In silico environmental risk assessment of fate and effects of pharmaceuticals and their TPs generated and treated by coupling tertiary processes in hospital wastewater†
Alexandre
Della-Flora
,
Davi
Scunderlick
,
Marcelo L.
Wilde
,
Adriano
de A. Gomes
,
Eder C.
Lima
 and
Carla
Sirtori
and
Carla
Sirtori
 *
*
Instituto de Química, Universidade Federal do Rio Grande do Sul, Av. Bento Gonçalves 9500, CEP 91501-970, Porto Alegre, RS, Brazil. E-mail: carla.sirtori@ufrgs.br
First published on 5th December 2022
Abstract
The solar photo-Fenton process leads to the formation of transformation products (TPs) that are new compounds with an unknown chemical, physical, and biological characteristics. There are no commercial analytical standards for these new TPs. Due to this difficulty, ecotoxicity studies have been able to provide only limited information, while intrinsic features of the samples (such as complexity and matrix effects), as well as issues related to the sensitivity of the test organism, can further hinder investigations in cases where TPs coexist in mixtures. However, a viable option is to carry out in silico studies that do not use analytical standards or living organisms and do not require long testing periods to reach endpoints. In the present work, 90 chemical compounds (pharmaceuticals and their TPs generated by the solar photo-Fenton process) were evaluated using freely available software to assess the following endpoints: biodegradability, carcinogenicity, mutagenicity, and PBT (persistence, bioaccumulation, and toxicity). Chemometric analyses (HCA and PCA) were also applied to enhance the interpretation of the results. The main findings were that flutamide, chloramphenicol, and nimesulide, together with their TPs, were mostly non-biodegradable compounds that could be potentially mutagenic and carcinogenic. Therefore, these drugs and their TPs should not be released into the environment. The results indicated that it could be important to improve the solar photo-Fenton process or to couple this Advanced Oxidation Process with additional physicochemical treatment in order to efficiently remove these organic microcontaminants.
Water impactThe present manuscript evaluates the both following aspects: – the release of transformation products (TPs) in treated effluents by AOPs could have negative impacts on aquatic organisms, causing adverse effects in ecosystems. In this situation, the use of in silico prediction models, employing (Q)SAR tools, can provide an initial assessment of TP ecotoxicity. This can reveal the ideal final point of the treatment process, as indicated by the formation or presence of TPs predicted to be nontoxic. Chemometric analysis (HCA and PCA) facilitates the interpretation of (Q)SAR results, since the compounds are organized in clusters. It is then possible to determine the best strategy to adopt (to extend the treatment time, to optimize more drastic treatment conditions, or to identify a suitable additional coupled process). |
1. Introduction
Due to the limitations of the solar photo-Fenton (SPF) treatment process, its association with adsorption employing activated carbon seems to be one way to reduce the release of undegraded parent compounds and their transformation products (TPs) into the environment.1 This is essential since TPs are new compounds with unknown physicochemical characteristics, whose toxicities may be equal to, or greater than, those of the parent compounds, and they may also be persistent and resistant towards degradation.2,3 The assessment of TPs and many drugs have been neglected because most of them are not considered in legislation worldwide. Treated effluents are commonly discharged without any qualitative or quantitative control of these organic microcontaminants, whether before, during, or after treatment.4,5 In addition, the presence of pharmaceuticals in effluents is worrying because some compounds, such as fluconazole, Furosemide, Losartan, and Nimesulide, are not easily removed by conventional wastewater treatments.6–12 Additionally, the TPs generated during the new treatment technologies can maintain characteristics, be resistant to conventional removal treatments, and even present other properties.13 It is evident that the characterization and identification of TPs are fundamental for predicting the environmental risks that they might present. Due to the lack of analytical standards, the difficulty of analysis makes this process more complicated, necessitating the use of sophisticated analytical equipment where chromatographic separation is coupled with high-resolution mass spectrometry (HRMS). Orbitrap, time-of-flight (TOF), and hybrid quadrupole-time-of-flight (QTOF) analyzers are often used to elucidate the proposed chemical structures of unknown compounds (such as TPs) since these systems offer high mass resolution capacity.14,15 Studies have been published with proposals for new TPs found during different Advanced Oxidation treatments, supported by these robust tools.16,17 However, it is very difficult to evaluate the toxicities of the TPs in vitro due to the lack of analytical standards and, often, the presence of “mixtures” of TPs.18 After an advanced treatment process, an ecotoxicity assessment of a mixture may indicate that the final mixture is less toxic than the initial mixture, giving a general indication of decreased effluent toxicity. This can sometimes lead to an incorrect conclusion since the concentrations of the chemical compounds, especially the initial compounds, have only been reduced or transformed by the treatment. Even if a mixture that contains TPs does not present toxicity in vitro, it is important to note that these TPs are unknown compounds and that their concentrations could be too low to alter the physiological functions of the test organisms. Therefore, a better approach would be to evaluate the toxicological effects individually (for each TP), using in silico prediction models that avoid any need for TP analytical standards and enable rapid prediction of different endpoints for each TP.18–20In silico (quantitative) structure–activity relationship ((Q)SAR) prediction models are mathematical models that describe the relationship between molecular structures and their related properties, comparing the chemical structure of each studied compound with those of compounds included in databases that present results for endpoints with a high degree of certainty.21 These in silico models offer easy applicability for the prediction of only one endpoint, such as biodegradability,22 or a range of endpoints, such as toxicity towards different organisms, physicochemical parameters, carcinogenicity, and mutagenicity.16,23–25 The main advantages of (Q)SAR models are that these tools do not require lengthy experiments or tests employing animals, plants, or other living organisms. In the case of TPs from AOPs, (Q)SAR tools have excellent potential to be used routinely, especially in laboratories with little infrastructure for ecotoxicity and molecular biology studies. Furthermore, the use of these models in a step prior to performing bioassays could assist in selecting appropriate experimental procedures and in vitro tests.
The present study aimed to perform a risk assessment and environmental fate analysis for selected pharmaceuticals and their TPs generated during a solar photo-Fenton (SPF) process coupled with adsorption, applied as a treatment for a hospital wastewater matrix. The following endpoints were analyzed using different (Q)SAR model software packages: biodegradability, carcinogenicity, mutagenicity, and PBT (persistence, bioaccumulation, and toxicity).
