Open Access Article
Open Access ArticleModulating the optical properties of carbon dots by peptide condensates†
Dor
Gaash
a,
Simran
Dewan
a,
Avigail Baruch
Leshem
a,
Kumar Sagar
Jaiswal
bc,
Raz
Jelinek
 bc and
Ayala
Lampel
bc and
Ayala
Lampel
 *adef
*adef
aShmunis School of Biomedicine and Cancer Research, George S. Wise Faculty of Life Sciences, Tel Aviv University, Tel Aviv 69978, Israel. E-mail: ayalalampel@tauex.tau.ac.il; Web: https://twitter.com/Lampel_Lab
bDepartment of Chemistry, Ben Gurion University of the Negev, 84105 Beer Sheva, Israel
cIlse Katz Institute for Nanoscale Science and Technology (IKI), Ben Gurion University of the Negen, Israel
dCenter for Nanoscience and Nanotechnology, Tel Aviv University, Tel Aviv 69978, Israel
eSagol Center for Regenerative Biotechnology, Tel Aviv University, Tel Aviv 69978, Israel
fCenter for the Physics and Chemistry of Living Systems, Tel Aviv University, Tel Aviv 69978, Israel
First published on 27th September 2023
Abstract
Here, we utilized designed condensates formed by liquid–liquid phase separation (LLPS) of cationic and aromatic peptide to sequester tyrosine-based carbon dots (C-dots). The C-dots fluorescence is quenched and retrieved upon partitioning and release from condensates, allowing a spatial regulation of C-dots fluorescence which can be utilized for biosensing applications.
Synthetic biomolecular condensates are supramolecular disordered compartments, that are inspired by cellular condensates such as stress granules and nucleoli.1 These designed compartments are formed via liquid–liquid phase separation (LLPS) of proteins,2 polypeptides,3 or peptides4–9 with or without nucleic acids,10 where a molecule-rich liquid phase is condensed within a molecule-poor phase. An attractive characteristic of synthetic biomolecular condensates is that they can be designed to assemble/disassemble or change their properties in response to stimuli. Examples of such systems include condensates which change their chemical composition upon enzymatic activation,11,12 oxidation,13 or addition of chemical fuel.14 Condensates can also be designed to change their material properties following a light trigger5,15 or disassemble upon stimuli such as pH16 and temperature.17
Owing to their responsiveness and ability to efficiently sequester (macro)molecules, synthetic condensates are being exploited1 for the regulation of biocatalytic reactions,13,18 protein synthesis in a cell-free transcription-translation system,19–21 and material synthesis.22 Other than being used as reactors, biomolecular condensates, particularly those formed by peptides, may serve as delivery vectors of therapeutic biomolecules8 including intracellular delivery.23 However, the design of condensates for biosensing applications is yet to be explored.
Building on the ability of designed condensates to sequester large payloads including nanoparticles,24 we sought to develop a biosensing system based on the sequestration of optical nanoparticles in synthetic condensates, including C-dots.16 Owing to the responsive nature of condensates and their ability to encapsulate nanoparticles, we envisioned that the encapsulation and release of C-dots from condensates can provide a means to regulate C-dots optical properties. We recently reported on minimalistic condensates that are formed by LLPS of designer 14-mer peptides containing 3 aromatic amino acids.4 These condensates showed high encapsulation efficiency towards aromatic small molecule dyes. Thus, we hypothesized that condensates formed by aromatic amino acid-containing peptides will sequester optical nanoparticles containing aromatic moieties by π–π interactions.
