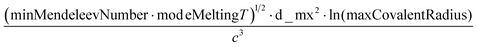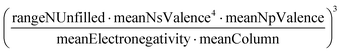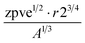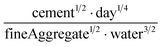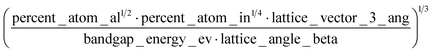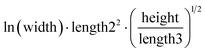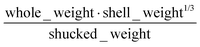Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Interpretable models for extrapolation in scientific machine learning†
Eric S.
Muckley
a,
James E.
Saal
 *a,
Bryce
Meredig
*a,
Bryce
Meredig
 a,
Christopher S.
Roper
b and
John H.
Martin
b
a,
Christopher S.
Roper
b and
John H.
Martin
b
aCitrine Informatics, Redwood City, CA 94063, USA. E-mail: jsaal@citrine.io
bHRL Laboratories, Malibu, CA 90265, USA
First published on 21st August 2023
Abstract
Data-driven models are central to scientific discovery. In efforts to achieve state-of-the-art model accuracy, researchers are employing increasingly complex machine learning algorithms that often outperform simple regressions in interpolative settings (e.g. random k-fold cross-validation) but suffer from poor extrapolation performance, portability, and human interpretability, which limits their potential for facilitating novel scientific insight. Here we examine the trade-off between model performance and interpretability across a broad range of science and engineering problems with an emphasis on materials science datasets. We compare the performance of black box random forest and neural network machine learning algorithms to that of single-feature linear regressions which are fitted using interpretable input features discovered by a simple random search algorithm. For interpolation problems, the average prediction errors of linear regressions were twice as high as those of black box models. Remarkably, when prediction tasks required extrapolation, linear models yielded average error only 5% higher than that of black box models, and outperformed black box models in roughly 40% of the tested prediction tasks, which suggests that they may be desirable over complex algorithms in many extrapolation problems because of their superior interpretability, computational overhead, and ease of use. The results challenge the common assumption that extrapolative models for scientific machine learning are constrained by an inherent trade-off between performance and interpretability.
Introduction
Machine learning has become a principal catalyst for scientific discovery, particularly in the design of novel functional materials.1,2 In efforts to build predictive models with state-of-the-art performance, researchers are employing increasingly complex black box algorithms, including large-scale ensembles and deep neural networks, because of their ability to approximate high-dimensional response surfaces with arbitrary precision.3 Recent availability of low-cost data storage and computing resources has unlocked model architectures capable of handling large numbers (105–107) of input features,4,5 enabling the development of deep learning models such as Elemnet6 and SchNet7 which can learn feature encodings directly from the elemental compositions of input materials. While the complexity of these models often leads them to outperform traditional regression techniques by standard cross-validation scoring metrics, they also suffer from notable disadvantages.8,9Increases in model complexity are generally accompanied by corresponding decreases in model portability and usability by non-experts. Many state-of-the-art black box algorithms require significant computational resources and hyperparameter optimization during training, which limits their usefulness in edge computing environments and necessitates experienced practitioners for managing model serialization, programming environments and version control systems, and input data compatibility.10,11 Growing complexity also inherently limits model interpretability by humans,12,13 which increases the likelihood of model overfitting, reduces researcher trust in predictions, and creates difficulty in troubleshooting.14 Perhaps most importantly, poor model interpretability impedes domain expert intuition by obscuring natural underlying patterns that often guide researchers toward novel insights and new physics.1,15,16
The use of simple interpretable models has garnered renewed attention amid mounting evidence that the trade-off between model performance and interpretability is often overstated.1,14 Interpretability aids in diagnosis of model biases, management of multi-objective trade-offs, and mitigation of unexpected results,8,17–20 which are essential considerations in the design of materials and chemicals for novel drugs, electronics, catalysts, and alloys.2,21 The primary challenge of materials informatics, accurate prediction of the physical properties of a material from its chemistry or other known characteristics, relies on the discovery of interpretable physics-informed input features: empirically or theoretically derived vectors of known quantities which can be used to predict the value of a target property using simple mappings, such as a linear transformations.6,22 Ideal input features are (i) general: they maintain predictive performance across a broad range of materials, (ii) extensible: they can be constructed from readily available data sources or simple calculations, and (iii) interpretable: they can provide insights into underlying physics or correlations which aid in predicting the material property of interest.23
The discovery of features meeting the aforementioned three criteria remains one of the primary challenges of scientific machine learning (SciML) and represents a principal roadblock on the path toward interpretable physics-informed models.24,25 Features for materials informatics models are often engineered to encode one or more material properties, including those derived from chemical composition, topology, electronic behavior, or structural fingerprints.18 The most commonly used features are composition-based vectors constructed from properties of the constituent elements of the material12,26 such as those included in the Magpie feature set,27 which provides descriptive statistics such as minimum, maximum, mean, and range of a set of tabulated element properties for a given chemical composition. Magpie has been used for building successful models across a broad range of materials classes22,28,29 and serves as the foundation for the widely-used Matminer suite of open-source materials informatics tools.30 While the standard Magpie library enables simple human-interpretable featurization of chemical composition, its strong reliance on elemental composition generally leads to a lack of robust encoding of underlying interactions beyond those of the material's constituent elements. Featurization based on elemental composition is increasingly being augmented with geometric and topological information such as crystal structure, lattice constants, bond types, and graph-based molecular representations,5,25,31–34 complex spatial descriptors such as the many-body tensor representation,35 and string-based molecular encodings such as SMILES,36 BigSMILES,37 and SELFIES.38 Structure-based encodings are widely utilized for featurization in academic studies,39–41 but obtaining details about crystallinity or molecular structure for large sets of candidate materials can be infeasible in practice if the structure of materials in the design space is not well known. Moreover, graph-based topological featurization methods are inherently difficult to implement for materials with poorly-defined structures such as glasses and multi-principal element alloys.