2. Materials and methods
2.1 In silico quantitative structure–activity relationship ((Q)SAR) model predictions
In order to perform in silico evaluation of the selected pharmaceuticals (chloramphenicol, fluconazole, flutamide, furosemide, gemfibrozil, ibuprofen, losartan, nimesulide, and paracetamol) and their TPs, the first step was to transform the chemical structure of each compound into computational language. For this, ChemBioDraw Ultra (v.12) was used to transform the structures into SMILES codes (Table S1, ESI†). All the possible structures were considered for those TPs that showed more than one possible chemical structure (such as constitutional isomers). In some cases, it was not possible to elucidate the chemical structure of the unknown TP (for example, the exact position of a double bond or the position where the hydroxyl group occurs in an aromatic ring) based only on the results provided by the LC-QTOFMS system. Fig. S1–S7 (ESI†) show all the possible structures of the TPs evaluated in this study.| Model | Endpoint | Description | Database |
|---|---|---|---|
| BIOWIN 1 (linear) | <0.5 not rapidly biodegradable≥0.5 fast biodegradability | The assessment of biological degradation for the generation of metabolites | Total 295 chemicals |
| BIOWIN 2 (nonlinear) | |||
| BIOWIN 3 (linear) | ≤1.000 longer than months1.000–1.999 months and longer 2.000 months 2.001–2.999 weeks to months 3.000 weeks 3.001–3.999 days to weeks 4.000 days 4.001–4.999 hours to days ≥5.000 hours |
The evaluation of mineralization by biological degradation and transformation of the parent compound into carbon dioxide, water, and inorganic salts, and generation of metabolites | Total 200 chemicals |
| BIOWIN 4 (nonlinear) | |||
| BIOWIN 5 (linear) | <0.5 not rapidly biodegradable≥0.5 fast biodegradability | Assessment of whether the compound is readily biodegradable or not | Total 884 chemicals |
| BIOWIN 6 (nonlinear) | |||
| BIOWIN 7 | <0.5 not rapidly biodegradable≥0.5 fast biodegradability | Evaluation of biodegradation under anaerobic conditions using the serum bottle test | Total 169 chemicals |
| Ready biodegradability model |
RB – readily biodegradablepRB – possibly readily biodegradable pnRB – possibly not readily biodegradable nRB – not readily biodegradable |
This model is based on the OECD TG 301C modified MITI I test. It assesses the possibility of the compound being biodegradable for 28 days in a bacteria-rich environment | Total 486 chemicals |
2.2 Chemometric analysis
Multivariate classification analysis was used to identify similarities among the endpoints evaluated for this study's (Q)SAR models. For this purpose, the PCA Toolbox (v.1.5) tool available in MatlabR2012 software30 was employed to perform hierarchical cluster analysis (HCA) and principal component analysis (PCA). The construction of the data matrix to perform HCA and PCA was achieved by assembling a spreadsheet with endpoints from all the (Q)SAR models evaluated in this work. The results were converted into binary numbers for the biodegradability analyses using the BIOWIN 1–7 models, with ‘1’ for non-biodegradable compounds and ‘0’ for biodegradable compounds. Similarly, for the mutagenicity and carcinogenicity models, the response was obtained using the sum of the numbers of the alerts. For each positive alert, a weight of ‘1’ was applied, while a weight of ‘0’ was applied for compounds that did not have alerts. This procedure was adopted because the mutagenicity and carcinogenicity models indicated which fragments could be potentially mutagenic or carcinogenic without responding to the form of numerical values. Accordingly, the results were presented in a 90 × 25 matrix, where the lines corresponded to the pharmaceuticals and the TPs, while the columns contained the endpoints of the ecotoxicity models (Table S3, ESI†). The data were scaled using the mean centering method for the HCA and PCA analyses. Construction of the HCA dendrogram used Euclidean distances and single links between clusters.3. Results and discussion
3.1 Biodegradability
The environmental fates of the pharmaceuticals and their TPs were evaluated considering their persistence and resistance to biodegradation, using the BIOWIN 1–7 models complemented with the Ready Biodegradability model (IRFMN). The BIOWIN 1–6 models consider aerobic conditions, while the BIOWIN 7 model assumes anaerobic conditions. It should be noted that the models are not directly comparable since they are each designed for a certain test or a specific type of biodegradation (applicability domain). For example, the BIOWIN 5 and IRFMN models are comparable with the OECD 301C biodegradation model.26 In turn, the BIOWIN 3 and 4 models provide final and partial biodegradation results, respectively, and the BIOWIN 7 model provides results under anaerobic conditions. The results for the predictions of persistence and biodegradability are shown in Table S2 (ESI†).When the antibiotic chloramphenicol (CLO) is ingested, around 5–10% is excreted unchanged. However, after ingestion, it is transformed into chloramphenicol glucuronide, reaching almost 90%. However, after being excreted in the metabolized form, chloramphenicol glucuronide can be transformed back to the parent compound.31 It has been reported that using this antibiotic can lead to resistance in bacteria.32,33 Furthermore, this pharmaceutical is not fully removed in sewage treatment plants, where the elimination efficiency is approximately 45%.34 The BIOWIN 5 and VEGA IRFMN models indicated that CLO was not biodegradable, which could have been because the deactivating aromatic nitro group prevents the action of enzymes.35 The same was observed for the TPs of CLO that contained the nitro group in the aromatic ring. However, CLO and its TPs possess electron-donating groups, such as hydroxyl groups, that are susceptible to enzymatic hydrolysis and can increase aerobic biodegradability.35
Nimesulide (NMS), a non-steroidal anti-inflammatory drug, is metabolized in the liver, with up to 3% of the parent compound being excreted in the urine.36 It has been reported that low NMS removal rates in biodegradation processes are due to the presence of two aromatic rings in its chemical structure.37 Boethling et al.35 considered that the presence of two functional groups (aromatic nitro and arylamino) increases the resistance of NMS to biodegradation because electron-withdrawing groups prevent the action of enzymes. Consequently, the BIOWIN 5 and VEGA IRFMN models indicated that NMS and its TPs were non-biodegradable compounds.
Losartan (LOS) has been reported to present low removal rates of around 33% in wastewater treatment plants (WWTPs).6 Another study reported that LOS is a compound that is difficult to remove in WWTPs.7 These difficulties in the removal of LOS using biological treatments were predicted by the BIOWIN 5 and VEGA IRFMN in silico models. The biodegradation may be hindered by the presence of chlorine in the aromatic ring since this substituent strongly removes electrons from the aromatic ring, hindering the action of enzymes.35 Since LOS TP14 did not possess chlorine in the aromatic ring, it could finally be biodegraded, as indicated by the BIOWIN 3 model. However, the BIOWIN 5 and VEGA IRFMN models found that LOS and all the LOS TPs observed in this study were not biodegradable.
Furosemide (FRS) is mainly excreted in the form of the parent compound (around 90%). Park et al.8 evaluated the efficiencies of FRS removal by membrane bioreactor treatment and biological processes commonly employed in WWTPs. The results indicated that FRS was not removed by conventional treatment, while a low removal rate was achieved using biological processes. This recalcitrance could be attributed to the presence of chlorine in the FRS aromatic ring.35 The in silico biodegradability predictions obtained for FRS and its TPs are presented in Table S2 (ESI†). The compounds containing chlorine in their structures (FRS and FRS TP3) were typically found to be non-biodegradable by all the models, while FRS TP12 and FRS TP14 were found to be readily biodegradable, according to the VEGA IRFMN model. As expected, these TPs did not possess chlorine in their structures.
Fluconazole (FCZ) is a pharmaceutical used to treat fungal infections. The literature reports that biological treatments provide low FCZ removal rates.9–12 The in silico biodegradability results indicated that FCZ and its TPs containing halogens in the aromatic rings were non-biodegradable molecules. Even after replacing an electron-withdrawing group with an electron donor moiety (eliminating the fluorine and adding a hydroxyl group), FCZ TP2 remained non-biodegradable.
Paracetamol (PCT) is an analgesic widely used globally, especially because it can be purchased without needing a medical prescription. As a consequence, it is commonly found in surface waters and wastewaters.38,39 PCT is easily degraded under aerobic conditions40 and readily eliminated in WWTPs, with high removal percentages.41–44 The in silico biodegradability predictions showed that PCT and its TPs were biodegradable compounds. This could be explained by the presence of the amide functional group, which facilitates enzymatic hydrolysis.35,45
Gemfibrozil (GFZ) is a lipid regulator that is not completely eliminated by WWTP processes.46–48In silico biodegradability predictions using BIOWIN 5 and VEGA IRFMN identified GFZ as being non-biodegradable. On the other hand, GFZ TP8 was classified as readily biodegradable by VEGA IRFMN and non-biodegradable by BIOWIN 5.
