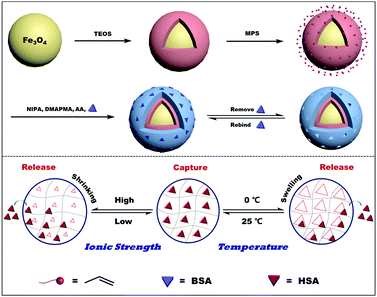
Multi-stimuli responsive molecularly imprinted nanoparticles with tailorable affinity for modulated specific recognition of human serum albumin†
Junjie
Zhang‡
a,
Yi
Hao‡
ab,
Xuemeng
Tian
a,
Yulu
Liang
c,
Xiwen
He
c,
Ruixia
Gao
*a,
Langxing
Chen
 *c and
Yukui
Zhang
cd
*c and
Yukui
Zhang
cd
aSchool of Chemistry, Xi’an Jiaotong University, Xi’an, Shaanxi 710049, China. E-mail: ruixiagao@xjtu.edu.cn
bSchool of Pharmacy, Health Science Center, Xi'an Jiaotong University, Xi’an, Shaanxi 710061, China
cCollege of Chemistry, Tianjin Key Laboratory of Biosensing and Molecular Recognition, Nankai University, Tianjin 300071, China. E-mail: lxchen@nankai.edu.cn
dDalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China
First published on 1st March 2022
Abstract
A kind of novel multi-stimuli responsive molecularly imprinted polymers with bovine serum albumin (BSA) as a dummy template (MSR-BSA-MIPs) was fabricated for specific recognition of human serum albumin (HSA) with modulated affinity. The MSR-BSA-MIPs were prepared through free radical polymerization using vinyl modified magnetic nanoparticles as substrates, bovine serum albumin (BSA), with high amino acid sequence similarity but low price compared to HSA, as the dummy template, N-(3-(dimethylamino)-propyl)-methacrylamide (DMAPMA) and N-isopropylacrylamide (NIPAm) as functional monomers with ionic strength and temperature response. The conditions of polymerization, adsorption and elution were systematically investigated. As expected, the obtained MSR-BSA-MIPs exhibited rapid dispersion or separation states under magnetic control, flexible conversion of adsorption and desorption for the target protein under temperature or ionic strength adjustment. Ten adsorption–desorption cycles were carried out with a little decrease in adsorption capacity under two different elution methods, which also inspired us to combine two elution methods while considering both the stability and adsorption capacity of MSR-BSA-MIPs. The adsorption capacity (Q) and imprinting factor (IF) of MSR-BSA-MIPs for HSA are 43.01 mg g−1 and 4.26, respectively. Furthermore, the blood was opted as a realistic specimen for evaluating the adsorption capability of the proposed adsorbent, emphasizing its good practicality for target protein recognition and enrichment.
1. Introduction
Human serum albumin (HSA) is a highly water-soluble spherical monomeric protein found abundantly in human blood plasma. It plays a vital role in numerous physiological activities, and is liable for transporting hormones, fatty acids, and other compounds such as drugs, and maintains osmotic pressure.1 The normal range of has in adults is 35–55 g L−1. Studies show that a lower/higher level of HSA is associated with various disorders, such as chronic hepatitis, burns, kidney disorders, and diabetes mellitus.2–4 Meanwhile, HSA has been the focus of attention in the pharmaceutical industry because of its large number of drug binding sites.5 Therefore, rapid purification and separation of HSA from complex samples is a prerequisite for subsequent analysis or function studies of HSA.Molecularly imprinted polymers (MIPs), as tailor-made artificial receptors, have exhibited great potential in a variety of fields6–8 due to their high specificity, mechanical and chemical stability, easy preparation, and low cost. To date, creating MIPs towards small molecules is successful and some even achieve an industrialization level,9 while imprinting biomacromolecules, especially proteins, still face a huge challenge resulting from the influence of the characteristics of the protein itself, such as large molecular size, structure complexity, conformational flexibility, solubility, and sensitivity to the environment.10 To solve this problem, several new imprinting approaches have been developed, such as surface imprinting, metal-chelating imprinting, and epitope imprinting.11–13 Among them, the surface imprinting technique, making the binding sites distribute on or close to the substrate surface, is considered a promising strategy to imprint proteins.
Surface imprinting can overcome the restricted mass transfer for protein, solving the problem of protein template elution to a large extent because of the accessible imprinted cavities of MIPs.14 Magnetic nanoparticles with large surface areas, good biocompatibility, and high chemical stability have become one of the most extensively used substrates for preparing surface MIPs.15,16 The synthesized magnetic MIPs can be separated within several seconds under an external magnetic field without a complicated isolation procedure. Additionally, although conventional eluents are acid, alkali, salt, and surfactants, such as sodium dodecyl sulfate (SDS), even a mixture of these solutions can effectively elute the protein,17,18 but in the process, the protein template tends to denature easily and the process is time-consuming. Therefore, milder approaches are required to replace traditional elution methods.
Stimuli-responsive MIPs, as a class of intelligent materials, can respond to special external stimuli such as temperature, magnetism, pH, solvent, and light,19–23 which becomes a good choice for protein imprinting. Among them, temperature-sensitive MIPs can change their structure and physical properties in response to the change of temperature, which is very similar to the recognition of proteins in natural systems.24 poly-(N-isopropylacrylamide) (pNIPAm) is the most commonly used thermo-responsive polymer with a lower critical solution temperature (LCST) of approximately 32–34 °C.25 Below LCST, the polymer chains expand and dissolve in water, while the polymer chains curl and are insoluble above LCST, making the swelling and shrinking transformation against temperature changes. This special feature integrated into MIPs is capable of controlling the uptake and release of target proteins resulting from the shape change of imprinted sites from expansion and contraction. More importantly, the elution conditions are relatively mild, which helps keep the structure and characteristics of the protein not being affected by the capture/elution conditions, providing a basis for the subsequent protein structure analysis.
