Open Access Article
Open Access ArticleColorimetric response in polydiacetylene at the single domain level using hyperspectral microscopy†
Jiali
Chen
,
Jianlu
Zheng
,
Yuge
Hou
and
Kaori
Sugihara
 *
*
Institute of Industrial Science, The University of Tokyo, 4-6-1 Komaba Meguro-Ku, Tokyo 153-8505, Japan. E-mail: kaori-s@iis.u-tokyo.ac.jp
First published on 10th March 2023
Abstract
The structural variance of polydiacetylene (PDA) at the nanoscale level, even under the same fabrication conditions, is one of the origins of its poor reproducibility in chemo/biosensing. In this work, we present a spatial map of such structural distributions within a single crystal by taking advantage of the recent development of hyperspectral microscopy at visible wavelengths. Hyperspectral microscopy provides the distribution of absorption spectra at the spatial resolution of standard optical microscopy. By tracking the blue-to-red transition via this technique, we found that heat or pH stimulation leaves a unique pattern in the transition pathways.
Polydiacetylenes (PDAs) are a class of mechanochromic polymers that present a blue-to-red color transition and emit fluorescence when triggered by various stimuli such as heat,1 pH,2 mechanical stress,3 biomolecules,4 and so on. Being encouraged by their customizable chemical structure and facile fabrication method,5 much effort has been devoted to developing PDA sensors6 involving biomolecular detection via receptor–ligand interactions,7 food deterioration via pH monitoring,8 and volatile organic compound identification.9 However, some challenges remain for bringing these technologies forward to real-life applications.10
One of them is the unwanted variation of PDA structures, where the fabrication procedures seem to yield a mixture of PDAs with slightly different structures. Such distribution can vary from batch to batch, worsening the reproducibility of chemo/biosensing.11 Many factors, such as the monomer distance during polymerization,12 the alkyl spacer properties,13 and the lipid composition and size,14 lead to subtle differences in the PDA structure, which ultimately affect the mechanochromic properties. Raman spectroscopy, infrared spectroscopy (IR), X-ray diffraction, and nuclear magnetic resonance (NMR) have been utilized to decipher the PDA structure during polymerization and the blue-to-red transition.15–19 However, they are all based on ensemble measurements, and fail to capture the distribution of different structures within a sample.
In this work, we introduce hyperspectral microscopy that operates in the visible wavelength range to visualize the distribution of different structures at the sub-crystal level. Hyperspectral microscopy provides an absorption spectral map with a spatial resolution that is comparable to standard optical microscopy. Hyperspectral imaging dates back to the 1970s when NASA mapped infrared (IR) spectra for remote geology sensing20 and mineral detection.21 Since its first commercialization in the late 1980s, recent improvements in the computing technology and the detector have significantly broadened its applications towards medical diagnosis,22 chemical identification,23 quality monitoring,24 agriculture,25 astronomy26 and geological studies.27 By applying this novel technique to PDA research, we will show that both blue and red PDA films contain various structures even within a single crystal, and that each structure responds differently to stimuli. Tracking the transformation between these blue and red structures yields a unique pattern of the transition pathways.
Blue PDA is made of microdomains with at least 10 different structures. PDA made of 10,12-tricosadiynoic acid (TRCDA) assembled on glass coverslips via drop-casting of the monomer solution in chloroform, followed by UV irradiation (see Materials and methods (ESI†) for details). Bright-field microscopy imaging captured the PDA crystals, which look uniformly blue (Fig. 1a). However, the mapping of their absorption spectra via hyperspectral microscopy revealed that the PDA sample consists of many different “blues”, where their spectra are slightly different from each other (Fig. 1c; note that the top 10 spectra were color-coded and their spatial distribution is shown in Fig. 1b).