Ibuprofen (IBP) is mainly used to treat pain, inflammation, and fever. After ingestion, excretion can account for up to 85% of the original compound.45,48–51 Biological treatments easily eliminate IBP, which can be explained by the presence of electron donor groups in itsstructure.35,45 Despite being readily removed in WWTPs, the predictions of the BIOWIN 5 and VEGA IRFMN models classified IBP as non-biodegradable. The VEGA IRFMN model associated this recalcitrance with the presence of the 1,4-diethylbenzene moiety.
Flutamide (FLT) is a drug used to treat prostate cancer. A previous study using in silico predictions reported the biodegradability of FLT and its TPs.25 Here, FLT and its TPs were classified as non-biodegradable compounds, according to the BIOWIN 5 and VEGA IFRMN models.
3.2 Mutagenicity
Mutagenicity predictions were obtained using the eleven available models, evaluating the structural alerts for each endpoint, as shown in Fig. 1. Table S4 (ESI†) presents the final results of each model, indicating whether the compounds were potentially mutagenic or not. Additionally, the model predictions obtained using QSAR Toolbox provided structural alerts (SAs).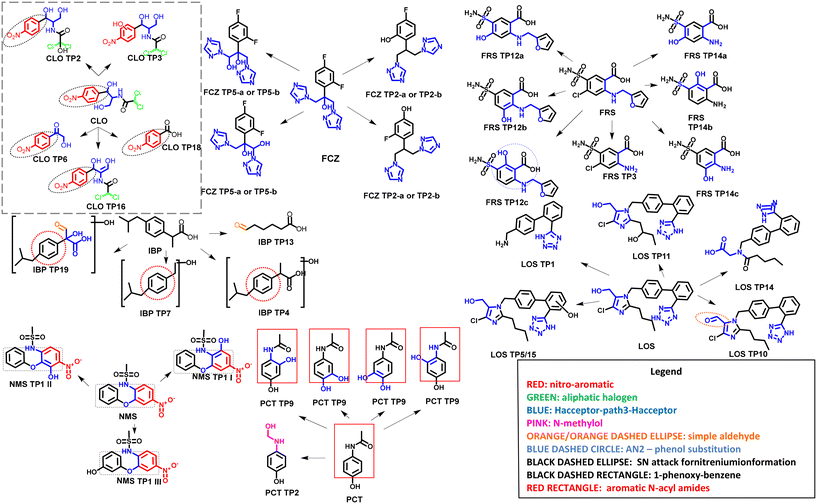 | ||
| Fig. 1 Main in silico structural alerts predicted by (Q)SAR toolbox, CASE ULTRA, and VEGA HUB software for the chemical compounds (pharmaceuticals and TPs) analyzed. | ||
Several compounds, such as CLO and its TPs, could have more than one SA. These compounds presented the aromatic nitro SA, Hacceptor-path3-Hacceptor, while TPs that retained chlorine in their structures presented the aliphatic halogens SA. The aromatic nitro SA represented 19% of the alerts found in this study, with the presence of this SA being observed for 7 of the 11 models.52 The mutagenic potential is associated with the metabolization of the aromatic nitro moiety, which is reduced to a nitronium radical and stabilized by resonance. This radical can reduce molecular oxygen, producing toxic oxygen species interacting with DNA and changing its structure. There are two mechanisms proposed for this type of SA. The SN1 reaction mechanism involves nucleophilic attack after the reduction and formation of the nitrene ion. The other possible mechanism is radical-mediated and involves indirect reactive oxygen species (ROS) formation in the p-nitrobenzene position.53,54
The aliphatic halogen SA was observed for 4% of the evaluated compounds in 7 of the 11 models. This SA is observed for compounds containing the halogens Cl, Br, and I, but not fluorine (F) since F is not biologically active.55,56 Although the mechanism of toxicity related to aliphatic halogen is unclear, this group can react with proteins and DNA. In addition, the presence of halogens can favor biotransformation with glutathione (GSH), which can increase toxicity. Aliphatic halogen compounds may also interact with cytochrome P450 to form aldehydes, resulting in an oxidizing metabolite that can cause cell damage.55,56
The Hacceptor-path3-Hacceptor SA indicates the potential of the compound to undergo interactions with DNA and/or proteins. This SA was found for a majority (67%) of the compounds evaluated here. For FCZ and its TPs, it was the only SA present.
The KNN/Read-Across model does not have SAs as endpoints. Instead, it compares the structure with a database containing 5770 compounds and indicates whether the compound is potentially mutagenic. For FCZ and its TPs, the KNN/Read-Across model indicated mutagenic potential. Although in silico evaluation showed the potential mutagenicity of FCZ, the minimum dose for this effect is unknown. In work by Silva et al.,57 using human mononuclear blood cells, it was found that DNA damage occurred above a concentration of 30 μg mL−1, while a mutagenic effect occurred above 6 μg mL−1.
The same Hacceptor-path3-Hacceptor SA observed for FCZ was also found for FRS and its TPs. In addition, FRS TP12(c) presented a possible nucleophilic addition SA (AN2) related to clastogenicity, due to substitutions by phenols in chromosomes.58
GFZ showed no SA for mutagenicity, and the same was observed for GFZ TP8 (they are not shown in Fig. 1). Leusch et al.59 performed in vivo mutagenicity tests for 39 chemical contaminants, among which GFZ presented a negative result.
For IBP and its TPs, SAs were only observed for some of the TPs. IBP TP13 and IBP TP19 presented the simple Aldehyde SA, while IBP TP19(a,c,e,f,g,h) also showed the H-acceptor-path3-H-acceptor SA. For the TPs IBP TP4(a,b,c,d), IBP TP7(c,d,e,f,j), and IBP TP19(a,c), there was the possibility of nucleophilic addition (AN2) when the hydroxyl moiety was found in the aromatic ring. The Simple Aldehyde SA was observed using 4 models for about 9% of the compounds evaluated in this study. This SA reflects the potential to form a Schiff base when reacting with a primary amine, which can bond to DNA and damage its structure.60
In the case of LOS and its TPs, the main SA found was Hacceptor-path3-Hacceptor. Additional SAs observed were nucleophilic addition (AN2) for LOS TP5 and LOS TP15 and Simple Aldehyde for LOS TP10. Although the in silico results for some of the models showed mutagenicity alerts for LOS and its TPs, previous in vivo and in vitro studies had reported an absence of genotoxic effects for LOS.61,62
Studies carried out using rats have evaluated the possible mutagenic effects of NMS, finding that this drug is potentially genotoxic.63,64 Here, the in silico results showed that NMS and its TPs presented SAs for mutagenicity, with the aromatic nitro SA being the most prevalent. In addition, the H-acceptor-path3-H-acceptor and 1-phenoxy-benzene SAs were also observed.