Meanwhile, dual and multi-stimuli responsive protein MIPs have been proposed such as temperature/light,26 temperature/pH,27 magnetic/pH,28 magnetic/light,29 magnetic/temperature,30 and temperature/pH/magnetic31 dual/multi-response MIPs, in order to achieve the double/multiple response control capture and release of the target protein. For example, Wang et al. developed a facile and reliable approach to prepare the protein imprinted polymers with photonic and magnetic dual responsive ability for specific recognition of BHb, and the prepared MIPs not only showed outstanding magnetic properties for rapid separation, but also exhibited a good selective photoreponsive ability towards template protein by alternating irradiation with 365 and 440 nm light. Fan et al. reported a route to prepare a multiple stimuli-responsive rattle-type magnetic hollow molecularly imprinted nanosphere for specific recognition of BSA using a magnetic nanosphere as carriers and N-isopropylacrylamide (NIPAm) and α-methacrylic acid (MAA) as co-monomers, which can be responsive to an external magnetic field, environmental temperature and pH, respectively. However, the multi-response MIPs are still limited to date, and in particular magnetic/temperature/ionic strength multi-responsive MIPs have not been reported so far.
In this study, a novel kind of intelligent molecularly imprinted polymers with temperature, ionic strength, and magnetic multi-responsive property was fabricated for specific recognition of human serum protein (HSA) with modulated affinity under external temperature or ionic strength stimulus and the rapid dispersion or separation state under magnetic control. The obtained MSR-MIPs were prepared through free radical polymerization using vinyl modified magnetic nanoparticles as substrates, bovine serum protein (BSA), with high amino acid sequence similarity but only about 0.75% price of HSA, as the dummy template, and N-isopropylacrylamide (NIPAm) and N-(3-(dimethylamino)-propyl)-methacrylamide (DMAPMA) as the temperature and ionic strength response functional monomer, respectively. The polymerization, adsorption, and elution conditions were systematically optimized. Meanwhile, the capture and release experiments were carried out under temperature and ionic strength adjustment to estimate the reusability of MSR-MIPs. Moreover, the practicability of the proposed MSR-MIPs was evaluated by purification of BSA and HSA in bovine and human blood samples.
2. Experimental
2.1 Reagents and materials
N-Isopropylacrylamide (NIPAm), acrylamide (AAm), 3-methacryloxypropy trimethoxysilane (MPS), N-(3-(dimethylamino)-propyl)-methacrylamide (DMAPMA), and tetraethoxysilane (TEOS) were purchased from Alfa Aesar. Tris base, N,N-methylene-bisacrylamide (MBAA), ammonium persulfate (APS), N,N,N′,N′-tetramethylethylenediamine (TEMED), bovine serum albumin (BSA; MW 66.0 kDa, pI 4.8), bovine hemoglobin (BHb; MW 64.0 kDa, pI 6.9), human serum albumin (HSA; MW 69.0 kDa, pI 4.8), and hemoglobin (Hb; MW 64.5 kDa, pI 6.8) were obtained from Sigma-Aldrich. All other chemicals used in this study were of analytical reagent grade and without further treatment.2.2 Characterization
The morphology and structure of materials were examined using a Tecnai G2 T2 S-TWIN transmission electron microscope (TEM). The infrared spectra were recorded on a Nicolet AVATAR-360 Fourier transform infrared (FT-IR) spectrometer. The identification of the crystalline phase was performed on a Rigaku D/max/2500v/pc (Japan) X-ray diffractometer with a Cu Ka source with 2θ range from 10° to 80° and a scan rate of 4° min−1. The magnetic properties were analyzed with a vibrating sample magnetometer (VSM) (LDJ 9600-1, USA). Electrophoretic analysis of proteins was performed using regular SDS-PAGE (Bio-Rad, Hercules, CA) with 12% running and 5% stacking gels. Proteins were stained with Coomassie Brilliant Blue R-250.2.3 Preparation of imprinted and non-imprinted polymers
The monodispersed Fe3O4 nanoparticles (NPs) and Fe3O4@SiO2 were prepared in the same way as described in our previous work.32 Briefly, FeCl3·6H2O (1.08 g) and NaOAc (4.0 g) were dissolved in ethylene glycol (14 mL) and diethylene glycol (26 mL) in a Teflon-lined stainless-steel autoclave, sealed and heated to 200 °C. After reacting for 15 h, the autoclave was cooled to room temperature. The obtained Fe3O4 NPs were washed several times with highly purified water and ethanol. Finally, the products were collected with a magnet and then dried under vacuum. Fe3O4 NPs (0.2 g) were dissolved in ethanol (100 mL) and highly purified water (20 mL) by stirring for 10 min, followed by the addition of ammonium hydroxide (3 mL) and TEOS (1 mL). The mixture was reacted for 12 h at room temperature with continuous stirring. The resultant silica-coated Fe3O4 NPs (denoted as Fe3O4@SiO2) were collected using an external magnetic field and rinsed repeatedly with highly purified water, and then dried at 40 °C. The resultant vinyl groups grafted onto the Fe3O4@SiO2 (denoted as Fe3O4@SiO2@MPS) were prepared according to previous literature.33 200 mg of Fe3O4@SiO2 and 2 mL of MPS were placed in 50 mL of anhydrous toluene. The solution was protected by nitrogen, and reacted for 24 hours. After the reaction, a magnetic field is applied to separate and washed with ultrapure water and ethanol, following drying at 40 °C. The multi-stimuli responsive molecularly imprinted polymers were prepared as follows. Typically, Fe3O4@SiO2@MPS (0.50 g), NIPAm (0.40 g), AAm (0.0050 g), DMAPMA (16.0 μL), MBAA (0.092 g), 10% APS (50.0 μL), and template protein BSA (0.10 g) were dissolved in 20.0 mL of Tris-HCl buffer solution (10.0 mM, pH 7.0). The solution was deoxygenated by purging with nitrogen for 10 min, and a volume of 5.0 μL of TEMED was added. The mixture was stirred at 300 rpm and 50 °C for 18 h under an inert atmosphere of nitrogen. After the reaction, the resultant polymers were eluted with 10.0 mL of NaCl (0.50 M) to remove the template protein at intervals of three hours with the help of shaking until an absorbance less than 0.001 was detected in the eluent using a UV-vis spectrophotometer at about 280 nm to assure that the template was almost removed. The residual amount of BSA after removal was roughly estimated by the amount of BSA in the supernatant after polymerization and the total amount of BSA in the eluents (Table S1, ESI†) and the residual BSA in the resultant products (denoted as MSR-BSA-MIPs) is about 1.2 mg. The MSR-HSA-MIPs were synthesized in the same way as that of the MSR-BSA-MIPs except for with HSA as the template instead of BSA. Non-imprinted polymers (denoted as NIPs) were prepared by following the same procedure but in the absence of the template.2.4 Measurements of the swelling ratio of imprinted and non-imprinted polymers
For the temperature and ionic strength response studies, the MSR-BSA-MIPs and NIPs were incubated in Tris-HCl buffer solution (10.0 mM, pH 7.0) at various predetermined temperatures ranging from 0 °C to 50 °C with 3.0 mM NaCl for 24 h, and with different NaCl concentrations ranging from 0–10.0 mM at 25 °C for 24 h. After equilibrium, the polymers were removed from the buffer solution by an external magnetic field and washed with ultrapure water, and the excess water on the polymer surface was blotted with filter paper and vacuum dried at room temperature. They were then weighed with a microbalance, and the swelling ratio (SR) was calculated using eqn (1):34| SR = (Ws − Wd)/Wd | (1) |
2.5 Binding experiments
In the binding experiment, 20.0 mg of MSR-BSA-MIPs and NIPs were added to 10.0 mL of BSA solutions and incubated on a reciprocating shaker at 25 °C. The solvent is 10.0 mM Tris-HCl (pH 7.0) containing 3.0 mM NaCl. Then, the supernatants and adsorbents were separated using an external magnetic field and the protein concentrations in the supernatant were measured by UV-vis spectrometry. The adsorption capacity (Q) of the protein bound to the adsorbents was calculated from eqn (2):| Q = (C0 − Ct) V/W | (2) |
In the kinetic binding experiment, the incubation time is changed from 0 to 40 min at regular intervals while keeping the initial concentration of BSA constantly at 2.0 mg mL−1. The pseudo-first-order and pseudo-second-order kinetic models are used to fit the kinetic data based on eqn (3) and (4) respectively.