An example of widely used fluorescent nanoparticles with tunable optical properties are quantum dots.25,26 The optical properties of quantum dots mainly depend on their size and chemical composition. For instance, quantum dots are known to emit light at a wavelength proportional to their diameter for a given excitation wavelength,27 thus their fluorescence can be tailored for specific applications. Quantum dots may be synthesized from inorganic or carbon-based molecules, where the latter are known as C-dots. The fluorescent properties of C-dots are dependent on the chemical content of their crystalline carbonaceous core.28 In addition, the fluorescence of C-dots is known to be excitation dependent. C-dots are also characterized by “structural memory”, the ability to present functional groups of their precursors on the surface of the carbon core.29 C-dots may be classified based on their structure, atom content, and heterogeneity in the crystalline core made mainly out of carbon.30 C-dots can also be synthesized from simple building blocks such as amino acids.29,30 The functional group presented on the synthesized C-dots, originating from the amino acid, determines the quantum yield in correlation to hydrophobicity and volume.31
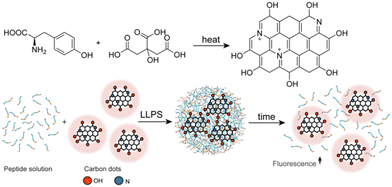
Here, we synthesized C-dots from the aromatic amino acid tyrosine. We showed that these C-dots partition in synthetic condensates that are formed by LLPS of an aromatic amino acid-containing peptide. The fluorescence of partitioned C-dots is quenched due to their confinement and retrieved upon disassembly of the condensates. Thus, the designed peptide-C-dots condensates can be further developed to respond to microenvironmental physical or biochemical cues for the detection of pH fluctuations or increased local enzyme expression.
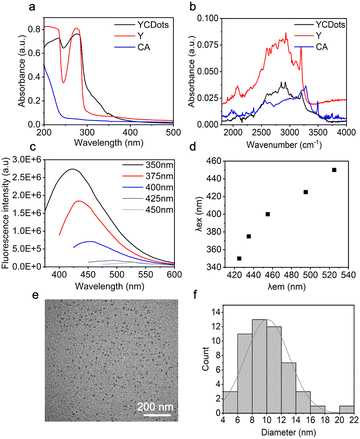
We first synthesized C-dots from citric acid (CA) and tyrosine (Y), designated for interaction with phase-separating peptides (Fig. 1). The tyrosine-based C-dots, termed YCDots, were synthesized via hydrothermal treatment, in an aqueous solution28 under conditions of continuous heat.32 The formation of the desired product was confirmed by using surface chemistry, optical properties, and morphology. First, we used UV-Vis absorption spectroscopy to analyze the resulting YCDots. Compared to its precursors, tyrosine and citric acid, the YCDots showed an absorbance peak at 230 nm (Fig. 2a). This absorbance maximum is attributed to a π–π* transition, which is characteristic of C-dots.33 In addition, the absorbance peak at 280 nm, present in both the YCDots and the tyrosine spectra, indicates the preservation of the phenol moiety on the surface of the conjugated C-dots (Fig. 2a). In contrast, the precursor citric acid exhibited low absorbance in the UV region.
To further analyze the functional groups on the YCDots surface, we performed FTIR analysis. The YCDots and tyrosine both exhibit common peaks at ∼2000 cm−1 and ∼3000 cm−1 (Fig. 2b), corresponding to the phenol group and the hydroxyl moiety within the phenol group, respectively.30 These findings coincide with the structural memory property of C-dots.,29 for the phenol group originating in tyrosine is preserved on the YCDots surface in a conjugated form (Fig. 1).
Next, we performed fluorescence spectroscopy analysis of the YCDots. For this, the emission of YCDots was measured at increasing excitation wavelengths at a range of 350–450 nm. The YCDots showed specific emission maxima up to excitation of 425 nm, where the emission maxima become broader and lower in intensity at higher excitation wavelengths (Fig. 2c). Moreover, the emission maxima redshift in an excitation-dependent manner29 (Fig. 2d).
To characterize the morphology and size of the YCDots, we used transmission electron microscopy (TEM) imaging (Fig. 2e). The TEM analysis confirmed the formation of nanometer-sized particles, with a size distribution which varies between 4–22 nm (Fig. 2f). The relatively wide size distribution is in line with a broad absorption and emission spectra,29 in terms of C-dots properties.34
To further analyze the functional groups on the surface of the YCDots, we performed XPS analysis (Fig. S1, ESI†). The formation of C-dots is validated by the peak at 289.8 eV of the XPS spectrum for the C 1s of the product (Fig. S1b, ESI†). Moreover, the peak at 401.72 eV in the N1 spectrum suggests that the YCDots core is doped with the nitrogen that originates from the amine group of the tyrosine (Fig. S1c, ESI†). The presence of doped nitrogen may lead to a redshift in the emission spectra,28 further contributing to the broad fluorescence of the C-dots.29 This coincides with the dependence of fluorescence on heteroatoms doped into the carbonaceous crystalline core of the C-dots.35 The peak at 532.67 eV for the O1 provides further indication of the presence of phenol groups on the surface of the YCDots (Fig. S1d, ESI†).36 Thus, the XPS results confirm the formation of YCDots and shed light on their optical properties.