The aromatic amine SA was found for PCT and PCT TP9, reflecting potential carcinogenicity following metabolization. Aromatic amines undergo N-oxidation mediated by cytochrome P-450, with a series of reactions leading to the formation of the nitrenium ion, which is highly reactive towards DNA.65,66 In addition to this alert, the Hacceptor-path3-Hacceptor SA was also observed for PCT TP9. PCT TP2 presented the N-methylol derivatives SA, reflecting hydrolysis with the release of formaldehyde, which can then attack DNA according to the Schiff base reaction mechanism.67,68
The compounds FLT, FLT TP6, FLT TP7, FLT TP9, FLT TP10, FLT TP11, and FLT TP13 presented the aromatic nitro and H-acceptor-path3-H-acceptor SAs. The presence of the aromatic nitro SA was indicative of the SN1 reaction, with radical alerts due to the probability of the formation of the nitrenium ion. In contrast, FLT TP8, which did not possess the nitro group, presented the H-acceptor-path3-H-acceptor SA and the AN2 addition mechanism SA. Despite the positive alerts for mutagenicity, previous studies using the Ames Salmonella/microsome mutagenicity assay did not find DNA alteration activity of FLT.69,70
3.3 Carcinogenicity
Studies to determine the in vivo carcinogenicity of certain chemicals in humans are carried out using rats of both sexes and last for 2 years.71In silico predictions can assist such studies by providing information concerning the potential carcinogenicities of drugs and their TPs.For CLO and its TPs, the endpoints of 4 models indicated the potential carcinogenicity of these compounds (Table S4, ESI†). The International Agency for Research on Cancer (IARC) classifies CLO as a probable carcinogen.72 According to Martelli et al.,73 a carcinogenic effect is possible when a dose 25 times higher than the therapeutic limit is applied. For this reason, the use of CLO in veterinary medicine has been banned, although it is still prescribed for humans.74
The in silico results for FCZ and its TPs indicated positive carcinogenicity, according to the models applied. Although studies indicate that FCZ can cause tumors, this is extremely unlikely to occur in humans.75 In recent work, Tamura et al.76 investigated liver hypertrophy and tumor development in mice induced by FCZ and two other triazoles. It was found that FCZ could induce hepatic hypertrophy and the development of tumors in the liver.
Four models showed positive alerts for the carcinogenicity of FRS and its TPs. It has been found that FRS can induce the development of breast cancer in rats,77 while other work reported that it is potentially carcinogenic.78 Furthermore, when FRS is metabolized, metabolites are formed that can be associated with the toxic effects of FRS.79
GFZ presented two alerts for carcinogenicity, while GFZ TP8 had no alerts. In contrast, Fitzgerald et al.80 evaluated the potential carcinogenic effect of GFZ in rats and concluded that it did not induce tumor formation in these animals.80
LOS and all its TPs presented alerts for carcinogenicity, with the exception of LOS TP1. Despite this, previous studies found that LOS has no genotoxic effect,61 indicating that it does not induce cancer and has antineoplastic effects.81 The IBP TPs (IBP TP4 and IBP TP7) only presented a single positive alert for carcinogenicity, while IBP TP13 and IBP TP19 showed between 2 and 4 positive alerts. According to the work of Marshall et al.,82 long-term daily use of IBP may be associated with an increased risk of developing breast cancer.
Carcinogenicity alerts for the compounds NMS, NMS TP1 II, and NMS TP1 III were found for five of the models, while NMS TP1 I presented carcinogenicity for four models. Although the in silico results indicated that NMS is a carcinogen, it has been reported that NMS can inhibit the development of breast cancer.83 In addition, NMS can inhibit cyclooxygenase-2 (COX-2), helping to prevent the proliferation of cancer cells.84,85
PCT and PCT TP2 had three alerts for carcinogenicity, PCT TP9(b,c) had two alerts, PCT TP9(d) had three alerts, and PCT TP9(a) had four alerts. The parent compound (PCT) has been reported to have no carcinogenic effects at concentrations that do not cause hepatotoxic damage (300 mg kg−1 day−1 in rats or 1000 mg kg−1 day−1 in mice).86 Furthermore, Kirkland et al.87 recently reviewed 69 studies concerning the genetic toxicology of PCT and concluded that PCT is incapable of causing any genetic damage, so it is not a compound with carcinogenicity risk.88
FLT and its TPs had positive alerts for carcinogenicity. In addition, FLT was found to have a carcinogenic effect in rats when an amount greater than 3 times the equivalent human dose was administered. Despite this, FLT is not considered carcinogenic, and this effect is only rarely seen when doses far above the therapeutic limit are administered.89
3.4 PBT assessment
Another in silico evaluation was performed using the PBT parameter applied to the pharmaceuticals and TPs in the effluent. In natural aqueous environments, the compounds may be persistent, bioaccumulative, and toxic (PBT), even when present at low concentrations on the order of ng L−1 to μg L−1. It should be noted that for most of the TPs, the concentrations are hypothetical since no analytical standards are available for the purpose of quantification, so their occurrence is based on the concentration of the parent compound. The values obtained using the PROMETHEUS v.1.0 software indicated whether the compounds were non-PBT (<0.5) or PBT (≥0.5). There is a region of uncertainty in the range from 0.475 to 0.525, where a compound could be considered PBT or non-PBT. Values within this uncertainty range were found for 11 compounds: LOS TP1, LOS TP10, LOS TP14, LOS TP15(d), NMS, NMS TP1 I, NMS TP1 II, NMS TP2 III, FLT TP6–7(a), FLT TP13(a), and FLT TP13(b). Only two TPs were identified as PBT: FLT TP6(b,c) and FLT TP7(b,c). Finally, 77 of the compounds evaluated were not considered PBT.3.5 Chemometric analysis
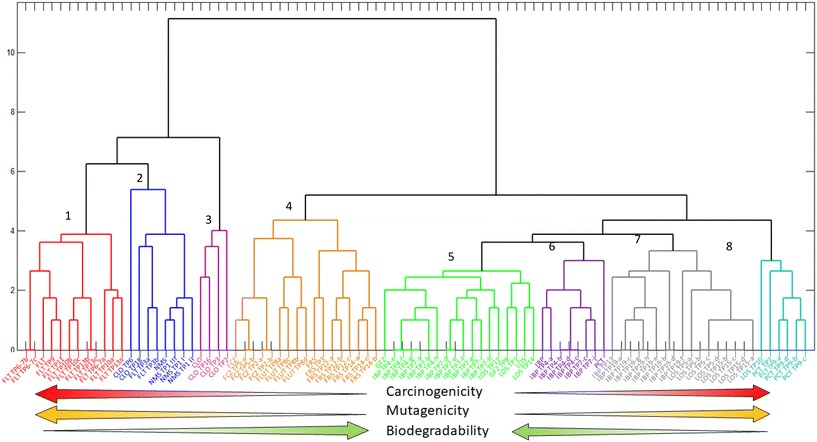
The diversity of the results obtained in this study made it difficult to identify any patterns among the results provided by the different models for all the endpoints. Therefore, multivariate statistical analyses were used to elucidate relationships among the (Q)SAR results for the pharmaceuticals and their TPs.The use of hierarchical cluster analysis (HCA) allowed an overview of data groupings according to their characteristics, such as carcinogenicity, biodegradability, or mutagenicity.90 The results of HCA application to the in silico results are shown in Fig. 2. The HCA dendrogram could be divided into two large groups, with the one on the left comprising three subdivisions (shown in red, blue, and pink colors) and the one on the right with 5 subdivisions (shown in orange, green, and purple, grey, and cyan colors). Therefore, it was possible to organize the TPs and pharmaceuticals into 8 clusters. The large group located on the left of Fig. 2 contained three clusters: the first (red color) comprised the compounds FLT and its TPs; the second (blue color) included NMS and its TPs, together with FLT TP3(a,b), CLO TP6, and CLO TP18; the third cluster (pink color) comprised CLO and its TPs. The FLT TP3(a,b) compounds layout side the FLT cluster since they had a lower number of alerts for biodegradability compared to the other FLT TPs. The FLT TP8(a,b,c,d) compounds lay outside the grouping because they had fewer alerts for mutagenicity and carcinogenicity. This was due to the loss of the nitro group of the aromatic ring, consequently decreasing the number of alerts. The compounds CLO TP6 and CLO TP18, which presented lower numbers of alerts for biodegradability, mutagenicity, and carcinogenicity, compared to CLO and its TPs, were not classified in the parent compound cluster. The compounds present in the large group on the left-hand side of the dendrograma had a common characteristic, namely the presence in their structures of the aromatic nitro moiety, which is associated with mutagenicity and carcinogenicity SAs. The compounds in this large group were classified as non-biodegradable, carcinogenic, and mutagenic.