| ln (Qe − Qt) = ln Qe − k1t | (3) |
| t/Qt = 1/k2Qe2 + t/Qe | (4) |
In the isotherms binding experiment, the initial concentration of BSA is adjusted from 0.10 to 4.0 mg mL−1 under the same incubation time of 20 min. The saturation binding data were processed with the Scatchard, Langmuir, and Freundlich isothermal models to further estimate the adsorption process. The equations are defined by eqn (5)–(7):
| Qe/Ce = (Qmax − Qe)/KD | (5) |
| Ce/Qe = 1/QmaxKL + Ce/Qmax | (6) |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Qe = m Qe = m![[thin space (1/6-em)]](https://www.rsc.org/images/entities/i_char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ce+ log Ce+ log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KF KF | (7) |
The recognition property of MSR-BSA-MIPs, MSR-HSA-MIPs, and NIPs to HSA, the mixed solution of BHb and BSA, and the mixed solution of Hb and HSA were also investigated in the optimized adsorption conditions.
2.6 Reusability of MSR-BSA-MIPs
The reusability of adsorbents was investigated by using the same batch of MSR-BSA-MIPs. Briefly, 20.0 mg of adsorbents were added to 10.0 mL of protein solutions at optimized adsorption conditions. Then, the adsorbents were separated using a magnet and eluted with 0.50 M NaCl for 6 h or incubated at 0 °C for 1 h in Tris-HCl buffer solution (10.0 mM, pH 7.0) to ensure complete removal of the residual BSA or HSA. The recovered product was then reused for adsorption of BSA or HSA for at least ten cycles.2.7 Real sample analysis
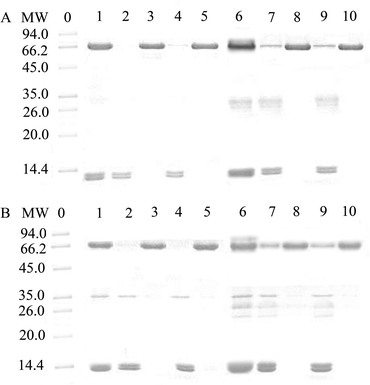
Bovine whole blood and human whole blood were chosen as the real samples to evaluate the practical application of adsorbents. Typically, 50 mg of MSR-BSA-MIPs and MSR-HSA-MIPs were incubated with 10 mL of the 250-fold diluted blood samples under gentle shaking for 60 min. After the adsorption process, the adsorbents were magnetically separated and eluted at 0 °C for 1 h in Tris-HCl buffer solution (10.0 mM, pH 7.0) to release the captured protein. SDS-PAGE was used to analyze the initial, adsorbed, and eluted solutions.3. Results and discussion
3.1 Synthesis and characterization of MSR-BSA-MIPs
The synthesis procedure for MSR-BSA-MIPs is illustrated in Scheme 1. Firstly, Fe3O4 nanoparticles were synthesized by a modified solvothermal reaction and coated with a silica shell, which can screen the magnetic dipolar attraction to favor the dispersion of magnetite nanoparticles in liquid media, protect the magnetite nanoparticles from leaching in an acidic environment, and make the magnetite nanoparticles be easily modified with diversified groups for bioconjugation purposes. Then, the vinyl groups were functionalized on the surface of Fe3O4@SiO2 with MPS using a sol–gel process. Then, under the guiding effect of the template, the imprinted layer was formed on the Fe3O4@SiO2 through the free radical polymerization with functional monomers (NIPAm, AAm, and DMAPMA), cross-linking agent (MBAA), initiator (APS), accelerator (TEMED), and dummy template molecule (BSA). Finally, after the polymerization, the obtained adsorbents were eluted with Tris-HCl buffer solution (10.0 mM, pH 7.0) containing 0.50 M NaCl or 0 °C to remove the templates.The template removal efficiency, adsorption capacity, and imprinting factor of MSR-BSA-MIPs and NIPs with different cross-linking degrees ranging from 1% to 10% are compared in Table 1. The removal efficiency decreases as the degree of cross-linking increases, which may result from the fact that the structure of MSR-BSA-MIPs becomes tighter and tighter to make template proteins come out more and more difficult. The removal efficiency of MSR-BSA-MIPs was best when the degree of cross-linking was 1%. However, due to the worse mechanical strength of MSR-BSA-MIPs at the degree of cross-linking 1%, the adsorption capacity and imprinting factor were not ideal. Considering these three factors, a 2% cross-linking degree was chosen in the polymer preparation.