 | ||
| Fig. 1 Structural mapping of blue and red PDA crystals. (a and e) Optical image of blue and red PDA, respectively. (b and f) Spectral map of blue and red PDA, respectively. (c and g) Absorption spectra of blue and red PDA, respectively. (d and h) Structure distribution of blue and red PDA, respectively. Details such as the error bars are shown in Fig. S1 (ESI†). Note that the spectra are color-coded, and the same color code is used for (b–d) and (f–h). | ||
The spectral angle mapper method (see Material and methods (ESI†) for the details) that we used for classification does not distinguish the spectra that can be superposed by the multiplication of their amplitudes. This avoids categorizing PDA by thickness. This suggests that each blue spectrum that we obtained (B1–B10) corresponds to a different structure of PDA. This is because each PDA absorption spectrum reflects the electronic structure at the conjugated backbone, which is influenced by the polymer length and its architecture. This inhomogeneity originates from the lipidic monomer self-assembly process. Lipid self-assembly produces structures that are energetically favorable under the experimental conditions, yet in reality it often yields a distribution of various architectures, such as those with different packing arrangements, lipid tail angle relative to the monolayer leaflet, etc. To obtain statistics, we analyzed 11 samples. Their average showed that the spectra B7, B8, and B2 correspond to 21%, 19%, and 18% of the entire PDA crystals, respectively, being the top three major structures in the blue PDA (Fig. 1d). These structures seem to form microdomains in the crystals (Fig. 1b), where some PDA crystals contain more than two structures. This implies that the structure at the nanoscale is not uniform, even within a single crystal.
Red PDA also consists of at least 10 different structures, but the domain size is smaller than that of blue PDA. Next, we heated these blue PDA samples at 240 °C for 1 min on the microscope (see Materials and methods (ESI†)) to convert them into red PDAs before characterizing them via hyperspectral microscopy. The sample appeared to be red in a relatively uniform manner (Fig. 1e), yet the spectral mapping visualized that it contains at least ten different “reds” (Fig. 1f and g). Interestingly, their domain size became smaller on average (Table S1, ESI†), which implies an increase in the entropy, as has previously been reported.28,29 As a result, a single crystal accommodates many sub-micrometer domains that have different structures. Averaging the results from 11 samples shows that there are also some major red structures (Fig. 1h, e.g., R2, R1, and R4 of 19%, 10%, and 10%, respectively). The 26% grey portion represents glass areas that are newly created during the blue-to-red transition because of the shrinkage of the crystals. This implies that the PDA contracted upon heating. A similar reduction of the area during the blue-to-red transition has been reported before.30
Hyperspectral dark-field microscopy has previously been applied to peptide-PDA films made of 10,12-pentacosadiynoic acid to confirm its homogeneity.31 Nevertheless, observed inhomogeneity and microdomains in our samples have been also previously reported with other types of PDA. Gaber's group observed that PDA vesicles made of 1,2-bis(tricosa-10,12-diynoyl)-sn-glycero-3-phosphocholine (DC89PC) yielded different sizes, as characterized using size exclusion chromatography, and that each of them presented unique properties.32 The addition of a surfactant has also been shown to alter the PDA vesicle size, where the smaller vesicles were more sensitive.33 Korostylev and co-workers reported that optimization of the distance and the spatial arrangement of diacetylene monomers are critical for efficient polymerization, and thus affect the final polymer structures and their functions.34 Jelinek's group studied TRCDA Langmuir films using a Brewster angle microscope and has found that monolayer and multilayer domains were mixed in single samples.35 Atomic force microscopy (AFM)36,37 and transmission electron microscopy (TEM)38 have also been used to visualize microdomains within PDA crystals. These different structures mixed in PDA samples were suspected to be one of the origins of the poor reproducibility in PDA bio/chemosensing.
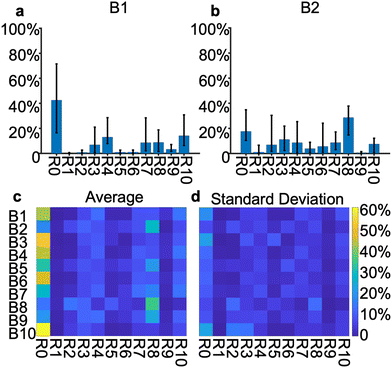
There is a unique pattern for the structural transformation from blue to red PDA. To decipher the transformation behavior from blue to red stimulated by heat, we studied, as a percentage, how much of B1 transformed into R0, R1, … and R10, respectively (Fig. 2a). On average, 43% of the pixels that had B1 spectra were transformed into R0 (glass area) after heating, whereas B1 → R1, B1 → R2, B1 → R5, and B1 → R6 were negligible pathways because their portions were less than 3%. Similarly, we analyzed the transition pathways from B2 (Fig. 2b), and up to B10 (Fig. S2, ESI†). All of the results are summarized in Fig. 2c and d for average and standard deviation data, respectively. R8 seems to be the preferred-endpoint red structure for many blue structures, besides R0 (glass area). By contrast, R1, R5, R6, and R9 seem to be unpreferred red structures as their transition portions are small for most of the blue structures. Overall, the standard deviation is not necessarily small, yet for some of the major pathways such as B8 → R8 the value (35%) is larger compared with the standard deviation (13%), suggesting that the blue-to-red transition induced by heat takes place with a certain pattern. Note that this pattern is visible regardless of the error in the spatial superposition between the blue and the red polymers due to shrinkage of the crystals.