The large group on the right-hand side of the dendrogram in Fig. 2 had two subdivisions and five clusters. The group of compounds in the fourth cluster (shown in orange) had two divisions, one for FCZ, its TPs, and FLT TP8(a,b,c,d), and the other for FRS and its TPs. As a characteristic of being non-biodegradable, the compounds in this cluster had low numbers of SAs for mutagenicity (from 1 to 3) and numbers of SAs for carcinogenicity ranging from 3 to 7. The fifth cluster (shown in green) included the compounds GFZ, GFZ TP8, IBP TP4(e,f,g,h), IBP TP7(a,b,d,e,f,g,h,i), LOS, LOS TP1, LOS TP11, and LOS TP14. The features of these compounds included a low number of SAs (ranging from 0 to 2) for carcinogenicity, and values of 0 for mutagenicity SAs, except for IBP TP7(d,e,f), LOS, LOS TP1, LOS TP11, and LOS TP14, which presented only one SA. The micropollutants associated with the sixth cluster (purple color) were IBP, IBP TP4(a,b,c,d), IBP TP7(c,j), and PCT. These compounds showed favorable biodegradability, with low numbers of SAs for mutagenicity (1 to 2) and carcinogenicity (0 to 2). The seventh cluster (grey color) consisted of the compounds IBP TP13, IBP TP19, LOS TP5, and LOS TP15. These compounds presented non-biodegradability, with three SAs for mutagenicity and 2 to 4 SAs for carcinogenicity. Finally, the eighth cluster (cyan color) consisted of LOS TP10, PCT TP2, and PCT TP9. These compounds were non-biodegradable, with high numbers of SAs for mutagenicity (from 7 to 9) and carcinogenicity (from 3 to 4).
The compounds' biodegradability increased from the extremes to the center of the dendrogram (Fig. 2), while the presence of compounds with substantial toxicity (considering mutagenicity and carcinogenicity) increased from the center to the extremes. Accordingly, the compounds in the fifth cluster (green color) were likely to be the least harmful to the environment. Conversely, compounds present at the extremes of the dendrogram, such as those in clusters 1, 2, 3, and 8, were probably the most harmful, as shown by the high numbers of mutagenicity and carcinogenicity alerts, in addition to non-biodegradability.
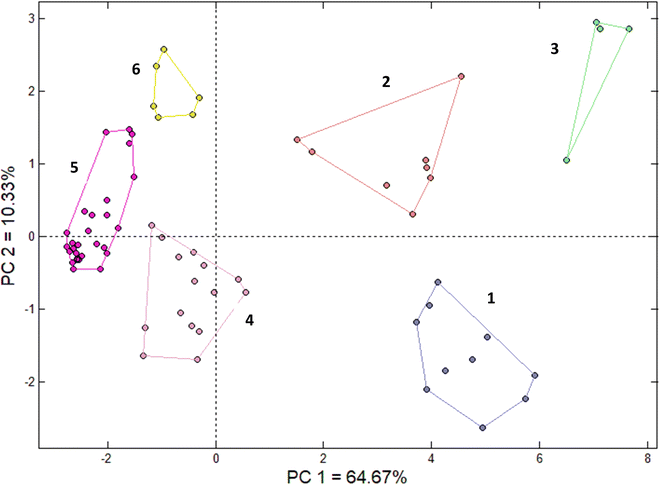
The same data and standardization methodology employed in the HCA was used for applying PCA. The results (Fig. 3) showed that two principal components (PCs) could explain 75% of the total variance, with PC1 explaining 64.67% and PC2 explaining 10.33%. Six well-defined clusters were obtained (Fig. 3), of which five (clusters 1, 2, 3, 4, and 6) were consistent with the HCA results. The compounds in the first three clusters presented the aromatic nitro group, were non-biodegradable and had mutagenicity and carcinogenicity alerts. The fourth cluster identified by the PCA included the compounds identified in the fourth and fifth HCA clusters (FCZ and its TPs, FRS and its TPs, FLT TP2, and FLT TP8). The compounds were non-biodegradable, with low numbers of alerts for mutagenicity and high numbers of alerts (from 3 to 7) for carcinogenicity. A general characteristic of this cluster was aromatic structures, with the presence of halogens in some compounds. The fifth PCA cluster contained compounds previously identified in HCA clusters 5–7. This cluster presented low numbers of alerts for mutagenicity and carcinogenicity, with an absence of these alerts for some compounds, while some compounds showed favorable biodegradability and others did not. Finally, the sixth cluster was the equivalent of the eighth cluster identified in the HCA analysis, consisting of LOS TP10, PCT TP2, and PCT TP9. These biodegradable compounds presented SAs for mutagenicity and were positive for carcinogenicity.
4. Conclusions
Biodegradability is affected by process conditions and also by the structures of chemical compounds. The in silico predictions considered the optimal conditions for biodegradation, with a nutrient-rich environment and an ideal temperature. Therefore, such in silico models do not replace experimental studies but can assist in elucidating biodegradation potential.The endpoints of in vivo/in vitro mutagenicity and carcinogenicity studies of pharmaceuticals reported in the literature are primarily aimed at humans. The concentrations at which drugs are found in effluents cause minimal effects in human beings since they are on the order of μg L−1 to ng L−1. There are no in vivo/in vitro studies for TPs since these are unknown compounds generated during treatments (in this case, the SPF process). However, releasing these TPs in effluents could negatively impact aquatic organisms, causing adverse effects on ecosystems. In this situation, using in silico prediction models, employing (Q)SAR tools can provide an initial assessment of TP ecotoxicity. This can reveal the ideal final point of the treatment process, as indicated by the formation or presence of TPs predicted to be non-toxic.
The results of this study showed that the vast majority of the TPs were non-biodegradable, with mutagenic and carcinogenic potentials. Such information obtained employing in silico predictions can be valuable in guiding decision-making. In addition, chemometric analysis, for example, using HCA and PCA, facilitates the interpretation of (Q)SAR results since the compounds are organized in clusters. It is then possible to determine the best strategy to adopt. For example, if the TPs are non-aggressive to the ecosystem (biodegradable, non-mutagenic, and non-carcinogenic), they could be released together with the effluents. However, their removal from the effluent would be important if their characteristics indicated that release would not be appropriate. In this case, options that could be considered include extending the treatment time, optimizing more drastic treatment conditions, or identifying a suitable additional coupled process.
