Open Access Article
Open Access ArticleSynthesis and functionalization of dendritic polyglycerol-based nanogels: application in T cell activation†
Felix
Reisbeck
a,
Stefanie
Wedepohl
 a,
Mathias
Dimde
a,
Ann-Cathrin
Schmitt
a,
Jens
Dernedde
a,
Mathias
Dimde
a,
Ann-Cathrin
Schmitt
a,
Jens
Dernedde
 b,
Miguel
Álvaro-Benito
b,
Miguel
Álvaro-Benito
 *c,
Christian
Freund
*c,
Christian
Freund
 *c and
Rainer
Haag
*c and
Rainer
Haag
 *a
*a
aInstitute of Chemistry and Biochemistry, Freie Universität Berlin, Takustr. 3, 14195 Berlin, Germany. E-mail: haag@chemie.fu-berlin.de
bCharité-Universitätsmedizin Berlin, Insitute of Laboratory Medicine, Clinical Chemistry, and Pathobiochemistry, CVK Augustenburger Platz 1, 13535 Berlin, Germany
cInstitute of Chemistry and Biochemistry, Freie Universität Berlin, Thielallee 63, 14195 Berlin, Germany. E-mail: chfreund@zedat.fu-berlin.de; m.alvaro@fu-berlin.de
First published on 25th November 2021
Abstract
The concept of multivalency finds various applications in the fields of chemistry and biology, relying on the principle that multiple weak interactions can lead to strong adhesive forces. Polymeric carriers are promising tools to translate these properties into the field of biomedicine, especially upon functionalization by active biomolecules, such as antibodies. In this study we report on the synthesis of dendritic polyglycerol (dPG) and dPG-based nanogels (NGs) as platforms for the multivalent display of molecules and their potential application as carrier units. Macromolecules based on dPG were synthesized and NGs were generated by strain-promoted azide–alkyne cycloaddition (SPAAC) by inverse nanoprecipitation under mild conditions. Scale-up screening rendered a reproducible method for a batch size of up to 50 mg for the formation of NGs in a size range of 150 nm with narrow dispersity. Dye-labelled bovine serum albumin (FITC-BSA) was chosen as a model protein and showed successful conjugation to the carriers, while the protein's secondary structure was not affected. Consequently, cyanine-5-amine (Cy5-NH2) and avidin (Av) were conjugated in order to exploit the strong avidin-biotin interaction, facilitating the directed attachment of a myriad of biotinylated (bio)molecules. As a proof-of-concept, the biotinylated monoclonal antibodies (mAbs) α-CD3 and α-CD28 were attached to the platforms and their capability to activate T cells was assessed. Experiments were performed with a Jurkat reporter cell line which expresses green fluorescent protein (GFP) upon activation, providing a rapid and reliable readout by flow cytometry. Carriers clearly outperformed conventional compounds for activation (i.e. antibodies crosslinked with anti-IgG antibody) at significantly lower dosages. These findings could be confirmed by confocal laser scanning microscopy (CLSM), showing accumulation of the functional nanoplatforms at the cell surface and cytoplasmic GFP expression (>95% activation of cells for the multivalent conjugates at 10 μg mL−1 compared to 37% activation with conventionally crosslinked mAbs at 25 μg mL−1), whereas carriers without mAbs could not activate cells. As the attachment of biotinylated molecules to the functional nanoplatforms is straightforward, the results obtained show the great potential of our platforms for a broad range of applications.
1. Introduction
Multivalency plays a key role in nature and describes interactions between a large number of binding partners, leading to improved binding as compared to the mere sum of its monovalent participants.1,2 This phenomenon is used for example by pathogens such as viruses and bacteria to enhance their cell binding strength and thus promote internalization. As an analogy, relevant examples that rely on multivalency are the burr or Velcro®, where relatively weak binding between hooks and loops results in powerful adhesion due to the sheer number of multivalent interactions.Biocompatible polymeric carriers are excellent candidates for introducing a variety of binding sites and generate multivalency, since they are usually easy to manufacture on an industrial batch scale, show little immunogenicity and are rarely prone to enzymatic degradation and opsonization in the body.3 Moreover, they can be manufactured in a variety of architectures including linear,4 star-like,5 and dendritic structures.6–8 These systems can have a particularly large beneficial impact for the multivalent display of biomolecules, such as sugars, proteins, peptides or antibodies, which are subject to very low availability upon administration and furthermore can be cleared by the body easily via enzymatic degradation or renal clearance.9–13
Besides natural polymers such as alginate, chitosan and hyaluronic acid, synthetic biocompatible polymers that have been explored include the polyesters N-(2-hydroxypropyl)methylacrylamide (HPMA),14,15 poly(lactic-co-glycolic acid) (PLGA),16 polyglutamic acid (PGA)17,18 and polyethers such as the FDA-approved poly(ethylene) glycol (PEG),19 which is considered to be the gold standard for biomedical applications.
Another excellent carrier for such applications is dendritic polyglycerol (dPG), a hyperbranched polymer which is synthesized by controlled anionic ring-opening polymerization of glycidol.20–23 Apart from its tuneable molecular weight and size, the free hydroxyl groups allow for high-precision chemical modifications as well as introduction of reactive groups, hydrophobic residues or surface charges. Besides its globular shape and polyether backbone, dPG is highly biocompatible and water-soluble, while the inertness of the polymer to biological components is retained.24 In order to enhance the surface area for multivalent display, larger nano-sized polymer-based carriers for example polymersomes, micelles or nanogels (NGs) can be synthesized. The latter are highly hydrated, crosslinked networks of amphiphilic or hydrophilic polymers in the size range of 1–1000 nm.25–27 NGs can be prepared in a variety of methods, including the dispersion of preformed polymers via solvent evaporation, salting out, dialysis, supercritical fluid technologies or by different polymerization techniques (e.g. mini- and microemulsion or nanoprecipitation).27–29 Many of these techniques, however, rely on the use of surfactants, potentially hampering their use in biomedical applications if not purified properly. The surfactant-free formation of nanogels using Cu(I)-catalyzed azide–alkyne cycloaddition, thiol-Michael addition, strain-promoted azide–alkyne cycloaddition (SPAAC) and inverse electron demand Diels–Alder (iEDDA) cyclization via inverse nanoprecipitation under mild conditions has been reported previously.30–34 When considering the display of biomolecules by polymeric carriers, one key factor is their accessibility and correct geometry after coupling. One way to achieve the site-specific coupling is the use of avidin–biotin interactions. Avidin is a highly stable, tetrameric glycoprotein and shows the strongest non-covalent interaction to biotin as a counterpart, showing orders of magnitude stronger affinity than antigen–antibody interaction.35 The use of multivalent structures exploiting these interactions could be particularly interesting for the display of prodrugs or the emerging field of drug-free macromolecular therapeutics, which aims at the treatment of diseases by crosslinking receptors without the need of low molecular weight drugs.36 Due to the versatility of nanomedicine, the development of novel multivalent platforms is still highly demanded. In this work, we report on the synthesis of dPG and dPG-based NGs as nanoplatforms for the multivalent display of biomolecules, using avidin–biotin interaction. Nanogels are formed via inverse nanoprecipitation by SPAAC and subsequently coupled to Av to allow for further directed coupling of any biotinylated biomolecule. We sought to compare an assembled NG with a size distribution around 150 nm to a monomeric dPG with a hydrodynamic diameter of 25 nm. As a proof-of-principle, Av was coupled and biotinylated antibodies (α-CD3 and α-CD28) were loaded. The functionality of the conjugates was tested in a cell-based assay. Activation of the TCR was visualized by microscopy and flow cytometry in a respective reporter cell line.
