Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
ESAMP: event-sourced architecture for materials provenance management and application to accelerated materials discovery†
Michael J.
Statt
*a,
Brian A.
Rohr
a,
Kris
Brown
a,
Dan
Guevarra
c,
Jens
Hummelshøj
b,
Linda
Hung
 b,
Abraham
Anapolsky
b,
John M.
Gregoire
b,
Abraham
Anapolsky
b,
John M.
Gregoire
 *c and
Santosh K.
Suram
*c and
Santosh K.
Suram
 *b
*b
aModelyst LLC, Palo Alto, CA 94303, USA. E-mail: michael.statt@modelyst.io
bAccelerated Materials Design and Discovery, Toyota Research Institute, Los Altos, CA 94040, USA. E-mail: santosh.suram@tri.global
cDivision of Engineering and Applied Science, California Institute of Technology, Pasadena, CA 91125, USA. E-mail: gregoire@caltech.edu
First published on 21st June 2023
Abstract
While the vision of accelerating materials discovery using data driven methods is well-founded, practical realization has been throttled due to challenges in data generation, ingestion, and materials state-aware machine learning. High-throughput experiments and automated computational workflows are addressing the challenge of data generation, and capitalizing on these emerging data resources requires ingestion of data into an architecture that captures the complex provenance of experiments and simulations. In this manuscript, we describe an event-sourced architecture for materials provenance (ESAMP) that encodes the sequence and interrelationships among events occurring in a simulation or experiment. We use this architecture to ingest a large and varied dataset (MEAD) that contains raw data and metadata from millions of materials synthesis and characterization experiments performed using various modalities such as serial, parallel, multi-modal experimentation. Our data architecture tracks the evolution of a material's state, enabling a demonstration of how state-equivalency rules can be used to generate datasets that significantly enhance data-driven materials discovery. Specifically, using state-equivalency rules and parameters associated with state-changing processes in addition to the typically used composition data, we demonstrated marked reduction of uncertainty in prediction of overpotential for oxygen evolution reaction (OER) catalysts. Finally, we discuss the importance of ESAMP architecture in enabling several aspects of accelerated materials discovery such as dynamic workflow design, generation of knowledge graphs, and efficient integration of simulation and experiment.
Introduction
Accelerating materials discovery is critical for a sustainable future and the practical realization of emergent technologies. Data-driven methods are anticipated to play an increasingly significant role in enabling this desired acceleration, which would be greatly facilitated by the establishment and community adoption of data structures and databases that capture data from the broad range of materials experiments. In computational materials science, automated workflows have been established to produce large and diverse materials datasets. While these workflows and associated data management tools can be improved to facilitate capturing of a materials' state and enable easy capture of re-configurable analysis methods, their current implementations have facilitated a host of materials discoveries,1–4 emphasizing the importance of continued development of materials data architectures. In case of experimental materials science, the majority of the data remains in human readable format and is not ingested into a database. In cases where the databases exist, they are either large with limited scope (ICSD, ICDD, which contains hundreds of thousands of X-ray diffraction patterns) or are diverse but have limited data.5–7 This has limited application of machine learning for acceleration of experimental materials discovery to specific datasets such as microstructure data, X-ray diffraction spectra, X-ray absorption spectra, or Raman spectra.8–11Recent application of high-throughput experimental techniques has resulted in two large, diverse experimental datasets: (a) High Throughput Experimental Materials (HTEM) dataset, which contains synthesis conditions, chemical composition, crystal structure, and optoelectronic property measurements (>150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 entries), and (b) Materials Experiment and Analysis Database (MEAD) that contains raw data and metadata from millions of materials synthesis and characterization experiments, as well as the corresponding property and performance metrics.12,13 These datasets contain thousands to millions of data entries for a given type of experimental process, but the experimental conditions or prior processing of the materials leading up to the process of interest can vary substantially. The multitude of process parameters and provenances results in datasets whose richness could be fully realized and utilized if the context and provenance of each experiment were appropriately modeled. In contrast to computational data, where the programmatic workflows facilitate provenance tracking, experimental workflows generally experience more variability from many on-the-fly decisions as well as environmental factors and evolution of the instrumentation. Sensitivity to historical measurements is generally higher in experiments since any measurement could conceivably alter the material, making any materials experiment a type of “processing.” Factors ranging from instrument contamination to drifting detector calibration also may play a role. Therefore, a piece of experimental data must be considered in the context of the parameters used for its generation and the entire experimental provenance.
000 entries), and (b) Materials Experiment and Analysis Database (MEAD) that contains raw data and metadata from millions of materials synthesis and characterization experiments, as well as the corresponding property and performance metrics.12,13 These datasets contain thousands to millions of data entries for a given type of experimental process, but the experimental conditions or prior processing of the materials leading up to the process of interest can vary substantially. The multitude of process parameters and provenances results in datasets whose richness could be fully realized and utilized if the context and provenance of each experiment were appropriately modeled. In contrast to computational data, where the programmatic workflows facilitate provenance tracking, experimental workflows generally experience more variability from many on-the-fly decisions as well as environmental factors and evolution of the instrumentation. Sensitivity to historical measurements is generally higher in experiments since any measurement could conceivably alter the material, making any materials experiment a type of “processing.” Factors ranging from instrument contamination to drifting detector calibration also may play a role. Therefore, a piece of experimental data must be considered in the context of the parameters used for its generation and the entire experimental provenance.
The importance of sample and process history of the experimental data makes it challenging to identify which measurement data can be aggregated to enable data-driven discovery. The standard practice for generating a shareable dataset is to choose data that match a set of process and provenance parameters and consider most or all other parameters to be inconsequential. This method is highly subjective to the individual researcher. For both human and machine users of the resulting dataset, the ground truth of the sample-process provenance is partially or fully missing. In addition, an injection of assumptions prior to ingestion into a database creates datasets that do not adhere to the Findability, Accessibility, Interoperability, and Reusability (FAIR) guiding principles14 resulting in lack of interoperability, creation of data silos that cannot be analyzed efficiently to generate new insights and accelerate materials discovery. As a result, the data's value is never fully realized, motivating the development of data management practices that closely link data ingestion to data acquisition.