Author contributions
Alexandre Della-Flora: investigation; formal analysis; writing – original draft; visualization. Marcelo L. Wilde: investigation; formal analysis; writing – original draft; visualization. Adriano de A. Gomes: investigation; formal analysis; writing – original draft; visualization. Davi Scunderlick: investigation. Eder C. Lima: supervision; project administration. Carla Sirtori: conceptualization; writing – original draft; supervision; project administration; funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are grateful for financial support provided by CNPq (403.051/2016-9; 303.622/2017-2; 310.717/2020-5; 402.450/2021-3; 303.612/2021-5; 155.905/2018-0) and FAPERGS (19/2551-0001865-7). This study was financed in part by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES, Finance Code 001).References
- A. Della-Flora, M. L. Wilde, P. S. Thue, D. Lima, E. C. Lima and C. Sirtori, J. Hazard. Mater., 2020, 396, 122699 CrossRef CAS.
- D. Ma, G. Liu, W. Lv, K. Yao, X. Zhang and H. Xiao, Environ. Sci. Pollut. Res., 2014, 21, 7797–7804 CrossRef CAS PubMed.
- G. Š. Parezanović, M. Lalic-Popovic, S. Golocorbin-Kon, V. Vasovic, B. Milijašević, H. Al-Salami and M. Mikov, Arch. Environ. Contam. Toxicol., 2019, 77, 155–161 CrossRef.
- M. la Farré, S. Pérez, L. Kantiani and D. Barceló, TrAC, Trends Anal. Chem., 2008, 27, 991–1007 CrossRef.
- B. Petrie, R. Barden and B. Kasprzyk-Hordern, Water Res., 2015, 72, 3–27 CrossRef CAS PubMed.
- H. Matsuo, H. Sakamoto, K. Arizono and R. Shinohara, Bull. Environ. Contam. Toxicol., 2011, 87, 31–35 CrossRef CAS.
- A. M. Botero-Coy, D. Martínez-Pachón, C. Boix, R. J. Rincón, N. Castillo, L. P. Arias-Marín, L. Manrique-Losada, R. Torres-Palma, A. Moncayo-Lasso and F. Hernández, Sci. Total Environ., 2018, 642, 842–853 CrossRef CAS PubMed.
- J. Park, N. Yamashita, C. Park, T. Shimono, D. M. Takeuchi and H. Tanaka, Chemosphere, 2017, 179, 347–358 CrossRef CAS.
- J. Casado, I. Rodríguez, M. Ramil and R. Cela, J. Chromatogr. A, 2014, 1339, 42–49 CrossRef CAS.
- Z.-F. Chen and G.-G. Ying, Environ. Int., 2015, 84, 142–153 CrossRef CAS.
- S. Arriaga, N. de Jonge, M. L. Nielsen, H. R. Andersen, V. Borregaard, K. Jewel, T. A. Ternes and J. L. Nielsen, Water Res., 2016, 107, 37–46 CrossRef CAS.
- W.-R. Liu, Y.-Y. Yang, Y.-S. Liu, L.-J. Zhang, J.-L. Zhao, Q.-Q. Zhang, M. Zhang, J.-N. Zhang, Y.-X. Jiang and G.-G. Ying, J. Hazard. Mater., 2017, 329, 310–320 CrossRef CAS.
- A. J. Li, O. J. Schmitz, S. Stephan, C. Lenzen, P. Y.-K. Yue, K. Li, H. Li and K. S.-Y. Leung, Water Res., 2016, 89, 68–75 CrossRef CAS.
- L. Lin, H. Lin, M. Zhang, X. Dong, X. Yin, C. Qu and J. Ni, RSC Adv., 2015, 5, 107623–107636 RSC.
- M. Ibáñez, J. V. Sancho, L. Bijlsma, A. L. N. van Nuijs, A. Covaci and F. Hernández, TrAC, Trends Anal. Chem., 2014, 57, 107–117 CrossRef.
- R. Wielens Becker, M. L. Wilde, D. Salmoria Araújo, D. Seibert Lüdtke and C. Sirtori, Water Res., 2020, 184, 116183 CrossRef CAS.
- E. Evgenidou, A. Ofrydopoulou, N. Malesic-Eleftheriadou, C. Nannou, N. M. Ainali, E. Christodoulou, D. N. Bikiaris, G. Z. Kyzas and D. A. Lambropoulou, Sci. Total Environ., 2020, 741, 140394 CrossRef CAS.
- T. Matsushita, S. Honda, T. Kuriyama, Y. Fujita, T. Kondo, Y. Matsui, N. Shirasaki, H. Takanashi and T. Kameya, Water Res., 2018, 129, 347–356 CrossRef CAS.
- J. Menz, A. P. Toolaram, T. Rastogi, C. Leder, O. Olsson, K. Kümmerer and M. Schneider, Environ. Int., 2017, 98, 171–180 CrossRef CAS.
- L. Yin, B. Wang, H. Yuan, S. Deng, J. Huang, Y. Wang and G. Yu, Emerging Contam., 2017, 3, 69–75 CrossRef.
- T. I. Netzeva, A. P. Worth, T. Aldenberg, R. Benigni, M. T. D. Cronin, P. Gramatica, J. S. Jaworska, S. Kahn, G. Klopman, C. A. Marchant, G. Myatt, N. Nikolova-Jeliazkova, G. Y. Patlewicz, R. Perkins, D. W. Roberts, T. W. Schultz, D. T. Stanton, J. J. M. van de Sandt, W. Tong, G. Veith and C. Yang, ATLA, Altern. Lab. Anim., 2005, 33, 155–173 CrossRef CAS PubMed.
- E. Cuervo Lumbaque, C. Sirtori and V. J. P. Vilar, Sci. Total Environ., 2020, 743, 140629 CrossRef CAS.
- I. González-Mariño, I. Rodríguez, L. Rojo and R. Cela, Sci. Total Environ., 2018, 644, 995–1005 CrossRef.
- R. A. Osawa, B. T. Barrocas, O. C. Monteiro, M. C. Oliveira and M. H. Florêncio, Sci. Total Environ., 2019, 692, 503–510 CrossRef CAS PubMed.
- A. Della-Flora, M. L. Wilde, I. D. F. Pinto, É. C. Lima and C. Sirtori, Environ. Res., 2020, 183, 109223 CrossRef CAS PubMed.
- F. Pizzo, A. Lombardo, A. Manganaro and E. Benfenati, Sci. Total Environ., 2013, 463–464, 161–168 CrossRef CAS PubMed.
- S. K. Chakravarti, R. D. Saiakhov and G. Klopman, J. Chem. Inf. Model., 2012, 52, 2609–2618 CrossRef CAS.
- R. Saiakhov, S. Chakravarti and G. Klopman, Mol. Inf., 2013, 32, 87–97 CrossRef CAS.
- F. Pizzo, A. Lombardo, A. Manganaro, C. I. Cappelli, M. I. Petoumenou, F. Albanese, A. Roncaglioni, M. Brandt and E. Benfenati, Environ. Res., 2016, 151, 478–492 CrossRef CAS.
- D. Ballabio, Chemom. Intell. Lab. Syst., 2015, 149, 1–9 CrossRef CAS.
