Open Access Article
Open Access ArticleA sensitive isothermal fluorescence biosensor for microRNA detection coupling primer exchange reaction with catalytic hairpin assembly†
Jiatong
Liu
,
Minzhe
Shen
,
Jadera
Talap
,
Xudan
Shen
 ,
Zihan
Song
,
Haihong
Hu
,
Su
Zeng
and
Sheng
Cai
,
Zihan
Song
,
Haihong
Hu
,
Su
Zeng
and
Sheng
Cai
 *
*
Institute of Drug Metabolism and Pharmaceutical Analysis, Zhejiang Province Key Laboratory of Anti-Cancer Drug Research, Zhejiang University, Hangzhou, Zhejiang 310058, China. E-mail: caisheng@zju.edu.cn
First published on 25th August 2022
Abstract
The miRNA, which has been proven to regulate the expression of many cancer-related genes, is expected to become a new type of biomarker for cancer diagnosis and prognosis research. Herein, a novel biosensor for sensitive detection of miRNAs was established based on coupling the primer exchange reaction (PER) with catalytic hairpin assembly (CHA). A self-gated hairpin was designed as the template hairpin of the PER. Benefiting from the cascade PER and CHA, the proposed fluorescence biosensor showed high sensitivity and excellent selectivity for the miR-200a detection. Additionally, the biosensor was successfully applied for the detection of miR-200a from a diluted human serum sample. To sum up, the proposed fluorescence biosensor we established could be used as a highly sensitive platform. By replacing the specific sequences in the self-gated hairpin, the biosensor was promising to be used for simultaneous detection of various miRNA biomarkers in bioanalysis.
Introduction
MicroRNAs (miRNAs), a class of endogenous and noncoding small RNAs with a length of 19–25 nucleotides, have been proven to play an important role in biological processes,1–6 including embryonic differentiation,7 apoptosis,8–10 metabolism,11 growth control,12 and immune reactions.13 Recent studies have proven that the expression of miRNAs may be involved in various human diseases, such as diabetes, cancer, AIDS, heart diseases, etc.14,15 Thus, the detection of concentration of miRNAs has attracted great attention due to its far-reaching significance. However, due to their high homology, low abundance, and small sequence, the development of accurate and sensitive detection of miRNAs is still challenging. Conventional detection methods include northern blotting,16–18 reverse transcription-quantitative PCR,19–22 rolling circle amplification,23–25 loop-mediated amplification,26,27 and miRNA chips;28 however, these techniques have some limitations, such as complicated operation, high requirements for equipment, and being time-consuming. To address these challenges, a detection method with high sensitivity, good specificity, and easy operation has been valued greatly.The Primer Exchange Reaction (PER) was firstly proposed in 2018.29 In the PER process, short primers were programmed to synthesize ssDNA. A single hairpin was used as a catalytic template and a user-defined sequence was attached to the short DNA primer under the action of a DNA polymerase. Through the toehold-mediated branch migration process, the synthesized ssDNA can be spontaneously isolated from the hairpin, and other primers re-participated to initiate the next round of ssDNA synthesis. Only one hairpin and one primer were required to perform a rapid and specific signal amplification. The PER has been used in a series of high-performance biosensors for proteins, DNA, and RNA in cells and tissues.30–33
Herein, we proposed a sensitive isothermal fluorescence biosensor for microRNA detection by coupling catalytic hairpin assembly (CHA) and the PER. CHA is an isothermal amplification technique based on the toehold-mediated strand displacement (TMSD) reaction, which has very bright application prospects due to its rapid response and simple operation.34–37 This strand displacement reaction not only can be used for signal amplification, but also provides an approach for DNA logic operations.38 In our fluorescence biosensor, a self-gated hairpin was designed as the template hairpin of the PER. The presence of miR-200a could lead to opening of the self-gated hairpin which would trigger the PER. Meanwhile, the nascent long DNA strand generated by the PER was used as the trigger sequence for CHA, bringing isothermal nucleic acid amplification. Thus, the combination of the PER and CHA achieved simple operation, rapid detection, good specificity, and high sensitivity, promising PER–CHA to become a powerful approach for miRNA detection.
