Open Access Article
Open Access ArticleCarbon dioxide to bio-oil in a bioelectrochemical system-assisted microalgae biorefinery process†
Silvia
Bolognesi
 ab,
Lluís
Bañeras
c,
Elisabet
Perona-Vico
c,
Andrea G.
Capodaglio
b,
Maria Dolors
Balaguer
a and
Sebastià
Puig
ab,
Lluís
Bañeras
c,
Elisabet
Perona-Vico
c,
Andrea G.
Capodaglio
b,
Maria Dolors
Balaguer
a and
Sebastià
Puig
 *a
*a
aLEQUiA, Institute of the Environment, Universitat de Girona, Carrer Maria Aurèlia Capmany 69, Girona 17003, Spain. E-mail: sebastia.puig@udg.edu; Fax: +34 972418150; Tel: +34 972418182
bDepartment of Civil Engineering and Architecture, University of Pavia, Via Ferrata 1, 27100 Pavia, Italy
cGroup of Molecular Microbial Ecology. Institute of Aquatic Ecology, Universitat de Girona, Carrer Maria Aurèlia Capmany 40, Girona 17003, Spain
First published on 24th November 2021
Abstract
Microbial electrosynthesis (MES) for bioelectro carbon dioxide (CO2) recycling is an interesting and sustainable opportunity to exploit off gases from industrial facilities and convert them into valuable energy sources. In the present study, a two-step process based on coupling a bioelectrochemical system (BES) and heterotrophic microalgae Auxenochlorella protothecoides is proposed to convert carbon dioxide into a biodiesel compatible oil. The MES effluent was further processed in a heterotrophic microalgae batch reactor, where the acetate previously synthesized from CO2 was converted into bio-oil in a subsequent, extraction-free step. Two MES reactors were operated in batch mode at an applied cathodic potential of −0.8 V vs. SHE (standard hydrogen electrode) for 95 days. The system reached a concentration of up to 13 g L−1 of acetate (at a maximum production rate of 0.29 g L−1 d−1). Microbial community analysis revealed the presence of Clostridium spp. in both reactors. In a second stage, the effluent from the biocathode was transferred to microalgae reactors containing A. protothecoides to assess oil production. The bio-oil content was up to 22% w/w (dry weight), sufficient to further explore the feasibility of microalgae-to-oil recovery in the future. According to our estimations, 7.59 kg CO2 can be converted into 1 kg acetate, which can be used to grow heterotrophically 1.11 kg dry algae; an overall balance of 0.03 kg bio-oil produced per kg CO2 captured was assessed. The oil obtained can be further processed to produce a biodiesel compatible with EU requirements for biofuels.
1. Introduction
Amongst many biological methods for carbon dioxide (CO2) sequestration and conversion, microbial electrosynthesis (MES) emerges as a promising technology in microbial electrochemistry,1–4 through bioelectro CO2 recycling into multi-carbon molecules, such as added value chemicals and biofuels.5 Bioelectrochemical systems (BES) are characterized by two electrodes working concurrently with a redox reaction and microorganisms catalyzing reactions happening at one or both electrodes. In MES, the reducing-power is provided by external application of a current, or by fixing the potential at the cathode of a BES to supply bacteria with the necessary reducing equivalents and perform the bioconversion of CO2 into medium and short-chain fatty acids.6 Many strategies have been evaluated to expand the portfolio of potential products since further chain elongation is hard to reach in single-step processes, acetic acid being the most abundant product.7 Control of operational parameters (such as pH and partial pressure of hydrogen, pH2) to steer production towards longer chain volatile fatty acids (VFAs) or alcohols is paramount.6,8–10 Genetic engineering of homoacetogens is currently at the centre of microbiologists' interest, to increase biomass tolerance to stress or toxicity conditions and to attempt new fermentation pathways.7 Purple phototrophic bacteria (PPB) are currently being studied for their capability to convert waste products into biofuels like hydrogen (H2), bioplastics such as PHA (polyhydroxyalkanoate) or single-cell protein.11,12 Electrical enhancement of hydrogenotrophic bacteria may lead to the establishment of resilient co-cultures, ideally composed of a H2 producing strain and a homoacetogen, improving MES processes and electro-fermentation.13,14Another interesting option is coupling MES with a second step to increase the value of the VFAs produced: for example, coupling an MES reactor to a fermenter to convert short-chain carboxylates (C2–C4) into medium-chain carboxylates (C6–C8).6,8 Pepè Sciarria et al. (2018) suggested conversion of C2–C4 VFAs produced through MES into bioplastics (PHA) in a three-step process (MES – VFA extraction – anaerobic digestion), obtaining 0.41 kg PHA per kg of carbon as CCO2 with a carbon fixation efficiency (into acetate and butyrate) of 73%.15
Coupling BES technology with microalgae is a strategy explored by several researchers so far. Integrated photo-bioelectrochemical (IPB) reactors with microalgae inserted in the cathode of a microbial fuel cell for electricity generation and nutrient removal from wastewater are extensively reported in the literature, with encouraging results and reduced use of chemicals as terminal electron acceptors.4,16 Chlorophyll from microalgae Chlorella vulgaris has also been used as a biocatalyst in carbon nanotube anodes to produce electricity in photo-bioelectrochemical cells.17
On the other hand, heterotrophic microalgae, such as Auxenochlorella protothecoides, can use acetate and other short chain volatile fatty acids for growth in the presence of oxygen, and require no direct light exposure; thus they can be used as oxygen scavengers in an MES and subsequently exploited for their properties.18 Several valuable products can be obtained from microalgae biorefinery.19–22 Some species of microalgae, among these A. protothecoides, can accumulate large amounts of lipids when cultivated heterotrophically, which can be converted into biodiesel through conventional transesterification processes after microalgal oil extraction.23,24 Liquid biofuels, e.g. biodiesel, bioethanol, biobutanol and jet fuels, are the most likely outcomes of algal biorefining.19,25 These are targeted by the EU's and other Nations' policies as priority fuels for transportation to reduce GHG emissions and fossil fuel use.26 When comparing biodiesel production yields from microalgae and vegetable crops, results seem to indicate a promising advantage of the former.27 Biodiesel produced using microalgal oil presents a higher heating value (41 MJ kg−1) and H/C ratio (1.81) fully compatible with ASTM biodiesel standards.28 However, heterotrophic algal growth is not linked directly to de novo CO2 fixation, and two-step fermentation processes should be explored to benefit from higher growth rates of heterotrophy, while maintaining net CO2 fixation levels for the whole process. In the present study, a two-step process based on coupling a bioelectrochemical system and heterotrophic microalgae A. protothecoides is proposed to convert carbon dioxide into a biodiesel compatible oil.18 The overall process from CO2 to bio-oils includes a step for VFA production via MES, and their subsequent conversion into microalgae biorefinery products by heterotrophic batch processing. Microalgae can exploit both the carbon compounds produced at the biocathode of the MES reactor and the oxygen produced from water splitting at the anode. Oil extraction was performed at the end of each one-week long batch experiment. The process was then assessed in terms of carbon mass balance and overall performance.