- R. Hirsch, T. Ternes, K. Haberer and K.-L. Kratz, Sci. Total Environ., 1999, 225, 109–118 CrossRef CAS PubMed.
- M. Galimand, G. Gerbaud, M. Guibourdenche, J.-Y. Riou and P. Courvalin, N. Engl. J. Med., 1998, 339, 868–874 CrossRef CAS PubMed.
- R. Szczepanowski, B. Linke, I. Krahn, K.-H. Gartemann, T. Gützkow, W. Eichler, A. Pühler and A. Schlüter, Microbiology, 2009, 155, 2306–2319 CrossRef CAS.
- W. Xu, G. Zhang, X. Li, S. Zou, P. Li, Z. Hu and J. Li, Water Res., 2007, 41, 4526–4534 CrossRef CAS.
- R. S. Boethling, E. Sommer and D. DiFiore, Chem. Rev., 2007, 107, 2207–2227 CrossRef CAS.
- A. Bernareggi, Inflammopharmacology, 2001, 9, 81–89 CrossRef CAS.
- I. Bragança, A. S. Danko, J. Pacheco, D. Frascari, C. Delerue-Matos and V. F. Domingues, Water, Air, Soil Pollut., 2016, 227, 227 CrossRef.
- Q. Yan, X. Gao, Y.-P. Chen, X.-Y. Peng, Y.-X. Zhang, X.-M. Gan, C.-F. Zi and J.-S. Guo, Sci. Total Environ., 2014, 470–471, 618–630 CrossRef CAS PubMed.
- J. L. Wilkinson, A. B. A. Boxall, D. W. Kolpin, K. M. Y. Leung, W. S. Lai, C. Galbán-Malagón, A. D. Adell, J. Mondon, M. Metian, R. A. Marchant, A. Bouzas-Monroy, A. Cuni-Sanchez, A. Coors, P. Carriquiriborde, M. Rojo, C. Gordon, M. Cara, M. Moermond, T. Luarte, V. Petrosyan, Y. Perikhanyan, C. S. Mahon, C. J. McGurk, T. Hofmann, T. Kormoker, V. Iniguez, J. Guzman-Otazo, J. L. Tavares, G. D. de Figueiredo, M. T. P. Razzolini, V. Dougnon, G. Gbaguidi, O. Traoré, J. M. Blais, L. E. Kimpe, M. Wong, D. Wong, R. Ntchantcho, J. Pizarro, G-G. Ying, C-E. Chen, M. Paéz, J. Martínez-Lara, J-P. Otamonga, J. Poté, S. A. Ifo, P. Wilson, S. Echeverría-Sáenz, N. Udikovic-Kolic, M. Milakovic, D. Fatta-Kassinos, L. Ioannou-Ttofa, V. Belušová, J. Vymazal, M. Cárdenas-Bustamante, B. A. Kassa, J. Garric, A. Chaumot, P. Gibba, I. Kunchulia, S. Seidensticker, G. Lyberatos, H. P. Halldórsson, M. Melling, T. Shashidhar, M. Lamba, A. Nastiti, A. Supriatin, N. Pourang, A. Abedini, O. Abdullah, S. S. Gharbia, F. Pilla, B. Chefetz, T. Topaz, K. M. Yao, B. Aubakirova, R. Beisenova, L. Olaka, J. K. Mulu, P. Chatanga, V. Ntulli, N. T. Blama, S. Sherif, A. Z. Aris, L. J. Looi, M. Niang, S. T. Traore, R. Oldenkamp, O. Ogunbanwo, M. Ashfaq, M. Iqbal, Z. Abdeen, A. O'Dea, J. M. Morales-Saldaña, M. Custodio, H. de la Cruz, I. Navarrete, F. Carvalho, A. B. Gogra, B. M. Koroma, V. C. Flajs, M. Gombac, M. Thwala, K. Choi, H. Kang, J. L. C. Ladu, A. Rico, P. Amerasinghe, A. Sobek, G. Horlitz, A. K. Zenker, A. C. King, J-J. Jiang, R. Kariuki, M. Tumbo, U. Tezel, T. T. Onay, J. B. Lejju, Y. Vystavna, Y. Vergeles, H. Heinzen, A. Pérez-Parada, D. B. Sims, M. Figy, D. Good and C. Teta, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2113947119 CrossRef CAS.
- D. Dionisi and C. C. Etteh, J. Environ. Chem. Eng., 2019, 7, 103282 CrossRef CAS.
- K. Kołecka, M. Gajewska, P. Stepnowski and M. Caban, Sci. Total Environ., 2019, 647, 149–157 CrossRef PubMed.
- W.-J. Sim, J.-W. Lee and J.-E. Oh, Environ. Pollut., 2010, 158, 1938–1947 CrossRef CAS PubMed.
- S. K. Behera, H. W. Kim, J.-E. Oh and H.-S. Park, Sci. Total Environ., 2011, 409, 4351–4360 CrossRef CAS.
- Y. Li, X. Niu, C. Yao, W. Yang and G. Lu, Int. J. Environ. Res. Public Health, 2019, 16 Search PubMed.
- N. H. Tran and K. Y.-H. Gin, Sci. Total Environ., 2017, 599–600, 1503–1516 CrossRef CAS PubMed.
- Y. Fang, A. Karnjanapiboonwong, D. A. Chase, J. Wang, A. N. Morse and T. A. Anderson, Environ. Toxicol. Chem., 2012, 31, 550–555 CrossRef CAS.
- R. R. Z. Tarpani and A. Azapagic, Sci. Total Environ., 2018, 622–623, 1417–1430 CrossRef CAS.
- Y. Tang, Z. Xu, Q. Guo, X. Li, L. Wang, C. Hong and Y. Bing, in 2012 International Conference on Biomedical Engineering and Biotechnology, 2012, pp. 1576–1581 Search PubMed.
- A. Joss, E. Keller, A. C. Alder, A. Göbel, C. S. McArdell, T. Ternes and H. Siegrist, Water Res., 2005, 39, 3139–3152 CrossRef CAS.
- C. I. Kosma, D. A. Lambropoulou and T. A. Albanis, J. Hazard. Mater., 2010, 179, 804–817 CrossRef CAS PubMed.
- I. Martínez-Alcalá, J. M. Guillén-Navarro and C. Fernández-López, Chem. Eng. J., 2017, 316, 332–340 CrossRef.
- S. D. Nelson, J. Med. Chem., 1982, 25, 753–765 CrossRef CAS.
- G. Sabbioni, Environ. Health Perspect., 1994, 102(Suppl), 61–67 CAS.
- M. Shimizu and E. Yano, Mutat. Res., Genet. Toxicol., 1986, 170, 11–22 CrossRef CAS.
- J. Kazius, R. McGuire and R. Bursi, J. Med. Chem., 2005, 48, 312–320 CrossRef CAS.
- A. B. Bailey, R. Chanderbhan, N. Collazo-Braier, M. A. Cheeseman and M. L. Twaroski, Regul. Toxicol. Pharmacol., 2005, 42, 225–235 CrossRef CAS PubMed.
- G. S. Silva, L. Zuravski, M. M. M. F. Duarte, M. M. Machado and L. F. S. Oliveira, Immunopharmacol. Immunotoxicol., 2019, 41, 123–129 CrossRef CAS.
- D. C. Thompson, J. A. Thompson, M. Sugumaran and P. Moldéus, Chem.-Biol. Interact., 1993, 86, 129–162 CrossRef CAS PubMed.