Given the complexity and variability in materials experimentation, several tailored approaches such as ARES, AIR-Chem, and Chem-OS have been developed to enable integration between data ingestion and acquisition for specific types of experiments.15–17 Recently, a more generalizable solution for facilitating experiment specification, capture, and automation called ESCALATE was developed.18 Such approaches aim to streamline and minimize information loss that occurs in an experimental laboratory. We focus on modeling the complete ground truth of materials provenances that could operate on structured data resulting either from a specialized in-house data management software or a more general framework such as ESCALATE.
Prior efforts such as The Materials Commons,19 GEMD,20 and PolyDAT21 have also focused on modeling materials provenances. GEMD uses a construction based on specs and runs for materials, ingredients, processes, and measurements. However, there isn't an explicit distinction between measurements and processes. Especially, in case of in-operando or in situ experiments, a single experiment corresponds to both a process and also a measurement. PolyDAT focuses on capturing transformations and characterizations of polymer species. Materials Commons focuses on creation of samples, datafiles, and measurements by processes. We acknowledge the efforts of these earlier works, here we aim to further simplify the data architecture such that it is easily generalizable for various data sources. We also simplify various terminologies such as materials, ingredients, processes, measurements, characterizations, transformations into three main entities – sample, process, and process data. We also introduce a concept called “state” that enables dynamic sample → process data mapping and demonstrate its value for machine learning.
We use an event-sourced architecture for materials provenances (ESAMP) to capture the ground truth of materials experimentation. This architecture is inspired by event-sourced architectures used in software design wherein the whole application state is stored as a sequence of events. This architecture maintains relationships among experimental processes, their metadata, and their resulting primary data to strive for comprehensive representation of the experiments. We believe that these attributes make ESAMP broadly applicable for materials experiments and beyond. We discuss database architecture decisions that enable deployment for a range of experiment throughput and automation levels. We also discuss the applicability of ESAMP to primary data acquisition modes such as serial, parallel, and multimodal experimentation. Finally, we present a specific instantiation of ESAMP for one of the largest experimental materials databases (MEAD) named Materials Provenance Store22 (MPS) consisting of more than 6 million measurements on 1.5 million samples. We demonstrate facile information retrieval, analysis, and knowledge generation from this database. The primary use case described herein involves training machine learning models for catalyst discovery, where different definitions of provenance equivalence yield different datasets for model training that profoundly impact the ability to predict catalytic activity in new compositions spaces. We also discuss the universality of our approach for materials data management and its opportunities for the adoption of machine learning in many different aspects of materials research.
ESAMP description
Overview
ESAMP is a database architecture designed to store experimental materials science data. It aims to capture all three of the types of aforementioned data: (1) information about the samples in the database including storing provenance regarding how they were created and what processes they have undergone, (2) raw data from processes run on the samples, and (3) information derived from analyses of these raw data.Altogether, this architecture enables users to use simple SQL queries to answer questions like:
• What is the complete history of a given sample and any other samples used to create this one?
• How many samples have had XRD run on them both before and after an electrochemistry experiment?
• What is the figure of merit resulting from a given set of raw data analyzed using different methods?
Identification of data to evaluate any scientific question requires consideration of the context of the data, motivating our design of the ESAMP structure to intuitively specify contextual requirements of the data. For example, if a researcher wishes to begin a machine learning project, creating a custom dataset for their project can be done by querying data in the ESAMP architecture. For example, training data for machine learning prediction of the overpotential in chronopotentiometry (CP) experiments from catalyst composition can be obtained via a query to answer questions such as
• Which samples have undergone XPS then CP?
• How diverse are the sample compositions in a dataset?
The researcher may further restrict the results to create a balanced dataset or a dataset with specified heterogeneity with respect to provenance and experiment parameters. The query provides transparent self-documentation of the origins of such a dataset; any other researcher wondering how the dataset was created can look at the WHERE clause in the SQL query to see what data was included and excluded.
To enable these benefits, we must first track the state of samples and instruments involved in a laboratory to capture the ground truth completely. In this article, we focus mainly on the state of samples and note that the architecture could capture the state of instruments or other research entities. A sample provenance can be tracked by considering three key entities: sample, process, and process_data, which are designed to provide intuitive ingestion of data from both traditional manual experiments and their automated or robotic analogues.
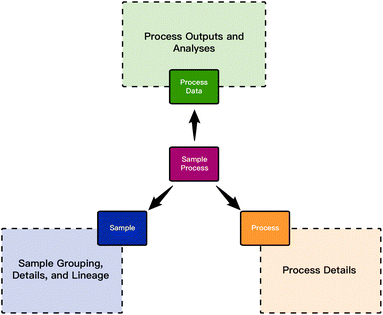
These three entities connected via a sample_process table form the framework's central structure. Fig. 1 shows these entities and their relationships. The three shaded boxes indicate the secondary tables that support the central tables by storing process details, sample details, and analyses. Each region is expanded upon below.
Samples, collections, and lineage
The trinity of sample, process, and process-data enable us to have a generalized framework that captures the ground truth associated with any given sample in an experimental dataset. However, interpretation of experimental data requires us to capture the provenance of a sample completely. That is, throughout the sample's lifetime, it is important to track three key things:• How was the sample created?
• What processes occurred to the sample?
• If the sample no longer exists, how was it consumed?
The middle question is directly answered by the sequence of entries in the sample_process table wherein each record in sample_process specifies the time that a sample underwent a process. This concept is complicated by processes that merge, split, or otherwise alter physical identification of samples. Such processes are often responsible for the creation and consumption of samples, for example the deposition of a catalyst onto an electrode or the use of the same precursor in many different molecule formulations. In these cases, the process history of the “parent” catalyst or precursor is an inherent part of the provenance of the “child” catalyst electrode or molecular material. These potentially-complex lineages are tracked through the sample_ancestor and sample_parent entities as shown in Fig. 2a.
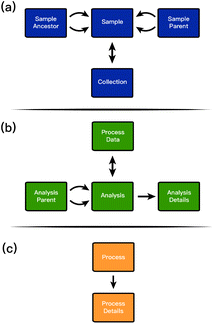 | ||
| Fig. 2 An overview of the three major areas of the framework as shown in Fig. 1. Each region is centered on one of the three entities connected to the central SampProc entity: (a) Samp (b) ProcData (c) Proc. | ||
Both the SampParent and SampAnc entities are defined by their connection to two sample entities, indicating a parent/ancestor and child/descendant relationship, respectively. The SampParent entity indicates that the child sample was created from the parent sample and should inherit its process history lineage. Each SampParent can be decorated with additional attributes to indicate its role in the parent–child relationship, such as labeling the anode and cathode when creating a battery. The SampAnc entity is nearly identical to SampParent with an additional attribute called “rank” that indicates the number of generations between the ancestor and the descendant. A rank of 0 indicates a parent–child relationship, while a rank of 2 indicates a great-grandparent type relationship. The parent and the ancestor tables are not essential to the database and are tables that can be derived from the materials provenance. However, these derived tables are extremely valuable for simplifying complex queries dependent on sample lineages.