The structural change during the blue to red transition has been extensively studied before. 13C NMR indicated the twist of the backbone, where the deplanarization of the alkyl side chains was detected during the blue-to-red transition.39 NMR and Raman spectroscopy, combined with theoretical calculations, have shown that the PDAs made of poly(1,6-bis(diphenylamino)-2,4-hexadiyne) (poly-THD) had twisted, slightly-twisted, and flat (planar) side chains for the red, intermediate, and blue forms, respectively, where the strongly-twisted red polymer was the most stable and the major red structure.40 By contrast, AFM and Fourier-transform infrared spectroscopy (FTIR) indicated an increase in the molecular-scale ordering and the near-perfect hexagonal packing of alkyl side chains as a response to the heat stimulus.41 As such, different blue structures have been suggested in the literature and a variety of methods have detected that each blue form has its distinct response against stimuli. Although we do not know what the exact structures of B1–B10 and R1–R10 are, their structural differences probably originate from a slight shift in the monomer distance, the carbon chain angle relative to the substrate or to the backbone, or different lipid phases such as lamellar and micelle architectures and their mixtures. Each form of PDA potentially has a different sensitivity to heat and transfers into different red structures. Fig. 2c shows the first comprehensive map of such transitions from various blue to various red structure for PDA made using TRCDA stimulated by heat. To study whether this transition map is unique in each stimulus, next we selected thin blue PDA areas, stimulated them using heat and pH, and obtained their blue-to-red transition patterns via the same analytical procedure (Fig. S3 and S4, ESI†). Note that new sets of 10 blue and 10 red spectra were used in this analysis; therefore, B1, for example, in Fig. 1 and 3 are not identical. The results show that each stimulus (heat or pH) leaves a unique (different) transition pattern (Fig. 3). This indicates the potential for using such a map to identify different stimuli. These transition maps were reproducible in crystal structures, but less so in amorphous PDAs (Fig. S5, ESI†).
 | ||
| Fig. 3 Blue-to-red transition map of thin PDAs stimulated using (a) heat and (b) pH. Note that a new set of B1–B10 were used in this analysis compared with those used in Fig. 1 and 2. | ||
We visualized the distribution of the microdomains from different blue and red structures by introducing hyperspectral microscopy that operates at visible wavelengths. This was enabled due to the spectral mapping ability of the technique with a spatial resolution that goes below the single crystal size, in contrast to conventional ensemble measurements such as UV-Vis, IR, Raman spectroscopy, NMR, X-ray diffraction, and other imaging methods that have the spatial resolution but not the spectral information such as AFM and TEM.42 Other spectral mapping based on scanning microscopy methods, such as infrared scattering scanning near-field optical microscopy (IR-s-SNOM)43,44 and tip-enhanced Raman spectroscopy (TERS), may be an alternative tool.45 We found that each blue structure undergoes a unique path to diverse types of red structure, where a pattern, e.g., a certain blue structure prefers to transform into a certain red structure, was detected. The result implies that the stimulus (heat, in this work) leaves a fingerprint to the PDA blue-to-red transition pathways. Deconvoluting the spectra at the sub-crystal domains and tracking their blue-to-red transitions separately instead of observing their average may help with tackling the sample inhomogeneity issue and the related reproducibility problem in PDA sensing. In addition, the hyperspectral microscopy that is used in this work, which can be also operated in fluorescence mode, can be applied to other (mechano)chromic materials in the future.46
Part of the research leading to these results received funding from the UTEC-UTokyo FSI Research Grant Program, the FY 2020 University of Tokyo Excellent Young Researcher, the Takeda Science Foundation, the Mitsubishi Foundation, the Inoue Foundation for Science, the Naito Foundation, Fuji Seal Foundation, JSPS KAKENHI Grant Number JP22K03544, JP22H04525, JST FOREST Program Grant Number JPMJFR211Q, and JPNP20004, subsidized by the New Energy and Industrial Technology Development.