Recent research efforts suggest that simple composition-based features may be used to build predictive SciML models without sacrificing human interpretability or relying on prior knowledge about material structure.25,42,43 Often this is accomplished through the use of feature engineering, the refinement of model inputs by pruning of existing features or construction of new features from simpler base features, which enables physics-based mapping of model inputs to target properties through the use of symbolic regression.23,24 Similar methods have been demonstrated for building interpretable classification models in health care and criminal justice,44–46 but quantitative comparisons between the performance of black box models and interpretable models enabled by symbolic regression have not been widely documented for SciML applications.20,24
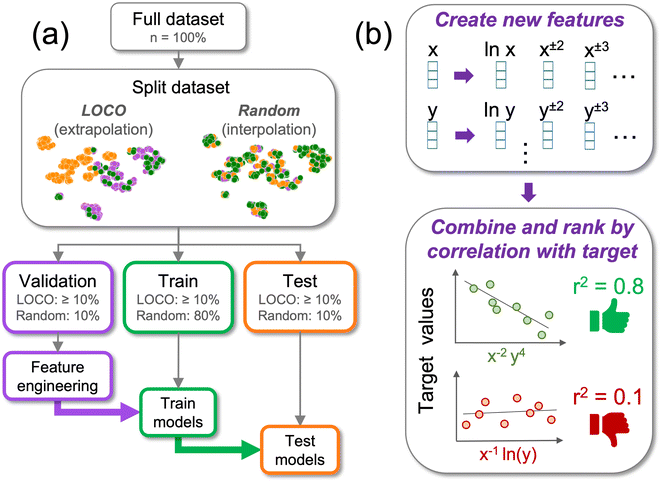
In this study we investigate the use of empirically-derived feature vectors for constructing interpretable predictive models. We screen models by their ability to extrapolate to new clusters in the input design space using leave-one-cluster-out (LOCO)47 cross-validation (CV), a specific implementation of leave-group-out (LOG)48 and other extrapolation-based CV techniques49 (Fig. 1). We compare the extrapolation performance of linear models which rely on a single interpretable feature to that of black box random forest and neural network models which utilize up to 102–103 different input features. As a case study, we examine model extrapolation on a set of 9 open datasets: (i) 3 materials datasets with Magpie featurization, (ii) 3 materials datasets without Magpie featurization, and (iii) 3 non-materials datasets in the physical sciences which are used to test generality of the results. The study highlights important considerations for balancing model performance and interpretability and demonstrates the potential for building interpretable single-feature linear models with extrapolation performance that is comparable to that of black box algorithms in many SciML problems.
Methods
Benchmark datasets were selected to span a broad range of materials informatics problems, using both Magpie- and non-Magpie-featurized materials, as well as non-materials scientific datasets to test generality of the results beyond materials. One variable in each dataset was selected as the regression target to simulate a real-world scientific modeling problem. Target variables were selected to sample a broad range of material properties, including electronic, mechanical, and thermodynamic properties, and to ensure diversity in target value distributions across each dataset. Future work may be necessary to consider additional dataset types not considered here (e.g., those with discrete and/or categorical values or those with complex underlying functions).Prior to partitioning into cross-validation (CV) sets, datasets were cleaned by removal of rows which contained at least one value which was greater than 3 interdecile ranges away from the median value of the column, removal of duplicate rows and columns, removal of non-numeric columns, removal of rows containing null values, and removal of columns with less than 5 unique values. The 9 cleaned benchmark datasets, along with a description of each target variable, dataset size, and data source, are shown in Table 1. Histograms of each target value are shown in Fig. S1† where each panel corresponds to one test dataset and is labeled by the name of the target variable in the dataset. Three materials datasets (colored green in Fig. S1†) were featurized based on chemical formula using the element_property, BandCenter, and AtomicOrbitals featurization methods in Matminer. Features generated from Matminer which contained non-numeric values or constant values across all rows were removed, resulting in slightly different sets of Matminer features in each dataset.
| Target | Description | Inputs | Type | Shape | Source |
|---|---|---|---|---|---|
| Melting temp | Melting temperature of AxBy compounds in K | Magpie chemistry (e.g. atomic numbers, atomic weights, valences) | Exp. | (243, 101) | Ref. 26 |
| Bulk modulus | Bulk modulus of M2AX compounds in GPa | Magpie chemistry (e.g. lattice constants, atomic numbers, valences) | Comp. | (223, 74) | Matminer50 |
| Band gap | Electronic band gap of double perovskites in eV | Magpie chemistry (e.g. atomic numbers, atomic weights, valences) | Comp. | (1306, 69) | Matminer51 |
| Heat capacity | Heat capacity of organic molecules at 298.15 K in cal mol−1 K−1 | Non-magpie chemistry (e.g. rotational constants, HOMO, LUMO, ZPVE) | Comp. | (1331, 11) | DeepChem52 |
| Compressive strength | Compressive strength of concrete formulations in MPa | Non-magpie formulations (e.g. water, fly ash, cement content) | Exp. | (985, 9) | UCI53 |
| Formation energy | Formation energy of transparent conductors in eV per atom | Non-magpie chemistry (e.g. lattice vector angles, bandgap, percent aluminum) | Comp. | (2137, 13) | Kaggle54 |
| Fish weight | Weight of fish in g | Physical measurements (e.g. length, height, width) | Exp. | (158, 6) | Kaggle55 |
| Airfoil sound | Intensity of sound in decibels created by airfoil flight | Physical measurements (e.g. airfoil angle, velocity, chord length) | Exp. | (1497, 5) | UCI56 |
| Abalone rings | Number of rings (corresponding to age) in an abalone shell | Physical measurements (e.g. length, diameter, weight) | Exp. | (4175, 8) | UCI57 |
To prevent data leakage during feature engineering and model testing, each dataset was partitioned into 3 splits: train (for training predictive models), test (for evaluating model performance), and validation (for assessing the quality of engineered features), as shown in Fig. 1a. For each dataset, data partitioning was repeated 10 times for each of 2 CV strategies: leave-one-cluster-out (LOCO CV), to simulate model extrapolation (Fig. S2†), and random CV, to simulate model interpolation (Fig. S3†), resulting in 20 total unique train-test-validation splits per dataset. For LOCO CV, splits were generated following the method of Meredig et al.47 Standard normalization of input features was performed, the data was shuffled, k-means clustering with random initialization was performed where the number of clusters was selected randomly between 3 (the minimum number of clusters required for train, test, and validation sets) and 10 (the largest traditional choice for clusters in cross-validation), and each cluster was randomly assigned to a train, test, or validation set such that no set contained less than 10% of the entire dataset. For random CV, the dataset was divided randomly so that 80% was used for training, 10% was used for testing, and 10% was used for validation. The uniqueness of each CV split was verified to ensure that no two splits contained identical partitioning of samples.