2. Materials and methods
2.1. Experimental set-up
Two H-type MES reactors (each comprising two 0.25 L glass bottles, Pyrex V-65231 Scharlab, Spain) were operated, indicated as HT1 and HT2. Cathodic and anodic chambers were separated by a cationic exchange membrane (2 cm2, CMI-1875T, Membranes International, USA). Carbon cloth (working surface 30 cm2, thickness 490 μm; NuVant's ELAT, LT2400 W, FuelCellsEtc., USA) connected to a stainless-steel wire was used as the cathode (working electrode) while a graphite rod was used as the anode electrode (counter electrode, length 25 cm, diameter 4 mm, EnViroCell, Germany). An Ag/AgCl reference electrode (+0.197 V vs. SHE, standard hydrogen electrode, model SE11-S, Sensotechnik Meinsberg, Germany) was placed in the cathodic chamber. Both MES reactors were operated in a three-electrode configuration with a potentiostat (NEV 3.2, Nanoelectra, Spain) controlling the cathode potential at −0.8 V vs. SHE. The net liquid volumes for both anodic and cathodic chambers were 220 mL. Complete mixing of the cathodic chamber was induced by magnetic stirring. The cathodic chambers of the two reactors were checked for gas leaks by performing pressure tests (up to 1 bar) independently before inoculation. Both cathodes were inoculated with 20 mL (10% v/v) of electroactive inoculum from a parent thermophilic (50 °C) MES reactor.29 The experiment was conducted at 25 ± 3 °C.2.2. MES reactor operation
The two MES reactors were operated in batch mode. In the first 25 days of operation, both anodic and cathodic chambers were filled with a low-buffered inorganic medium (modified ATCC1754 PETC medium adjusted to pH 6, as reported by Blasco-Gómez et al.8). After 25 days, ATCC1754 medium in the anodic chamber was substituted by BG-11 medium, typical growth medium used for microalgae Chlorella, to simulate microalgae renewal from an autotrophic cultivation unit in an integrated system.30 CO2 (99.9%, Praxair, Spain) was the only carbon source, and it was supplied to the systems every 3 days for 5 minutes, saturating the cathodic chamber. pH was monitored throughout the experimentation and eventually corrected up to 5.50 with NaOH 1 M when below 4.70. The anodic pH dropped down to highly acidic values (up to 2), and the medium was replenished or substituted when necessary. Before feeding CO2, gas and liquid samples were collected and analysed to monitor the gas composition and the production of volatile fatty acids (VFAs) and alcohols.2.3. Microalgae cultivation
Two algal strains, Chlorella vulgaris 211-11b and Auxenochlorella protothecoides 211-7a (SAG, Culture Collection of Algae, Göttingen, Germany) were maintained under autotrophic conditions in BG-11 medium until use. A. protothecoides has been extensively used to produce good amounts of lipids18,28,31 and thus tested throughout the experimentation, while C. vulgaris was only operated in preliminary tests and as a comparison for microalgae growth. Two methacrylate tubular photobioreactors (d = 0.04 m, H = 1 m, 1 L each) were built to preserve the cultures from external contamination. Air was provided through a diffuser from the bottom of the photobioreactor to prevent sedimentation of the microalgae and keep the culture in suspension. BG-11 medium (pH 7) was added periodically to replenish the column.2.4. Heterotrophic tests
Heterotrophic batch tests were performed to evaluate the possibility of growing microalgae using VFAs (acetate) produced in biocathodes. Starting parameters were assessed according to preliminary tests and values found in the literature.18 Erlenmeyer flasks (100 mL) were filled with microalgae and cathode effluent at different ratios (Table 1). They were continuously mixed by magnetic stirrers and kept in the dark. Each test lasted one week, in replicate. Tests A, B, and C with Chlorella vulgaris were performed with the same biocathode (HT1) effluent at different dilution rates. In tests D and E, previously autotrophically cultivated A. protothecoides was diluted up to a concentration of OD540 of 1.3 (corresponding to an algal dry weight 0.80 g L−1), by addition of the inorganic medium operated in the MES systems to the concentrated microalgae solution. When cathodic effluent was fed to the microalgae, the amount of effluent to be added was calibrated to properly reach a concentration of initial VFAs between 1.5 and 2.0 g L−1 in each flask. 3 mL samples were taken every day from each flask to monitor variations in pH, optical density (OD540), conductivity and chlorophyll a content (Table 1). VFA consumption was monitored through GC analysis. At the end of each test, the remaining mixture was stored to perform oil extraction.| Test | Algae![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) biocathode effluent ratiob biocathode effluent ratiob |
Volume | Samples | Microalgae species | Time of biocathode sample collection (days) |
|---|---|---|---|---|---|
| a Preliminary batch tests were performed in synthetic medium with both inorganic and BG-11 media, under heterotrophic (2 g L−1 acetate) and autotrophic conditions, and at different pH starting values (4, 5.5, 7, 9). Results of preliminary batch tests are reported in the ESI (Fig. S2–S5). N.A.: not available. b The algae/biocathode effluent ratio of each test was chosen according to values of OD540 and VFA content of the sample. | |||||
| PB1 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
80 mL | 1a | C. vulgaris | N.A.a |
| PB2 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
80 mL | 1a | A. protothecoides | N.A.a |
| A | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
60 mL | 2 | C. vulgaris | 25 |
| B | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
60 mL | 2 | C. vulgaris | 25 |
| C | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
60 mL | 2 | C. vulgaris | 25 |
| D | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
70 mL | 2 | A. protothecoides | 80 |
| E | 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
70 mL | 2 | A. protothecoides | 80 |
Chlorophyll a analysis was performed at the beginning of each test and at least every second day. Being a primary pigment directly connected to phototrophic metabolism, chlorophyll a was measured as a parameter to evaluate the change in microalgae metabolism (from phototrophic to heterotrophic).32 2 mL of sample was taken and put in a clean tube and centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min) to pellet cells. The supernatant was discarded, and 2 mL of acetone was added to the pellet and mixed. Tubes were left overnight in the freezer (−20 °C) to facilitate complete extraction of the pigments. Samples were centrifuged again (10
000 rpm, 10 min) to pellet cells. The supernatant was discarded, and 2 mL of acetone was added to the pellet and mixed. Tubes were left overnight in the freezer (−20 °C) to facilitate complete extraction of the pigments. Samples were centrifuged again (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min) and the supernatant OD was measured at different wavelengths (400–850 nm). Chlorophyll a content was calculated as reported in eqn (1) (modified from Lorenzen (1967)).33
000 rpm, 10 min) and the supernatant OD was measured at different wavelengths (400–850 nm). Chlorophyll a content was calculated as reported in eqn (1) (modified from Lorenzen (1967)).33
| Chl a = (1.56(OD665 − OD830) − 2.0(OD645 − OD830) − 0.8(OD636 − OD830)) × Vac/Vsam | (1) |
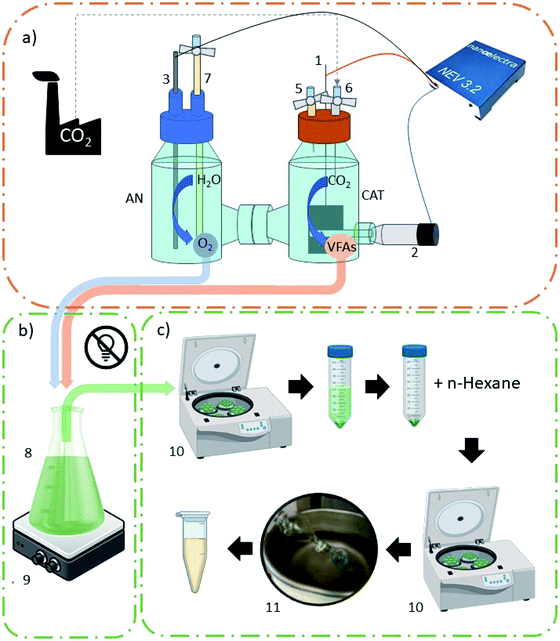
Oil extraction was performed at the end of every batch test. Samples were put in falcon tubes and centrifuged to separate the solid fraction from water. Algal cells were resuspended in distilled water (DW) to remove impurities and centrifuged again (6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min). 10 mL n-hexane (purity ≥ 99%, Sigma Aldrich) per gram of wet algae was added, and cell disruption was achieved mechanically at first by friction and then using an ultrasonic bath (40 kHz, 10 min). Tubes were centrifuged again to separate the different phases (pellet, residual water and n-hexane with lipids dissolved in it). The supernatant was then separated, and oil was extracted using a rotary evaporator (bath temperature 35 °C, 20 rpm). The experimental setup is represented in Fig. 1.