Once YCDots formation was validated, we sought to encapsulate them in peptide condensate, which are formed by peptide LLPS in the presence of the YCDots. For this, we used a 14-mer LLPS-promoting peptide WGRGRGRGWPGVGY noted as WGR-1 (Fig. S2, ESI†). The peptide contains three glycine-arginine (RG) dyads, where the glycine promotes flexibility of the backbone and the arginine promotes electrostatic and π–π interactions, which overall promote phase separation. The peptide also contains three aromatic amino acids: two tryptophans (W) and one tyrosine (Y), and elastin-like polypeptide (ELP) domain, which also promotes phase separation.37 Peptide condensates containing YCDots were formed by first dissolving the YCDots (0.5 mg mL−1) in phosphate buffer with 0.2 M NaCl at pH 7.5, and thereafter dissolving the WGR-1 peptide (20 mM) in the YCDots solution. Upon increasing the pH to 7.5, the solution became turbid, and condensates were formed due to the deprotonation of the terminal amine and decrease of electrostatic repulsion between the cationic peptide molecules, which facilitates peptide–peptide interaction and phase separation.4
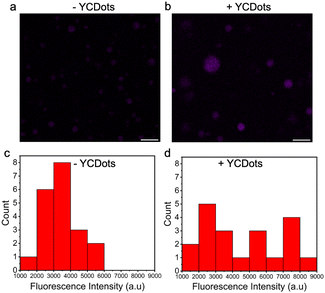
The encapsulation efficiency (%EE) of the YCDots in the condensates was analyzed spectroscopically by measuring the emission of YCDots in the supernatant following centrifugation of the peptide condensates containing YCDots (see Method section in ESI†). The calculated encapsulation efficiency is ∼65.6 ± 5.8% (n = 5), indicating the YCDOts preferentially partition within the condensed peptide phase compared to the dilute phase. The YCDots encapsulation in the condensates is also supported by the increase in the fluorescence intensity of peptide condensates which were prepared in the presence of YCDots compared to that of condensates which were prepared without YCDots, as shown by confocal microscopy analysis using λex = 405 nm (Fig. 3). Measuring the fluorescence intensity of the condensates (Fig. 3c) shows that the peptide condensates themselves have intrinsic autofluorescence, presumably due to the presence of aromatic residues within the peptide sequence. Yet, the presence of YCDots increased the fluorescence intensity of the peptide condensates, as shown by the fluorescence intensity histograms (Fig. 3c and d). Thus, these results suggest that the YCDots partition in the WGR-1 peptide condensates. Based on the surface chemistry analysis, it is plausible that the YCDots partitioning is a result of π–π stacking interactions between the YCDots and the aromatic side chains of the WGR-1 peptide. Yet, the relatively low %EE of the YCDots in the condensates compared with the reported values for other vehicles (Table S1, ESI†) might be a result of electrostatic repulsion between the arginine side chains4 of the peptide and the positively charged nitrogen (Fig. S1c, ESI†) in the YCDots core.
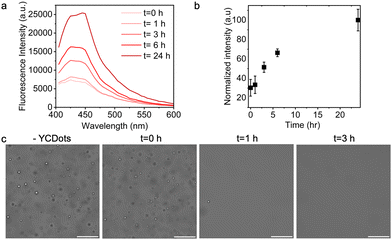
We envisioned that the YCDots fluorescence intensity will decrease upon their encapsulation in condensates due to the tendency of C-dots to quench upon aggregation when partitioned in the condensed phase,38 and that their signal will increase over disassembly of the condensates. Since the condensates are dynamic assemblies that disassemble over time yet are stable at high temperature or high ionic strength (Fig. S3 and S4, ESI†), we monitored the fluorescence of the YCDots in condensates as a function of incubation time by fluorescence spectroscopy. The fluorescence intensity of partitioned YCDots is 34% lower than that of free particles (Fig. S5, ESI†). This partial quenching can be attributed to the presence of free YCDots in the dilute phase or might be a result of YCDots diffusion and mobility in the condensates. The fluorescence signal of the partitioned YCDots increases over incubation time, with a 5-fold increase in intensity after 24 h (Fig. 4a and b). The abundance of the condensates significantly decreases after 1 h of incubation, and the condensates are completely dissociated following 3 h of incubation (Fig. 4c).