After datasets were partitioned into train, test, and validation sets, feature engineering was performed with the goal of creating interpretable input features built from the set of existing base features, as described in Fig. 1b. To enable a variety of interpretable functional forms for engineered features, base input features were raised to varying powers: from feature x, the features x−1, x±1/2, x±1/3, x±1/4, x±2, x±3, x±4, and ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) x were generated when possible (for example, x−1 was not generated for feature x when feature x contained values equal to 0). Next, between 2 and 5 of the newly-constructed features were selected at random. The selected features were multiplied together to generate a new feature. This process was repeated 5 × 105 times at each CV split for each dataset, and each resulting feature was ranked by its Pearson r2 correlation to the target values in the validation set. The engineered feature which exhibited the highest r2 correlation to the target values of the validation set at a given CV split was selected as the best engineered feature (BE) for that CV split. While recent efforts have demonstrated more advanced feature engineering methods for materials informatics,20,24 the random search approach described here was used to demonstrate how feature engineering can be performed using a simple procedure without the need for dedicated software packages or computationally-expensive genetic algorithms. No constraints were applied to enforce dimensionality of the resultant features. The number of generated features scales linearly as O(n) with the number of distinct random combinations of input features. The 5 × 105 cap in iterations is intended to ensure linear scaling when adding new datasets and input features.
x were generated when possible (for example, x−1 was not generated for feature x when feature x contained values equal to 0). Next, between 2 and 5 of the newly-constructed features were selected at random. The selected features were multiplied together to generate a new feature. This process was repeated 5 × 105 times at each CV split for each dataset, and each resulting feature was ranked by its Pearson r2 correlation to the target values in the validation set. The engineered feature which exhibited the highest r2 correlation to the target values of the validation set at a given CV split was selected as the best engineered feature (BE) for that CV split. While recent efforts have demonstrated more advanced feature engineering methods for materials informatics,20,24 the random search approach described here was used to demonstrate how feature engineering can be performed using a simple procedure without the need for dedicated software packages or computationally-expensive genetic algorithms. No constraints were applied to enforce dimensionality of the resultant features. The number of generated features scales linearly as O(n) with the number of distinct random combinations of input features. The 5 × 105 cap in iterations is intended to ensure linear scaling when adding new datasets and input features.
After the BE feature was selected for each CV split, it was used for fitting single-feature linear regressions which were compared to black box models in both interpolative and extrapolative regimes. At each CV split, 3 regression models were compared: a random forest, a neural network with stochastic gradient-descent optimizer, and a linear regression, all implemented using default Scikit-learn methods (RandomForestRegressor, MLPRegressor, and LinearRegression, respectively) with default hyperparameters (100 estimators and no maximum tree depth for the random forest, and 1 hidden layer with 100 neurons, ReLU activation, Adam solver, and 200 maximum iterations for the neural network).58 Default hyperparameters were used to investigate the performance of models with standard architectures for both interpolation and extrapolation tasks, as optimization of hyperparameters often yields improvements in model interpolation at the potential expense of performance in extrapolative settings, which may lead researchers to overestimate model utility for real-world problems.47 Stochastic processes in the training of the neural network and random forest models were avoided by the use of fixed seeds in the published results.
Before fitting the models, each dataset was scaled using the Scikit-learn RobustScaler method. Model performance was quantified using non-dimensional model error (NDME),59 calculated as the ratio between root-mean-square error (RMSE) between predicted values and ground truth values in the test set, and the standard deviation in the ground-truth values of the test set. NDME was used as a performance metric to ensure that model performance could be compared across datasets and target properties with varying units and magnitudes, where NDME of 0 corresponds to a perfect model, and NDME of 1 corresponds to a model with average prediction error which is equal to standard deviation in the ground truth values. Models were evaluated on their ability to predict target values in the test set of each CV split for 3 different featurization strategies: (1) best engineered (BE), where the single best engineered feature at the given CV split was used as the only model input, (2) BE + original, where all the original dataset input columns, in addition to the single best engineered feature at the given CV split, were used as input features, and (3) original, where the original dataset input columns were used as input features. Covariance confidence intervals of model NDMEs were calculated by excluding extreme model outliers with NDME > 5.