2. Experimental
2.1 Materials and methods
Anhydrous solvents were chemically dried by conventional methods prior to use or commercially purchased from Acros Organics™ in septum-sealed bottles. All reactions which involved air- or water-sensitive compounds were carried out in a dried flask under an argon atmosphere. Water was used from a Milli-Q station from Millipore (Merck, Millipore).Dendritic Polyglycerol (dPG) with a molecular weight of 10 kDa, dPG-N3(10%) and dPG-NH2(10%) were synthesized according to an established procedure.20,37 Since dPG is a hygroscopic polymer, it was dried (50 °C, 10−3 mbar, 24 h) prior to reactions. dPG1.4 MDa-N3(7%) was kindly provided by Matthias Wallert.
Methane sulfonylchloride, diethylene glycol, propargyl bromide, potassium tert-butoxide, tert-butyl acrylate, sodium azide, N,N,N′,N′-tetramethyl-O-(N-succinimidyl)uronium hexafluorophosphate (HSTU), triphenyl phosphine, bovine serum albumine (BSA), fluorescein-labeled BSA (FITC-BSA) and benzoylated dialysis tubing with a molecular weight cut-off (MWCO) of 2 kDa were purchased from Sigma-Aldrich. Pierce™ BCA Protein Quantification Kit was purchased from ThermoFisher and used according to the manufacturer's instructions. Spectra/POR dialysis membrane (100–500 Da, 50 kDa and 100 kDa MWCO) were purchased from Carl Roth. Moreover, the following compounds were acquired: trifluoroacetic acid (Acros), cyanine-5-amine (Lumiprobe), avidin (Pierce, ThermoFisher), biotinylated human anti CD3 antibody clone OKT3, biotin anti human CD28 antibody clone CD28.2 (Biolegend), goat anti-mouse IgG H&L (abcam).
1H and 13C NMR spectra were recorded with a Bruker DPX (400 MHz) or a Bruker AVANCE III (500 MHz; 700 MHz) instrument. As deuterated solvents, CDCl3, D2O and DMSO-d6 were used. Chemical shifts δ are given in ppm relative to TMS as an internal standard or relative to the resonance of the solvent (1H NMR: Chloroform: δ = 7.26 ppm, DMSO: δ = 2.50 ppm; D2O: δ = 4.79 ppm; 13C NMR: chloroform: δ = 77.00 ppm, DMSO: δ = 39.70 ppm). All spectra were recorded at room temperature.
2.2 Synthesis of dendritic polyglycerol (dPG)-based macromolecules
Dendritic polyglycerol (dPG), dPG-(NH2)10% and dPG-(N3)10% were synthesized according to established protocols.20,37 Their corresponding coupling partners for the formation of the macromolecules Bicyclo[6.1.0]non-4-yn-9-ylmethyl-(4-nitrophenyl) carbonate (BCN) and azide-linker (3) were synthesized according to Dommerholt et al. and Kumar et al., respectively.38,39 The final dPG-(BCN)10% and dPG-azide-linker macromonomers were synthesized according to modified reported procedures.40,41Analogously to 6, the large dPG1.4 MDa-(N3)7% was decorated with the azide linker 3, aiming for 200 groups of carboxylic acids on the surface.
2.3 Preparation of dPG nanogels (NGs) by nanoprecipitation
dPG-azide-linker 6 (20 mg, 16.8 μmol azide groups) and dPG-BCN 8 (30 mg, 31.9 μmol alkyne) were dissolved separately in Milli-Q water (5 mL each). The solutions were unified, vortexed and injected into acetone (800 mL) under vigorous stirring. The reaction was quenched after 45 min by addition of 3-azidopropanol (100 μg). After 24 h, Milli-Q water (50 mL) was added, the non-solvent was removed in vacuo in order to obtain blue shimmering dispersions of the nanoparticles. Nanogels were dialyzed against Milli-Q water with regular exchange of the dialysate for 3 d using a MWCO 50 kDa membrane and were characterized by means of DLS, AFM and NTA.2.4 Preparation of active esters
Nanogels were transferred into DMF with subsequent evaporation of water using a vacuum line. All carboxylic acids in the nanogel were converted with 1.5 equiv. of DIPEA as well as HSTU into NHS esters for amide coupling. Active esters were formed in situ under an argon atmosphere and left overnight. Protein coupling was further investigated in solution.2.5 FITC-BSA coupling
We coupled NGs in different ratios to FITC-BSA. The protein (1 mg mL−1 stock solution in PBS (2.0, 0.3 and 0.1 equiv. per NHS ester) was added dropwise under cooling on ice. The reaction mixture was stirred for 24 h and then dialyzed against phosphate buffer (pH 7.4) for 72 h with a MWCO 100 kDa membrane. Conjugates were analysed by means of DLS and CD. The coupling efficiency was determined based on UV/vis measurements against a calibration curve (Fig. S1, ESI†).2.6 Avidin coupling
For decoration of the carriers with avidin, we chose to couple each, dPG-azide-linker 6 in the NG network and dPG1.4 MDa-(N3)-COOH with 1 equivalent avidin. The reaction was performed similar to the one using the model protein. However, since Av was not dye-labelled itself, Cy5-NH2 (1 mg mL−1 stock solution in DMSO; 0.1 equiv. per NHS ester) was added to the reaction mixture two hours after the addition of the protein. Avidin-coupled nanogels were dialyzed against phosphate buffer (pH 7.4, MWCO 100 kDa) for 72 h and their volume was reduced in vacuo for further analysis. Here, coupling efficiency was determined using a BCA protein quantification kit (Pierce). The integrity of the secondary structure was determined using CD spectroscopy. Similar modifications were performed with dPG1.4 M-(COOH)200. A detailed description is given in the ESI.†2.7 Coupling of α-CD3 and α-CD28 to the architectures
Based on the values obtained by BCA assay, carriers bearing Av were incubated with 4 equiv. of either only one antibody or a combination of both antibodies (50/50 mol%) at room temperature for 1 h. Conjugates were dialyzed against Milli-Q water (MWCO 100 kDa) for 24 h and the dialysate was exchanged regularly. Finally, the volume of reduced in vacuo and the sample was stored in PBS. As negative controls, bare carriers were processed in the same fashion.2.8 Physical characterization of dPG-based NGs