The final entity connected to a sample is the collection. It is common for researchers to group samples. For example, in high throughput experiments many samples may exist on the same chip or plate, or researchers may include in a collection all samples synthesized for a single project. In these cases, researchers need to be able to keep track of and make queries based on that information. It is clear from the previously-mentioned example that many samples can (and almost always do) belong to at least one collection. It is also important that we allow for the same sample to exist in many collections. For example, a researcher may want to group samples by which plate or wafer they are on, which high-level project they are a part of, and which account they should be billed to all at the same time. The corresponding many-to-many relationships are supported by ESAMP.
Processes & process details
A process represents one experimental procedure (e.g. a synthesis or characterization) that is applied to a sample. The only requirement imposed on a process is that it must be possible to sort them chronologically. Chronological sorting is essential for accurately representing a sample's process history. Therefore, each process is uniquely associated with a timestamp and machine/user. There is an underlying assumption that for a given timestamp and a given machine/user, only 1 process is occurring, although that process may involve multiple samples.While single-step experiments on machine-based workflows can easily provide a precise timestamp for each process, it is cumbersome and error-prone for researchers to provide these at the timescale of seconds or even hours. Additionally, some multi-step processes may reuse the initial timestamp throughout each step, associating an initiation timestamp with a closely-coupled series of experiments whose ordering is known but whose individual timestamps are not tracked. It is important to add a simple ordering parameter to represent the chronology when the timestamp alone is insufficient. For tracking manual experiments, this ordering parameter allows researchers to record the date and a counter for the number of experiments they have completed that day. In multi-step processes, each step can be associated with an index to record the order of steps.
Processes indicate that an experimental event has occurred to one or more samples. However, it is important to track information describing the type of process that occurred and the process parameters used, or generally any information that would be required to reproduce the experiment. A given research workflow may comprise many different types of experiments, such as electrochemical, XPS, or deposition processes. Each of these types of processes will also be associated with a set of input parameters. The ProcDet entity and its associated process-specific tables are used to track this important metadata for each process. A more comprehensive discussion on the representation of process details for various relational database management system (RDMS) implementations is provided in the ESI.†
Process data & analysis
While ProcDet tracks inputs to a Proc, ProcDet tracks the output of a Proc. For reproducibility, transparency, and ability to continue experiments without reliance on an active database connection, it is prudent to store process outputs as raw files independent from the data management framework. Therefore, while ProcData may include relevant data parsed from the raw files, it should also always include a raw file path. Additionally, attributes can be added to specify the location to search for the file, such as an Amazon S3 bucket or local storage drive. A single file may also contain multiple pieces of data that each refers to different samples. This complexity motivates the inclusion of the start and end line numbers for a file identifying information for ProcData. If an entire file should be consumed as a single piece of process data, null values can be provided for those attributes. As a significant amount of scientific data is stored as comma-separated values (CSV) files, it can also be beneficial to parse these files directly into values in the database utilizing flexible column data types, such as JavaScript Object Notation (JSON) that is supported by modern RDMS's. For large datasets, storing data using efficient binary serializations such as Messagepack could be beneficial.23The relationship between process outputs and their associated processes and samples can be complex. The most straightforward relationship is one piece of process data is generated for a single sample, which is typically the case for serial experimentation and traditional experimentation performed without automation. In parallel experimentation, a single process involves many samples, and if the resulting data is relevant to all samples, SampProc has a many-to-one relationship to ProcData. In multi-modal experiments, multiple detectors can generate multiple pieces of data for a single sample in a single process, where SampProc has a one-to-many relationship to ProcData. Parallel, multi-model experimentation can result in many-to-many relationships. To model these different types of experimentation in a uniform manner, ESAMP manages many-to-many relationships between SampProc and ProcData.
The raw output of scientific processes may require several iterative analytical steps before the desired results can be obtained. As the core tenet of this framework design is tracking the full provenance of scientific data, analytical steps must have their lineage tracked similarly to that of samples and processes. This is achieved by the analysis, analysis_details, and analysis_parent tables. The analysis table represents a single analytical step and, similar to Proc, is identified by inputs, outputs, and associated parameters. Just as Proc has a many-to-many relationship with sample, analysis has a many-to-many relationship with process_data; a piece of process data can be used as an input to multiple analyses and a single analysis can have multiple pieces of process data as inputs. The type of analysis and its input parameters are stored in the analysis_detail entity. The analysis type should define the analytical transformation function applied to the inputs, while the parameters are fed into the function alongside the data inputs.
An important difference between analysis and Proc is that an analysis can use the output of multiple ProcData and analysis entities as inputs. This is analogous to the parent–child relationship as that modeled by SampParent. The introduction of analysis_parent table allows for this complex lineage to be modeled. This allows for even the most complex analytical outputs to be traced back to the raw ProcData entities and the intermediate analyses on which they are based.