000 rpm, 10 min). 10 mL n-hexane (purity ≥ 99%, Sigma Aldrich) per gram of wet algae was added, and cell disruption was achieved mechanically at first by friction and then using an ultrasonic bath (40 kHz, 10 min). Tubes were centrifuged again to separate the different phases (pellet, residual water and n-hexane with lipids dissolved in it). The supernatant was then separated, and oil was extracted using a rotary evaporator (bath temperature 35 °C, 20 rpm). The experimental setup is represented in Fig. 1.
2.5. Analyses and calculations
VFAs and alcohols in the liquid phase were analysed with a gas chromatograph (GC) (Agilent 7890A, Agilent Technologies, USA) equipped with a DB-FFAP column and a flame ionization detector (FID). Unless otherwise stated, the concentration of organic compounds in the liquid phase is expressed throughout the manuscript in mg L−1. Conductivity (EC), pH and optical density (OD600) were also measured for the liquid sample. The gas pressure in the headspace of the reactor was measured before sampling and after feeding with a differential manometer (Model-Testo-512; Testo, Germany). Gas samples were taken using a glass syringe before taking liquid samples and analysed using a Micro-GC (Agilent 490 Micro GC system, Agilent Technologies, USA) equipped with two columns: a CP-molsieve 5A for methane (CH4), carbon monoxide (CO), hydrogen (H2), oxygen (O2) and nitrogen (N2) analysis, and a CP-Poraplot U for carbon dioxide (CO2) analysis. Both columns were connected to a thermal conductivity detector (TCD). The partial pressure of hydrogen (pH2) was calculated from the total pressure measured with the differential manometer before taking the gas samples and the composition of the gas detected in the headspace of the biocathode. The concentrations of dissolved H2 and CO2 were calculated according to Henry's law at 25 °C. Both MES reactors were flushed and saturated with CO2 after the sampling phase, assuming conditions of CO2 saturation and absence of H2 in the liquid phase after each feeding. Coulombic efficiency (CE) was calculated according to Rovira-Alsina et al.29Carbon conversion efficiency was finally calculated according to the maximum acetate production rate achieved. The final product (bio-oil) conversion efficiency from CO2 was calculated according to the amount of acetate fed to the microalgae, and the ratio between the weight of the dry sample collected and the weight of the oil sample obtained at the end of the extraction procedure.
2.6. Extraction of DNA and microbial community structure determination
Samples of carbon cloth and bulk liquid were taken to assess the microbial community composition at the end of the experiments. Before DNA extraction, bulk liquid cells were pelleted by centrifugation, whereas carbon cloth samples were used directly. DNA was extracted using the FastDNA® SPIN kit for Soils (MP Biomedicals, USA) following the manufacturer's instructions. The extracts were distributed in aliquots and stored at −20 °C, and the DNA concentration was measured using a Nanodrop™ 1000 spectrophotometer (Thermo Fisher Scientific, USA). The quality of DNA extracts for downstream molecular applications was checked after PCR detection of 16S rRNA using the universal bacterial primers 515F and 806R.The hypervariable V4 region of the 16S rRNA gene for all the samples was amplified using the primers 515F and 806R following the method described by Kozich and Schloss, which was adapted to produce dual-indexed Illumina compatible libraries in a single PCR step.34 First, PCR was performed using fusion primers with target-specific portions,35 and Fluidigm CS oligos at their 5′ ends. Second, PCR targeting the CS oligos was used to add sequences necessary for Illumina sequencing and unique indexes. PCR products were normalized using Invitrogen SequalPrep DNA normalization plates and the pooled samples were sequenced using an Illumina MiSeq flow cell (v2) in a 500-cycle reagent kit (2 × 250 bp paired-end reads). Finally, sequencing was done at the RTSF Core facilities at the Michigan State University, USA (https://rtsf.natsci.msu.edu/).
Raw sequences from the MiSeq platform were analyzed and treated through DADA2 software from the open-source Bioconductor project as mentioned by Callahan and co-workers.36 Output demultiplex sequences obtained in the fastq format were sorted to ensure reads were in the same order, quality-filter with a maximum expected error of 2 to filter out low-quality sequencing reads and trimmed to a consistent length. After trimming, average sequencing read lengths were 220 and 200 nt for forward and reverse reads, respectively. Sequences were then dereplicated by combining the identical sequences into unique sequences to remove redundancy, denoised to remove sequences errors and identify the biological sequence in the reads. A sequence table was obtained with the inferred amplicon sequence variant (ASVs), distributed according to the length. ASVs with a length between 220 and 265 nt were kept and checked for chimera removal by comparing each inferred sequence with the others in the table and removing sequences that can be reproduced by stitching two more abundant sequences. Taxonomy was assigned to each ASV sequence using the SILVA 132 database (https://www.arb-silva.de/) as a reference to obtain an ASV relative abundance table. A phylogenetic tree was constructed using DECIPHER and phangorn R packages to perform downstream analyses, especially for making comparisons between microbial communities and determine phylogenetic diversities. A phyloseq object was created with the data for its further analysis using the phyloseq R package. Sequences identified as chloroplasts and mitochondria were removed. Diversity analyses were carried out using different indicators such as species richness (observed number of ASVs) and α-diversity (Shannon index). To compare the community structure between sample groups principal coordinate analysis (PCoA) plots employing weighted UniFrac distance matrices were obtained.37 All indices were determined using the R package phyloseq38 and plotted using ggplot (https://www.r-project.org/).