Here, we developed a peptide condensate system that enables C-dot encapsulation and fluorescence control. The C-dots design based on the amino acid tyrosine as a precursor, relies on phenol group functionalization that promotes attractive forces with the aromatic peptide side chains. The efficiency of C-dots encapsulation in the condensates is however constrained by repulsive forces caused by doping with positively charged nitrogen from the tyrosine amine groups. This limitation may be resolved by designing C-dots from different precursors i.e., acidic amino acids, and changing the composition of the LLPS-promoting peptide building blocks accordingly. We demonstrated that the release of partitioned C-dots from deconstructed condensates regulates their fluorescence signal. This system can be further developed as a stimuli-responsive platform for applications in precision biosensing, by leveraging the optical characteristics of C-dots and the capacity of condensates to respond to physical or biochemical stimuli. For instance, condensates formed mainly through electrostatic interactions can disassemble and release C-dots upon phosphorylation by specific cancer marker kinases for detection of diseased tissues.
This research was supported by the Israel Science Foundation (ISF) (grant no. 2589/21). We thank Dr V. Holdengreber for the help with the TEM imaging, S. Veretnik for the assistance with the schematic illustration, P. Shekhter for the help with XPS analysis, Dr S. Ben-Zichri N. Shauloff and Dr E. Arad for the help with carbon dots synthesis. We thank the Chaoul Center for Nanoscale Systems of Tel Aviv University for the use of instruments and staff assistance.
Conflicts of interest
There are no conflicts to declare.References
- Y. Dai, L. You and A. Chilkoti, Nat. Rev. Bioeng., 2023, 1, 466–480 CrossRef PubMed.
- X. Wang, C. Shi, J. Mo, Y. Xu, W. Wei and J. Zhao, Angew. Chem., 2020, 132, 2701–2705 CrossRef.
- W. A. Scott, E. G. Gharakhanian, A. G. Bell, D. Evans, E. Barun, K. N. Houk and T. J. Deming, J. Am. Chem. Soc., 2021, 143, 18196–18203 CrossRef CAS PubMed.
- A. Baruch Leshem, S. Sloan-Dennison, T. Massarano, S. Ben-David, D. Graham, K. Faulds, H. E. Gottlieb, J. H. Chill and A. Lampel, Nat. Commun., 2023, 14, 421 CrossRef CAS PubMed.
- I. Katzir, E. Haimov and A. Lampel, Adv. Mater., 2022, 34, 2206371 CrossRef CAS PubMed.
- R. V. Ulijn and A. Lampel, Isr. J. Chem., 2020, 60, 1129–1140 CrossRef CAS.
- M. Abbas, W. P. Lipiński, J. Wang and E. Spruijt, Chem. Soc. Rev., 2021, 50, 3690–3705 RSC.
- J. Liu, E. Spruijt, A. Miserez and R. Langer, Nat. Rev. Mater., 2023, 8, 139–141 CrossRef CAS.
- C. Yuan, Q. Li, R. Xing, J. Li and X. Yan, Chem, 2023, S2451929423002413 Search PubMed.
- W. A. Wee, H. Sugiyama and S. Park, iScience, 2021, 24, 103455 CrossRef CAS PubMed.
- W. M. Aumiller and C. D. Keating, Nat. Chem., 2016, 8, 129–137 CrossRef CAS PubMed.
- K. K. Nakashima, J. F. Baaij and E. Spruijt, Soft Matter, 2018, 14, 361–367 RSC.
- M. Abbas, J. O. Law, S. N. Grellscheid, W. T. S. Huck and E. Spruijt, Adv. Mater., 2022, 34, 2202913 CrossRef CAS PubMed.
- C. Donau, F. Späth, M. Sosson, B. A. K. Kriebisch, F. Schnitter, M. Tena-Solsona, H.-S. Kang, E. Salibi, M. Sattler, H. Mutschler and J. Boekhoven, Nat. Commun., 2020, 11, 5167 CrossRef CAS PubMed.
- N. Martin, L. Tian, D. Spencer, A. Coutable-Pennarun, J. L. R. Anderson and S. Mann, Angew. Chem., 2019, 131, 14736–14740 CrossRef.
- B. Saini, R. R. Singh, D. Nayak and T. K. Mukherjee, ACS Appl. Nano Mater., 2020, 3, 5826–5837 CrossRef CAS.