Atomic Force Microscopy (AFM) was performed on a Multimode 8 microscope which was equipped with a Nanoscope V controller (Bruker Corporation). 15 μL of highly diluted sample (10 μg mL−1) was deposited on the substrate and incubated at room temperature for 15 minutes. Cantilever A, type SNL-10 from Bruker with a nominal tip radius and nominal spring constant of 0.35 N m−1 were used in these experiments to map the samples. Prior to imaging, cantilevers were calibrated on the naked mica surface. A compression on mica was taken to extract the cantilever sensitivity and then, the thermal noise method was used to derive its spring constant.42 Imaging in soft tapping mode (sTM) was performed at room temperature in air. Imaging in PeakForce mode was carried out with a maximal applied force of 200 pN using a 512 points per line at a scan rate of 0.7 Hz. Image analysis was performed using Nanoscope Analysis 1.5 software. All images were processed by the Plane fit program and the flatten tool (order 1) to correct from effects of bowing from the piezo movement.Circular Dichroism (CD) spectra were recorded using a JASCO J-810 spectropolarimeter with a HAAKEWKL recirculating chiller (ThermoFisher Scientific). The cuvettes were set to a constant temperature of 25 °C with a JASCO PTC-423S Peltier-type thermocouple. Measurements were performed in quartz cuvettes (Suprasil Hellma) with a path length of 2 mm.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× and an acceleration voltage of 200 kV. Images were recorded by a Falcon 3CE direct electron detector (48 aligned frames) at full size (4k). The defocus was chosen to be 4 μm in all cases to create sufficient phase contrast.
000× and an acceleration voltage of 200 kV. Images were recorded by a Falcon 3CE direct electron detector (48 aligned frames) at full size (4k). The defocus was chosen to be 4 μm in all cases to create sufficient phase contrast.
DLS and zeta potential measurements were performed on a Malvern zetasizer nano ZS 90 with a He–Ne laser (λ = 633 nm) at 25 °C. Size and zeta potential were measured in triplicates at a concentration of 1 mg mL−1 in phosphate buffer at pH 7.4.
Nanoparticle Tracking Analysis (NTA) was performed in triplicates using a Nanosight NS500 (Malvern Instruments). As compared to DLS, samples were diluted 1000 times in Milli-Q water.
UV/vis measurements were performed on a Agilent Cary 8654 UV-visible spectrophotometer using half-micro quartz cuvettes.
2.9 Cytotoxicity measurements
The cytotoxic effect of NGs on three cancer cell lines (HeLa, A549, and MCF-7) as well as the Jurkat reporter cell line was determined using the cell viability assay Cell Counting Kit 8 (CCK-8) from Sigma Aldrich according to the manufacturer's instructions. HeLa (DSMZ no.: ACC 57), A549 (DSMZ no.: ACC107), and MCF-7 (DSMZ no.: ACC115) were obtained from Leibniz-Institute DSMZ – Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH and cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% (v/v) FBS, 100 U mL−1 penicillin and 100 μg mL−1 streptomycin (all from Gibco/ThermoFisher Scientific). Cells were regularly cultured at least twice a week when they reached 70–90% confluency. For the cytotoxicity measurements, 90 μL of a cell suspension in DMEM containing 50 × 104 cells per mL were seeded in each inner well of a 96-well plate and incubated overnight at 37 °C and 5% CO2. In the outer wells, 90 μL of DMEM without any cell were added. The next day, serial dilutions of all samples were prepared and 10 μL each were added in triplicates to the cells, as well as to one outer well for background correction. SDS (1%) and untreated cells were used as controls. After another 48 h at 37 °C and 5% CO2, the CCK-8 solution (10 μL per well) was added and absorbance was measured after approximately 3 h incubation at a wavelength of 450 nm and a reference wavelength of 650 nm with a microplate reader (TECAN infinite M200Pro). Measurements were performed in triplicate and the cell viability was calculated by setting the untreated controls to 100% after subtracting the background using Excel software. All cell experiments were conducted according to German genetic engineering laws and German biosafety guidelines in the laboratory (safety level 1).2.10 Sample preparation for T cell activation and flow cytometry
A Jurkat JE6.1 NFκB-eGFP T cell line engineered to express a T cell receptor (TCR) specific for a immunodominant T cell epitope (Art v 125-36 presented on HLA-DRB1*01:01) was used as reporter for T cell activation. The cells had been previously reported by Rosskopf et al., were obtained directly from the authors and were cultured and maintained as described in the original publication.43 The cell line was routinely cultivated in RPMI medium (Gibco/ThermoFisher Scientfic) supplemented with 10% FBS and 1% Penicillin/Streptomycin at 37 °C and 5% CO2. Cell clumps were dispersed by pipetting up and down and the cell density was kept between 0.5 × 106 and 2 × 106 cells per mL by dilution with fresh medium twice a week. One day before the experiments, cells were adjusted to 1 × 106 cells per mL in fresh medium. The next day, cells were counted and distributed in 96 well plates at 3 × 105 cells per well. Test compounds were added at different concentrations and incubated for 6 h at 37 °C and 5% CO2. After incubation, cells were transferred to microcentrifuge tubes and centrifuged for 4 minutes at 140 × g. The supernatant was removed and cells were washed 2× with 1 mL PBS supplemented with 1% (v/v) BSA at 4 °C and centrifugation at 140 × g for 4 minutes. After washing, cells were resuspended in PBS-BSA (200 μL) and kept on ice.Flow cytometry was performed on an Accuri C6 (BD Biosciences) flow cytometer. Data were acquired by the BD Accuri C6 software and further processed with the FlowJo V10 analysis software.