State
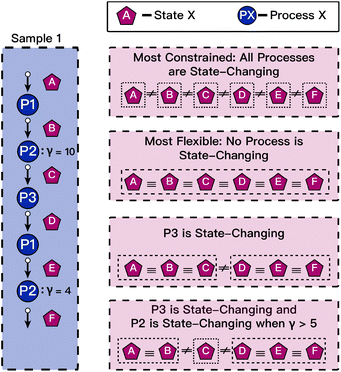
During experiments a sample may be intentionally or unintentionally altered. For example, a researcher could measure the composition of a sample, perform an electrochemical process that unknowingly changes the composition, and finally perform a spectroscopic characterization. Even though the sample label is preserved throughout these three processes, directly associating the composition measurement with the spectroscopic measurement can lead to incorrect analysis because the intervening process altered the link between the two. This example motivates the need for the final entity in the framework, state. The ESAMP model for state assumes that every process irreversibly changes the sample. A state is defined by two sample_process entities that share the same sample and have no sample_process chronologically between them. By managing state under the most conservative assumption that every process alters the sample's state, any state equivalency rules (SERs), i.e. whether a certain type of process alters the state or not, can be applied in a transparent manner. A new state table can be constructed from these SERs, which may be easily modified either by a human or a machine.As state essentially provides a link between the input and output of a process, it is best visualized as a graph. Fig. 3 shows an example state graph. Sample 1 undergoes a series of five processes that involve three distinct types of processes. A new state is created after each process. If no relaxation assumptions are applied, all processes are assumed to be state-changing, and since all states are non-equivalent, it might be invalid to share process data or derived analysis amongst them. Under the most relaxed constraint, no processes are state-changing. However, the utility of state is the ability to apply domain and use-specific rules to model SERs. For example, consider process 3 (P3) to be a destructive electrochemical experiment that changes the sample's composition, while the other processes are innocuous characterization experiments. By designating only P3 as state-changing, the sample can be considered to have only 2 unique states. SERs can be further parameterized by utilizing the ProcDet's of the process to determine state-changing behavior. For example, if P2 is an anneal step, we might only consider it state-changing if the temperature rises above a certain level. By defining simple rules, merging equivalent states yields simpler state graphs that serve as the basis for dataset curation. This powerful concept of state is enabled by the core framework's ability to track the process provenance of samples throughout their lifetime.
Database implementation
The framework (Fig. 4) so far has been defined using standard entity relationship language. It is important to note that this framework can be instantiated in most or all RDMS's and is not tied to a specific implementation. However, the specific implementation details of the framework may change slightly depending on the RDMS used. These changes are vital in deciding the RDMS system that is appropriate for a particular use case.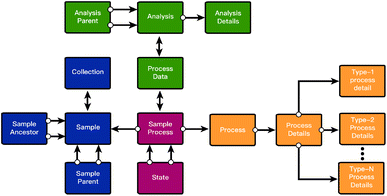 | ||
| Fig. 4 A full graphical representation of the framework described in Fig. 1 and 2. Single headed arrows indicate a many-to-one relationship in the direction of the arrow. Double-headed arrows indicate a many-to-many relationship. | ||
Fig. S1† shows the framework in its entirety. All double-sided arrows indicate a many-to-many relationship. The implementation of many-to-many relationships differs between SQL, NoSQL, and graph databases. In a SQL RDMS such as PostgreSQL, the standard practice uses a “mapping” table where a row is defined simply by its relationship to the two tables with the many-to-many relationship. In graph databases, many-to-many relationships can be represented simply as an edge between two nodes. Additionally, entities that track lineages, such as SampParent, state, and analysis_parent, can also be represented simply as edges between two nodes of the same type. The cost of this simplicity is the reduced constraints on column datatypes as well as a less standardized query functionality.
If complicated process provenance and lineages are expected to exist along with a need to query those lineages, then a graph database may be the right choice. However, if simpler lineages with large amounts of well-structured data are used, a standard SQL RDMS would be more advantageous. Data can even be migrated quite easily between implementations of this framework in two RDMS's if the slight differences noted above are carefully considered. In this implementation we used a postgreSQL database due to the presence of a large amount of reasonably well-structured data. In addition, the postgreSQL database allows us to build a graph database on top of it, which can be used for complex provenance queries.
Results
Implementation of the ESAMP framework is demonstrated via ingestion and modeling of MEAD, the database resulting from high throughput experimental investigation of solar fuels materials in the Joint Center for Artificial Photosynthesis (JCAP).24 MEAD contains a breadth and depth of experiments that make it representative of a broad range of materials experiments. For example, the 51 types of processes include serial, parallel, and multi-modal experiments.Using the most conservative rule that every process is state-changing, the database contains approximately 17 million material states. This dataset contains many compositions in high-order composition spaces, particularly metal oxides with three or more cation elements. For electrocatalysis of the oxygen evolution reaction (OER), the high throughput experiments underlying MEAD have led to the discovery of catalysts with nanostructured mixtures of metal oxides in such high-order composition spaces.25–27 Given the vast number of unique compositions in these high-dimensional search spaces, a critical capability for accelerating catalyst discovery is the generation of machine learning models that can predict composition-activity trends in high-order composition spaces, motivating illustration of ESAMP for this use case.
Catalyst discovery use case
To demonstrate the importance and utility of the management of process provenance and parameters, we consider a use case where data is curated to train a machine learning model and predict the catalytic activity of new catalyst compositions. We commence by considering all MEAD measurements of metal oxides synthesized by inkjet printing and evaluated as OER electrocatalysts, particularly the OER overpotential for an anodic electrochemical current density of 3 mA cm−2. This overpotential is the electrochemical potential above 1.23 V vs. RHE required to obtain the current density, so smaller values correspond to higher, desirable catalytic activity. Measurement of this overpotential can be made by cyclic voltammogram (CV) or chronopotentiometry (CP) measurements.Querying MEAD for all measurements of this overpotential and identifying the synthesis composition for each sample produces a dataset of composition and activity regardless of each sample's history prior to the CP experiment and the electrochemical conditions of the measurement. This dataset is referred to as dataset A in Fig. 5a and contains 660![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 260 measurements of overpotential. Considering a provenance to be the ordered set of process types that occurred up to the overpotential measurement, this dataset contains 19
260 measurements of overpotential. Considering a provenance to be the ordered set of process types that occurred up to the overpotential measurement, this dataset contains 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 129 unique provenances. To increase the homogeneity in provenance and materials processing, the SERs can require that the catalyst samples have been annealed at 400 °C. Additionally, to generate a single activity metric for each sample, the SERs can also require only the most recent or “latest” measurement of activity, which results in a dataset B containing 66
129 unique provenances. To increase the homogeneity in provenance and materials processing, the SERs can require that the catalyst samples have been annealed at 400 °C. Additionally, to generate a single activity metric for each sample, the SERs can also require only the most recent or “latest” measurement of activity, which results in a dataset B containing 66![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 653 measurements, corresponding to 304 unique provenances. To further increase the homogeneity, the SERs can also require the electrolyte pH to be within 0.5 of pH 13 and require those catalysts to have been operated for at least 100 minutes before catalyst activity measurement, resulting in dataset C containing 20
653 measurements, corresponding to 304 unique provenances. To further increase the homogeneity, the SERs can also require the electrolyte pH to be within 0.5 of pH 13 and require those catalysts to have been operated for at least 100 minutes before catalyst activity measurement, resulting in dataset C containing 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 012 measurements. This dataset contains only 29 unique provenances that differ in their sequence of electrochemical experiments that preceded the overpotential measurement.
