
.
- M. Al-Sarayreh, M. M. Reis, W. Q. Yan and R. Klette, Detection of red-meat adulteration by deep spectral–spatial features in hyperspectral images, J. Imaging, 2018, 4(5), 63 CrossRef
.
- H. A. Neto, W. L. F. Tavares, D. C. S. Z. Ribeiro, R. C. O. Alves, L. M. Fonseca and S. V. A. Campos, On the utilization of deep and ensemble learning to detect milk adulteration., BioData Min., 2019, 12(1), 1–13 CrossRef PubMed
.
- P. Visciano and M. Schirone, Food frauds: global incidents and misleading situations, Trends Food Sci. Technol., 2021, 114, 424–442 CrossRef CAS
.
- D. Mao, F. Wang, Z. Hao and H. Li, Information asymmetry, blockchain and food safety, Int. J. Environ. Res. Public Health, 2016, 11(1), 53–56 Search PubMed
.
- Y. Liu, H. Pu and D. W. Sun, Efficient extraction of deep image features using convolutional neural network (CNN) for applications in detecting and analysing complex food matrices, Trends Food Sci. Technol., 2021, 113, 193–204 CrossRef CAS
.
-
FAO, Climate Change and Food Systems: Global Assessments and Implications for Food Security and Trade, 2015 Search PubMed
.
- K. D. Hall, J. Guo, M. Dore and C. C. Chow, The progressive increase of food waste in America and its environmental impact, PLoS One, 2009, 4(11), e7940 CrossRef PubMed
.
- V. Guillard, S. Gaucel, C. Fornaciari, H. Angellier-Coussy, P. Buche and N. Gontard, The next generation of sustainable food packaging to preserve our environment in a circular economy context, Front. Nutr., 2018, 5, 121 CrossRef PubMed
.
- C. Batello, O. Jan, C. Tostivint, A. Turbé, C. O’Connor, P. Lavelle, A. Flammini, N. El-H. Scialabba, J. Hoogeveen, M. Iweins and F. Tubiello, Food Wastage Footprint: Impacts on Natural Resources, 2013 Search PubMed.
- Y.-C. Wang, Biosensors and intelligent packaging to improve food safety, J. Anim. Sci., 2020, 98, 64 CrossRef
.
- Estimating the burden of foodborne diseases, https://www.who.int/activities/estimating-the-burden-of-foodborne-diseases, accessed 17 August 2022.
- J. Coronel-Reyes, I. Ramirez-Morales, E. Fernandez-Blanco, D. Rivero and A. Pazos, Determination of egg storage time at room temperature using a low-cost NIR spectrometer and machine learning techniques, Comput. Electron. Agric., 2018, 145, 1–10 CrossRef
.
| This journal is © The Royal Society of Chemistry 2023 |