Open Access Article
Open Access ArticleApplying deep learning to iterative screening of medium-sized molecules for protein–protein interaction-targeted drug discovery†
Yugo
Shimizu
 a,
Tomoki
Yonezawa
a,
Yu
Bao
a,
Junichi
Sakamoto
b,
Mariko
Yokogawa
a,
Toshio
Furuya
c,
Masanori
Osawa
a,
Tomoki
Yonezawa
a,
Yu
Bao
a,
Junichi
Sakamoto
b,
Mariko
Yokogawa
a,
Toshio
Furuya
c,
Masanori
Osawa
 a and
Kazuyoshi
Ikeda
a and
Kazuyoshi
Ikeda
 *ad
*ad
aDivision of Physics for Life Functions, Keio University Faculty of Pharmacy, 1-5-30 Shibakoen, Minato-ku, Tokyo 105-8512, Japan. E-mail: ikeda-kz@pha.keio.ac.jp
bAxcelead Drug Discovery Partners, Inc., 26-1, Muraoka-Higashi 2-chome, Fujisawa, Kanagawa 251-0012, Japan
cDrug Discovery Department, Research & Development Division, PharmaDesign, Inc., Hatchobori 2-19-8, Chuo-ku, Tokyo 104-0032, Japan
dHPC- and AI-driven Drug Development Platform Division, Center for Computational Science, RIKEN, Yokohama 230-0045, Japan
First published on 11th May 2023
Abstract
We combined a library of medium-sized molecules with iterative screening using multiple machine learning algorithms that were ligand-based, which resulted in a large increase of the hit rate against a protein–protein interaction target. This was demonstrated by inhibition assays using a PPI target, Kelch-like ECH-associated protein 1/nuclear factor erythroid 2-related factor 2 (Keap1/Nrf2), and a deep neural network model based on the first-round assay data showed a highest hit rate of 27.3%. Using the models, we identified novel active and non-flat compounds far from public datasets, expanding the chemical space.
Protein–protein interactions (PPIs) are important targets in drug discovery research.1 Given the low hit rate in primary screening with small-molecule libraries for PPI targets, it is difficult to obtain new active compounds based on the commonality of structures and properties of hit compounds (i.e., hit expansion).2 Compounds with relatively large molecular weights, multiple aromatic rings, and non-flat structures are suitable for inhibiting PPI targets.3,4 We previously successfully identified hit compounds that inhibit the Kelch-like ECH-associated protein 1/nuclear factor erythroid 2-related factor 2 (Keap1/Nrf2) PPI using a combination of machine learning (ML) and a PPI-oriented library (DLiP1).5 Keap1/Nrf2 PPI is an important target that regulates antioxidant defense system and is involved in various diseases such as neurodegenerative and metabolic disorders.6 DLiP1 consists of 12
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 593 medium-sized compounds suitable for the PPI interface. Recently, we developed a new PPI library called DLiP2 containing 2722 of these compounds. The DLiP1/DLiP2 library compounds were synthetic medium-sized molecules with molecular weights between 400 and 650, designed with consideration for further structural expansion in the hit-to-lead process to molecules with molecular weights ≥ 500.
593 medium-sized compounds suitable for the PPI interface. Recently, we developed a new PPI library called DLiP2 containing 2722 of these compounds. The DLiP1/DLiP2 library compounds were synthetic medium-sized molecules with molecular weights between 400 and 650, designed with consideration for further structural expansion in the hit-to-lead process to molecules with molecular weights ≥ 500.
In recent years, ML including deep learning (DL), has been increasingly applied during the early stages of drug discovery research.7 However, some limitations of using ML for hit and lead identification exist. For example, the applicability of models is limited when the training and test data are located in distant chemical spaces.8 Although we showed that ligand-based virtual screenings (LBVSs) using ML methods can slightly increase the hit rate even when the chemical space is far apart, they are still not efficient enough for drug discovery to reach the next stages such as hit expansion.5 To solve this problem, an iterative screening method is considered a promising approach.9,10 Iterative screening methods involve repeating a cycle of assays using a relatively small number of compounds selected based on predictive models, adding new experimental results from the assays into the training data of the models, and then reconstructing the models. Reker et al. used iterative screening combined with active learning to efficiently identify PPI inhibitors from a large, high-throughput screening (HTS) compound collection of small molecules.11 However, PPIs are challenging targets, and adaptation of iterative screening for PPI targets with medium-sized molecules has not yet been demonstrated.