3. Results and discussion
3.1. Biocathode performance
The biocathodes inoculated with 20 mL of a parent MES operated under thermophilic conditions (50 °C)29 produced acetate from carbon dioxide (max production rate 28 g m−2 d−1, with a maximum titre of 5250 mg L−1). Adaptation to mesophilic conditions (25 °C) lasted 25 days, when low VFA production was observed and, once stabilized, modified ATCC1754 PETC medium was replaced by BG-11 in the anode (Section 2.2). Fig. 2 presents VFA production over time after the acclimatization period for the systems HT1 (a) and HT2 (b). | ||
| Fig. 2 VFA production, pH and current density profile throughout the experimentation: (a) HT1, (b) HT2. | ||
During the first 5 days of operation no significant VFA production was detected in either HT1 or HT2. At day 10, HT1 started producing acetate (126 L−1 d−1) and butyrate (5 mg L−1 d−1), maintaining a constant trend up to the end of the experimentation. The reduction of CO2 to acetate occurs by the well-known Wood–Ljungdahl pathway.39 On day 30, pH decreased from 6.3 to 5.6 and partial pressure of hydrogen (pH2) values increased up to 0.86 bar, resulting in the inception of ethanol production (15 mg L−1 d−1). The pH was maintained in the range 5.1–5.6 up to day 60, leading to increased ethanol production, as expected according to Blasco-Gómez et al. (2019).8 Subsequently, the pH dropped below 5, due to increased acetogenic activity, and residual pH2 detected was negligible switching again to the main metabolic pathway acted upon by the biomass. Between days 53 and 73 the sampling routine was changed, by adding a third sampling point in between the feeding points. This increased renewal of the medium and nutrients, together with favorable pH conditions, led to fast acetate production increase at day 77. An increase in acetate production associated with augmented nutrient availability was already reported by Rovira-Alsina and co-workers.29 At day 80 the maximum acetic acid production rate (289 mg L−1 d−1) and its highest concentration in the whole experimentation were obtained in HT1 (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 063 mg L−1; Table 2). Comparable values in terms of concentration have seldom been reported in the literature. Jiang et al. achieved a maximum acetate concentration of 13.4 g L−1 using a slurry electrode composed of a stainless steel brush and powdered activated carbon (5 g L−1), almost threefold the concentration obtained with a stainless steel brush alone.40 Mohanakrishna et al., (2020) achieved a maximum acetate production of 260 mg L−1 d−1 by adding a supplementary inorganic carbon source (15 g HCO3− per L) to the solution. These previous results showed the relevance of those obtained in the present study with CO2 as the only carbon source.41
063 mg L−1; Table 2). Comparable values in terms of concentration have seldom been reported in the literature. Jiang et al. achieved a maximum acetate concentration of 13.4 g L−1 using a slurry electrode composed of a stainless steel brush and powdered activated carbon (5 g L−1), almost threefold the concentration obtained with a stainless steel brush alone.40 Mohanakrishna et al., (2020) achieved a maximum acetate production of 260 mg L−1 d−1 by adding a supplementary inorganic carbon source (15 g HCO3− per L) to the solution. These previous results showed the relevance of those obtained in the present study with CO2 as the only carbon source.41
| HT1 | HT2 | |||||
|---|---|---|---|---|---|---|
| Acetate | Butyrate | Ethanol | Acetate | Butyrate | Ethanol | |
| mg L−1 d−1 (average) | 126.33 | 4.98 | 15.02 | 71.67 | 1.76 | 4.43 |
| gprod melectrode−2 d−1 | 9.26 | 0.28 | 1.19 | 4.97 | 0.14 | 0.20 |
| mg L−1 d−1 (peak) | 289.41 | 60.48 | 59.58 | 220.60 | 15.39 | 56.41 |
| gprod melectrode−2 d−1 | 21.22 | 4.43 | 4.37 | 16.18 | 1.13 | 4.14 |
On day 80, one third (60 mL) of the cathodic volume was extracted to perform microalgae heterotrophic tests and replaced with fresh medium. Production rates for acetic acid and butyric acid were not affected by the extraction. HT2 followed similar trends in production rates and product spectrum.
At the end of the experimentation, both reactors were stopped on day 95, and microbiological samples from bulk solution and biofilm were collected to perform DNA analysis.
In terms of CE, the two reactors behaved differently. Since almost no H2 was found in the headspace of HT1 at any time, it could be assumed that all the hydrogen produced was consumed in the acetate production process, while the lower VFA production in HT2 consumed less H2, which was always found in the headspace gas composition (even at high overpressure, over 1 bar). However, the two systems were not equally efficient. HT1 consumed less coulombs, and approximately 2/3 of them were mostly used up in acetate production, while HT2 used up most of the coulombs for hydrogen production, but with overall lower coulombic efficiency (Fig. S1, ESI†). The maximum CE for acetate production was close to 100% and 92% for HT1 and HT2, respectively, between days 72 and 75, while the average observed CE was 41% for HT1, and 21% for HT2. Such low CE for HT2 could be imputed to undetected fugitive hydrogen emissions due to the high overpressure at the biocathode, or to the use of electrons in producing undesired compounds (such as methane). Increased internal resistance due to membrane/electric circuit connection deterioration could explain the difficulty in maintaining the cathode potential fixed at −0.997 V vs. Ag/AgCl in HT2 during the latter phase of the experimentation.