Results and discussion
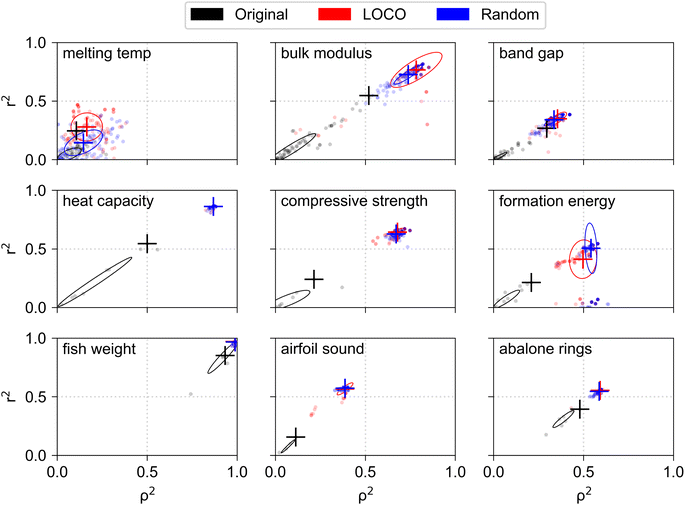
The primary challenge of creating interpretable physics-informed models lies in the discovery of features which provide a linear mapping between input values and target values. We performed feature engineering with the goal of finding new features which were linearly correlated with the target variable of each test dataset (Fig. S4†). Fig. 2 summarizes the input features associated with each test dataset before and after the feature engineering process. Each panel corresponds to one dataset and is labeled by the target variable name. Each scatter point corresponds to a single input feature, where the original features of each dataset are shown in black, the top 100 features engineered using LOCO CV (top 10 features for each of 10 CV splits) are shown in red, and the top 100 features engineered using random CV are shown in blue. Each feature is plotted by its Pearson r2 correlation with the target variable on the vertical axis, for quantifying linearity with the target, and its and its Spearman ρ2 correlation on the horizontal axis, for quantifying monotonicity with the target. Large cross symbols show the median location of engineered points in the LOCO and random groups, and the single feature with the highest r2 value in the original feature set. While top features were selected during the feature engineering process based on their r2 correlation to the validation set, the correlations shown in Fig. 2 represent correlations between input features and the target value of the entire dataset. Points which lie near the origin (0, 0) represent features which are generally non-informative for linear models, as they lack linearity and monotonicity with the target variable. Features near the coordinate (0, 1) exhibit high linearity but poor monotonicity, which suggests that high degeneracy in target values may be predicted for a single value of the input feature. Features near coordinate (1, 0) exhibit high monotonicity but poor linearity, which makes them informative inputs for nonlinear regression models. Features near coordinates (1, 1) exhibit both high linearity and monotonicity with the target values, which makes them suitable for use in simple linear regression models.In every dataset, the feature engineering process uncovered new features with r2 correlation to the target variable which was higher than that of any of the original features. In 4 of 9 datasets (heat capacity compressive strength, formation energy, airfoil sound), feature engineering resulted in discovery of new features which achieved significant (>100%) increases in r2 correlation to the target, which suggests that simple algebraic manipulation and combination of existing features may be used for creating highly informative input features in many SciML problems. A noteworthy result is that higher correlations between original features and the target variable do not necessarily enable engineering of new features with proportionally higher correlations. For example, the best original input feature for abalone rings exhibits r2 correlation of ∼0.45, while that of compressive strength exhibits r2 of ∼0.2. However, feature engineering for compressive strength results in new features with median r2 of ∼0.7, whereas r2 for engineered abalone rings features reaches a maximum value at ∼0.6. The results demonstrate that the return on investment in feature engineering is highly dependent on characteristics of the specific dataset and may be difficult to predict from correlations between the original input features and the target variable alone.
A sample of the best engineered feature discovered per dataset (that which exhibited the highest r2 correlation to the full dataset, after being evaluated only on its correlation with the validation set during feature engineering) is shown in Table 2. The variable names present in each feature represent the corresponding column names in the original datasets (including the feature names created from Magpie featurization in the first 3 test datasets). Variable names follow the standard Matminer naming convention: minimum (min), maximum (max), range, mean, mode, and average deviation (avgdev). The Pearson r2 correlation between the engineered feature and the target variable is shown in the BE r2 column, while the highest r2 correlation between the target variable and any original input features is shown in the Orig. r2 column. Both r2 values were calculated using the full dataset, even though features were engineered based on their correlation to the validation set only.
The human-readability of each engineered feature enables rapid identification of the input variables which are predictive of the given target variable, in addition to whether input variables are directly or inversely proportional to the target variable, and whether they exhibit higher correlation to the target variable when raised to a specific power or after taking the natural logarithm. For example, heat capacity is highly correlated (r2 = 0.87) with the square root of zero-point vibrational energy (zpve) divided by the cube root of A, a rotational constant. This is consistent with general thermodynamic considerations, as vibrational and rotational degrees of freedom give rise to heat capacity,60,61 and heat capacity is proportional to a factor with dependence on zpve.62 Fish weight is strongly predicted (r2 = 0.98) by the square of the length2 property multiplied by factors related to width and the ratio of height to length3, which differs from the basic intuition that weight scales linearly with volume, which in a first-order approximation is the simple product of length, width, and height. The feature for predicting the compressive strength of concrete includes roughly half of the variables available in the original dataset, which suggests that several ingredients in the concrete formulation (water, cement, aggregates) are significantly more important for predicting compressive strength than others (fly ash, furnace slag, and superplasticizers) which are not present in the engineered feature. These insights are not easily obtained through the use of black box models, which encode the relationships between different input variables using large arrays of coefficients that are generally not human-readable due to their high dimensionality.
The results in Table 2 demonstrate how model complexity may be offloaded from the internal model structure to the input features. In black box models, the relationships between inputs are generally represented by complex data structures (tree ensembles in random forests, hidden layer node interconnections in neural networks), which are difficult to interpret by human domain experts. Using symbolic regression for feature engineering, these relationships can be represented in human-readable form.46 The human-readable features exhibit higher correlations with the target variable than any of the original features in the dataset, which unlocks the potential for using simple linear regression models instead of black box algorithms to achieve the same predictive power. Transfer of the model complexity to the input features significantly enhances model portability, as the resulting linear models can be fully represented by just 2 coefficients: slope and intercept, which makes them easy to deploy in a broad range of settings without the need for specialized machine learning experts.