| Degree of cross-linkingb (%) | 1 | 2 | 3 | 4 | 5 | 10 |
|---|---|---|---|---|---|---|
| a In these experiments, 10.0 mL of 2.0 mg mL−1 BSA in Tris-HCl (10.0 mM, pH 7.0) containing 3.0 mM NaCl incubated by 20.0 mg of MIPs and NIPs synthesized at different cross-linking degrees at 25 °C for 20 min. b The degree of cross-linking was defined as the mass fraction of the cross-linking agent. c The imprinting factor (IF) was defined as the ratio of QMIPs to QNIPs. d The removal efficiency was the ratio of template protein to be washed to polymerize. | ||||||
| Q MIPs (mg g−1) | 41.46 | 54.76 | 43.94 | 33.48 | 25.68 | 13.12 |
| Q NIPs (mg g−1) | 9.174 | 10.18 | 9.816 | 8.257 | 6.881 | 3.854 |
| IFc | 4.519 | 5.379 | 4.476 | 4.055 | 3.732 | 3.404 |
| Removal efficiencyd (%) | 97.85 | 95.79 | 90.68 | 85.36 | 80.05 | 56.62 |
Different mass ratios of template to thermo-sensitive monomer were also investigated, ranging from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 1
1 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4. As we can see (Fig. 1A), low mass ratios (1
4. As we can see (Fig. 1A), low mass ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3–1
3–1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) mean many excessive monomers may embed recognition sites, while a high mass ratio (1
4) mean many excessive monomers may embed recognition sites, while a high mass ratio (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) indicates that fewer monomers could not create enough recognition sites; both extreme conditions are not conducive to adsorption capacity. Therefore, the mass ratio of template to thermo-sensitive monomer was fixed at 1
1) indicates that fewer monomers could not create enough recognition sites; both extreme conditions are not conducive to adsorption capacity. Therefore, the mass ratio of template to thermo-sensitive monomer was fixed at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.
2.
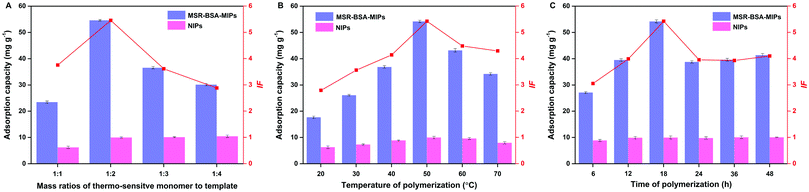 | ||
| Fig. 1 Effect of the mass ratio of the thermo-sensitive monomer to template (A), polymerization temperature (B), and polymerization time (C) on the imprinting performance of MSR-BSA-MIPs. | ||
The polymerization temperature was investigated varying from 20–70 °C. As presented in Fig. 1B, the adsorption capacity of MSR-BSA-MIPs increase with the increment of polymerization temperature from 20–50 °C, inferring that a higher polymerization temperature will result in shorter polymer chains and thus a more rigid polymer network, helping to maintain the spatial integrity after template proteins removal. Continuing to increase the temperature from 50–70 °C, the adsorption capacity of MSR-BSA-MIPs declines obviously as the imprinted layer becomes very crispy, also not suitable for practical applications. As a comparison, the adsorption capacity of NIPs fluctuates little under temperature changes, resulting from the nonspecific adsorption of the NIPs. Therefore, 50 °C was adopted as the polymerization temperature in the subsequent experiments.
The influence of the polymerization time was examined varying from 6–48 h. As shown in Fig. 1C, the best adsorption capacity and imprinting factor are demonstrated at a polymerization time of 18 h, as the optimal recognition sites and spatial network of polymers were formed. When the polymerization time is over 18 h, too many monomers were polymerized onto the MSR-BSA-MIPs to block the recognition sites, while the polymeric level was defective when the polymerization time was less than 18 h. The polymerization time is selected as 18 h.
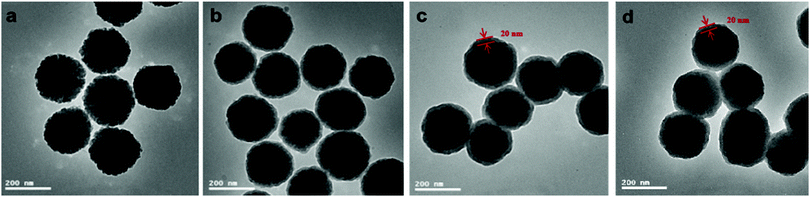
Fig. 2 displays the TEM images of Fe3O4, Fe3O4@SiO2, MSR-BSA-MIPs, and NIPs. The pure Fe3O4 is uniform with a diameter of about 200 nm (Fig. 2a). After coating the silica shell, the core–shell structure of Fe3O4@SiO2 is distinct and the thickness of the shell is about 20 nm (Fig. 2b). Compared with the morphology of Fe3O4@SiO2, MSR-BSA-MIPs and NIPs also exhibit obvious core–shell structures with an increased layer thickness (Fig. 2c and d), corresponding to a 20 nm thick imprinted layer on the Fe3O4@SiO2, which was sufficient for mass transport between the solution and the surface of MSR-BSA-MIPs.
FT-IR spectra of Fe3O4, Fe3O4@SiO2, Fe3O4@SiO2@MPS, and MSR-BSA-MIPs are shown in Fig. 3A. The peak at 587 cm−1, attributed to the stretching vibration of Fe–O, appears in all the materials. In comparison with the curve of pure Fe3O4 (Fig. 3A-a), the characteristic peak of the Si–O–Si group at approximately 1100 cm−1 (Fig. 3A-b) indicates the formation of a silica shell on the surface of Fe3O4. After the modification of MPS, the peculiar peak of the vibration of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C at 1710 cm−1 represented in Fig. 3A-c, demonstrates that vinyl groups are grafted on the surface of Fe3O4@SiO2. In Fig. 3A-d, the characteristic peaks of –NH2 at ∼3300 cm−1, –C
C at 1710 cm−1 represented in Fig. 3A-c, demonstrates that vinyl groups are grafted on the surface of Fe3O4@SiO2. In Fig. 3A-d, the characteristic peaks of –NH2 at ∼3300 cm−1, –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O at ∼1650 cm−1, and the isopropyl group at ∼2970 cm−1 derived from NIPAm, AAm, and DMAPMA indicate that all monomers were successfully incorporated into the backbone of the MSR-BSA-MIPs.
O at ∼1650 cm−1, and the isopropyl group at ∼2970 cm−1 derived from NIPAm, AAm, and DMAPMA indicate that all monomers were successfully incorporated into the backbone of the MSR-BSA-MIPs.