2.11 Confocal laser scanning microscopy
After flow cytometry, the remaining cells were fixed by addition of formaldehyde to a final concentration of 10% (v/v) for 30 minutes. Fixed cells were washed 3× with PBS and transferred to a 8-well slide (ibidi μ-slide 8) for microscopy on a Leica SP8 confocal laser scanning microscope. Images were acquired using LASX software and the preset settings for GFP and Cy5, as well as transmission.3. Results and discussion
3.1 Synthesis of nanogel precursors
Inverse nanoprecipitation relies on fast reaction kinetics, hence click reactions such as [3+2] cycloadditions or inverse electron demand Diels–Alder (iEDDA) reactions can be used.30,44 Here, we chose the strain-promoted azide alkyne cycloaddition (SPAAC) for the synthesis of the macromolecular precursors of the nanogels (Scheme 1).This reaction is performed without the addition of any catalyst, and in water at room temperature.
In this process, 10% of the hydroxyl groups of a 10 kDa dPG 4 were converted in two reaction steps into azide groups according to a well-established protocol to obtain dPG-N3(10%)5.37 In order to allow for post-functionalization via amide coupling, four azide groups of dPG-N3(10%) were converted with 3-(2-(2-(prop-2-yn-1-yloxy)ethoxy)ethoxy)propanoic acid 3, in a copper(I)-catalysed azide–alkyne cycloaddition to macromolecule dPG-azide-linker 6 according to a modified procedure.45 This step leaves about ten remaining groups for the nanogel crosslinking. Then, dPG-N3(10%)5 was reduced in a Staudinger reduction with PPh3 to the corresponding amine 7, which was then decorated with bicyclo[6.1.0.]non-4-yne (BCN) by using its activated carbonate to the second macromolecule dPG-BCN 8.38 Carboxylated NGs 9 were obtained by inverse nanoprecipitation of an aqueous precursor solution into acetone.
3.2 Nanogel synthesis and characterization
To find a method for the reproducible preparation of nanogels, different nanoprecipitation variables such as ratio of macromonomers, amount of solvent and non-solvent, reaction and quenching times were screened. Our hypothesis is that medium-sized flexible NGs can attach to the cell surface in a multivalent fashion. Therefore, we sought after conditions that reliably yield NGs in a size range of 100–200 nm. Macromonomers were dissolved separately in Milli-Q water, briefly mixed, and then injected into acetone under vigorous stirring. The reaction was quenched by the addition of 3-azidopropanol after 40 minutes. The resulting nanogel sizes, PDIs and zeta potentials are summarized in Table 1. We found that for this system, a ratio of a slight excess (1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 w/w) of dPG-BCN 8 to dPG-azide-linker 6 is favourable.
1 w/w) of dPG-BCN 8 to dPG-azide-linker 6 is favourable.
| Batch size (mg mL−1) | H2O (mL) | Acetone (mL) | Azidopropanol (μg) | Size (d, nm) | PDI | ξ (mV) |
|---|---|---|---|---|---|---|
| 5 | 1 | 80 | 10 | 185 ± 2 | 0.16 | −16 ± 1 |
| 25 | 5 | 400 | 50 | 149 ± 6 | 0.22 | −13 ± 1 |
| 50 | 10 | 800 | 100 | 143 ± 2 | 0.02 | −19 ± 1 |
We could implement a scale-up of the nanoprecipitation and up to 50 mg batch size, yielding NGs with a narrow size distribution. It is worth noting that all gels have a negative zeta potential, indicating the presence of carboxylic acids on the surface of NGs and not entangled in the polymer network (Table 1).
This facilitated surface coupling for the next steps, in which proteins and/or antibodies need to be conjugated. The particle size and morphology of NG 9 from a 50 mg batch was characterized by means of DLS, nanoparticle tracking analysis (NTA), atomic force microscopy (AFM) (Fig. 1).
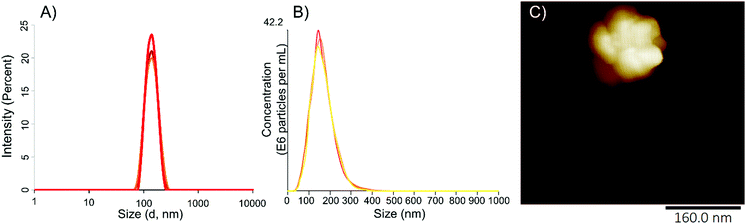 | ||
| Fig. 1 Physical characterization of NG 9 from a 50 mg batch. (A) Particle size by DLS or (B) NTA; and (C) particle morphology as determined by AFM. | ||
DLS indicated an average size of the NG of 170 nm, which was corroborated by NTA with 155 nm. Microscopic methods could demonstrate the globular shape of the nanogel, consisting of crosslinked dPG macromolecules, and their narrow size distribution. We could further confirm this by AFM measured in soft tapping mode.