Experimental
Reagents and materials
All DNA sequences that were used in the experiment were synthesized and purified by Sangon Biotech Co., Ltd (Shanghai, China). The Bst DNA polymerase, large fragment (8 U μL−1, the following text is referred to as Bst DNA polymerase), Klenow fragment (3′ → 5′ exo-), and phi29 DNA polymerase were purchased from New England Biolabs, Inc (America). RNase-free water and dHTP were purchased from Takara Bio. Co., Ltd (Dalian, China). miRNA sequences were synthesized and purified by Genepharma, Ltd (Shanghai, China). Prior to use, all DNAs were heated at 95 °C for 5 min and gradually cooled down to 25 °C at a speed of 0.1 °C s−1 (PCR instrument). Related oligonucleotide sequences are shown in Table S1.†Agarose gel electrophoresis
Agarose gel electrophoresis was utilized to verify the amplification of the PER and the feasibility of PER–CHA. The DNA solutions were mixed with 10× loading buffer and analyzed by 2% agarose gel electrophoresis, which was executed in 1× TAE at an electrophoretic voltage of 170 V for 30 min. The gels were stained with nucleic acid gel stain DuRed to image non-labeled DNA.Fluorescence measurements of the PER
All fluorescence measurements were conducted with a 20 μL reaction solution system containing 8 μL target miRNA, 2 μL primer a (1 μM), 2 μL self-gated hairpin (500 nM), 1 μL dHTP (2.5 mM), 3 μL MgSO4 (100 mM) and 2 U Bst DNA polymerase. After mixing, the system was incubated at 37 °C for 1 h, and then incubated at 80 °C for 20 min to inactivate the enzyme. Following that, 2 μL of fluorescence dye 20× EvaGreen was added. The fluorescence intensities were identified on a microplate detector (Synergy H1, BioTek, America) with an excitation wavelength of 492 nm and an emission wavelength of 520 nm.Fluorescence measurements of PER–CHA
As for the fluorescence measurements of PER–CHA, the 20 μL reaction solution should contain 8 μL target miRNA, 2 μL primer a (1 μM), 2 μL self-gated hairpin (500 nM), 1 μL dHTP (2.5 mM), 3 μL MgSO4 (100 mM) and 8 U Bst DNA polymerase. After incubation at 31 °C for 150 minutes, the PER was quenched by heating at 80 °C for 20 min. The CHA was initiated by adding 2 μL of CHA probe mixture (H1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2
H2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H3 = 1
H3 = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the concentration of each probe was 900 nM) to the reaction system, and 20 μL was taken to obtain fluorescence intensities on the microplate detector.
1, the concentration of each probe was 900 nM) to the reaction system, and 20 μL was taken to obtain fluorescence intensities on the microplate detector.