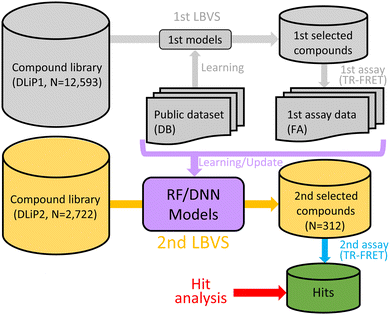
In this study, we applied a model update approach using DL (Fig. 1) to identify Keap1/Nrf2 PPI inhibitors from the DLiP2 library to expand the chemical space of hit compounds from our first assay5 against the DLiP1 library using time-resolved fluorescence resonance energy transfer (TR-FRET). We propose a new approach using libraries of medium-sized molecules and demonstrate the effectiveness of this method in expanding hits against PPI targets to which small molecular libraries are difficult to adapt.
Whereas the DLiP1 library was designed based on structure-based drug design approach (docking calculations for the interfaces of known PPI targets), the DLiP2 library was designed as containing compounds that had non-flat (sphere-like) shapes or new scaffolds (spiro) suitable for PPI inhibitors. For the retrieval of sphere-like compounds, the criterion for the sum of the two normalized principal moments of inertia ratios (NPR1 + NPR2) > 1.3 was used, resulting in 48% (1294/2722) of the DLiP2 compounds meeting this criterion. In our first assay5 for Keap1/Nrf2 using DLiP1, fifteen compounds were identified as hit compounds and 47% (7/15) of those had NPR1 + NPR2 values > 1.3, which was higher than the 11% (1382/12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 492) of the DLiP1. Therefore, the DLiP2 library was considered as prospective for discovering new Keap1/Nrf2 PPI inhibitors.
492) of the DLiP1. Therefore, the DLiP2 library was considered as prospective for discovering new Keap1/Nrf2 PPI inhibitors.
To improve our previous models for predicting Keap1/Nrf2 PPI inhibitory activities of medium-sized molecules, we created new training datasets combining public datasets and our first inhibitory assay (FA) results (i.e., the initial batch of iterative screening). We used two datasets obtained from public data sources: one was a database (DB) dataset (also used in our previous work)5 including 108 active and 106 inactive compounds against the Keap1/Nrf2 PPI and the other was a miscellaneous putative inactive (MISC-PI) dataset including 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 973 active compounds against PPI targets other than Keap1/Nrf2. The MISC-PI dataset was created based on the assumption that compounds that inhibit a target other than Keap1/Nrf2 rarely inhibit Keap1/Nrf2 as well. We created two deep neural network (DNN) models from molecular descriptor feature sets: one was a DNN-hybrid model from the DB and FA datasets, and the other was a DNN-hybrid-PI model from the DB, FA, and MISC-PI datasets. Two random forest (RF) models using the fingerprint feature set were also created: one was from the FA dataset only (RF-FA), and the other was from both the FA and DB datasets (RF-hybrid). In addition, two RF-DB models (RF-true inactive, RF-DB-TI and RF-putative inactive, RF-DB-PI) created in our previous work5 without the FA dataset were also used for comparison. We performed LBVSs using the models to identify medium-sized molecular inhibitors of the Keap1/Nrf2 PPI and investigated the performance of iterative screening.
973 active compounds against PPI targets other than Keap1/Nrf2. The MISC-PI dataset was created based on the assumption that compounds that inhibit a target other than Keap1/Nrf2 rarely inhibit Keap1/Nrf2 as well. We created two deep neural network (DNN) models from molecular descriptor feature sets: one was a DNN-hybrid model from the DB and FA datasets, and the other was a DNN-hybrid-PI model from the DB, FA, and MISC-PI datasets. Two random forest (RF) models using the fingerprint feature set were also created: one was from the FA dataset only (RF-FA), and the other was from both the FA and DB datasets (RF-hybrid). In addition, two RF-DB models (RF-true inactive, RF-DB-TI and RF-putative inactive, RF-DB-PI) created in our previous work5 without the FA dataset were also used for comparison. We performed LBVSs using the models to identify medium-sized molecular inhibitors of the Keap1/Nrf2 PPI and investigated the performance of iterative screening.