3.2. Bacterial community analysis
Microbial communities for both MES on day 95 were dominated by Firmicutes bacteria, accounting for 93–99% in biofilm and 60–72% in the bulk. Proteobacteria were also present, being more abundant in liquid (26–39%) than in biofilm samples (1–6%). At the order level, the microbial community was composed mainly of Clostridiales, Betaproteobacteriales and Selenomonadales, the former being the most abundant (Fig. 3). At the genus level, both reactors were clearly dominated by Clostridium spp. Further analyses performed via BLASTn searches revealed the presence of different Clostridium species in reactors HT1 and HT2.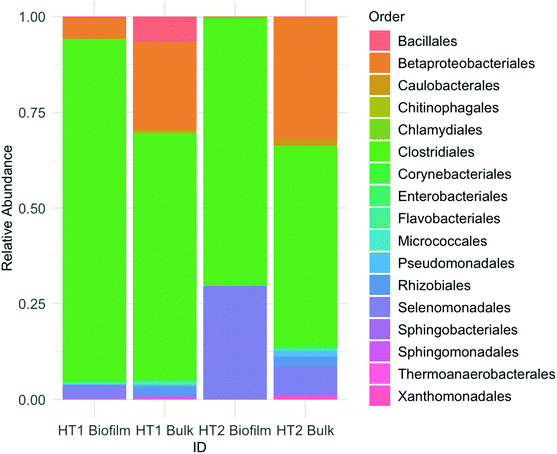 | ||
| Fig. 3 Microbial community composition in biofilm and bulk samples. The bar chart shows relative abundances of main bacterial orders. | ||
HT1 was characterized by the presence of Clostridium autoethanogenum and Clostridium ljungdahlii (Table 3). Both species are model homoacetogens, and have been consistently reported to be able to accept electrons from a cathode via the Wood–Ljungdahl pathway.42,43Clostridium autoethanogenum also has a role in autotrophic ethanol production from industrial waste gases.44,45
| ASV | Most probable identification | Similarity (%) | Relative number of sequences (%) | |||
|---|---|---|---|---|---|---|
| HT 1 | HT 2 | |||||
| Biofilm | Bulk | Biofilm | Bulk | |||
| 1 | Clostridium autoethanogenum DSM 10061 NR_121758.1 | 100 | 66.2 | 53.4 | < 0.01 | <0.01 |
| Clostridium ljungdahlii DSM 13528 NR_074161.1 | 100 | |||||
| 2 | Clostridium aciditolerans strain JW/YJL-B3 | 96.4 | 7.5 | 1.8 | 70.3 | 32.2 |
| Clostridium nitrophenolicum strain 1D | 96.4 | |||||
Sequences retrieved from HT2 showed a higher similarity to Clostridium aciditolerans and Clostridium nitrophenolicum (96% identity), so far still not reported as typical species in MES processes. Although additional tests should be performed to confirm the identity of the most abundant ASVs in HT2, the absence of true homoacetogenic clostridia is intriguing in view of the acetate production. It should be noted that sequence similarity of ASV2 to cultured bacterial species was below 97% which could determine the presence of a previously undescribed species in the reactor. In fact, 100% similar sequences to those retrieved here have been found in anaerobic reactors inoculated with rumen fluids and hydrogen producing reactors both reporting similar pH as stated here for HT2.46,47Clostridium aciditolerans has been proven to survive acid stress conditions, and to play an indispensable role in biohydrogen production and accumulation, potentially through butyrate/H2 fermentation of sugars;48 however, no evidence of autotrophic growth and acetate production exists for isolates of this species.49 Homoacetogenesis took place at similar rates in the two reactors, and measured state variables were also comparable during the entire experimentation, except for cell voltage, which was higher in HT2 (by 1–2 V) than in HT1. Collectively, it is likely that a change in operating conditions in the two reactors resulted in the selection of two different homoacetogenic populations as predominant species, although one of them could not be correctly identified with the methods used herein. Nevertheless, observed sequence differences may denote the presence of slightly different homoacetogenic clostridia, which together with variations in the total bacterial abundance, could have contributed to differences in acetate production in the two reactors.
3.3. Heterotrophic A. protothecoides growth tests with effluents of biocathodes
Several one-week long heterotrophic tests with cathode effluents were performed with different volumes of microalgae/biocathode effluent ratios (Table 1). The results of the preliminary tests with the synthetic effluent at different pHs are reported in the ESI (Fig. S2–S5†). A. protothecoides can grow heterotrophically using both ATCC1754 and BG-11 media plus acetate (2 g L−1), with a maximum peak at day 5. Autotrophic growth instead was evident only when BG-11 at pH 7 was used. In contrast, C. vulgaris responded well to any autotrophic condition tested, while under heterotrophic conditions after a peak at day 3 microalgae concentration in the batch reactor rapidly decreased, except for the test conducted with inorganic medium at pH 5.5.Tests with the biocathode effluent were performed on both strains, considering that for C. vulgaris in preliminary tests the synthetic medium most similar to the biocathode effluent composition was the one yielding the most promising results. Fig. 4 reports results from tests A, B and C with C. vulgaris. Different initial concentrations of VFAs (due to the different microalgae/biocathode effluent ratios) were provided to the microalgae reactor. It was found that higher VFA concentrations significantly increase biomass production, as shown in tests A, B, and C, where a biomass production rate of 0.137 ± 0.016 g L−1 d−1 (corresponding to a biomass yield of 0.576 g L−1) was obtained in test B (4 g L−1 acetate). The OD540 profile showed that maximum growth was achieved in the sample collected on day 4 under all conditions tested, confirming the peak rates previously detected in preliminary batch tests. Comparing values presented in this study with literature data for open pond cultivation, similar values to those obtained in test B with C. vulgaris were found in Griffiths and Harrison's study50 reporting an average biomass productivity of 0.17 g L−1 d−1.
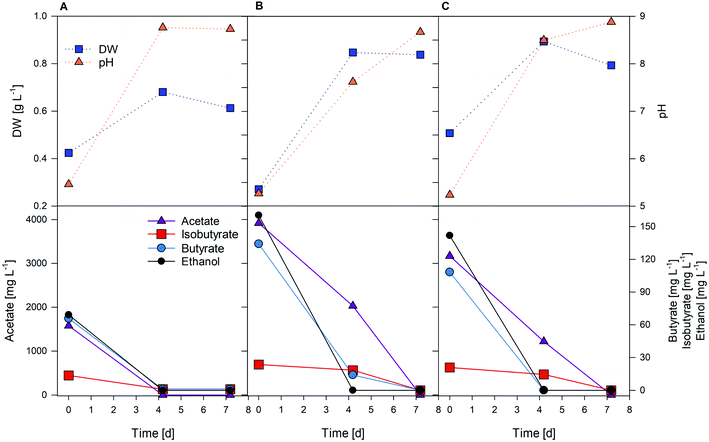 | ||
| Fig. 4 Heterotrophic tests (A, B and C) with the cathode effluent at different dilution percentages (C. vulgaris). | ||
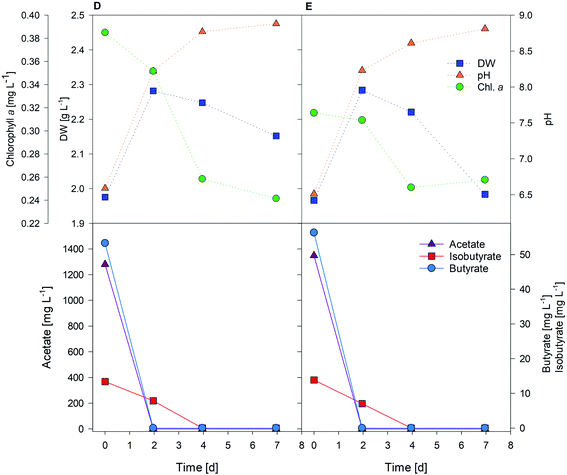
Fig. 5 shows two replicates obtained from one of the tests performed with A. protothecoides. Both proved that this strain was capable of rapidly switching from autotrophic to heterotrophic metabolism, and of using biocathode-produced VFAs for growth. Acetic acid was fully consumed after two days, while more complex molecules (i.e., isobutyric acid) took longer to be assimilated by microalgae.