Conflicts of interest
There are no conflicts to declare.References
- C. Girard-Reydet, R. D. Ortuso, M. Tsemperouli and K. Sugihara, J. Phys. Chem. B, 2016, 120, 3511–3515 CrossRef CAS PubMed.
- S. J. Kew and E. A. H. Hall, Anal. Chem., 2006, 78, 2231–2238 CrossRef CAS PubMed.
- L. Juhasz, R. D. Ortuso and K. Sugihara, Nano Lett., 2021, 21, 543–549 CrossRef CAS PubMed.
- J. Zhao and K. Sugihara, J. Phys. Chem. B, 2021, 125, 12206–12213 CrossRef CAS PubMed.
- D. J. Ahn, S. Lee and J.-M. Kim, Adv. Funct. Mater., 2009, 19, 1483–1496 CrossRef CAS.
- B. Yoon, S. Lee and J.-M. Kim, Chem. Soc. Rev., 2009, 38, 1958–1968 RSC.
- D. H. Charych, J. O. Nagy, W. Spevak and M. D. Bednarski, Science, 1993, 261, 585–588 CrossRef CAS PubMed.
- M. Weston, R. P. Kuchel, M. Ciftci, C. Boyer and R. Chandrawati, J. Colloid Interface Sci., 2020, 572, 31–38 CrossRef CAS PubMed.
- S. Dolai, S. K. Bhunia, S. S. Beglaryan, S. Kolusheva, L. Zeiri and R. Jelinek, ACS Appl. Mater. Interfaces, 2017, 9, 2891–2898 CrossRef CAS PubMed.
- M. Weston, A. D. Tjandra and R. Chandrawati, Polym. Chem., 2020, 11, 166–183 RSC.
- Y. K. Jung, T. W. Kim, H. G. Park and H. T. Soh, Adv. Funct. Mater., 2010, 20, 3092–3097 CrossRef CAS.
- S. Okada, S. Peng, W. Spevak and D. Charych, Acc. Chem. Res., 1998, 31, 229–239 CrossRef CAS.
- R. Potai, K. Faisadcha, R. Traiphol and N. Traiphol, Colloids Surf., A, 2018, 555, 27–36 CrossRef CAS.
- A. Nopwinyuwong, T. Kitaoka, W. Boonsupthip, C. Pechyen and P. Suppakul, Appl. Surf. Sci., 2014, 314, 426–432 CrossRef CAS.
- D. J. Sandman and Y. J. Chen, Polymer, 1989, 30, 1027–1031 CrossRef CAS.
- D. N. Batchelder, N. J. Poole and D. Bloor, Chem. Phys. Lett., 1981, 81, 560–564 CrossRef CAS.
- D. N. Batchelder and D. Bloor, J. Phys. C: Solid State Phys., 1982, 15, 3005 CrossRef CAS.
- E. Armando Zaragoza-Contreras, C. A. Hernández-Escobar and I. Rodríguez-Sánchez, Spectrochim. Acta, Part A, 2022, 267, 120506 CrossRef CAS PubMed.
- R. D. Ortuso, N. Ricardi, T. Bürgi, T. A. Wesolowski and K. Sugihara, Spectrochim. Acta, Part A, 2019, 219, 23–32 CrossRef CAS PubMed.
- S. F. Alexander, H. Goetz, G. E. Vane Jerry Solomon and B. N. Rock, Science, 1985, 228, 1147–1153 CrossRef PubMed.
- G. A. Roth, S. Tahiliani, N. M. Neu-Baker and S. A. Brenner, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2015, 7, 565–579 CAS.
- G. Lu and B. Fei, J. Biomed. Opt., 2014, 19, 010901 CrossRef PubMed.
- V. Farley, M. Chamberland, P. Lagueux, A. Vallières, A. Villemaire and J. Giroux, SPIE Proc., 2007, 6739, 673918 CrossRef.
- H. Huang, L. Liu and M. O. Ngadi, Sensors, 2014, 14, 7248–7276 CrossRef CAS PubMed.