- F. D. L. Leusch, S. J. Khan, S. Laingam, E. Prochazka, S. Froscio, T. Trinh, H. F. Chapman and A. Humpage, Water Res., 2014, 49, 300–315 CrossRef CAS.
- G. Speit, P. Schütz, J. Högel and O. Schmid, Mutagenesis, 2007, 22, 387–394 CrossRef CAS.
- R. G. Silva-Oliveira, P. C. Orsolin and J. C. Nepomuceno, Food Chem. Toxicol., 2016, 95, 211–218 CrossRef CAS PubMed.
- M. F. de M. Leão, J. A. Duarte, P. D. Sauzen, J. C. E. da Piccoli, L. F. S. de Oliveira and M. M. Machado, Environ. Toxicol. Pharmacol., 2018, 63, 1–5 CrossRef PubMed.
- D. Borkotoky, S. K. Panda, G. R. Sahoo and S. C. Parija, Drug Chem. Toxicol., 2014, 37, 178–183 CrossRef CAS.
- R. Tripathi, P. Tripathi, S. S. Pancholi and C. N. Patel, Drug Chem. Toxicol., 2014, 37, 255–260 CrossRef CAS.
- R. Benigni, A. Giuliani, R. Franke and A. Gruska, Chem. Rev., 2000, 100, 3697–3714 CrossRef CAS.
- Y.-T. Woo and D. Y. Lai, Patty's Toxicol., 2012, pp. 1–96 Search PubMed.
- H. M. Coley, N. Brooks, D. H. Phillips, A. Hewer, T. C. Jenkins, M. Jarman and I. R. Judson, Biochem. Pharmacol., 1995, 49, 1203–1212 CrossRef CAS PubMed.
- G. Cheng, Y. Shi, S. J. Sturla, J. R. Jalas, E. J. McIntee, P. W. Villalta, M. Wang and S. S. Hecht, Chem. Res. Toxicol., 2003, 16, 145–152 Search PubMed.
- BC Cancer Agency Cancer, Flutamide Monograph, 2014 Search PubMed.
- Merck, EUFLEX* MONOGRAPH, Quebec, 2011 Search PubMed.
- P. Steinberg, In Vitro–In Vivo Carcinogenicity, in In vitro Environmental Toxicology - Concepts, Application and Assessment, ed. G. Reifferscheid and S. Buchinger, Advances in Biochemical Engineering/Biotechnology, Springer, Cham, 2016, vol. 157, DOI:10.1007/10_2015_5013.
- INTERNATIONAL AGENCY FOR RESEARCH ON CANCER, IARC Monographs on the Evaluation of Carcinogenic Risks to Humans Volume 50, 1990 Search PubMed.
- A. Martelli, F. Mattioli, G. Pastorino, L. Robbiano, A. Allavena and G. Brambilla, Mutat. Res., Genet. Toxicol., 1991, 260, 65–72 Search PubMed.
- J. C. Hanekamp and A. Bast, Environ. Toxicol. Pharmacol., 2015, 39, 213–220 CrossRef CAS.
- G. Paulus, L. Longeart and A. M. Monro, Teratog., Carcinog., Mutagen., 1994, 14, 251–257 CrossRef CAS PubMed.
- K. Tamura, K. Inoue, M. Takahashi, S. Matsuo, K. Irie, Y. Kodama, T. Gamo, S. Ozawa and M. Yoshida, Food Chem. Toxicol., 2015, 78, 86–95 CrossRef CAS PubMed.
- J. R. Bucher, J. Huff, J. K. Haseman, S. L. Eustis, W. E. Davis Jr. and E. F. Meierhenry, J. Appl. Toxicol., 1990, 10, 369–378 CrossRef CAS.
- S. C. Mondal, D. N. Tripathi, A. Vikram, P. Ramarao and G. B. Jena, Fundam. Clin. Pharmacol., 2012, 26, 383–392 CrossRef CAS.
- D. P. Williams, D. J. Antoine, P. J. Butler, R. Jones, L. Randle, A. Payne, M. Howard, I. Gardner, J. Blagg and B. K. Park, J. Pharmacol. Exp. Ther., 2007, 322, 1208–1220 CrossRef CAS PubMed.
- J. E. Fitzgerald, J. L. Sanyer, J. L. Schardein, R. S. Lake, E. J. McGuire and F. A. de la Iglesia, J. Natl. Cancer Inst., 1981, 67, 1105–1116 CAS.
- S. Kim, H. Toyokawa, J. Yamao, S. Satoi, H. Yanagimoto, T. Yamamoto, S. Hirooka, S. Yamaki, K. Inoue, Y. Matsui and A.-H. Kwon, Pancreas, 2014, 43, 886–890 CrossRef CAS PubMed.
- S. F. Marshall, L. Bernstein, H. Anton-Culver, D. Deapen, P. L. Horn-Ross, H. Mohrenweiser, D. Peel, R. Pinder, D. M. Purdie, P. Reynolds, D. Stram, D. West, W. E. Wright, A. Ziogas and R. K. Ross, J. Natl. Cancer Inst., 2005, 97, 805–812 CrossRef CAS PubMed.
- S. Nakatsugi, T. Ohta, T. Kawamori, M. Mutoh, T. Tanigawa, K. Watanabe, S. Sugie, T. Sugimura and K. Wakabayashi, Jpn. J. Cancer Res., 2000, 91, 886–892 CrossRef CAS PubMed.
- B. Zhong, X. Cai, S. Chennamaneni, X. Yi, L. Liu, J. J. Pink, A. Dowlati, Y. Xu, A. Zhou and B. Su, Eur. J. Med. Chem., 2012, 47, 432–444 CrossRef CAS PubMed.
- M. Afzal, D. P. Bhardwaj, R. Khan, I. Kazmi, S. Saleem, F. A. Al-Abbasi and F. Anwar, Inflammopharmacology, 2019, 27, 89–98 CrossRef CAS PubMed.
- K. Bergman, L. Müller and S. W. Teigen, Mutat. Res., Fundam. Mol. Mech. Mutagen., 1996, 349, 263–288 CrossRef.
- D. Kirkland, M. Kovochich, S. L. More, F. J. Murray, A. D. Monnot, J. V. Miller, H. Jaeschke, D. Jacobson-Kram, M. Deore, S. K. Pitchaiyan, K. Unice and G. Eichenbaum, Regul. Toxicol. Pharmacol., 2021, 122, 104892 CrossRef CAS.
- F. J. Murray, A. D. Monnot, D. Jacobson-Kram, S. M. Cohen, J. F. Hardisty, S. B. Bandara, M. Kovochich, M. Deore, S. K. Pitchaiyan, C. K. Gelotte, J. C. K. Lai, E. Atillasoy, A. Hermanowski-Vosatka, E. Kuffner, K. M. Unice, K. Yang, Y. Gebremichael, B. A. Howell and G. Eichenbaum, Regul. Toxicol. Pharmacol., 2020, 118, 104801 CrossRef CAS PubMed.
- L. C. Zacharia, Toxicol. Sci., 2017, 159, 279–289 CrossRef CAS.
- N. P. Kalogiouri, R. Aalizadeh, M. E. Dasenaki and N. S. Thomaidis, Anal. Chim. Acta, 2020, 1134, 150–173 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ew00518b |
| This journal is © The Royal Society of Chemistry 2023 |