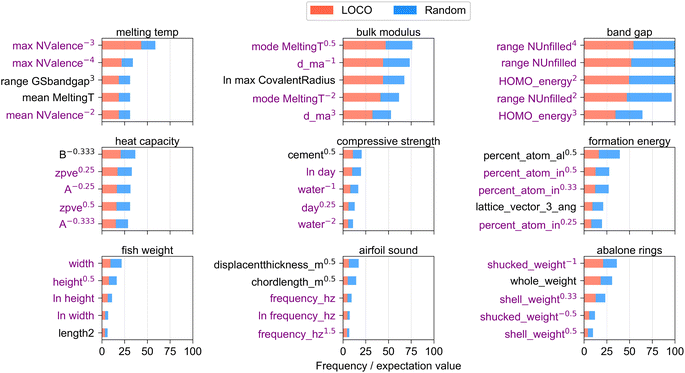
Fig. 3 shows the 5 variable functional forms which were most frequently included in the top 10 engineered features per CV split of a given dataset. The horizontal axis is scaled by the expectation value of the frequency of each variable in the top 10 engineered features if it had been selected randomly. The functional form of each variable, i.e. whether they were raised to powers or a natural logarithm was taken, is shown in each variable label. On average, ∼3.7 of the top 5 variables appeared in the top 5 variables more than once in different functional forms (shown by an average of 3.7 purple variable labels per panel), which suggests that a small number of informative features were used frequently during the feature engineering process due to their high correlation to the target value compared to other variables in the dataset. The results enable key diagnostics about which input variables, and which functional forms of those input variables, contributed to informative inputs for feature engineering. Unlike standard feature importance rankings which are commonly reported for ensemble regression methods58 or make use of game-theoretic formulations such as Shapley,63 the methodology demonstrated here enables ranking of the important input features while additionally highlighting the corresponding functional forms (i.e. power, logarithm) which yield highest correlations to target values, and highlights the resiliency of certain input variables to remain informative over different regions (CV splits) of the dataset.
It is worth noting that variables in the first 3 datasets in Fig. 3 generally appear in higher frequency than those in subsequent datasets. The large number of Magpie columns in the first 3 datasets (see dataset size in Table 1) lowers the expectation value that a single variable will appear in high-performing engineered features across multiple CV splits, which increases the value of frequency/expectation plotted on the horizontal axis. The occurrence of some properties in multiple functional forms, particularly NValence in the melting temp dataset and range NUnfilled in the band gap dataset, attests to the powerful utility of those properties for predicting the given target variable across a broad range of extrapolative CV splits.
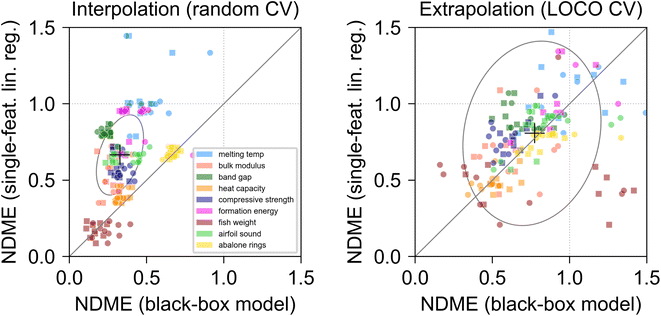
To investigate the trade-off between model performance and interpretability, we compared the prediction error of single-feature linear models to that of black box models in both interpolation and extrapolation problems. The BE feature at each CV split was used as the input for a single-feature linear regression. The prediction accuracy of the linear regression was compared to that of two black box algorithms: one random forest (RF) and one feed-forward back-propagating neural network (NN). Both black box algorithms were fitted using all original input features as inputs. Results from the model comparison are shown in Fig. 4.
For interpolation problems (random CV, left panel), the median NDME across all CV splits was ∼0.67 for single feature linear models and ∼0.33 for black box models, and black box models outperformed linear regressions in 87% of CV splits. This is consistent with the understanding that complex algorithms can effectively capture subtleties of the response surface in the immediate neighborhood of training data with arbitrary precision.3 In three datasets (fish weight, heat capacity, and abalone rings), the linear models performed comparably to the black box models (median NDME for linear models was similar to that of black box models). In the band gap, formation energy, and melting temp datasets, linear models exhibited NDME which was roughly twice as high as that of corresponding black box models. These datasets exhibited some of the lowest BE r2 values of any datasets in Table 2, which supports the intuition that higher r2 correlations between engineered features and target variables enable the construction of higher-performing linear models.
In extrapolation problems (LOCO CV, right panel), the performance difference between single-feature linear regressions and black box models was much less pronounced. The median NDME for linear models, ∼0.81, was an average of ∼5% higher than that of the NDME for black box models (∼0.77). Black box models outperformed linear regressions in 60% of CV splits, with roughly ∼40% of the area of the covariance confidence ellipse, shown by the gray curve, lying inside the region below the y = x line, which suggests that the performance difference between linear models and black box models is highly dependent on the particular dataset and extrapolation region (CV split), and difficult to predict and generalize across different datasets. For extrapolation in the fish weight and heat capacity datasets, single-feature linear regressions almost always outperformed black box models. The engineered features discovered for those datasets exhibited very high median correlations to the target values (r2 > 0.85, Fig. 2), which indicates that the r2 correlation between input features and target variables may serve as a rough guide for predicting whether single feature linear models will outperform black box models in extrapolation problems.
We also examined the effect of including BE features as inputs to the black box models. For each dataset, the performance of linear regression models was compared to that of RF and NN models using 3 different featurization strategies: (1) original: the model was trained using only the original input features in the dataset, including those calculated using Magpie, (2) best engineered (BE): the model was trained using the single best engineered input feature for the given CV split as ranked by its r2 correlation to the validation set, and (3) BE + original: the model was trained using all original input features in addition to the single best engineered feature. Model performance was quantified by the median NDME across all train-test splits for a given dataset and extrapolation or interpolation task (Fig. S5†).