012 measurements. This dataset contains only 29 unique provenances that differ in their sequence of electrochemical experiments that preceded the overpotential measurement.
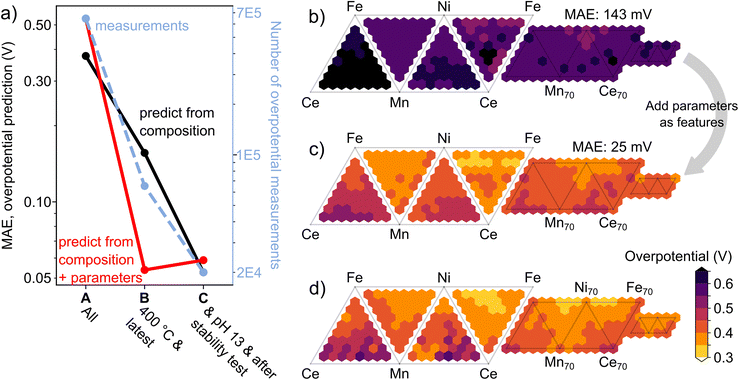
Dataset C contains 63 unique 4-cation composition spaces. To demonstrate machine learning prediction of catalyst activity in new composition spaces, each of these 63 combinations of 4-cation elements is treated as an independent data instance in which the test set is taken to be all catalyst measurements from dataset C where the catalyst composition contains three or all four of the respective 4-cation elements. Keeping the test set consistent, three independent eXtreme Gradient Boosting (XGB) random forest regression models, one for each of the three datasets, were trained to predict over-potential from composition, wherein each case the composition spaces that comprise the test are held out from training. Repeating these exercises for all 63 data instances enables calculation of the aggregate mean absolute error (MAE) for predicting catalyst activity, as shown in Fig. 5a for the three different datasets. The MAE improves considerably when increasing the homogeneity of provenance and experimental parameters from dataset A to B and from dataset B to C, demonstrating the value of using appropriate SERs to curate materials databases with specific provenance and property conditions to generate suitable training data for a specific prediction task.
The parameters used for creating the SERs can also be considered as properties of the catalyst measurements, enabling the training of machine learning models that not only use composition as input but also additional parameters, in the present case the maximum annealing temperature, the number of previous measurements of the catalyst activity, the electrolyte pH, the duration of prior catalyst stability measurements, and whether the measurement occurred by CV or CP. Fig. 5a shows the corresponding results for the same exercise described above wherein the aggregate MAE is calculated for each dataset A, B, and C. This more expressive input space enables a substantial decrease in the MAE when using the dataset B. Whereas, for dataset A this expressive input space marginally increased the MAE, highlighting the importance of combining SER based data classification with regression using richer expressions of the input space.
For the Ce–Fe–Mn–Ni data instance, Fig. 5b shows the prediction using dataset B and only composition as model input, resulting in an MAE of 143 mV. Using the same dataset but expanding the model input to include the experiment and catalyst parameters lowers the MAE to 25 mV, which is the approximate measurement uncertainty (Fig. 5c). Comparison to the ground truth values in Fig. 5d reveals that the prediction in Fig. 5c captures the broad range in activity and the composition-activity trends in each of the four 3-cation and 4-cation composition spaces. Overall, these results demonstrate that curation of data to accelerate materials discovery via machine learning requires management of experiment provenance and parameters.
Discussion
Automated ML pipelines
In the catalyst discovery use case described above, we identified that the choice of state-changing processes had a significant effect in predicting OER overpotential. To avoid making such decisions a priori, which is often not possible in experimental research, all distinguishable processes should be reflected in the data management. For instance, a sample storage event is typically assumed to be non state-changing, which may not be the case. The simplest example is air-sensitive materials whose sample handling between experiments should be documented as “sample handling” processes. The ESAMP framework allows for every event in the laboratory to be defined as a process. However, in practice, capturing every event in a laboratory is infeasible in a typical laboratory setting. There may always exist “hidden” processes that altered a material's state but were not tracked, which compounds the issues discussed above with human-made decisions about what processes are state-changing and whether that designation varies with either sample or process parameters. By liberally defining what constitutes a process and aggregating data from many experimental workflows, ESAMP will ultimately enable machine learning to identify hidden processes and which tracked processes are indeed state-changing.Recently, several research works have focused on developing closed-loop methods to identify optimal materials and processing conditions for several applications such as carbon nanotube synthesis,28 halide perovskite synthesis,29 and organic thin film synthesis.30 The workflows of these experiments are typically static. Similarly, several high-throughput experimental systems deploy static workflows or utilize simple if-then logic to choose amongst a set of pre-defined workflows. Machine learning on data defined using ESAMP that contain various process provenances along with definitions of state-changing processes will enable dynamic identification of workflows that maximize knowledge extraction.
Generality for modeling other experimental workflows
While the breadth of process provenances and the dynamic range of depth within each type of provenance makes the MEAD database an excellent demonstrator of ESAMP, the provenance management and the database schema are intended to be general to all experimental and computational workflows. A given type of experiment may be considered equivalent when performed in 2 different labs, although differences in the process parameters and data management have created hurdles to universal materials data management. Such differences may require lab-specific ingestion scripts and tables, but custom development of these components of ESAMP comprise a low-overhead expansion of the database to accept data from new labs as well as new types of processes. One of the most widely used experimental inorganic crystal structural and diffraction databases (ICDD) was generated by manual curation and aggregation over several decades of X-ray diffraction data generated in many laboratories. We anticipate that ESAMP's universal data management will result in a more facile generation of several large experimental datasets with full provenance that enables data-driven accelerated materials discoveries.In addition to the generation of new insights from provenance management and acceleration of research via more effective incorporation of machine learning, we envision materials provenance management to profoundly impact the integrity of experimental science. In the physical sciences, the complexity of modern experimentation contributes to issues with reproducing published results.31 However, the complexity itself is not the issue, but rather the inability of the Methods sections in journal articles to adequately describe the materials provenance, for example, via exclusion of parameters or processing steps that were assumed to be unimportant, which is exacerbated by complex, many-process workflows. Provided an architecture for provenance management such as ESAMP, data can ultimately determine what parameters and processes are essential for reproducible materials experiments.