Clinical sample preparation
Serum samples were obtained from healthy volunteers at the Second Affiliated Hospital of Medical College of Zhejiang University. The study was implemented after being approved by the ethics review committee. Whole blood was incubated with a coagulant for 30 min and then centrifuged at 3000 rpm at 4 °C for 5 min in a low-temperature centrifuge. The faint yellow supernatant was collected as the required serum sample. Different concentrations of miRNAs were added to the prepared serum to authenticate the resistance of our established method to biological matrix interference.Results and discussion
Overall principle of the developed strategy
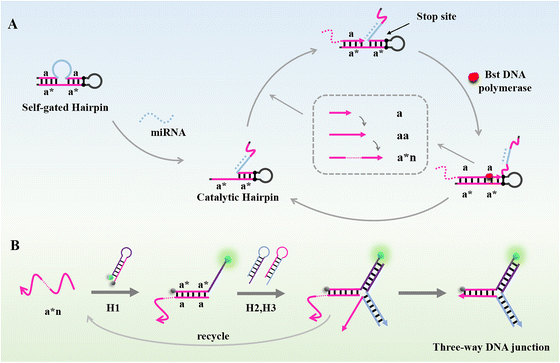
The principle of the PER–CHA detection is shown in Scheme 1. Due to the lack of dGTP in raw materials, the hairpin probe was designed with a G–C pair as a stop site.39 In the presence of the target miR-200a, the delicately designed self-gated hairpin was initiated into the catalytic hairpin, which contained the complementary domain of primer a. With the efforts of the Bst DNA polymerase, primer a could be used as a trigger to generate ssDNA “aa” and halted at the stop site. Under the competitive displacement, the extended sequence was released from the catalytic hairpin. With the same sequence of primer a, the extended ssDNA aa could trigger the next amplification cascade, consequently forming an enlarged DNA strand containing a repetitive “a” sequence, also mentioned as “a*n”. The combination of the PER and CHA was conducted by adding CHA hairpins into the PER system. The existence of the PER product could trigger the dynamic assembly of H1, H2, and H3 to produce a three-way DNA junction (H1–H2–H3 complexes) accompanying the release of the trigger. And the released PER product could further trigger the next CHA cascade, resulting in an accumulated “turn-on” fluorescence signal.Investigation of the PER amplification and PER–CHA strategy
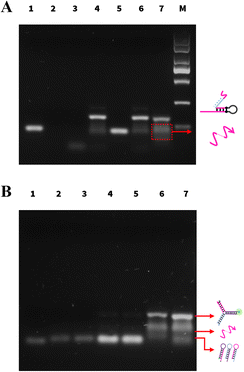
The feasibility of the PER amplification and PER–CHA strategy was verified by agarose gel electrophoresis. As shown in Fig. 1A, clear bands were noted in lane 1 and lane 3 representing the self-gated hairpin and the target miRNA, respectively. A new band appeared after the self-gated hairpin was mixed with the target miRNA indicating the formation of the catalytic hairpin (lane 4).When the self-gated hairpin was mixed with primer a (lane 5), no hybridization occurred. In the absence of the enzyme (lane 6), the mixture of the self-gated hairpin, primer a, and the target miRNA showed no new band, implying that no amplification occurred. In the presence of the enzyme, the PER system produced a ladder of different lengths of the product (lane 7), which was a new band longer than primer a, verifying the feasibility of the PER.
The activation of CHA was also exhibited by agarose gel electrophoresis in Fig. 1B. Lanes 1–3 showed the bands of H1, H2, and H3, respectively. Although containing complementary segments, there was almost no band position change when the CHA probes (H1, H2, and H3) were mixed (lane 4), indicating no spontaneous interactions between three hairpins. The same performance was also observed after the addition of primer a (lane 5), showing that primer a had no capability to trigger CHA. Upon introducing CHA hairpins to the PER, the bands of the CHA hairpins were significantly reduced (lanes 4 and 7); meanwhile, the brightness of the three-way DNA junction increased obviously (lane 7). The above result successfully demonstrated the capability of the PER amplification strategy and the feasibility of CHA induced by the PER product.
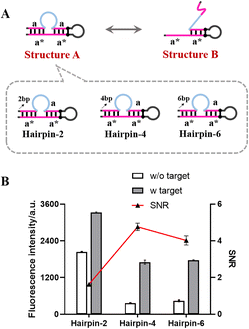
Design of the self-gated hairpin
As shown in Fig. 2A, the self-gated hairpin existed in two structures (structure A and structure B) that could be converted into each other under certain conditions. In the presence of the target miR-200a, the principle of least energy shifted the equilibrium reaction to the right, exposing the complementary sequence of primer a and initiating the signal amplification. In the absence of miR-200a, the more the self-gated hairpin existed as structure B, the higher the fluorescence intensity of the blank group was. To ensure the stability of structure A, we added complementary base pairs to the end of the self-gated hairpin to blockade structure A. Corresponding to the figure of complementary base pairs (2, 4, and 6), the hairpins were named as Hairpin-2, Hairpin-4, and Hairpin-6 (Fig. 2A), respectively.The performance of the self-gated hairpin with different blocking base pairs was investigated by comparing the signal/noise ratio (SNR) and fluorescence intensity. As shown in Fig. 2B, the fluorescence signal of Hairpin-2 without the target was significantly higher than those of the other groups without the target, which demonstrated the low blocking ability of the existence of two blocking base pairs. The blocking performances of Hairpin-4 and Hairpin-6 were almost the same level, both lower than the previous one. Hairpin-4 with the best SNR and fluorescence intensity was selected for further experiments.