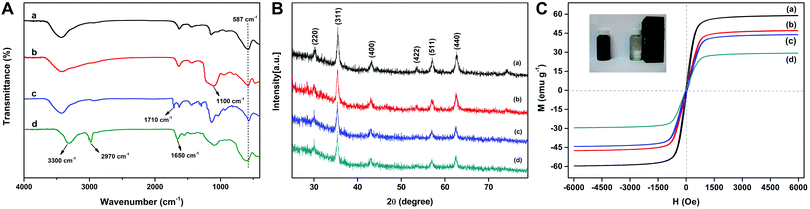 | ||
| Fig. 3 FT-IR spectra (A), XRD (B), and VSM (C) of Fe3O4 (a), Fe3O4@SiO2 (b), Fe3O4@SiO2@MPS (c), and MSR-BSA-MIPs (d). | ||
Fig. 3B shows the XRD patterns of the synthesized Fe3O4, Fe3O4@SiO2, Fe3O4@SiO2@MPS, and MSR-BSA-MIPs. In the 2θ range of 20–70°, six characteristic peaks for Fe3O4 (2θ = 30.38°, 35.58°, 43.14°, 53.48°, 57.08°, and 62.66°) were observed for the four samples. The peak positions at the corresponding 2θ values were indexed as (220), (311), (400), (422), (511), and (440), respectively, which matched well with the database for magnetite in the JCPDS-International Center for Diffraction Data (JCPDS Card: 19-629) file, indicating that the crystal form has not changed during the preparation process.
Fig. 3C displays the VSM curves to study the magnetic properties of the synthesized nanomaterials. Notably, Fe3O4, Fe3O4@SiO2, Fe3O4@SiO2@MPS, and MSR-BSA-MIPs exhibit no hysteresis and zero coercivity, reflecting their superparamagnetic properties, which implied that no remanence remained when the applied magnetic field is removed. The saturation magnetization values were 58.79, 46.98, 43.83, and 29.29 emu g−1 for Fe3O4, Fe3O4@SiO2, Fe3O4@SiO2@MPS, and MSR-BSA-MIPs, respectively. The saturation magnetization of MSR-BSA-MIPs was reduced to 29.29 emu g−1 in comparison with 58.79 emu g−1 of the pure Fe3O4, but remained strongly magnetic at room temperature and allowed for effective magnetic separation as shown in the inner image of Fig. 3C.
3.2 Binding Properties of MSR-BSA-MIPs
MSR-BSA-MIPs were synthesized adopting NIPAm and DMAPMA as functional monomers, which will exhibit the sensitive responses to both temperature and ionic strength. Fig. 4A shows the effect of temperature on the adsorption capacity, from which the adsorption capacity of MSR-BSA-MIPs increases firstly and then decreases as the temperature increases from 0 to 50 °C. The maximum adsorption capacity and the best imprinting factor is obtained at 25 °C, inferring the best imprinting state of MSR-BSA-MIPs whose shape of imprinted cavity and distribution of the charged groups tailor with the protein accurately. Correspondingly, the swelling ratio exhibits a continuous shrinking trend from 0 to 50 °C, accompanied by changes in the size of the imprinted cavity. MSR-BSA-MIPs has a value of 6.22 at 25 °C, which is an appropriate size to recognize the template protein, neither too big nor small (Fig. 4B). When the temperature is higher than 25 °C, the collapse of the polymer networks led to the destruction of the spatial integrity of the recognition site and the specific binding of MSR-BSA-MIPs becomes worse. The NIPs showed a slight increase in adsorption capacity with increasing temperature, which was possibly attributed to the enhancement of the hydrophobic interactions between the proteins and the polymer chains.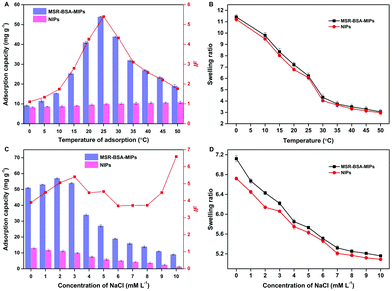 | ||
| Fig. 4 Effect of the adsorption temperature (A and B) and ionic strength (C and D) on the adsorption capacity and swelling ratio of MSR-BSA-MIPs and NIPs. | ||
Fig. 4C displays the effect of ionic strength on the adsorption capacity with varying the concentrations of NaCl from 0 to 10.0 mM. The polymer chains with tertiary amine groups were positively charged while the template protein BSA (pI 4.8) was negatively charged at a pH of 7.0, which led to the electrostatic attraction between the polymer and the protein, and also the electrostatic repulsion between the polymer chains. The adsorption of MSR-BSA-MIPs and NIPs for BSA is quite different with the increase of ionic strength. For NIP, the adsorption capacity gradually decreases as NIPs do not have specific recognition sites, their functional monomers are arranged in disorder and the adsorption force mainly relies on electrostatic attraction. For MSR-BSA-MIPs, when the ionic strength is 3 mM, the adsorption effect of MSR-BSA-MIPs is optimal considering both the adsorption capacity and imprinting factor, inferring that the formed imprinted cavity is just appropriate both in shape, size, and functional arrangement. In addition, NIPs exhibit a faster decrease of affinity than the MSR-BSA-MIPs because of less effective binding sites. Accordingly, as the ionic strength increases, the mutual repulsion between polymer chains gradually decreases, and both MSR-BSA-MIPs and NIPs show a state of gradual shrinkage (Fig. 4D).
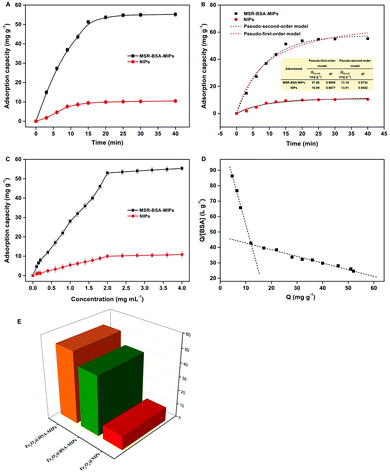
Fig. 5A presents the binding kinetics of MSR-BSA-MIPs. It can be seen that the adsorption capacity increases quickly in the first 15 min, and changes slowly with continually increasing the incubation time, and at last approaches equilibrium after 20 min. The fast adsorption equilibrium time in our case suggests that MSR-BSA-MIPs have good mass transport making BSA molecules reach the surface imprinted cavities of MSR-BSA-MIPs easily and rapidly. The adsorption kinetics of BSA onto MSR-BSA-MIPs is fitted by the two kinetics models and illustrated in Fig. 5B. The pseudo-first-order model is more adaptable for MSR-BSA-MIPs.35 In addition, the adsorption capacity (Qe, cal) value calculated from the pseudo-first-order kinetic model is in line with the experimental values (Qe, exp).