3.3 Synthesis of dye-labelled nanogel–protein conjugates
Prior to coupling of our carriers, NHS ester formation was verified for dPG-azide-linker 6 by 1H NMR (see ESI†). We evaluated the ability and capacity of the dPGs and NGs to bind different biomolecules. As coupling strategy, we chose unspecific amide coupling of the activated carboxylic acids on the NGs to amines (either N-terminal or lysine side chains) on the proteins. An overview of the process can be seen in Scheme 2.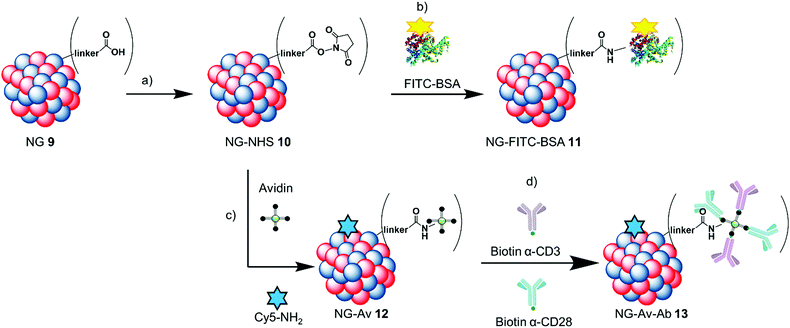 | ||
| Scheme 2 NG–protein conjugation scheme: (a) HSTU, DIPEA, DMF, r.t., 24 h; (b) FITC-BSA, DMF/PBS, 0 °C-r.t., 24 h; (c) avidin, Cy5-NH2, DMF/PBS, 0 °C-r.t., 24 h; (d) biotin-α-CD3/CD28, r.t., 1 h. | ||
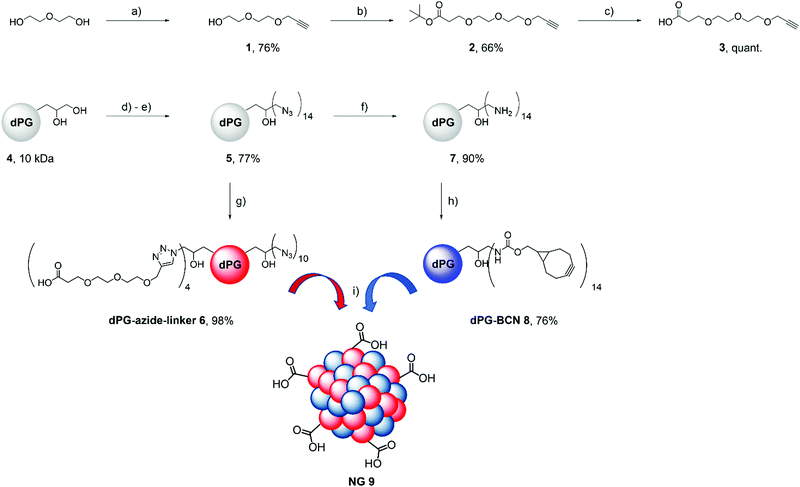
The carboxylic acids on NG 9 were first converted to form NG-NHS 10in situ, and subsequently the model protein was coupled to them resulting in NG-FITC-BSA 11. We established protein coupling to the nanogels using FITC-labeled bovine serum albumin (FITC-BSA) as a model protein as it is readily available, inexpensive and has a similar monomeric size compared to avidin (FITC-BSA 65 kDa and Av 67 kDa). We used the dye label as a convenient reporter for the coupling efficiency, as it can be followed by UV/Vis spectroscopy. Fig. 2 shows the evaluation of the coupling as well as stability of FITC-BSA coupled to the NG scaffold by DLS and CD spectroscopy. For the evaluation of the NG's NHS ester formation, they were transferred from water into DMF and DLS measurements were performed in order to determine any size difference (Fig. 2A).
NG 9 showed no difference in size or zeta potential upon storage for 1 year at 4 °C, indicating that they are stable under these conditions. There seemed to be no impact concerning the solvent either and FITC-BSA was coupled to the activated esters in DMF in different ratios. We calculated the coupling efficiency using a calibration curve (Fig. S1, ESI†) of the dye-labeled protein, and determined a range between 51 and 62%. DLS measurements showed that after work-up no free protein was contained within the mixture and that NG-FITC-BSA 11 increased very little in size compared to the unconjugated NG (Fig. 2B). This is further supported by the number average size distribution (Fig. S2, ESI†). In order to analyse the structural integrity of the protein after coupling, we performed CD spectroscopy of the blank NG 9, the free protein and NG-FITC-BSA conjugate 11. The resulting curves indicate that the α-helical secondary structure of FITC-BSA was still intact after coupling (Fig. 2C).46
3.4 Functionalization via avidin
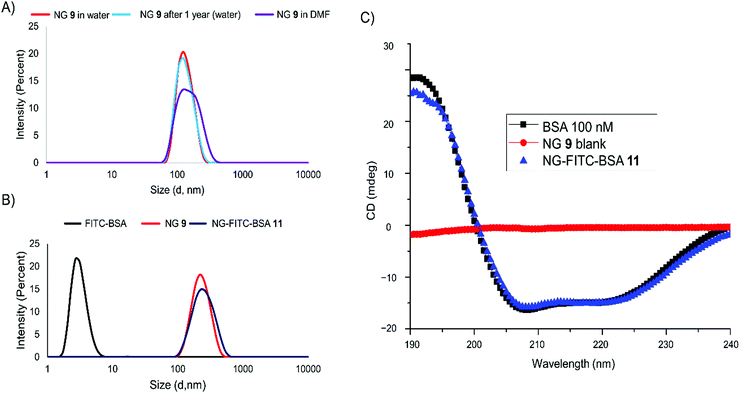
After successful coupling of the model protein, we proceeded with using avidin (Av) for the further development of this system as a platform for the multivalent display of biotinylated biomolecules. We chose to couple Av to 10% of the activated NHS esters on the nanogel and add Cy5-NH2. Addition of his dye label will allow for localization of the carriers for the evaluation of their performance. Fig. 3 shows the evaluation of NG-avidin coupling by protein quantification assay, DLS, CD spectroscopy and cryo-TEM.In contrast to the NG-FITC-BSA evaluation, here we used Av without any dye label. Therefore, the coupling efficiency was evaluated by a protein quantification kit (BCA assay) (Fig. 3A) and calculated to be in the range between 30% and 40% (for exemplary calibration see Fig. S3, ESI†). DLS indicated that the coupling leads to a small increase in the hydrodynamic radius from 140 nm (NG 9) to 160 nm (NG-Av 12), but also to a broadening of the size distribution after coupling. The hydrodynamic diameter of Av was determined to be around 6 nm (Fig. 3B). CD spectroscopy revealed that the secondary structure of Av was still intact after coupling (Fig. 3C).47Cryo-TEM images of NG-Av 12 (Fig. 3D) show spherical particles with a rather narrow size distribution. Just as for the measurements for the nanogel 9 (Fig. 1D), a smaller diameter as compared to DLS can be observed, which again can be attributed to the missing hydration shell.