Each test achieved the production of 1–2 g wet algae, further processed to extract microalgal oil, at a net biomass yield of 0.061 ± 0.006 g L−1 d−1 for C. vulgaris (test A) and 0.064 ± 0.010 g L−1 d−1 for A. protothecoides (tests D and E) fed with 1.5 g L−1 VFA solution. Chlorophyll a content decreased over time as expected, confirming the fast development of a heterotrophic regime. While light exposure undoubtedly induced chlorophyll synthesis,51 during the first 48 h of heterotrophic growth, the total amount of chlorophyll slowly decreased, afterwards the chlorophyll profile switched to a phase of net degradation, to finally reach a plateau after day 4 (0.25 ± 0.02 mg L−1), as evident from tests D and E (Fig. 5). The steady state trend of chlorophyll content in the last days of the test meant that the biosynthetic and the chlorophyll degradation reactions were finally evolving at the same rate, with green algae still synthesizing chlorophyll under dark conditions. Gene expression for chlorophyll metabolism, along with those for photosynthesis and carotenoid biosynthesis, is downregulated in the heterotrophic metabolism of C. protothecoides, while glycolysis, TCA cycle, and fatty acid synthesis processes increase.23,52 Microalgae concentration instead increased by 35 ± 3% in the first two days in both flasks according to DW values. Once the organic substrate was depleted, after day 3, microalgae concentration decreased again: when microalgae are exposed to nutrient starvation, their metabolic activity switches again, enhancing lipid accumulation in their cell wall.24 In a possible future optimization of the system, microalgae feeding cycles could be optimized to last three to five days, thereby allowing the maintenance of the highest growth rates. Another option for process optimization could be to evaluate the ideal feed length to maximize the lipid content, rather than for achieving the highest biomass concentration. At any rate, microalgae A. protothecoides demonstrated their ability to use an acetate-enriched biocathode effluent to grow and replicate heterotrophically.
3.4. Oil extraction and perspectives
After microalgae collection and concentration, oil extraction was performed, obtaining an average yield of 20 ± 2% w/w (algae dry weight). This value is in line with other results available in the literature,53 but higher values have also been reported (over 60% lipid content, using glucose as the substrate), showing that there is ample scope to improve the process.52 Maximum oil yield was obtained in test D, where the observed lipid/biomass ratio was 0.221 g g−1, in line with previous results from Fei et al. (0.19 g g−1) for A. protothecoides grown on a 2 g L−1 VFA solution.18 However, it was not possible to perform further characterization of the bio-oil produced due to the low amount of oil collected in each test (0.1–0.2 g), too small to return reliable enough results in the NMR/GC-MS analysis. Considering the values achieved in this study, 7.59 kg CO2 and 12.9 kW h electricity are necessary to produce 1 kg of acetate (corresponding to a production cost for acetate of 0.83 € per kg), in line with other values reported in the literature for biocathode-produced acetate (12 kW h per kg acetate).7 Starting from that, and without extraction and purification, heterotrophic microalgae can be grown directly on the biocathode effluent. Approximately 1.11 kg dry algae per kg acetate was obtained, from which 0.207 kg of lipids could be extracted and further processed for biodiesel production.Following optimization of the extraction process, biodiesel may be obtained from a subsequent transesterification process, with product properties complying with EU fuel specifications.54 Further bio-oil characterisation via GC-MS or NMR analysis is needed to identify the long chain fatty acid profile (myristic acid (C14:0), palmitic acid (C16:0), stearic acid (C18:0), oleic acid (C18:1), linoleic acid (C18:2), linolenic acid (C18:3)) and evaluate the compatibility of the oils with EU biodiesel standards.55,56
Microalgae biomass concentration is strictly dependent on the cultivation technique and strain chosen, with values ranging from 0.1 to 8 g L−1 in dry weight (where the higher range of values is only reached with heterotrophic cultivation).57 Autotrophic microalgae can capture CO2 directly to synthesize new biomass: 1 kg (dry) microalgae Chlorella vulgaris may capture 1.83 kg CO2 with a fixation rate of 0.73 to 2.22 g L−1 d−1 but, when cultivated in open air raceway ponds, part of the fed CO2 is lost to the atmosphere.27,58 The combined process herein presented instead allowed almost 100% CO2 capture and conversion to be reached (Table 4). It has to be noted that heterotrophic cultures can generate biomass in larger cultivation volumes (cultivation ponds exceeding 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 L are reported),59 are not strictly dependent on the presence of sunlight, and can yield hundreds of kilograms of biomass, making the heterotrophic strategy less expensive than the autotrophic one in the long haul, including the possibility to use a waste stream as the organic carbon source. A similar approach to the one presented in this study was tested by LanzaTech and Indian Oil on a larger scale, who developed an integrated carbon capture and utilization (CCU) process that converts CO2 into commercial-grade docosahexaenoic acid esters (DHAs) and biodiesel, yielding 0.08 kg biodiesel and 0.08 kg DHAs per kg CO2.60
000 L are reported),59 are not strictly dependent on the presence of sunlight, and can yield hundreds of kilograms of biomass, making the heterotrophic strategy less expensive than the autotrophic one in the long haul, including the possibility to use a waste stream as the organic carbon source. A similar approach to the one presented in this study was tested by LanzaTech and Indian Oil on a larger scale, who developed an integrated carbon capture and utilization (CCU) process that converts CO2 into commercial-grade docosahexaenoic acid esters (DHAs) and biodiesel, yielding 0.08 kg biodiesel and 0.08 kg DHAs per kg CO2.60
| Carbon source | CO2 capture efficiency | Carbon capture potential [kg CO2 per kg] | Raw material cost | Biomass weight [g L−1] | Lipid yield | Cost (lipid) [€ per kg] | Ref. | |
|---|---|---|---|---|---|---|---|---|
| a The same processing cost calculated by Fei et al. was applied. | ||||||||
| Autotrophic (open pond) | Atmospheric CO2 + sunlight | 39–60% | 1.83 | 0 | 0.14 | 0.15–0.30 w/w | 0.52 | 58 and 61 |
| Heterotrophic | VFAs | N.A. | N.A. | 0.36 | 0.39 | 0.19 g g−1 | 0.48 | 18 |
| MES + heterotrophic | CO2 | 99% | 6.86 | 0.83 | 2.48 | 0.21 g g−1 | 0.91a | This study |
This study demonstrates the feasibility of a microalgae electro-biorefinery proof-of-concept. H-type MES is characterized by small volumes and optimal conditions for batch operation mode (high pH2 in a controlled environment, but difficulty in pH and CO2 on-line monitoring), thus the main limitation of this experiment lies in the small volumes of biocathode effluent collected to perform heterotrophic tests with microalgae. The first step to further the application value of this study will therefore be the design of a setup compatible with continuous mode operation. Direct reuse of acetic acid produced in MES reactors waives the costs of its extraction and purification, lowering the overall economic passive balance items of the process.62,63 Furthermore, in an integrated system, the use of heterotrophic microalgae may also enhance the reactor performance by capturing the oxygen produced at the anode side and preventing its passage through the membrane, avoiding the presence of an alternative (easier to reduce) electron acceptor at the biocathode.
HT1 and HT2 reached maximum acetate concentrations of 13 g L−1 and 8 g L−1, respectively, and production rates up to 289 mg L−1 d−1. Such a value is lower than A. protothecoides carbon need, since according to our experimental data 1.4 g L−1 acetate was consumed in only two days out of seven; however, for a future implementation of A. protothecoides inside the MES reactor, nutrient starvation plays a fundamental role in lipid accumulation in microalgae, thus this value can be considered compatible with microalgae heterotrophic growth for biodiesel production and encouraging for further investigation.