The binding isotherms of BSA to MSR-BSA-MIPs and NIPs are depicted in Fig. 5C. The amount of BSA bound to MSR-BSA-MIPs increases quickly with increasing initial concentration from 0.010 to 1.8 mg mL−1, and becomes slow to reach saturation at the initial concentration of 2.0 mg mL−1. The maximum adsorption capacity of MSR-BSA-MIPs towards BSA is 52.99 mg g−1, dramatically higher than that of NIPs (9.99 mg g−1). Scatchard analysis for MSR-BSA-MIPs was performed and the Scatchard plots are found to not be a single straight line, but instead consist of two distinct linear sections with different slopes (Fig. 5D). This observation indicates that two kinds of binding sites were populated on MSR-BSA-MIPs. The linear regression equations for the linear regions were Qe/Ce = 114.4–5.998Qe (R2 = 0.997) and Qe/Ce = 46.31–0.4082Qe (R2 = 0.977). From the slope and the intercept of the straight line obtained, the values of KD and Qmax are 0.1667 mg L−1 and 19.07 mg g−1, 2.449 mg L−1 and 113.4 mg g−1, respectively. What is more, Langmuir and Freundlich adsorption models are also used to well explain the adsorption process. The fittings of the adsorption data into Langmuir and Freundlich adsorption models are summarized in Table S2 (ESI†). It could be clearly seen that both cases are more suitable for Freundlich (R2 > 0.979), implying that the adsorption of BSA on MSR-BSA-MIPs and NIPs are likely a heterogeneous adsorption process.
To investigate the adsorption ability of MSR-BSA-MIPs for HSA, the other two adsorbents MSR-HSA-MIPs and NIPs are used as comparisons. As shown in Fig. 5E, the adsorption capacities of MSR-HSA-MIPs and MSR-BSA-MIPs to HSA are 51.95 mg g−1 and 43.01 mg g−1 at adsorption equilibrium, and the imprinting factor (IF) is 5.15 and 4.26, respectively. The difference in the two adsorbents in adsorption capacity is less than 20%, indicating that the dummy protein selected can produce the desired imprinting effect to recognize HSA. The desired effect results from the highly similar amino acid sequence of 78% for BSA and HSA along with approximately the same isoelectric point. However, BSA is much cheaper and easier to get, as the price of BSA is only about 0.75% that of HSA. HSA is relatively expensive and has important clinical significance. The results indicated that BSA is an ideal dummy template for HSA.
3.3 Selectivity of MSR-BSA-MIPs
To investigate the selectivity of MSR-BSA-MIPs and MSR-HSA-MIPs, BHb or Hb were used as a competitor as it is another highly abundant protein in the blood. Two kinds of imprinted polymers, along with NIPs, were challenged with the mixture of BSA and BHb and the mixture of HSA and Hb to achieve full cross-reaction characterization. The selectivity properties of the MIPs and NIPs toward template proteins and other proteins was estimated by the imprinting factor (IF) and selectivity coefficient (SC) as defined by eqn (8) and (9):| IF = QMIPs/QNIPs | (8) |
| SC = IFTEM/IFOTHER | (9) |
As listed in Table 2, the adsorption capacity and imprinting factor of MSR-BSA-MIPs and MSR-HSA-MIPs on BSA or HSA are much better than that of BHb or Hb, and the selectivity coefficient is above 2.3. The main reason is that the existed specific recognition sites tailored well for the template protein in shape, size, and location of functional groups. In addition, the functional monomer DMAPMA contains tertiary amino groups and is positively charged in the adsorption solution (Tris-HCl, pH = 7.0),36 the template protein BSA or HSA (pI 4.8) is negatively charged, leading to the electrostatic attraction between the polymer and the template protein. Whereas the isoelectric point of BHb and Hb is 6.9 and 6.8, there is almost no charge in the adsorption solution. Due to the similar structure of BSA and HSA, two types of imprinted polymers both have satisfactory selective ability for BSA and HSA compared with that of BHb and Hb, further verifying that BSA is a suitable dummy template for HSA. The above results verified that the imprinting effect plays a crucial part during selective recognition.
| MSR-BSA-MIPs | MSR-HSA-MIPs | NIPs | ||
|---|---|---|---|---|
| Mixture of BHb and BSA | Q BSA (mg g−1) | 43.97 | 37.36 | 7.192 |
| Q BHb (mg g−1) | 13.06 | 12.93 | 6.143 | |
| IFBSA | 6.114 | 5.195 | — | |
| IFBHb | 2.126 | 2.105 | — | |
| SC | 2.876 | 2.468 | — | |
| Q HSA (mg g−1) | 33.64 | 40.67 | 7.241 | |
| Mixture of Hb and HSA | Q Hb (mg g−1) | 11.28 | 12.54 | 5.728 |
| IFHSA | 4.646 | 5.617 | — | |
| IFHb | 1.969 | 2.189 | — | |
| SC | 2.359 | 2.566 | — |
3.4 Thermo-modulated removal of the template
MSR-BSA-MIPs obtained in this experiment have a thermo-responsive property because the NIPAm as the main monomer was introduced as a temperature-sensitive element that allowed for swelling and shrinking in response to temperature changes to realize recognition and release of proteins. We studied the effect of temperature and time on the protein release from polymers. As exhibited in Fig. 6A and B, the release rate of MSR-BSA-MIPs to BSA and HSA increase with decreasing the temperature from 20 °C to 0 °C. In this process, MSR-BSA-MIPs have a continuous swelling trend as the temperature decreases, the distance of the target protein and recognition sites become larger, weakening the interaction of the target protein and recognition sites, causing the spatial configuration to be unsuitable for the target protein anymore, so BSA and has are gradually released from the polymer network. With comprehensive consideration of the release rate and manipulability, we chose 0 °C as the release temperature. In comparison, the release rates of BSA and HSA from NIPs are far less than that from MSR-BSA-MIPs (Fig. 6C and D). This may be that the adsorption and release are non-specific adsorption processes for NIPs. In addition, the release time of MSR-BSA-MIPs for adsorbed protein almost reach equilibrium at 240 min.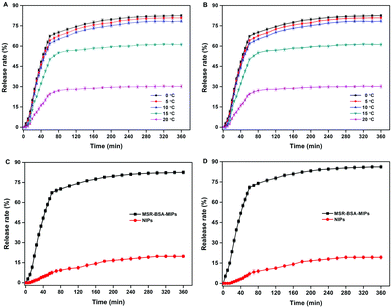 | ||