3.5 Evaluating the performance of Ab conjugates in T cell activation
We first investigated whether cultured cells incubated with the NGs would be affected in terms of their viability. For this, we performed cell viability assays with three well-established cell lines (HeLa, A459 and MCF-7). We could confirm the biocompatibility of dPGs, as it has been previously described.48 Up to the highest concentration tested (1 mg mL−1, see Fig. S4, ESI†), we did not observe any detrimental effects of the NGs to the cell viability of all three cell lines, supporting their safe use in biomedical applications.With these promising results regarding the method for particle formation, protein coupling and cell viability at hand, we then decided to evaluate the use of dPGs and NGs for the display of molecules. For this, biotinylated mAbs (α-CD3 and α-CD28) were coupled to our platforms and T cell receptor (TCR) activation was investigated. T cell activation can be achieved via simultaneous ligand binding to the co-stimulatory cell surface molecules CD3 and CD28 via monoclonal antibodies (mAbs). Usually, mAbs in solution fail to trigger T cell activation on their own. Essential is either a multivalent display on beads or crosslinking secondary antibodies (polyclonal) targeting the Fc domains of the primary antibodies.49
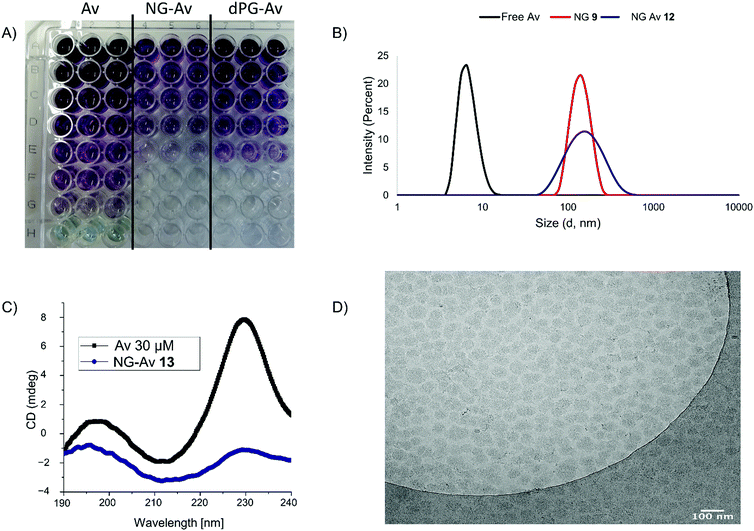
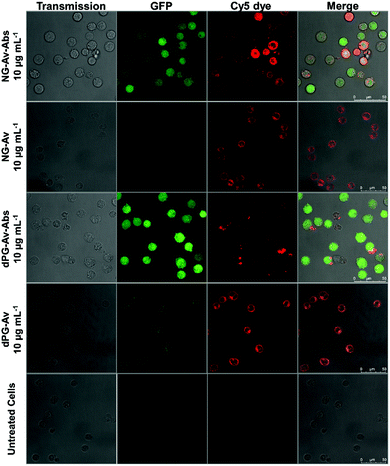
First, we investigated the T cell activation by confocal laser scanning microscopy (CLSM), using the Jurkat-NFκB-GFP reporter cell line that expresses green fluorescent protein (GFP) upon NFκB pathway activation. This signal pathway is initiated as result of TCR assembly, providing a rapid and reliable readout (Fig. 4).
Images were taken of non-treated cells as control as well as cells incubated with NG-Av, dPG-Av, NG-Av-Ab and dPG-Av-Ab, respectively, at a concentration of 10 μg mL−1 after 6 h. The images show that carriers (red) clearly bind to cells and accumulate along the cell surface for all formats. Moreover, strong cytoplasmic GFP expression can be observed for those carriers bearing conjugated mAbs (NG-Av-Ab and dPG-Av-Ab). This qualitatively demonstrates the ability of our platforms bearing both mAbs to activate T cells (as compared to carriers with only avidin).
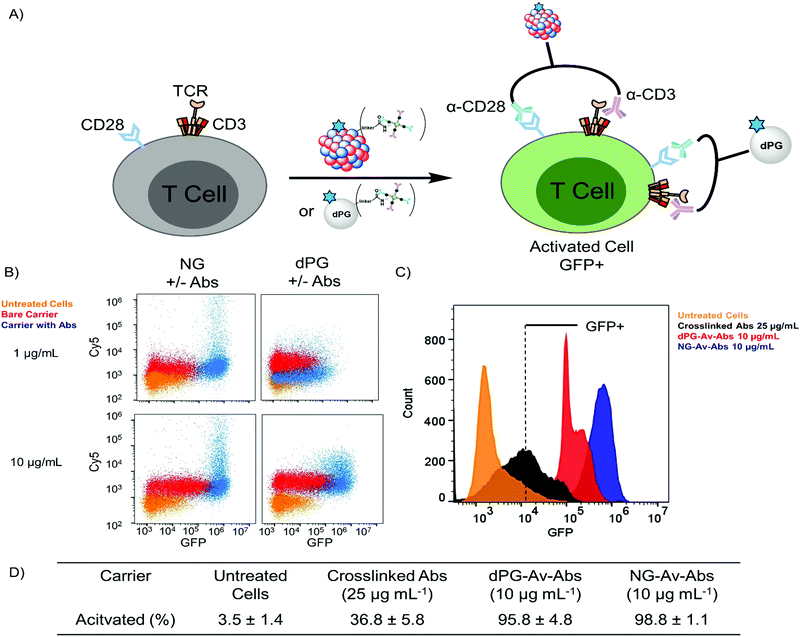
To complement our findings, we chose to quantify the T cell activation via flow cytometry and compare them to conventional methods for antibody crosslinking (Fig. 5). Under normal conditions, although a single conjugated antibody could trigger activation, a complete bulk activation will only occur if both molecules can target one single cell at once (Fig. 5A). Activation with PMA/Ionomycin, a conventional mix that bypasses cell surface activation, is achieved with 12.5 ng mL−1 within 6 h (see Fig. S6, ESI†).50 We evaluated different conditions used regularly in T cell activation based on mAbs α-CD3 and α-CD28. These conditions include crosslinking antibodies, which create CD3/CD28 clusters including the TCR. Cells were incubated with all different formats for 6 h and subsequently analysed via flow cytometry.
After 6 h incubation of dPG or NG conjugates bearing both α-CD3 and α-CD28 with the cells, we see a large proportion of the cell population to shift to higher fluorescence values in the GFP channel compared to untreated cells, which clearly demonstrates successful activation (Fig. 5B). Activation was possible with carrier concentrations as low as 1 μg mL−1, while the unconjugated carriers seem to contact the cell surface but are not competent to mediate significant pathway activation. On the y-axis that shows fluorescence intensity in the Cy5 channel, we see a shift of the cell populations that were treated with both the conjugated and unconjugated carriers, with the conjugates shifting markedly to higher fluorescence values. The shift in the fluorescence intensity of the cells treated with the bare carriers suggests that the carriers were taken up by the cells. The clear shift to higher GFP expression levels that can be seen in the histograms in Fig. 5C achieved with our carriers indicates that they outperform conventional methods based on soluble or conjugated mAbs, especially at lower dosages. This can be attributed to the multivalency effect. We determined the fraction of the gated main populations that can be activated within 6 h at a concentration of 10 μg mL−1 and found activation of more than 95% of the gated cells as compared to roughly 37% activation with the conjugated mAbs (Fig. 5D). When taking a more detailed look on the functionalized nanoplatforms bearing either one of the antibodies, slight activation can also be seen for these constructs at a concentration of 10 μg mL−1 (Fig. S8, ESI†).