Microalgal oil production was assessed in this study, however different conversion possibilities of the algal effluent should be explored in the future, including other recovery options. For example, the solid residue from the extraction process (solid fraction) can be pyrolyzed to biochar, a valuable recovery material,30 but also other valuable compounds such as proteins, pigments or other commodity chemicals may be considered to improve the economics of the process and overcome microalgae separation and oil extraction costs, representing relevant components of this approach.19
4. Conclusions
This study shows that MET-produced acetate can be used to achieve bio-oil production in algae cultures. This proof-of-concept study demonstrates the direct reuse of acetate produced in biocathodes through MES from CO2 as a feed for heterotrophic microalgae, aimed at bio-oil production in an integrated microalgae electro-biorefinery. Acetate (up to 13 g L−1) was produced from CO2 and electricity (7.59 kg CO2 and 12.9 kW h per kg acetate) in the MES reactors reaching a maximum production rate of 0.29 g L−1 d−1. Microbial community composition was different in the two reactors used, but dominated by Clostridium spp., synthesizing acetate through the Wood–Ljungdahl pathway.Microalgae integration with MES could lead to several advantages, amongst which direct reuse of acetate produced by the latter, thus avoiding the costs of its extraction and purification, and consequently lowering the overall economic footprint of the process is significant. Oil yield from microalgae of up to 22% w/w was assessed. According to the results, 0.03 kg bio-oil per kg CO2 captured could be recovered. Further studies will focus on process optimization to enhance bio-oil production and characterization, and to evaluate strategies to assess a multi-compound microalgae electro-biorefinery.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by the Spanish Ministry of Science through the grant RTI2018-098360-B-I00 and the Agency for Business Competitiveness of the Government of Catalonia (ACCIÓ) through the DigesTake Project (COMRDI16-1-0061). E. P.-V. is grateful for the Research Training grant from the University of Girona (IFUdG2018/52). S. P. is a Serra Húnter Fellow (UdG-AG-575) and acknowledges the funding from the ICREA Acadèmia award. LEQUiA and Ecoaqua have been recognized as consolidated research groups by the Catalan Government with codes 2017-SGR-1552 and 2017SGR-548, respectively.References
- A. ElMekawy, H. M. Hegab, G. Mohanakrishna, A. F. Elbaz, M. Bulut and D. Pant, Bioresour. Technol., 2016, 215, 357–370 CrossRef CAS
.
- K. P. Nevin, T. L. Woodard, A. E. Franks, Z. M. Summers and D. R. Lovley, Am. Soc. Microbiol., 2010, 1, 1–4 Search PubMed
.
- K. Rabaey and R. A. Rozendal, Nat. Rev. Microbiol., 2010, 8, 706–716 CrossRef CAS
.
- S. Bolognesi, D. Cecconet, A. Callegari and A. G. Capodaglio, Environ. Res., 2021, 192, 110263 CrossRef CAS PubMed
.
- P. Dessì, L. Rovira-Alsina, C. Sánchez, G. K. Dinesh, W. Tong, P. Chatterjee, M. Tedesco, P. Farràs, H. M. V. Hamelers and S. Puig, Biotechnol. Adv., 2021, 46, 107675 CrossRef
.
- I. Vassilev, P. A. Hernandez, P. Batlle-Vilanova, S. Freguia, J. O. Krömer, J. Keller, P. Ledezma and B. Virdis, ACS Sustainable Chem. Eng., 2018, 6, 8485–8493 CrossRef CAS
.
- A. Prévoteau, J. M. Carvajal-Arroyo, R. Ganigué and K. Rabaey, Curr. Opin. Biotechnol., 2020, 62, 48–57 CrossRef PubMed
.
- R. Blasco-Gómez, S. Ramió-Pujol, L. Bañeras, J. Colprim, M. D. Balaguer and S. Puig, Green Chem., 2019, 21, 684–691 RSC
.
- L. Jourdin, M. Winkelhorst, B. Rawls, C. J. N. Buisman and D. P. B. T. B. Strik, Bioresour. Technol., 2019, 7, 100284 CrossRef
.
- B. Bian, S. Bajracharya, J. Xu, D. Pant and P. E. Saikaly, Bioresour. Technol., 2020, 302, 122863 CrossRef CAS
.
- M. Grattieri, Photochem. Photobiol. Sci., 2020, 19, 424–435 CrossRef CAS
.
- I. A. Vasiliadou, A. Berná, C. Manchon, J. A. Melero, F. Martinez, A. Esteve-Nuñez and D. Puyol, Front. Energy Res., 2018, 6, 1–12 CrossRef
.
- E. Perona-Vico, L. Feliu-Paradeda, S. Puig and L. Bañeras, Sci. Rep., 2020, 10, 1–11 CrossRef
.
- S. Saheb-Alam, F. Persson, B. M. Wilén, M. Hermansson and O. Modin, Sci. Rep., 2019, 9, 1–13 Search PubMed
.
- T. Pepè Sciarria, P. Batlle-Vilanova, B. Colombo, B. Scaglia, M. D. Balaguer, J. Colprim, S. Puig and F. Adani, Green Chem., 2018, 20, 4058–4066 RSC
.
- Z. Wang, Z. He and E. B. Young, Curr. Opin. Chem. Biol., 2020, 59, 130–139 CrossRef CAS
.
- M. Christwardana, A. A. Septevani and L. A. Yoshi, Sol. Energy, 2021, 227, 217–223 CrossRef CAS
.
- Q. Fei, R. Fu, L. Shang, C. J. Brigham and H. N. Chang, Bioprocess Biosyst. Eng., 2015, 38, 691–700 CrossRef CAS PubMed
.
- Y. Liang, T. Kashdan, C. Sterner, L. Dombrowski, I. Petrick, M. Kröger and R. Höfer, Algal Biorefineries, 2015, 61–131 Search PubMed
.
- K. W. Chew, J. Y. Yap, P. L. Show, N. H. Suan, J. C. Juan, T.
C. Ling, D. J. Lee and J. S. Chang, Bioresour. Technol., 2017, 229, 53–62 CrossRef CAS
.
- S. H. Ho, S. W. Huang, C. Y. Chen, T. Hasunuma, A. Kondo and J. S. Chang, Bioresour. Technol., 2013, 135, 191–198 CrossRef CAS
.
- H.-H. Cheng, L.-M. Whang, K.-C. Chan, M.-C. Chung, S.-H. Wu, C.-P. Liu, S.-Y. Tien, S.-Y. Chen, J.-S. Chang and W.-J. Lee, Bioresour. Technol., 2015, 184, 379–385 CrossRef CAS PubMed
.
- C. Gao, Y. Wang, Y. Shen, D. Yan, X. He, J. Dai and Q. Wu, BMC Genomics, 2014, 15, 1–14 CAS
.
- M. Vitova, K. Bisova, S. Kawano and V. Zachleder, Biotechnol. Adv., 2014, 33, 1204–1218 CrossRef PubMed
.
- Y. Kanna Dasan, M. Kee Lam, S. Yusup, J. Wei Lim and K. Teong Lee, Sci. Total Environ., 2019, 688, 112–128 CrossRef
.