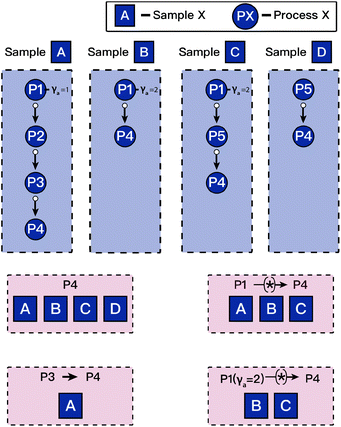
Generation of knowledge graphs and data networks
As discussed above, we anticipate ESAMP to provide the framework that enables the curation of large and diverse datasets with full provenance. Such large datasets are a great starting point for machine learning applications. However, ESAMP is quite general, and adapting a more specific data framework to one's use case can make knowledge extraction easier. These frameworks may extract subsets of the data stored in the main framework and apply simplifying assumptions that apply to the specific use case. However, as long as a link exists between the higher-level framework and ESAMP, then the complete provenance information will still be preserved and queryable. Machine learning datasets, such as those described in datasets A, B, and C in the above use case, are examples of a practical higher-level extraction. See (Fig. 6) for extraction of datasets based on process provenance constraints.One example of a higher-level framework enabled by ESAMP is that of knowledge graphs. Knowledge graphs are a powerful abstraction for storing, accessing, and interpreting data about entities interlinked by an ontology of classes and relations.32 This allows for formal reasoning, with reasoning engines designed for queries like “Return all triples (x1, x2, x3) where ϕ(x1, x2) and ϕ(x1, x3) and (ψ(x2, x3) if and only if θ(x3))”. Beyond direct queries which produce tabular results suited for traditional machine learning applications, machine learning models can be applied directly to relational databases33,34 and knowledge graphs.35 Applications involving knowledge graphs and ontologies have been explored in the space of chemistry and materials science research.36,37
The population of knowledge graphs is mainly facilitated by ESAMP in two ways. Firstly, data within a relational database structure is straightforwardly mappable into the data structure of knowledge graph triples.38 Secondly, a solid grasp of how to resolve distinct entities can be achieved through ESAMP before populating the nodes of the knowledge graph. Alternative approaches of merging all samples with the same label or considering every possibly-distinct sample to be a unique material are too coarse- and fine-grained, respectively. Beyond knowledge graphs, other high-level frameworks specialize in the migration and merging of data between research groups that structure their experiments and analyses differently,39 and these demand structured data such as ESAMP data as their initial input.
Better simulation-experiment integration
While the initial focus on experimental materials data is motivated by the historical lack of concerted effort to manage and track experimental provenance compared to computational materials science, we also envision that ESAMP has the inherent flexibility and expressiveness to model computational materials provenance as well and assist in the holy grail of combining simulation, experiment, and machine learning systematically.In the computational context, simulation details are recorded from automated workflows, input and output files, and code documentation, so the provenance and parameters involved in computations are simpler to track and ingest than experiments. To ingest computational data into ESAMP, we would consider the physically relevant aspects of a simulation, such as atomic positions, composition, micro-structure, or device components and dimensions, to comprise a sample. The simulation itself would be the process, with numerical parameters, simulation approximations, and the compute hardware and software, potentially being relevant process details. Output files and logs would be the process data. Just as samples in experiments can undergo multiple processes, a simulation “sample” can start in a specific configuration, undergo optimization during a “process,” and the new configuration, still associated with the same “sample,” can be passed on for further processing. Computational samples could be combined – results of a simulation are mixed in a new simulation – or partitioned into new samples. The ESAMP framework allowing analyses to be built on multiple process data components is relevant when post-processing simulation data.
Integrating simulation and experimental workflows has long been pursued in materials research. If a computational simulation indicates a material has desirable properties, it is advantageous to directly query all of the experimental data associated with that material to validate the prediction. Similarly, connecting a physical material to its computational counterpart can provide key insight into the fundamental source of its properties.
In general, the significant differences in metadata associated with simulation and experimental workflows have resulted in databases that have significantly different architecture, increasing the barrier for integration of experimental and simulation datasets. Since the key entities of ESAMP are independent of the type of samples, processes, and process data, it allows representation of various forms of data including simulation and experiments using similar architectures. This reduces the accessibility and queryability barrier for integrating experimental and simulation datasets.
As long as the experimental and simulation databases have a single common key (for example: composition, polymer ID) the barrier for initial comparison between simulation and experimental data is significantly reduced because of the increased accessibility and queryability enabled by ESAMP. However, complex queries that depend on the metadata that enable more detailed experiment to simulation comparison may not be obvious. We hope that experts who have experience in simulation-experiment integration will publicly share the specific queries used for comparison in addition to publishing simulation and experimental databases that use similar architecture. For example, an initial comparison of band gap derived from simulation vs. experiment could be based on a query that depends on common composition. A more detailed comparison could be to compare experimental measurements obtained on materials that have been annealed in air within a certain temperature range with simulated band gaps for compositions wherein the corresponding crystal structure is on the thermodynamic convex hull for specific ranges of oxygen chemical potential. Transparent publication of the queries that share similar language for simulation vs. experiment comparison will open the doors for more data-driven integration between theory and experiment. Wherein, simply comparing the findings from theory and experiment can help shed light on where the computational simulations are valid. Additionally, one could train machine learning models to map simulation values to experimental values and use that to make predictions about future experiments. The use of similar architecture for experimental and simulation databases is also likely to aid in development of an interface for simulation assisted autonomous experimentation.
Computational models are often benchmarked against experimentally obtained values. However, this mapping relies upon the common keys used for comparison between simulation and experiment to be valid for the measurement associated with the property. If an intervening process changes the material's state, the mapping between the simulation and experimental dataset would be incorrect. Therefore, it is advantageous to use ESAMP to define state equivalency rules similar to those described earlier, to ensure a more relevant comparison of simulation-experiment data.