| Fig. 6 Effect of temperature on the BSA (A) and (HSA) (B) release from MSR-BSA-MIPs; Effect of time on the BSA (C) and HSA (D) release from MSR-BSA-MIPs and NIPs at 0 °C. | ||
3.5 Reusability of MSR-BSA-MIPs
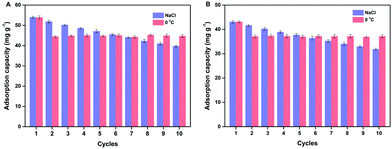
Reusability is one of the most important properties for the application of polymers. The obtained MSR-BSA-MIPs respond to temperature and ionic strength, so the same MSR-BSA-MIPs to BSA or HSA were washed by Tris-HCl buffer solution (10.0 mM, pH 7.0) containing 0.50 M NaCl or changed temperature to 0 °C in Tris-HCl buffer solution (10.0 mM, pH 7.0) ten times. As shown in Fig. 7, two washing methods can both elute BSA and HSA from MSR-BSA-MIPs. Interestingly, there is a trend in these ten adsorption–desorption cycles. Firstly, after the first washing process, the re-adsorption capacity of MSR-BSA-MIPs for two proteins is reduced by less than 5% for salt washing, however, it was reduced by over 15% for temperature washing. Then, with increasing the adsorption–desorption cycles, the re-adsorption capacities of MSR-BSA-MIPs to BSA or HSA become close after two kinds of washing methods. Finally, after seven washing cycles, the re-adsorption capacities of MSR-BSA-MIPs to BSA or HSA after washing with a low temperature are significantly higher than by salt washing. It is possible that some recognition sites in the network of MSR-BSA-MIPs could be blocked after regeneration or destroyed after rewashing by salt, and thus they were no longer fit for the template protein anymore. However, MSR-BSA-MIPs only underwent a swelling and shrinking process in response to temperature changes, and the effect on the recognition sites was negligible. Therefore, the adsorption ability of MSR-BSA-MIPs was stable after the second temperature washing, except for the incapable released proteins in the first washing cycle. In practical application, we can use the combination of two elution methods. In the early stage, we use salt elution to expose the imprinted sites as much as possible, and in the later stage, we use low-temperature elution to make the material more stable.3.6 Real sample analysis
The practical applicability of MSR-BSA-MIPs and MSR-HSA-MIPs was evaluated by using the binary standard protein mixture (BSA and BHb, HSA and Hb), 250-fold diluted bovine blood and human blood (buffer solution: Tris-HCl, pH = 7.0, 10 mM) as practical samples. As presented in Fig. 8, BSA or HSA is almost completely removed from the mixture solution of BSA and BHb or HSA and Hb (lane 1) after absorbing by MSR-BSA-MIPs (lane 2) and MSR-HSA-MIPs (lane 4) while the change in BHb or Hb was insignificant. After eluting the absorbed MSR-BSA-MIPs and MSR-HSA-MIPs through low temperature, the BSA or HSA band appears in lane 3 and lane 5. Similarly, for the bovine blood sample and human blood sample (lane 6), it could be clearly seen that the intensity of the BSA or HSA band was significantly weaker after treatment with MSR-BSA-MIPs (lane 7) and MSR-HSA-MIPs (lane 9), whereas other bands changed inconspicuously. The band of BSA or HSA reappears for the eluent of adsorbed MSR-BSA-MIPs (lane 8) and MSR-HSA-MIPs (lane 10) through low-temperature washing. These results suggest that MSR-BSA-MIPs and MSR-HSA-MIPs can specifically absorb BSA or HSA either in the binary protein mixture or in the diluted blood samples, further exhibiting the potential in practical applications.4. Conclusions
In this work, a kind of novel multi-stimuli responsive molecularly imprinted polymers was successfully fabricated using a surface imprinting technique for recognition and enrichment of HSA. The obtained MSR-BSA-MIPs not only possess a magnetic response reflected in rapid dispersion or separation state transformation, but also exhibit a temperature or ionic strength response for capture or release conversion for the target protein. Meanwhile, MSR-BSA-MIPs reveal fast adsorption kinetics with easy accessibility to the recognition sites, high adsorption capacity, and favorable selectivity for HSA. In addition, MSR-BSA-MIPs show good reusability only through controlling the temperature or ionic strength. Furthermore, the resultant MSR-BSA-MIPs could specifically capture BSA or HSA easily from real samples of bovine blood or human blood, suggesting that the proposed MSR-BSA-MIPs are a cheap and feasible alternative material with prospects in specific recognition and enrichment of proteins from complex samples.Author contributions
Junjie Zhang and Yi Hao: conceptualization, validation, investigation, writing – original draft. Xuemeng Tian: formal analysis, writing – reviewing. Yulu Liang: writing – reviewing and editing. Xiwen He: validation, supervision. Ruixia Gao: conceptualization, writing – reviewing and editing, project administration, funding acquisition. Langxing Chen: conceptualization, writing – reviewing and editing, project administration, funding acquisition. Yukui Zhang: supervision.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 21475067), the Natural Science Foundation of Shaanxi Province (No. 2020JM-066, 2020JQ-019), the Fundamental Research Funds for the Central Universities (No. xjh012020011), and the China Postdoctoral Science Foundation (No. 2020M673428).Notes and references
- G. J. Quinlan, G. S. Martin and T. W. Evans, Hepatology, 2005, 41, 1211–1219 CrossRef CAS PubMed.
- V. Arroyo, R. García-Martinez and X. Salvatella, J. Hepatol., 2014, 61, 396–407 CrossRef CAS PubMed.
- S. Bhat, M. G. Jagadeeshaprasad, V. Venkatasubramani and M. J. Kulkarni, Expert Rev. Proteomics, 2017, 14, 677–689 CrossRef CAS PubMed.
- A. Shibata, Y. Ishima, M. Ikeda, H. Sato, T. Imafuku, V. T. G. Chuang, Y. Ouchi, T. Abe, H. Watanabe, T. Ishida, M. Otagiri and T. Maruyama, Biochem. Biophys. Res. Commun., 2016, 479, 578–583 CrossRef CAS PubMed.