4. Conclusions
We present the synthesis of a platform for the multivalent display of biomolecules on the surface of biocompatible polymers in the size range of 25 and 150 nm. Macromonomers based on dendritic polyglycerol for the NG formation were successfully synthesized and the screening of the inverse nanoprecipitation rendered a reproducible method at a batch size of up to 50 mg. A 1.4 MDa dPG was functionalized in a similar fashion for comparison. FITC-BSA was coupled to NGs as a model protein, yielding coupling efficiencies between 51–60%, which were determined by UV/vis spectroscopy. DLS showed little impact of the conjugation on the carrier's size. The integrity of the protein's secondary structure was confirmed by CD spectroscopy, indicating successful coupling and formation of a stable polymer–protein conjugate. We chose avidin as a protein for conjugation, as it has four binding sites for biotin, allowing for a directed conjugation of biotinylated biomolecules to the surface of our carriers. In order to detect the carriers themselves, an additional Cy5-NH2 dye was incorporated in a one-pot reaction. The resulting conjugates were analysed by DLS, CD and cryo-TEM. Avidin coupling efficacy was determined using a BCA assay, indicating a ratio between 30–40%, and the structural integrity of the protein was confirmed by CD. To demonstrate further functionalization, biotinylated antibodies α-CD3 and α-CD28 were conjugated to dPG-Av and NG-Av, respectively. Bioactivation by the complete carriers was demonstrated on a Jurkat reporter cell line visualized by CLSM and quantified by flow cytometry. We determined 95.8% and 98.8% activation of the gated cells after incubation with 10 μg mL−1 of dPG-Av-Ab and NG-Av-Ab, respectively. This shows that our Ab-conjugated nanoplatforms perform significantly better than standard assays applying crosslinking antibodies and at lower dosages with 36.8% activation after incubation with 25 μg mL−1. Due to their facile synthesis and high biocompatibility, these polymeric nanoplatforms are promising candidates for the multivalent display of biotinylated compounds and their downstream applications.Author contributions
Conceptualization, methodology, synthesis, in vitro studies, validation, formal analysis, data interpretation, investigation, writing – original draft, visualization: F. R. Conceptualization, methodology, in vitro studies, validation, formal analysis, data interpretation, investigation, writing – review and editing: S. W. and M. A.-B. Conceptualization, data interpretation, writing – review and editing: M. D. Investigation: A.-C. S. Conceptualization, methodology, writing – review and editing, supervision: J. D. Conceptualization, methodology, writing – review and editing, supervision, project administration, funding acquisition: C. F. and R. H.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank Matthias Wallert for providing dPG1.4 MDa-N3(7%) and Jay Kizhakkedathu for the supervision. We are grateful to Katharina Achazi, Ernesto Rafael Osorio Blanco and Suvrat Chowdhary for helping with CLSM, AFM, and CD measurements, respectively. Elisa Quaas is acknowledged for performing cytotoxicity studies. The authors would like to acknowledge Winfried F. Pickl and Peter Steinberger (Medical University of Vienna) for providing the Jurkat cell line. This study was supported from the Dahlem Research School (DRS), Collaborative Research Centers SFB 765 and SFB 1449, a DFG grant to C.F. (FR1325/17-1 and TRR186, project A21N) and the core facility BioSupraMol (http://www.biosupramol.de), all funded by the Deutsche Forschungsgemeinschaft (DFG).Notes and references
- C. Fasting, C. A. Schalley, M. Weber, O. Seitz, S. Hecht, B. Koksch, J. Dernedde, C. Graf, E.-W. Knapp and R. Haag, Angew. Chem., Int. Ed., 2012, 51, 10472–10498 CrossRef CAS PubMed.
- M. Mammen, S.-K. Choi and G. M. Whitesides, Angew. Chem., Int. Ed., 1998, 37, 2754–2794 CrossRef PubMed.
- I. Ekladious, Y. L. Colson and M. W. Grinstaff, Nat. Rev. Drug Discovery, 2019, 18, 273–294 CrossRef CAS PubMed.
- T. X. Viegas, M. D. Bentley, J. M. Harris, Z. Fang, K. Yoon, B. Dizman, R. Weimer, A. Mero, G. Pasut and F. M. Veronese, Bioconjugate Chem., 2011, 22, 976–986 CrossRef CAS PubMed.
- D. Wang, P. Kopečková, T. Minko, V. Nanayakkara and J. Kopeček, Biomacromolecules, 2000, 1, 313–319 CrossRef CAS PubMed.
- Y. Zheng, S. Li, Z. Weng and C. Gao, Chem. Soc. Rev., 2015, 44, 4091–4130 RSC.
- P. M. Levine, T. P. Carberry, J. M. Holub and K. Kirshenbaum, MedChemComm, 2013, 4, 493–509 RSC.
- S. Chen, S. Huang, Y. Li and C. Zhou, Front. Chem., 2021, 9 DOI:10.3389/FCHEM.2021.659304.
- T. Nie, W. Wang, X. Liu, Y. Wang, K. Li, X. Song, J. Zhang, L. Yu and Z. He, Biomacromolecules, 2021, 22, 2299–2324 CrossRef CAS PubMed.
- S. Bhatia, M. Dimde and R. Haag, MedChemComm, 2014, 5, 862–878 RSC.
- P. A. Venter, A. Dirksen, D. Thomas, M. Manchester, P. E. Dawson and A. Schneemann, Biomacromolecules, 2011, 12, 2293–2301 CrossRef CAS PubMed.
- H. M. Yassine, J. C. Boyington, P. M. McTamney, C.-J. Wei, M. Kanekiyo, W.-P. Kong, J. R. Gallagher, L. Wang, Y. Zhang, M. G. Joyce, D. Lingwood, S. M. Moin, H. Andersen, Y. Okuno, S. S. Rao, A. K. Harris, P. D. Kwong, J. R. Mascola, G. J. Nabel and B. S. Graham, Nat. Med., 2015, 21, 1065–1070 CrossRef CAS PubMed.
- S. Hussain, J. Joo, J. Kang, B. Kim, G. B. Braun, Z.-G. She, D. Kim, A. P. Mann, T. Mölder, T. Teesalu, S. Carnazza, S. Guglielmino, M. J. Sailor and E. Ruoslahti, Nat. Biomed. Eng., 2018, 2, 95–103 CrossRef CAS.
- J. Yang and J. Kopeček, J. Controlled Release, 2016, 240, 9–23 CrossRef CAS PubMed.
- J. Yang and J. Kopeček, Curr. Opin. Colloid Interface Sci., 2017, 31, 30–42 CrossRef CAS PubMed.
- F. Danhier, E. Ansorena, J. M. Silva, R. Coco, A. Le Breton and V. Préat, J. Controlled Release, 2012, 161, 505–522 CrossRef CAS PubMed.