- S. Sahoo, S. Bera, S. Maiti and D. Dhara, ACS Omega, 2017, 2, 7946–7958 CrossRef CAS PubMed.
- A. M. Küffner, M. Prodan, R. Zuccarini, U. Capasso Palmiero, L. Faltova and P. Arosio, ChemSystemsChem, 2020, 2, e2000001 CrossRef.
- E. Sokolova, E. Spruijt, M. M. K. Hansen, E. Dubuc, J. Groen, V. Chokkalingam, A. Piruska, H. A. Heus and W. T. S. Huck, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 11692–11697 CrossRef CAS PubMed.
- Y. Hayashi, L. K. Ford, L. Fioriti, L. McGurk and M. Zhang, J. Neurosci., 2021, 41, 834–844 CrossRef CAS PubMed.
- Y. Dai, M. Farag, D. Lee, X. Zeng, K. Kim, H. Son, X. Guo, J. Su, N. Peterson, J. Mohammed, M. Ney, D. M. Shapiro, R. V. Pappu, A. Chilkoti and L. You, Nat. Chem. Biol., 2023, 19, 518–528 CrossRef CAS PubMed.
- T. Massarano, A. Baruch Leshem, M. Weitman and A. Lampel, ACS Appl. Mater. Interfaces, 2022, 14, 20520–20527 CrossRef CAS PubMed.
- Y. Sun, S. Y. Lau, Z. W. Lim, S. C. Chang, F. Ghadessy, A. Partridge and A. Miserez, Nat. Chem., 2022, 14, 274–283 CrossRef CAS PubMed.
- K. Kojima, S. Tomita and M. Kamimura, ChemPlusChem, 2023, 88, e202300207 CrossRef CAS PubMed.
- J. Zhou, Y. Yang and C. Zhang, Chem. Rev., 2015, 115, 11669–11717 CrossRef CAS PubMed.
- Q. Zhang, X. Zhang, F. Ma and C. Zhang, Coord. Chem. Rev., 2022, 469, 214674 CrossRef CAS.
- A. M. Wagner, J. M. Knipe, G. Orive and N. A. Peppas, Acta Biomater., 2019, 94, 44–63 CrossRef CAS PubMed.
- L. Đorđević, F. Arcudi, M. Cacioppo and M. Prato, Nat. Nanotechnol., 2022, 17, 112–130 CrossRef PubMed.
- D. N. Bloch, M. Sandre, S. Ben Zichri, A. Masato, S. Kolusheva, L. Bubacco and R. Jelinek, Nanoscale Adv., 2023, 5, 1356–1367 RSC.
- D. N. Bloch, S. Ben Zichri, S. Kolusheva and R. Jelinek, Nanoscale Adv., 2020, 2, 5866–5873 RSC.
- S. Pandit, P. Behera, J. Sahoo and M. De, ACS Appl. Bio Mater., 2019, 2, 3393–3403 CrossRef CAS PubMed.
- J. Zhou, Y. Yang and C. Zhang, Chem. Commun., 2013, 49, 8605 RSC.
- L. Yang, W. Jiang, L. Qiu, X. Jiang, D. Zuo, D. Wang and L. Yang, Nanoscale, 2015, 7, 6104–6113 RSC.
- S. Ben-Zichri, S. Rajendran, S. K. Bhunia and R. Jelinek, Bioconjugate Chem., 2022, 33, 1663–1671 CrossRef CAS PubMed.
- L. Efremushkin, S. K. Bhunia, R. Jelinek and A. Salomon, J. Phys. Chem. Lett., 2017, 8, 6080–6085 CrossRef CAS PubMed.
- D. Rosenthal, M. Ruta, R. Schlögl and L. Kiwi-Minsker, Carbon, 2010, 48, 1835–1843 CrossRef CAS.
- S. Roberts, T. S. Harmon, J. L. Schaal, V. Miao, K. Li, A. Hunt, Y. Wen, T. G. Oas, J. H. Collier, R. V. Pappu and A. Chilkoti, Nat. Mater., 2018, 17, 1154–1163 CrossRef CAS PubMed.
- N. Shauloff, S. Bhattacharya and R. Jelinek, Carbon, 2019, 152, 363–371 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc03945e |
| This journal is © The Royal Society of Chemistry 2023 |