- L. C. Y. Yu, Z. L. Hua, X. Y. Luo, T. Zhao and Y. Y. Liu, Biochim. Biophys. Acta, Rev. Cancer, 2022, 1877, 188655 CrossRef CAS PubMed.
- X. W. Li, M. Zangiabadi and Y. Zhao, J. Am. Chem. Soc., 2021, 143, 5172–5181 CrossRef CAS PubMed.
- Z. K. Gu, Y. R. Dong, S. X. Xu, L. S. Wang and Z. Liu, Angew. Chem., Int. Ed., 2021, 60, 2663–2667 CrossRef CAS PubMed.
- L. B. Wan, H. Liu, C. X. Huang and X. T. Shen, J. Mater. Chem. A, 2020, 8, 25931–25940 RSC.
- J. Wackerlig and R. Schirhagl, Anal. Chem., 2016, 88, 250–261 CrossRef CAS PubMed.
- K. C. Chen, R. He, X. Y. Luo, P. Z. Qin, L. Tan, Y. W. Tang and Z. C. Yang, Biosens. Bioelectron., 2017, 94, 609–615 CrossRef CAS PubMed.
- Q. Ding, Z. Y. Guo, W. Chen, H. Yu, X. X. Zhu, Q. Y. Liu and M. Fu, J. Colloid Interface Sci., 2021, 596, 225–232 CrossRef CAS PubMed.
- Z. T. Yang, Y. F. Zhang, J. Q. Ren, Q. Y. Zhang and B. L. Zhang, ACS Appl. Mater. Interfaces, 2021, 13, 34829–34842 CrossRef CAS PubMed.
- S. Dietl, H. Sobek and B. Mizaikoff, TrAC, Trends Anal. Chem., 2021, 143, 116414 CrossRef CAS.
- E. Turana, A. Zengin, Z. Suludere, N. Ö. Kalkan and U. Tamer, Talanta, 2022, 237, 122926 CrossRef PubMed.
- S. Rajpal, S. Bhakta and P. Mishra, J. Mater. Chem. B, 2021, 9, 2436–2446 RSC.
- C. Boitard, A. Bée, C. Ménager and N. Griffete, J. Mater. Chem. B, 2018, 6, 1563–1580 RSC.
- W. J. Lu, S. Wang, R. Liu, Y. Guan and Y. J. Zhang, Acta Biomater., 2021, 126, 249–258 CrossRef CAS PubMed.
- R. R. Xing, Y. Y. Ma, Y. J. Wang, Y. R. Wen and Z. Liu, Chem. Sci., 2019, 10, 1831–1835 RSC.
- Y. H. Cui, Z. Y. He, Y. Xu, Y. Su, L. Ding and Y. Li, Chem. Eng. J., 2021, 405, 126608 CrossRef CAS.
- R. T. Ma, X. Y. Sun, W. Ha, J. Chen and Y. P. Shi, J. Mater. Chem. B, 2017, 5, 7512–7518 RSC.
- R. R. Xing, S. S. Wang, Z. J. Bie, H. He and Z. Liu, Nat. Protoc., 2017, 12, 964–987 CrossRef CAS PubMed.
- M. J. Chen, H. L. Yang, Y. M. Si, Q. Tang, C. F. Chow and C. B. Gong, Food Chem., 2021, 355, 129656 CrossRef CAS PubMed.
- Y. Ma, Y. M. Yin, L. Ni, H. H. Miao, Y. J. Wang, C. Pan, X. H. Tian, J. M. Pan, T. Y. You, B. Li and G. Q. Pan, Bioact. Mater., 2021, 6, 1308–1317 CrossRef CAS PubMed.
- N. Zhang, N. Zhang, Y. R. Xu, Z. L. Li, C. R. Yan, K. Mei, M. L. Ding, S. C. Ding, P. Guan, L. W. Qian, C. B. Du and X. L. Hu, Macromol. Rapid Commun., 2019, 40, 1900096 CrossRef PubMed.
- L. Tang, L. Wang, X. Yang, Y. Y. Feng, Y. Li and W. Feng, Prog. Mater. Sci., 2021, 115, 100702 CrossRef CAS.
- C. X. Li, Y. Ma, H. Niu and H. Q. Zhang, ACS Appl. Mater. Interfaces, 2015, 7, 27340–27350 CrossRef CAS PubMed.
- X. Z. Dong, Y. Ma, C. P. Hou, B. L. Zhang, H. P. Zhang and Q. Y. Zhang, Polym. Int., 2019, 68, 955–963 CrossRef CAS.
- C. M. Hu, F. Peng, F. Mi, Y. Wang, P. F. Geng, L. Pang, Y. H. Ma, G. X. Li, Y. J. Li and M. Guan, Anal. Chim. Acta, 2021, 339289 Search PubMed.
- X. Y. Xie, Q. Hu, R. F. Ke, X. Y. Zhen, Y. S. Bu and S. C. Wang, Chem. Eng. J., 2019, 371, 130–137 CrossRef CAS.
- R. Zhang, T. J. Zhang, Y. Q. Lv, P. Y. Qin, H. M. Li, J. P. Li and T. W. Tan, Talanta, 2019, 201, 441–449 CrossRef CAS PubMed.
- J. P. Fan, J. X. Yu, X. M. Yang, X. H. Zhang, T. T. Yuan and H. L. Peng, Chem. Eng. J., 2018, 337, 722–732 CrossRef CAS.
- R. X. Gao, X. R. Mu, J. J. Zhang and Y. H. Tang, J. Mater. Chem. B, 2014, 2, 783–792 RSC.
- A. Kamgar and S. Hassanajili, J. Mol. Liq., 2020, 315, 113709 CrossRef CAS.
- A. G. B. Pereira, C. S. Nunes, A. F. Rubira, E. C. Muniz and A. R. Fajardo, Carbohydr. Polym., 2021, 266, 118116 CrossRef CAS PubMed.
- Y. H. Sun, C. Yao, Z. X. Xie and Y. G. Zhang, Colloids Surf., B, 2021, 197, 111446 CrossRef CAS PubMed.
- D. Keskin, L. Tromp, O. Mergel, G. Y. Zu, E. Warszawik, H. C. van der Mei and P. van Rijn, ACS Appl. Mater. Interfaces, 2020, 12, 57721–57731 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2tb00076h |
| ‡ These authors contributed equally to this paper. |
| This journal is © The Royal Society of Chemistry 2022 |