Effect of the DNA polymerase
The temperature of the PER, which was frequently held below 40 °C, had a certain gap with the optimal temperature of the Bst DNA polymerase at 65 °C. Due to the effect of temperature on the amplification efficiency of the enzyme, two DNA polymerases (Phi29 and Klenow) whose optimal temperature was below 40 °C were selected to verify the possibility for better amplification performance.The results are shown in Fig. 3. Compared to the Phi29 and Klenow polymerases, the Bst DNA polymerase showed a considerably high SNR level and satisfactory fluorescence intensity. The Bst DNA polymerase with the largest difference between the optimum temperature and the actual reaction temperature had the best performance. This may be due to the amplification efficiency of the PER being determined not only by the polymerization activity of the enzymatic reaction, but also by the strand displacement activity. The Bst DNA polymerase was much better at strand displacement than the other two polymerases. It could be speculated that the determinant of the amplification efficiency of the PER might be the strand displacement activity of the enzyme. According to the experimental results, the Bst DNA polymerase was chosen as the optimum DNA polymerase.
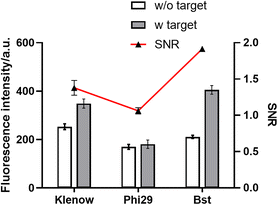 | ||
| Fig. 3 The effect of the DNA polymerase. Error bars: the standard deviation of triplicate independent measurements. | ||
Optimization of key parameters
Some relevant parameters, such as the concentration of the self-gated hairpin, the concentration of primer a, the amount of Bst DNA polymerase, the concentration of Mg2+, the concentration of dHTP, the incubation temperature, the incubation time, and the concentration of CHA probes, were known to influence the performance of the biosensor. The self-gated hairpin was not only a crucial factor to detect miR-200a, but also an amplification template in the amplification process of the PER, with a significant influence on the amplification efficiency. As shown in Fig. S1A,† with increasing self-gated hairpin concentration, the relative intensity and SNR increased. Hence, an optimized self-gated hairpin concentration of 500 nM was employed. Optimization of the concentration of primer a is critical as a lower concentration may lead to reduced amplification efficiency and a higher concentration may lead to nonspecific amplification. As shown in Fig. S1B,† with an increase in the concentration of primer a, the relative intensity increased, and then reached a plateau. At the same time, the SNR reached a peak. Thus, 1 μM primer a was chosen for the following experiment.As depicted in Fig. S1C,† we estimated the effect of the amount of Bst DNA polymerase on the elaboration capability using 5 different enzyme amounts (2, 4, 6, 8, and 10 U, and 2 U was selected as an optimal enzyme amount). The Mg2+ concentration, which influenced the activity of the Bst DNA polymerase, was the cofactor for the proposed biosensor. As shown in Fig. S1D,† the peak value of the SNR was acquired at 100 mM, therefore, 100 mM was chosen as the optimum concentration of Mg2+. dHTP containing dATP, dCTP, and dTTP without dGTP was a necessary raw material for amplification. As depicted in Fig. S1E,† the SNR showed an increase as the concentration of dHTP increased. Consequently, 2.5 mM dHTP with a maximum SNR was selected for the subsequent experiments. In addition, the optimized incubation temperature was 31 °C (Fig. S1F†) and the incubation time was 150 min (Fig. S1G†). As Fig. S1H† suggests, the concentration of CHA probes was selected to be 300 nM.