Adoption
To accelerate adoption of FAIR usage of experimental data, we believe that other aspects of data management such as data ingestion and data parsing need to be streamlined along with the use of generalizable database architectures such as ESAMP. The generalizable framework and language of our database architecture lends itself to development of simple user-interface modules that will assist in the data ingestion step. However, parsing data even after ingestion can be particularly challenging due to the presence of various file types such as files for X-ray diffraction, electrochemistry, X-ray photoemission spectroscopy etc. We believe that community sourcing of these parsers and their association with process types could be greatly beneficial to our ecosystem, and we particularly point to the effort undertaken by the MaRDA extractors working group.40We also point out that many prior efforts focus on static mapping of samples to attributes derived from process data. Our architecture in conjunction with the concept of “state” enables state equivalency rule based mapping of samples to process data attributes, which expands the utility of this database architecture to analysis of materials workflows that include state altering processes.
Another key barrier for adoption is inconsistencies in the nomenclature used for variables in the database. For example, various databases might use anneal_temperature or heating_temp to describe the same variable. In cases where the type of process (such as characterization, machining etc.) determines the database schema, inconsistent nomenclatures could result in inconsistencies in the database architecture increasing the barrier for interoperability. Whereas, in the case of ESAMP, these variables are present in details tables such as process_details. Therefore, defining sets of equivalent terms for terms used in the details tables can support in achieving interoperability amongst various databases.
Conclusions
In this work, we present a database architecture, called ESAMP, designed for storing materials science research data. We demonstrate that the database architecture captures each material sample's provenance, the data derived from experiments run using each sample, and high-level results derived from the raw data. We further demonstrate how this database can be used to enable material state-aware machine learning datasets. Finally, we discuss the role of ESAMP architecture in accelerated materials discovery via dynamic workflow design, generation of knowledge graphs, and efficient integration of simulation and experiment.Data availability
The entire MEAD data stored in ESAMP provenance is available in a PostgreSQL database. This format requires three steps to make use of: download the compressed SQL database dump file (.tar.gz format) from https://data.caltech.edu/records/hjfx4-a8r81; install PostgreSQL by following the instructions here; extract the .tar.gz file, which will yield a .sql file; follow the PostgreSQL documentation to create a new database from the .sql file. This will create a local copy of the database that we present in this work. The data can be browsed using the DBeaver user. Our docker container scripts to setup the database are provided here: https://github.com/modelyst/mps-docker. Database generation code: the database discussed in this manuscript was generated using the custom built DBgen tool: https://github.com/modelyst/dbgen/. Code to generate Fig. 5: all the scripts used to generate this figure are available at https://github.com/TRI-AMDD/ESAMP-usecase. The notebook ‘query_and_modeling.ipynb’ was used to generate the results and visualizations. The associated database queries are made available in eche_forms_query.sql and eche_pets_query.sql. In addition helper scripts such as myquaternaryulitity.py, myternaryutility.py, quaternary_faces_shells.py are provided to aid in visualization.Conflicts of interest
Modelyst LLC implements custom data management systems in a professional context.Acknowledgements
The development and implementation of the architecture were supported by the Toyota Research Institute through the Accelerated Materials Design and Discovery program. Generation of all experimental data was supported by the Joint Center for Artificial Photosynthesis, a US Department of Energy (DOE) Energy Innovation Hub, supported through the Office of Science of the DOE under Award Number DE-SC0004993. The development of the catalyst discovery use case was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Award DESC0020383. The authors thank Dr Edwin Soedarmadji for stewardship of MEAD and all members of the JCAP High Throughput Experimentation group for the generation of the data. The authors thank Daniel Schweigert for providing insights into standard database management practices. The authors thank Thomas E. Morell for facilitating implementation of DOI-based linkages between MPS and CaltechDATA.Notes and references
- A. Jain, S. P. Ong, G. Hautier, W. Chen, W. D. Richards, S. Dacek, S. Cholia, D. Gunter, D. Skinner, G. Ceder and K. a. Persson, APL Mater., 2013, 1, 011002 CrossRef.
- S. Kirklin, J. E. Saal, B. Meredig, A. Thompson, J. W. Doak, M. Aykol, S. Rühl and C. Wolverton, npj Comput. Mater., 2015, 1, 1–15 CrossRef.
- S. Curtarolo, W. Setyawan, S. Wang, J. Xue, K. Yang, R. H. Taylor, L. J. Nelson, G. L. W. Hart, S. Sanvito, M. Buongiorno-Nardelli, N. Mingo and O. Levy, Comput. Mater. Sci., 2012, 58, 227–235 CrossRef CAS.
- A. Jain, Y. Shin and K. A. Persson, Nat. Rev. Mater., 2016, 1, 15004 CrossRef CAS.
- P. Paufler, Cryst. Res. Technol., 1983, 18, 1318 CrossRef.
- M. Hellenbrandt, Crystallogr. Rev., 2004, 10, 17–22 CrossRef CAS.
- Y. Xu, M. Yamazaki and P. Villars, Jpn. J. Appl. Phys., 2011, 50, 11RH02 CrossRef.
- Y. C. Yabansu, A. Iskakov, A. Kapustina, S. Rajagopalan and S. R. Kalidindi, Acta Mater., 2019, 178, 45–58 CrossRef CAS.
- C. P. Gomes, J. Bai, Y. Xue, J. Björck, B. Rappazzo, S. Ament, R. Bernstein, S. Kong, S. K. Suram and R. B. van Dover, et al. , MRS Commun., 2019, 9, 600–608 CrossRef CAS.
- S. E. Ament, H. S. Stein, D. Guevarra, L. Zhou, J. A. Haber, D. A. Boyd, M. Umehara, J. M. Gregoire and C. P. Gomes, npj Comput. Mater., 2019, 5, 1–7 CrossRef.
- S. B. Torrisi, M. R. Carbone, B. A. Rohr, J. H. Montoya, Y. Ha, J. Yano, S. K. Suram and L. Hung, npj Comput. Mater., 2020, 6, 1–11 CrossRef.
- A. Zakutayev, N. Wunder, M. Schwarting, J. D. Perkins, R. White, K. Munch, W. Tumas and C. Phillips, Sci. Data, 2018, 5, 1–12 CrossRef PubMed.
- E. Soedarmadji, H. S. Stein, S. K. Suram, D. Guevarra and J. M. Gregoire, npj Comput. Mater., 2019, 5, 1–9 CrossRef.