In all cases, the best performing model configuration for a given dataset exhibited lower NDME when performing interpolation than when performing extrapolation, which is consistent with the common understanding that extrapolation outside of the range of training data is more difficult than interpolation of data which was used during the training process.47 Model extrapolation performance either improved or stayed unchanged upon addition of the single best engineered feature in the majority of all test cases, which indicates that the feature engineering process for addition of input variables generally provides performance benefits without risk of decreasing the prediction accuracy. This result suggests that when time and computational resources allow it, the feature engineering process should almost always be used prior to model training. In 4 of 9 test datasets (bulk modulus, heat capacity, fish weight, and abalone rings), single-feature linear models achieved extrapolation performance which was comparable (within error bars, which represent the standard deviation in NDME values cross the CV splits) to that of the best-performing black box models.
The results are noteworthy because they demonstrate that the generally-assumed trade-off between model interpretability and performance is often overstated, especially in extrapolation contexts. Linear regressions which utilize a single input feature in human-readable functional form can often extrapolate just as well as black box models which utilize 102–103 inputs. The linear models exhibit superior portability, decreased computational overhead, and improved ease of use by non-experts over black box models, which highlights their potential for deployment in edge computing applications and other resource-constrained environments without the need for expert operators.
Conclusions
Selection of predictive models for SciML problems is often driven by the assumption that an implicit trade-off exists between model performance and complexity. We investigated this trade-off by engineering human-readable input features for a set of 9 open datasets using a simple random search algorithm, and then compared the performance of single-feature linear regressions to that of black box random forest and neural network machine learning models in interpolative and extrapolative settings across different SciML tasks. For interpolation tasks, the linear models exhibited average prediction error roughly twice as high as that of the black box models, and outperformed black box models in just 13% of cross-validation splits. However, when extrapolating to new regions of the dataset which were not present in the training set, linear models exhibited average prediction error ∼5% higher than that of black box models, and outperformed black box models in ∼40% of cross-validation splits, while enabling faster model training times and improved model portability. Our results suggest that a possible indicator for extrapolation performance of these linear models is the r2 correlation between the engineered feature and the target variable. Further analysis of this effect may warrant further investigation. The use of human-readable input features provided insight into the functional forms of underlying variables which were most predictive of a given set of target values. The results suggest that extrapolation in many SciML problems may be performed using simple linear models which maintain human interpretability without sacrificing performance. When to utilize different models across interpolation and extrapolation regimes is a worthy topic for future work. Increased use of simple interpretable models may help unlock the full potential of machine learning for light-weight edge computing, rapid model portability, and generation of data-driven insights which augment existing human domain knowledge.Data availability
Data and code related to this paper are available at https://github.com/CitrineInformatics-ERD-public/linear-vs-blackbox.Author contributions
ESM, JES, BM, CSR, and JHM conceptualized the work. ESM and JES developed the code and wrote the manuscript.Conflicts of interest
The authors declare no competing financial or non-financial interests.Acknowledgements
This research was developed with funding from the Defense Advanced Research Projects Agency (DARPA) and material is based upon work supported by the United States Air Force under Contract No. FA8650-20-C-7002. Any opinions, findings and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the United States Air Force.References
- Y. Iwasaki, R. Sawada, V. Stanev, M. Ishida, A. Kirihara and Y. Omori, et al., Identification of advanced spin-driven thermoelectric materials via interpretable machine learning, npj Comput. Mater., 2019, 5(1), 1–6 CrossRef.
- J. Wei, X. Chu, X. Y. Sun, K. Xu, H. X. Deng and J. Chen, et al., Machine learning in materials science, InfoMat, 2019, 1(3), 338–358 CrossRef CAS.
- A. Agrawal and A. Choudhary, Deep materials informatics: Applications of deep learning in materials science, MRS Commun., 2019, 9(3), 779–792 CrossRef CAS.
- M. Maniruzzaman, M. J. Rahman, M. Al-MehediHasan, H. S. Suri, M. M. Abedin and A. El-Baz, et al., Accurate diabetes risk stratification using machine learning: role of missing value and outliers, J. Med. Syst., 2018, 42(5), 1–17 CrossRef PubMed.
- I. V. Tetko, D. M. Lowe and A. J. Williams, The development of models to predict melting and pyrolysis point data associated with several hundred thousand compounds mined from PATENTS, J. Cheminf., 2016, 8(1), 1–18 Search PubMed.
- D. Jha, L. Ward, A. Paul, Wk Liao, A. Choudhary and C. Wolverton, et al., Elemnet: Deep learning the chemistry of materials from only elemental composition, Sci. Rep., 2018, 8(1), 1–13 CAS.
- K. Schutt, P. Kessel, M. Gastegger, K. Nicoli, A. Tkatchenko and K. R. Muller, SchNetPack: A deep learning toolbox for atomistic systems, J. Chem. Theory Comput., 2018, 15(1), 448–455 CrossRef PubMed.
- R. Guidotti, A. Monreale, S. Ruggieri, F. Turini, F. Giannotti and D. Pedreschi, A survey of methods for explaining black box models, ACM Comput. Surv., 2018, 51(5), 1–42 CrossRef.
- Z. Yang, Y. C. Yabansu, R. Al-Bahrani, Wk Liao, A. N. Choudhary and S. R. Kalidindi, et al., Deep learning approaches for mining structure-property linkages in high contrast composites from simulation datasets, Comput. Mater. Sci., 2018, 151, 278–287 CrossRef CAS.
- L. Baier, F. Jöhren and S. Seebacher, Challenges in the deployment and operation of machine learning in practice, 2019 Search PubMed.