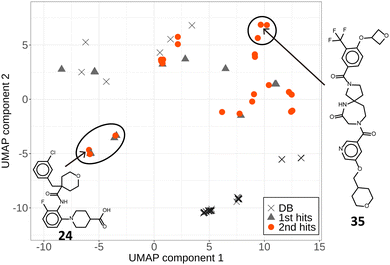
We selected 312 compounds using the LBVSs against 2722 newly synthesized library compounds (DLiP2) and tested the inhibitory activity of them against the Keap1/Nrf2 PPI using the TR-FRET assay (second assay). Forty-five compounds had an inhibition rate > 15% at a 100 μM concentration against the Keap1/Nrf2 PPI, among which five compounds were also active against the Bcl6/F1325 PPI. Therefore, 40 (12.8%) of the 312 compounds were considered specific hits for the Keap1/Nrf2 PPI (Fig. 2 and Table S1, ESI†). Most specific hits had molecular weights between 600 and 650 (Fig. S1, ESI†). The plot of the principal moments of inertia (PMI) clearly shows that the hits exist in a region apart from the rod-and-disc plane, indicating that most of them have non-flat structures that are less common in the DLiP1 (Fig. 3). Approximately 31% of the active compounds in the first assay had the common substructure of an ortho-substituted aromatic amide with a carboxylated piperidine.5 Compound 24 also had the common substructure, demonstrating the effectiveness of this substructure in inhibiting the Keap1/Nrf2 PPI. Fig. 4 (and Fig. S2 and S3, ESI†) visualizes the chemical space of the hit and library compounds of the first and second assays and the active compounds of the DB dataset. Some of the hits of both assays coexisted in a cluster due to the common active substructure (i.e., additionally acquired new cluster members by the second assay). Interestingly, the second hits not only existed around clusters of the first hits, but also formed DLiP2-specific clusters, including new structures with novel spiro-ring scaffolds (e.g., compound 35). Thus, Fig. 3 and 4 show that our iterative screening with ML models clearly extended the initial chemical space.
 | ||
| Fig. 2 Structures and inhibition rates at 100 μM concentration of compounds 1–5 that had the top five inhibition rates among the specific hit compounds. | ||
 | ||
| Fig. 3 PMI plot of DLiP1 compounds (gray dots) and the 40 specific hit compounds (magenta triangles for NPR1 + NPR2 ≥ 1.3 and orange triangles for NPR1 + NPR2 < 1.3). | ||
In the LBVSs, compounds were ranked based on their prediction scores for “active” and approximately 4% of the top-ranked compounds were extracted as candidates for the following experimental validation using the TR-FRET assay. The RF-FA and RF-hybrid models showed nearly identical molecules among the top 4% of predicted compounds. The RF models were created using structural fingerprints as the features, and the maximum structural similarities of the DLiP2 compounds against the DB dataset were much less than those against the FA dataset (Fig. S4, ESI†). These results suggest that the top 4% prediction of the RF-hybrid model was performed mainly by the contribution of the FA dataset in the training. Therefore, we used the LBVS results of the DNN-hybrid, DNN-hybrid-PI, RF-FA, RF-DB-TI, and RF-DB-PI models to select 320 compounds for the experimental assay. In fact, 312 available compounds, including 110, 67, 100, 96, and 100 compounds from the LBVS results of the DNN-hybrid, DNN-hybrid-PI, RF-FA, RF-DB-TI, and RF-DB-PI models, respectively, were selected and used in the following assays. The 312 compounds were not selected based only on their structural similarities to the known actives (Fig. S5, ESI†). The maximum structural similarities of the 312 compounds against known active compounds were not high (<0.31 against the DB dataset and <0.71 against the FA dataset, Fig. 5), indicating their novelty.
The hit rates of the models are listed in Table 1. Among the five models, the DNN-hybrid model had the highest hit rate of 27.3%. While the DNN-hybrid-PI model had the second highest hit rate of 23.9%, it showed no new hits compared with the DNN-hybrid model. Although the DNN-hybrid model hits included three non-specific hits, those of the DNN-hybrid-PI model included no non-specific hits, suggesting that the inclusion of compounds with inhibitory activity against targets other than Keap1/Nrf2 as putative inactives in the training contributed to the exclusion of non-specific actives in the DNN-hybrid-PI model. The hit rates of the RF-FA, RF-DB-TI, and RF-DB-PI models were 20.0, 8.0, and 14.6%, respectively. The models using our first assay data in the training data (DNN-hybrid, DNN-hybrid-PI, and RF-FA) exhibited much higher hit rates than the previous models (RF-DB-TI and RF-DB-PI), indicating the effectiveness of the iterative screening.
| Assay | Models | Selected | Hits | Specific hits |
|---|---|---|---|---|
| First | RF-DB-PI | 202 | 12 (5.9%) | 11 (5.4%) |
| First | RF-DB-TI | 223 | 6 (2.7%) | 5 (2.2%) |
| First | Random | 291 | 3 (1.0%) | 3 (1.0%) |
| Second | DNN-hybrid | 110 | 30 (27.3%) | 27 (24.5%) |
| Second | DNN-hybrid-PI | 67 | 16 (23.9%) | 16 (23.9%) |
| Second | RF-FA | 100 | 20 (20.0%) | 19 (19.0%) |
| Second | RF-DB-PI | 96 | 14 (14.6%) | 13(13.5%) |
| Second | RF-DB-TI | 100 | 8 (8.0%) | 6 (6.0%) |
Furthermore, the DNN models exhibited higher hit rates than the RF-FA model did, indicating the effectiveness of combining DNN and iterative screening. The hit rate of the DNN-hybrid model was extremely high compared with that of conventional HTS (<0.1%). Notably, the hit rates of the RF models were lower than those of the DNN models, and the hits of the RF models also differed slightly from those of the DNN models (Table S1, ESI†). The results show that the use of models with different algorithms or features is effective for obtaining diverse hits. The hit rates of the RF-DB-TI and RF-DB-PI models, (8.0% and 14.6%, respectively), were higher than those in the first assay (2.7% and 5.9%, respectively), suggesting that the new library compounds may have more three-dimensional conformational diversity than the previous library compounds and thus be applicable to Keap1/Nrf2 PPI inhibitors.