- G. Córdoba-David, A. Duro-Castano, R. C. Castelo-Branco, C. González-Guerrero, P. Cannata, A. B. Sanz, M. J. Vicent, A. Ortiz and A. M. Ramos, Sci. Rep., 2020, 10, 2056 CrossRef PubMed.
- A. Duro-Castano, R. M. England, D. Razola, E. Romero, M. Oteo-Vives, M. A. Morcillo and M. J. Vicent, Mol. Pharmaceutics, 2015, 12, 3639–3649 CrossRef CAS PubMed.
- P. L. Turecek, M. J. Bossard, F. Schoetens and I. A. Ivens, J. Pharm. Sci., 2016, 105, 460–475 CrossRef CAS.
- M. Wallert, J. Plaschke, M. Dimde, V. Ahmadi, S. Block and R. Haag, Macromol. Mater. Eng., 2021, 2000688 CrossRef CAS.
- A. Sunder, R. Mülhaupt, R. Haag and H. Frey, Adv. Mater., 2000, 12, 235–239 CrossRef CAS.
- A. Sunder, R. Mülhaupt, R. Haag and H. Frey, Macromolecules, 2000, 33, 253–254 CrossRef CAS.
- S. Abbina, S. Vappala, P. Kumar, E. M. J. Siren, C. C. La, U. Abbasi, D. E. Brooks and J. N. Kizhakkedathu, J. Mater. Chem. B, 2017, 5, 9249–9277 RSC.
- J. Khandare, M. Calderón, N. M. Dagia and R. Haag, Chem. Soc. Rev., 2012, 41, 2824–2848 RSC.
- M. Karg, A. Pich, T. Hellweg, T. Hoare, L. A. Lyon, J. J. Crassous, D. Suzuki, R. A. Gumerov, S. Schneider, I. I. Potemkin and W. Richtering, Langmuir, 2019, 35, 6231–6255 CrossRef CAS.
- K. S. Soni, S. S. Desale and T. K. Bronich, J. Controlled Release, 2016, 240, 109–126 CrossRef CAS PubMed.
- A. V. Kabanov and S. V. Vinogradov, Angew. Chem., Int. Ed., 2009, 48, 5418–5429 CrossRef CAS PubMed.
- N. Wang, X. Cheng, N. Li, H. Wang and H. Chen, Adv. Healthcare Mater., 2019, 8, 1801002 CrossRef.
- J. P. Rao and K. E. Geckeler, Prog. Polym. Sci., 2011, 36, 887–913 CrossRef CAS.
- D. Steinhilber, M. Witting, X. Zhang, M. Staegemann, F. Paulus, W. Friess, S. Küchler and R. Haag, J. Controlled Release, 2013, 169, 289–295 CrossRef CAS PubMed.
- A. Oehrl, S. Schötz and R. Haag, Colloid Polym. Sci., 2020, 298, 719–733 CrossRef CAS.
- M. Dimde, F. Neumann, F. Reisbeck, S. Ehrmann, J. L. Cuellar-Camacho, D. Steinhilber, N. Ma and R. Haag, Biomater. Sci., 2017, 5, 2328–2336 RSC.
- M. Giulbudagian, M. Asadian-Birjand, D. Steinhilber, K. Achazi, M. Molina and M. Calderón, Polym. Chem., 2014, 5, 6909–6913 RSC.
- M. Giulbudagian, G. Yealland, S. Hönzke, A. Edlich, B. Geisendörfer, B. Kleuser, S. Hedtrich and M. Calderón, Theranostics, 2018, 8, 450–463 CrossRef CAS PubMed.
- A. Jain and K. Cheng, J. Controlled Release, 2017, 245, 27–40 CrossRef CAS.
- J. Kopeček and J. Yang, Adv. Drug Delivery Rev., 2020, 156, 40–64 CrossRef PubMed.
- S. Roller, H. Zhou and R. Haag, Mol. Diversity, 2005, 9, 305–316 CrossRef CAS PubMed.
- J. Dommerholt, S. Schmidt, R. Temming, L. J. A. Hendriks, F. P. J. T. Rutjes, J. C. M. van Hest, D. J. Lefeber, P. Friedl and F. L. van Delft, Angew. Chem., Int. Ed., 2010, 49, 9422–9425 CrossRef CAS PubMed.
- A. Kumar, U. J. Erasquin, G. Qin, K. Li and C. Cai, Chem. Commun., 2010, 46, 5746–5748 RSC.
- D. Steinhilber, T. Rossow, S. Wedepohl, F. Paulus, S. Seiffert and R. Haag, Angew. Chem., Int. Ed., 2013, 52, 13538–13543 CrossRef CAS PubMed.
- T. Pecchioli, M. K. Muthyala, R. Haag and M. Christmann, Beilstein J. Org. Chem., 2015, 11, 730–738 CrossRef CAS PubMed.
- J. E. Sader, J. A. Sanelli, B. D. Adamson, J. P. Monty, X. Wei, S. A. Crawford, J. R. Friend, I. Marusic, P. Mulvaney and E. J. Bieske, Rev. Sci. Instrum., 2012, 83, 103705 CrossRef PubMed.
- S. Rosskopf, S. Jutz, A. Neunkirchner, M. R. Candia, B. Jahn-Schmid, B. Bohle, W. F. Pickl and P. Steinberger, Sci. Rep., 2016, 6, 31580 CrossRef CAS PubMed.
- A. Oehrl, S. Schötz and R. Haag, Macromol. Rapid Commun., 2020, 41, 1900510 CrossRef CAS PubMed.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596–2599 CrossRef CAS PubMed.
- A. Sanchez-Fernandez, K. J. Edler, T. Arnold, D. Alba Venero and A. J. Jackson, Phys. Chem. Chem. Phys., 2017, 19, 8667–8670 RSC.
- L. Bui-Le, A. P. S. Brogan and J. P. Hallett, Biotechnol. Bioeng., 2021, 118, 592–600 CrossRef CAS.
- P. Anilkumar, T. B. Lawson, S. Abbina, J. T. A. Mäkelä, R. C. Sabatelle, L. E. Takeuchi, B. D. Snyder, M. W. Grinstaff and J. N. Kizhakkedathu, Nat. Commun., 2020, 11, 2139 CrossRef CAS PubMed.
- K. Thümmler, N. Häntzschel, A. Skapenko, H. Schulze-Koops and A. Pich, Bioconjugate Chem., 2010, 21, 867–874 CrossRef PubMed.
- J. E. Kay, Immunol. Lett., 1991, 29, 51–54 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1tb02144c |
| This journal is © The Royal Society of Chemistry 2022 |