- M. Raboni, P. Viotti and A. G. Capodaglio, Rev. Ambiente Agua, 2015, 10, 10–21 Search PubMed
.
- Y. Chisti, Trends Biotechnol., 2008, 26, 126–131 CrossRef CAS
.
- X. Miao and Q. Wu, Bioresour. Technol., 2006, 97, 841–846 CrossRef CAS
.
- L. Rovira-Alsina, E. Perona-Vico, L. Bañeras, J. Colprim, M. D. Balaguer and S. Puig, Green Chem., 2020, 22, 2947–2955 RSC
.
- S. Bolognesi, G. Bernardi, A. Callegari, D. Dondi and A. G. Capodaglio, Biomass Convers. Biorefin., 2021, 11, 289–299 CrossRef CAS
.
- A. Wei, X. Zhang, D. Wei, G. Chen, Q. Wu and S. T. Yang, J. Ind. Microbiol. Biotechnol., 2009, 36, 1383–1389 CrossRef CAS
.
- K. J. M. Mulders, P. P. Lamers, D. E. Martens and R. H. Wijffels, J. Phycol., 2014, 50, 229–242 CrossRef CAS
.
- C. J. Lorenzen, Limnol. Oceanogr., 1967, 12, 343–346 CrossRef CAS
.
- J. J. Kozich, S. L. Westcott, N. T. Baxter, S. K. Highlander and P. D. Schloss, Appl. Environ. Microbiol., 2013, 79, 5112–5120 CrossRef CAS
.
- T. Stoeck, D. Bass, M. Nebel, R. Christen, M. D. M. Jones, H.-W. Breiner and T. A. Richards, Mol. Ecol., 2010, 19(Suppl 1), 21–31 CrossRef CAS
.
- B. J. Callahan, K. Sankaran, J. A. Fukuyama, P. J. McMurdie and S. P. Holmes, F1000Research, 2016, 5, 1–48 Search PubMed
.
- C. Lozupone and R. Knight, Appl. Environ. Microbiol., 2005, 71, 8228–8235 CrossRef CAS
.
- P. J. McMurdie and S. Holmes, PLoS One, 2013, 8(4), e61217 CrossRef CAS
.
- J. Wenzel, E. Fiset, P. Batlle-Vilanova, A. Cabezas, C. Etchebehere, M. D. Balaguer, J. Colprim and S. Puig, Front. Energy Res., 2018, 6, 15 CrossRef
.
- Y. Jiang, Q. Liang, N. Chu, W. Hao, L. Zhang and G. Zhan, Sci. Total Environ., 2020, 741, 140198 CrossRef CAS
.
- G. Mohanakrishna, I. M. Abu, K. Vanbroekhoven and D. Pant, Sci. Total Environ., 2020, 715, 137003 CrossRef CAS
.
- P. Batlle-Vilanova, R. Ganigué, S. Ramió-Pujol, L. Bañeras, G. Jiménez, M. Hidalgo, M. D. Balaguer, J. Colprim and S. Puig, Bioelectrochemistry, 2017, 117, 57–64 CrossRef CAS
.
- M. Köpke, C. Held, S. Hujer, H. Liesegang, A. Wiezer, A. Wollherr, A. Ehrenreich, W. Liebl, G. Gottschalk and P. Dürre, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 13087–13092 CrossRef
.
- F. Liew, A. M. Henstra, M. Köpke, K. Winzer, S. D. Simpson and N. P. Minton, Metab. Eng., 2017, 40, 104–114 CrossRef CAS
.
- H. Liu, T. Song, K. Fei, H. Wang and J. Xie, Bioresour. Bioprocess., 2018, 5, 7 CrossRef
.
- K. Li, H. Zhu, Y. Zhang and H. Zhang, Sustainability, 2017, 9, 243 CrossRef
.
- C. Etchebehere, E. Castelló, J. Wenzel, M. del Pilar Anzola-Rojas, L. Borzacconi, G. Buitrón, L. Cabrol, V. M. Carminato, J. Carrillo-Reyes, C. Cisneros-Pérez, L. Fuentes, I. Moreno-Andrade, E. Razo-Flores, G. R. Filippi, E. Tapia-Venegas, J. Toledo-Alarcón and M. Zaiat, Appl. Microbiol. Biotechnol., 2016, 100, 3371–3384 CrossRef CAS
.
- X. Lin, Y. Xia, Q. Yan, W. Shen and M. Zhao, Bioresour. Technol., 2014, 155, 98–103 CrossRef CAS PubMed
.
- Y. J. Lee, C. S. Romanek and J. Wiegel, Int. J. Syst. Evol. Microbiol., 2007, 57, 311–315 CrossRef CAS PubMed
.
- M. J. Griffiths and S. T. L. Harrison, J. Appl. Phycol., 2009, 21, 493–507 CrossRef CAS
.
- M. M. Maroneze, L. Q. Zepka, E. J. Lopes, A. Pérez-Gálvez and M. Roca, Antioxidants, 2019, 8, 600 CrossRef CAS
.
- A. Patel, L. Matsakas, U. Rova and P. Christakopoulos, Biotechnol Biofuels, 2018, 11, 1–16 CrossRef CAS PubMed
.
- F. Zhang and Z. He, Environ. Technol., 2013, 34, 1905–1913 CrossRef CAS PubMed
.
- A. Callegari, S. Bolognesi, D. Cecconet and A. G. Capodaglio, Crit. Rev. Environ. Sci. Technol., 2020, 50, 384–436 CrossRef CAS
.
- L. C. Meher, D. Vidya Sagar and S. N. Naik, Renewable Sustainable Energy Rev., 2006, 10, 248–268 CrossRef CAS
.
- B. Diehl and G. Randel, Lipid Technol., 2007, 19, 258–260 CrossRef CAS
.
- E. M. Valdovinos-García, J. Barajas-Fernández, M. de los Ángeles Olán-Acosta, M. A. Petriz-Prieto, A. Guzmán-López and M. G. Bravo-Sánchez, Energies, 2020, 13, 1–19 CrossRef
.
- J. C. M. Pires, M. C. M. Alvim-Ferraz, F. G. Martins and M. Simões, Renewable Sustainable Energy Rev., 2012, 16, 3043–3053 CrossRef CAS
.
- O. Perez-Garcia, F. M. E. Escalante, L. E. de-Bashan and Y. Bashan, Water Res., 2011, 45, 11–36 CrossRef CAS PubMed
.
- IndianOil and LanzaTech poised to scale-up CO2 to food, feed and fuel, https://bioenergyinternational.com/biofuels-oils/indianoil-and-lanzatech-poised-to-scale-up-co2-to-food-feed-and-fuel, accessed November 2021.
- Y. Chisti, Biotechnol. Adv., 2007, 25, 294–306 CrossRef CAS PubMed
.
- J. Xu, J. J. L. Guzman, S. J. Andersen, K. Rabaey and L. T. Angenent, Chem. Commun., 2015, 51, 6847–6850 RSC
.
- E. V. LaBelle and H. D. May, Front. Microbiol., 2017, 8, 1–9 Search PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1se01701b |
| This journal is © The Royal Society of Chemistry 2022 |