- M. D. Wilkinson, M. Dumontier, I. J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J.-W. Boiten, L. B. da Silva Santos, P. E. Bourne, J. Bouwman, A. J. Brookes, T. Clark, M. Crosas, I. Dillo, O. Dumon, S. Edmunds, C. T. Evelo, R. Finkers, A. Gonzalez-Beltran, A. J. G. Gray, P. Groth, C. Goble, J. S. Grethe, J. Heringa, P. A. C. 't Hoen, R. Hooft, T. Kuhn, R. Kok, J. Kok, S. J. Lusher, M. E. Martone, A. Mons, A. L. Packer, B. Persson, P. Rocca-Serra, M. Roos, R. van Schaik, S.-A. Sansone, E. Schultes, T. Sengstag, T. Slater, G. Strawn, M. A. Swertz, M. Thompson, J. van der Lei, E. van Mulligen, J. Velterop, A. Waagmeester, P. Wittenburg, K. Wolstencroft, J. Zhao and B. Mons, Sci. Data, 2016, 3, 1–9 Search PubMed.
- P. Nikolaev, D. Hooper, F. Webber, R. Rao, K. Decker, M. Krein, J. Poleski, R. Barto and B. Maruyama, npj Comput. Mater., 2016, 2, 1–6 CrossRef.
- J. Li, Y. Lu, Y. Xu, C. Liu, Y. Tu, S. Ye, H. Liu, Y. Xie, H. Qian and X. Zhu, J. Phys. Chem. A, 2018, 122, 9142–9148 CrossRef CAS PubMed.
- L. M. Roch, F. Häse, C. Kreisbeck, T. Tamayo-Mendoza, L. P. E. Yunker, J. E. Hein and A. Aspuru-Guzik, PLoS One, 2020, 15, e0229862 CrossRef CAS PubMed.
- I. M. Pendleton, G. Cattabriga, Z. Li, M. A. Najeeb, S. A. Friedler, A. J. Norquist, E. M. Chan and J. Schrier, MRS Commun., 2019, 9, 846–859 CrossRef CAS.
- B. Puchala, G. Tarcea, E. A. Marquis, M. Hedstrom, H. Jagadish and J. E. Allison, JOM, 2016, 68, 2035–2044 CrossRef.
- GEMD: Graphical Expression of Materials Data, https://citrineinformatics.github.io/gemd-docs/.
- T.-S. Lin, N. J. Rebello, H. K. Beech, Z. Wang, B. El-Zaatari, D. J. Lundberg, J. A. Johnson, J. A. Kalow, S. L. Craig and B. D. Olsen, J. Chem. Inf. Model., 2021, 61, 1150–1163 CrossRef CAS PubMed.
- M. J. Statt, B. A. Rohr, D. Guevarra, S. K. Suram, T. E. Morrell and J. M. Gregoire, Sci. Data, 2023, 10, 184 CrossRef PubMed.
- MessagePack: it’s like JSON. But fast and small, 2021, https://msgpack.org, online, accessed 17 March 2021.
- J. M. Gregoire, C. Xiang, S. Mitrovic, X. Liu, M. Marcin, E. W. Cornell, J. Fan and J. Jin, J. Electrochem. Soc., 2013, 160, F337–F342 CrossRef CAS.
- J. A. Haber, E. Anzenburg, J. Yano, C. Kisielowski and J. M. Gregoire, Adv. Energy Mater., 2015, 5, 1402307 CrossRef.
- M. Favaro, W. S. Drisdell, M. A. Marcus, J. M. Gregoire, E. J. Crumlin, J. A. Haber and J. Yano, ACS Catal., 2017, 7, 1248–1258 CrossRef CAS.
- J. A. Haber, Y. Cai, S. Jung, C. Xiang, S. Mitrovic, J. Jin, A. T. Bell and J. M. Gregoire, Energy Environ. Sci., 2014, 7, 682–688 RSC.
- P. Nikolaev, D. Hooper, N. Perea-López, M. Terrones and B. Maruyama, ACS Nano, 2014, 8, 10214–10222 CrossRef CAS PubMed.
- Z. Li, M. A. Najeeb, L. Alves, A. Z. Sherman, V. Shekar, P. Cruz Parrilla, I. M. Pendleton, W. Wang, P. W. Nega, M. Zeller, J. Schrier, A. J. Norquist and E. M. Chan, Chem. Mater., 2020, 32(13), 5650–5663 CrossRef CAS.
- B. P. MacLeod, F. G. L. Parlane, T. D. Morrissey, F. Häse, L. M. Roch, K. E. Dettelbach, R. Moreira, L. P. E. Yunker, M. B. Rooney, J. R. Deeth, V. Lai, G. J. Ng, H. Situ, R. H. Zhang, M. S. Elliott, T. H. Haley, D. J. Dvorak, A. Aspuru-Guzik, J. E. Hein and C. P. Berlinguette, Sci. Adv., 2020, 6, eaaz8867 CrossRef CAS PubMed.
- D. R. Baer and I. S. Gilmore, J. Vac. Sci. Technol., A, 2018, 36, 068502 CrossRef.
- H. van den Berg, in Current Issues in Mathematical Linguistics, ed. C. Martín-Vide, Elsevier, 1994, vol. 56 of North-Holland Linguistic Series: Linguistic Variations, pp. 319–328 Search PubMed.
- J. A. F. M. Van Gael, R. Herbrich and T. Graepel, Machine learning using relational databases, US Pat., 8,364,612, 2013 Search PubMed.
- M. Cvitkovic, arXiv preprint arXiv:2002.02046, 2020.
- F. Bianchi, G. Rossiello, L. Costabello, M. Palmonari and P. Minervini, arXiv preprint arXiv:2004.14843, 2020.
- K. R. Taylor, R. J. Gledhill, J. W. Essex, J. G. Frey, S. W. Harris and D. C. De Roure, J. Chem. Inf. Model., 2006, 46, 939–952 CrossRef CAS PubMed.
- A. Menon, N. B. Krdzavac and M. Kraft, Curr. Opin. Chem. Eng., 2019, 26, 33–37 CrossRef.
- J. F. Sequeda, S. H. Tirmizi, O. Corcho and D. P. Miranker, Knowledge Engineering Review, 2011, 26, 445–486 CrossRef.
- K. S. Brown, D. I. Spivak and R. Wisnesky, Comput. Mater. Sci., 2019, 164, 127–132 CrossRef.
- MaRDA Extractors, https://github.com/marda-alliance/metadata_extractors.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed schema discussion for relational database implementation of ESAMP. See DOI: https://doi.org/10.1039/d3dd00054k |
| This journal is © The Royal Society of Chemistry 2023 |