Docking calculations were performed on the hit compounds to inspect their interactions with Keap1. Our observations of the docking poses of the top five specific active compounds (Fig. 2) in this study showed that these compounds were bound at a position surrounded by arginine residues (Arg415 and Arg483) and aromatic residues (Tyr334, Tyr572, and Phe577) (Fig. S6 and S7, ESI†). The ligand binding site of the Keap1/Nrf2 PPI inhibitor is divided into five subpockets (P1–P5 in Fig. S6B, ESI†) and the number of subpockets occupied by the ligand has a significant influence on its inhibitory activity.6 We found that the top five hit compounds mainly interacted with a polar subpocket, P1, and hydrophobic subpockets, P3 and P4 (Fig. S6 and S7, ESI†). Of these, the top hit compound (compound 1) had aromatic ring interactions with the side chains of Tyr572 and Phe577, hydrogen bonds with the side chains of Gln530 and Ser555, while the bromophenyl group was located at the P1 subpocket just next to Arg415, and the fluorophenyl group occupied the P3 subpocket in the inner cavity of the Keap1 Kelch domain (Fig. S6B, ESI†). These hit compounds had a branched structure near the piperidine (Fig. 2), contributing to the binding of multiple subpockets.
By combining DL with iterative screening, we obtained training data with a chemical space similar to that of the candidate compounds and significantly increased hit rates in the subsequent assay. Hit analysis showed that our method succeeded in expanding the initial chemical space, leading to the discovery of novel inhibitors of the Keap1/Nrf2 PPI. This method is expected to contribute to efficient lead acquisition in assays for PPI drug discovery using medium-sized molecule compounds, which are difficult to screen on a large scale.
This work was supported by AMED under Grant Number JP19ak0101039h0005 and JP22ama121028. The authors appreciate Miyako Shibazaki for carrying out the bioassay. K. I. conceived and managed the research. Y. S. designed and performed in silico analysis. K. I. and T. Y. performed docking calculations. Y. B. created DL models. J. S. conducted the bioassay. M. O. and M. Y. contributed to the discussion of experimental results. Y. S. and K. I. drafted the manuscript.
Conflicts of interest
There are no conflicts to declare.Notes and references
- M. P. H. Stumpf, T. Thorne, E. de Silva, R. Stewart, H. J. An, M. Lappe and C. Wiuf, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 6959–6964 CrossRef CAS PubMed.
- W. Guo, J. A. Wisniewski and H. Ji, Bioorg. Med. Chem. Lett., 2014, 24, 2546–2554 CrossRef CAS PubMed.
- X. Morelli, R. Bourgeas and P. Roche, Curr. Opin. Chem. Biol., 2011, 15, 475–481 CrossRef CAS PubMed.
- O. Sperandio, C. H. Reynès, A.-C. Camproux and B. O. Villoutreix, Drug Discovery Today, 2010, 15, 220–229 CrossRef CAS.
- Y. Shimizu, T. Yonezawa, J. Sakamoto, T. Furuya, M. Osawa and K. Ikeda, Sci. Rep., 2021, 11, 7420 CrossRef CAS PubMed.
- S. Lee and L. Hu, Med. Chem. Res., 2020, 29, 846–867 CrossRef CAS PubMed.
- H. Chen, O. Engkvist, Y. Wang, M. Olivecrona and T. Blaschke, Drug Discovery Today, 2018, 23, 1241–1250 CrossRef PubMed.
- R. Liu, H. Wang, K. P. Glover, M. G. Feasel and A. Wallqvist, J. Chem. Inf. Model., 2019, 59, 117–126 CrossRef CAS PubMed.
- S. Paricharak, A. P. IJzerman, A. Bender and F. Nigsch, ACS Chem. Biol., 2016, 11, 1255–1264 CrossRef CAS PubMed.
- G. H. S. Dreiman, M. Bictash, P. V. Fish, L. Griffin and F. Svensson, SLAS Discovery, 2021, 26, 257–262 CrossRef PubMed.
- D. Reker, P. Schneider and G. Schneider, Chem. Sci., 2016, 7, 3919–3927 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc01283b |
| This journal is © The Royal Society of Chemistry 2023 |