- L. M. Dale, A. Thewis, C. Boudry, I. Rotar, P. Dardenne, V. Baeten and J. A. F. Pierna, Appl. Spectrosc. Rev., 2013, 48, 142–159 CrossRef CAS.
- M. Pisani and M. Zucco, SPIE-Int. Soc. Opt. Eng., 2018, 10677, 1067706 Search PubMed.
- S. Peyghambari and Y. Zhang, J. Appl. Remote Sens., 2021, 15, 31501 Search PubMed.
- S. J. Kew and E. A. H. Hall, J. Mater. Chem., 2006, 16, 2039–2047 RSC.
- J. Lee, S. Seo and J. Kim, ACS Appl. Mater. Interfaces, 2018, 10, 3164–3169 CrossRef CAS PubMed.
- J. Nuck and K. Sugihara, Macromolecules, 2020, 53, 6469–6475 CrossRef CAS.
- Y. Kuang, Z.-F. Yao, S. Lim, C. Ngo, H. Ann and M. ArdoñaSteric and Hydrophobic Control Over Sequence-Defined Peptide Templates to Modulate the Properties of Polydiacetylene Nanomaterials, ChemRxiv, 2022 DOI:10.26434/chemrxiv-2022-7zf4p.
- B. M. Peek, J. H. Callahan, K. Namboodiri, A. Singh and B. P. Gaber, Macromolecules, 1994, 27, 292–297 CrossRef CAS.
- A. Nopwinyuwong, T. Kitaoka, W. Boonsupthip, C. Pechyen and P. Suppakul, Appl. Surf. Sci., 2014, 314, 426–432 CrossRef CAS.
- R. v Vizgert and A. P. Korostylev, Polym. Sci. U.S.S.R., 1989, 31, 380–387 CrossRef.
- F. Gaboriaud, R. Volinsky, A. Berman and R. Jelinek, J. Colloid Interface Sci., 2005, 287, 191–197 CrossRef CAS PubMed.
- R. W. Carpick, D. Y. Sasaki and A. R. Burns, Tribol. Lett., 1999, 7, 79–85 CrossRef CAS.
- A. Lio, A. Reichert, J. O. Nagy, M. Salmeron and D. H. Charych, J. Vac. Sci. Technol., B, 1996, 14, 1481–1485 CrossRef CAS.
- Q. Ye, G. Zou, X. You, X. Yu and Q. Zhang, Mater. Lett., 2008, 62, 4025–4027 CrossRef CAS.
- H. Tanaka, M. Thakur, M. A. Gomez and A. E. Tonelli, Polymer, 1991, 32, 1834–1840 CrossRef CAS.
- J.-S. Filhol, J. Deschamps, S. G. Dutremez, B. Boury, T. Barisien, L. Legrand and M. Schott, J. Am. Chem. Soc., 2009, 131, 6976–6988 CrossRef CAS PubMed.
- A. Lio, A. Reichert, D. J. Ahn, J. O. Nagy, M. Salmeron and D. H. Charych, Langmuir, 1997, 13, 6524–6532 CrossRef CAS.
- A. Szerlauth, E. Balog, D. Takács, S. Sáringer, G. Varga, G. Schuszter and I. Szilágyi, Colloid Interface Sci. Commun., 2022, 46, 100564 CrossRef CAS.
- Q. Zhu, R. Zhou, J. Liu, J. Sun and Q. Wang, Nanomaterials, 2021, 11, 1353 CrossRef CAS PubMed.
- F. Zhong, H. Wang, Z. Wang, Y. Wang, T. He, P. Wu, M. Peng, H. Wang, T. Xu, F. Wang, P. Wang, J. Miao and W. Hu, Nano Res., 2021, 14, 1840–1862 CrossRef CAS.
- K. Jajcevic, A. M. Sequeira, J. Kalbacova, D. R. T. Zahn and K. Sugihara, Nanoscale, 2021, 13, 6927–6933 RSC.
- H. Traeger, Y. Sagara, D. J. Kiebala, S. Schrettl and C. Weder, Angew. Chem., Int. Ed., 2021, 60, 16191–16199 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods; Fig. S1 and S2. See DOI: https://doi.org/10.1039/d2cc06803f |
| This journal is © The Royal Society of Chemistry 2023 |