- A. Paleyes, R. G. Urma and N. D. Lawrence, Challenges in deploying machine learning: a survey of case studies, arXiv, 2020, preprint, arXiv:2011.09926, DOI:10.48550/arXiv.2011.09926.
- R. J. Murdock, S. K. Kauwe, A. Y. T. Wang and T. D. Sparks, Is domain knowledge necessary for machine learning materials properties?, Integr. Mater. Manuf. Innov., 2020, 9(3), 221–227 CrossRef.
- K. T. Butler, D. W. Davies, H. Cartwright, O. Isayev and A. Walsh, Machine learning for molecular and materials science, Nature, 2018, 559(7715), 547–555 CrossRef CAS PubMed.
- C. Rudin, Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead, Nat. Mach. Intell., 2019, 1(5), 206–215 CrossRef PubMed.
- N. Wagner and J. M. Rondinelli, Theory-guided machine learning in materials science, Front. Mater. Sci., 2016, 3, 28 Search PubMed.
- K. Lei, H. Joress, N. Persson, J. R. Hattrick-Simpers and B. DeCost, Aggressively optimizing validation statistics can degrade interpretability of data-driven materials models, J. Chem. Phys., 2021, 155(5), 054105 CrossRef CAS PubMed.
- C. B. Azodi, J. Tang and S. H. Shiu, Opening the black box: Interpretable machine learning for geneticists, Trends Genet., 2020, 36(6), 442–455 CrossRef CAS PubMed.
- P. Mikulskis, M. R. Alexander and D. A. Winkler, Toward interpretable machine learning models for materials discovery, Adv. Intell. Syst., 2019, 1(8), 1900045 CrossRef.
- F. Doshi-Velez and B. Kim, Considerations for evaluation and generalization in interpretable machine learning, in Explainable and interpretable models in computer vision and machine learning, Springer, 2018, pp. 3–17 Search PubMed.
- Z. Xiang, M. Fan, G. V. Tovar, W. Trehem, B. J. Yoon and X. Qian, et al., Physics-constrained Automatic Feature Engineering for Predictive Modeling in Materials Science, in Proceedings of the AAAI Conference on Artificial Intelligence, vol. 35, 2021, pp. 10414–10421 Search PubMed.
- T. Mueller, A. G. Kusne and R. Ramprasad, Machine learning in materials science: Recent progress and emerging applications, Rev. Comput. Chem., 2016, 29, 186–273 CAS.
- Z. Cao, Y. Dan, Z. Xiong, C. Niu, X. Li and S. Qian, et al., Convolutional neural networks for crystal material property prediction using hybrid orbital-field matrix and magpie descriptors, Crystals, 2019, 9(4), 191 CrossRef CAS.
- S. R. Kalidindi, Feature engineering of material structure for AI-based materials knowledge systems, J. Appl. Phys., 2020, 128(4), 041103 CrossRef.
- R. Ouyang, S. Curtarolo, E. Ahmetcik, M. Scheffler and L. M. Ghiringhelli, SISSO: A compressed-sensing method for identifying the best low-dimensional descriptor in an immensity of offered candidates, Phys. Rev. Mater., 2018, 2(8), 083802 CrossRef CAS.
- A. Seko, H. Hayashi, K. Nakayama, A. Takahashi and I. Tanaka, Representation of compounds for machine-learning prediction of physical properties, Phys. Rev. B, 2017, 95(14), 144110 CrossRef.
- A. Seko, T. Maekawa, K. Tsuda and I. Tanaka, Machine learning with systematic density-functional theory calculations: Application to melting temperatures of single-and binary-component solids, Phys. Rev. B, 2014, 89(5), 054303 CrossRef.
- L. Ward, A. Agrawal, A. Choudhary and C. Wolverton, A general-purpose machine learning framework for predicting properties of inorganic materials, npj Comput. Mater., 2016, 2(1), 1–7 CrossRef.
- J. Ling, M. Hutchinson, E. Antono, S. Paradiso and B. Meredig, High-dimensional materials and process optimization using data-driven experimental design with well-calibrated uncertainty estimates, Integr. Mater. Manuf. Innov., 2017, 6(3), 207–217 CrossRef.
- V. Stanev, C. Oses, A. G. Kusne, E. Rodriguez, J. Paglione and S. Curtarolo, et al., Machine learning modeling of superconducting critical temperature, npj Comput. Mater., 2018, 4(1), 1–14 CrossRef.
- L. Ward, A. Dunn, A. Faghaninia, N. E. Zimmermann, S. Bajaj and Q. Wang, et al., Matminer: An open source toolkit for materials data mining, Comput. Mater. Sci., 2018, 152, 60–69 CrossRef.
- A. A. Emery and C. Wolverton, High-throughput DFT calculations of formation energy, stability and oxygen vacancy formation energy of ABO3 perovskites, Sci. Data, 2017, 4(1), 1–10 Search PubMed.
- V. Venkatraman, S. Evjen, H. K. Knuutila, A. Fiksdahl and B. K. Alsberg, Predicting ionic liquid melting points using machine learning, J. Mol. Liq., 2018, 264, 318–326 CrossRef CAS.
- G. Sivaraman, N. E. Jackson, B. Sanchez-Lengeling, A. Vazquez-Mayagoitia, A. Aspuru-Guzik and V. Vishwanath, et al., A machine learning workflow for molecular analysis: application to melting points, Mach. Learn.: Sci. Technol., 2020, 1(2), 025015 Search PubMed.
- T. Xie and J. C. Grossman, Crystal graph convolutional neural networks for an accurate and interpretable prediction of material properties, Phys. Rev. Lett., 2018, 120(14), 145301 CrossRef CAS PubMed.
- H. Huo and M. Rupp, Unified representation of molecules and crystals for machine learning, arXiv, 2017, preprint, arXiv:1704.06439, DOI:10.48550/arXiv.1704.06439.
- D. Weininger, SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules, J. Chem. Inf. Model., 1988, 28(1), 31–36 CrossRef CAS.
- T. S. Lin, C. W. Coley, H. Mochigase, H. K. Beech, W. Wang and Z. Wang, et al., BigSMILES: a structurally-based line notation for describing macromolecules, ACS Cent. Sci., 2019, 5(9), 1523–1531 CrossRef CAS PubMed.
- M. Krenn, F. Häse, A. Nigam, P. Friederich and A. Aspuru-Guzik, Self-Referencing Embedded Strings (SELFIES): A 100% robust molecular string representation, Mach. Learn.: Sci. Technol., 2020, 1(4), 045024 Search PubMed.
- N. E. Jackson, M. A. Webb and J. J. de Pablo, Recent advances in machine learning towards multiscale soft materials design, Curr. Opin. Chem. Eng., 2019, 23, 106–114 CrossRef.
- S. Kearnes, K. McCloskey, M. Berndl, V. Pande and P. Riley, Molecular graph convolutions: moving beyond fingerprints, J. Comput.-Aided Mol. Des., 2016, 30(8), 595–608 CrossRef CAS PubMed.
- G. H. Gu, J. Noh, I. Kim and Y. Jung, Machine learning for renewable energy materials, J. Mater. Chem. A, 2019, 7(29), 17096–17117 RSC.
- E. Perim, D. Lee, Y. Liu, C. Toher, P. Gong and Y. Li, et al., Spectral descriptors for bulk metallic glasses based on the thermodynamics of competing crystalline phases, Nat. Commun., 2016, 7(1), 1–9 Search PubMed.
- J. Cheney and K. Vecchio, Prediction of glass-forming compositions using liquidus temperature calculations, Mater. Sci. Eng., A, 2007, 471(1–2), 135–143 CrossRef.
- C. Rudin and B. Ustun, Optimized scoring systems: Toward trust in machine learning for healthcare and criminal justice, Interfaces, 2018, 48(5), 449–466 CrossRef.
- J. Zeng, B. Ustun and C. Rudin, Interpretable classification models for recidivism prediction, arXiv, 2015, preprint, arXiv:1503.07810, DOI:10.48550/arXiv.1503.07810.
- E. Angelino, N. Larus-Stone, D. Alabi, M. Seltzer and C. Rudin, Learning certifiably optimal rule lists for categorical data, arXiv, 2017, preprint, arXiv:1704.01701, DOI:10.48550/arXiv.1704.01701.
- B. Meredig, E. Antono, C. Church, M. Hutchinson, J. Ling and S. Paradiso, et al., Can machine learning identify the next high-temperature superconductor? Examining extrapolation performance for materials discovery, Mol. Syst. Des. Eng., 2018, 3(5), 819–825 RSC.
- H. J. Lu, N. Zou, R. Jacobs, B. Afflerbach, X. G. Lu and D. Morgan, Error assessment and optimal cross-validation approaches in machine learning applied to impurity diffusion, Comput. Mater. Sci., 2019, 169, 109075 CrossRef.
- D. R. Roberts, V. Bahn, S. Ciuti, M. S. Boyce, J. Elith and G. Guillera-Arroita, et al., Cross-validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure, Ecography, 2017, 40(8), 913–929 CrossRef.
- M2AX dataset, https://hackingmaterials.lbl.gov/matminer/dataset_summary.html#m2ax.
- Double perovskites gap dataset, https://hackingmaterials.lbl.gov/matminer/dataset_summary.html#double-perovskites-gap.
- QM9 dataset, https://deepchemdata.s3-us-west-1.amazonaws.com/datasets/qm9.csv.
- Concrete dataset, https://archive.ics.uci.edu/ml/datasets/Concrete+Compressive+Strength.
- Transparent conductors dataset, https://www.kaggle.com/c/nomad2018-predict-transparent-conductors/data.
- Fish market dataset, https://www.kaggle.com/aungpyaeap/fish-market?ref=hackernoon.com.
- Airfoil self noise dataset. https://archive.ics.uci.edu/ml/datasets/Airfoil+Self-Noise.
- Abalone age dataset, https://archive.ics.uci.edu/ml/datasets/abalone.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion and O. Grisel, et al., Scikit-learn: Machine learning in Python, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- K. Liu, A. L. Nazarova, A. Mishra, Y. Chen, H. Lyu and L. Xu, et al., Dielectric Polymer Genome: Integrating Valence-Aware Polarizable Reactive Force Fields and Machine Learning, in Advances in Parallel & Distributed Processing, and Applications, Springer, 2021, pp. 51–64 Search PubMed.
- N. Sebbar, H. Bockhorn and J. W. Bozzelli, Structures, thermochemical properties (enthalpy, entropy and heat capacity), rotation barriers, and peroxide bond energies of vinyl, allyl, ethynyl and phenyl hydroperoxides, Phys. Chem. Chem. Phys., 2002, 4(15), 3691–3703 RSC.
- R. S. Grev, C. L. Janssen and H. F. Schaefer III, Concerning zero-point vibrational energy corrections to electronic energies, J. Chem. Phys., 1991, 95(7), 5128–5132 CrossRef CAS.
- E. A. Gomaa and R. T. Rashad, Thermal and thermodynamic parameters for glycine (GL) solvation in water theoretically, Biomed. j. sci. technol. res., 2019, 23(2), 17345–17348A Search PubMed.
- I. E. Kumar, S. Venkatasubramanian, C. Scheidegger and S. Friedler. Problems with Shapley-value-based explanations as feature importance measures, in International Conference on Machine Learning, PMLR, 2020, pp. 5491–500 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3dd00082f |
| This journal is © The Royal Society of Chemistry 2023 |