Analytical performance of the miRNA detection
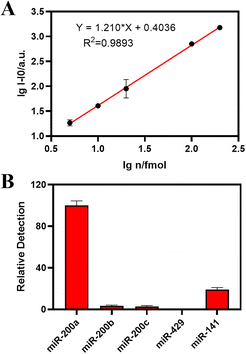
Under the optimal experimental conditions, we confirmed the sensitivity and selectivity of the two established methods for miRNA-200a. As shown in Fig. 4A, the log value of the miR-200a (lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n) concentration was taken as the abscissa, and the absolute fluorescence intensity I − I0 was taken as the ordinate to fit the standard curve. As for the biosensor cascading the PER and CHA, the standard curve line showed a strong linear relationship with an equation of lg
n) concentration was taken as the abscissa, and the absolute fluorescence intensity I − I0 was taken as the ordinate to fit the standard curve. As for the biosensor cascading the PER and CHA, the standard curve line showed a strong linear relationship with an equation of lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) I − I0 = 1.1210* lg
I − I0 = 1.1210* lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n + 0.4036 (R2 = 0.9893) from 5 to 200 fmol.
n + 0.4036 (R2 = 0.9893) from 5 to 200 fmol.
According to the 3σ rule, the LOD of PER–CHA was calculated to be 287 amol (14.35 pM), which showed superior sensitivity and stability compared to the previously reported method using CHA for miRNA detection (Table S1†). As shown in Fig. S3A,† the linear range of the established PER detection method was from 32 fmol to 512 fmol with an equation of lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) I − I0 = 0.8451* lg
I − I0 = 0.8451* lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) n + 0.9843 (R2 = 0.9474). Hence, the PER–CHA strategy showed a higher sensitivity and a better correlation coefficient than the pure PER strategy.
n + 0.9843 (R2 = 0.9474). Hence, the PER–CHA strategy showed a higher sensitivity and a better correlation coefficient than the pure PER strategy.
To investigate the selectivity of the proposed biosensor, the target miR-200a sequence was replaced with different subtypes in the same family of miR-200 like miR-200b and miR-200c, and a set of unrelated and non-complementary sequences such as miR-429 and miR-141. As shown in Fig. 4B, the biosensor exhibited excellent performance in the presence of miR-200a, while other miRNA sequences showed a negligible response under the same conditions. In addition, the selectivity of the pure PER strategy is presented in Fig. S3B.† All the results showed that the proposed biosensor exhibited a satisfactory selectivity for the detection of miRNA-200a.
Real sample analysis
To examine the practicality of the proposed biosensor for miRNA detection in human fluid samples, 20% human serum was chosen as a real biological sample. We prepared standard samples by adding different amounts (20, 40, and 80 fmol) of miR-200a into the 20% human serum. As shown in Table 1, the proposed biosensor provided miRNA amounts of 20, 40, and 80 fmol with recovery rates of 104.8%, 101.6%, and 99.1%, respectively, which indicates the satisfactory anti-interference performance of PER–CHA. In addition, the practicality of the pure PER strategy is presented in Table S3.†| miRNA | Added (fmol) | Detected (fmol) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| miR-200a | 20 | 21 | 104.8 | 1.14 |
| 40 | 40.6 | 101.6 | 1.55 | |
| 80 | 79.3 | 99.1 | 1.05 |
Conclusions
In summary, we constructed a convenient and efficient fluorescence biosensor for the detection of miRNA based on the PER cascading CHA. A self-gated hairpin was designed ingeniously to specifically bind to the target miR-200a. Under the primer exchange reaction, an efficient cyclic amplification was performed. By combining with CHA, the stability and sensitivity of miRNA detection were greatly promoted. With the modification of the complementary sequence of the target miRNA in the self-gated hairpin, the biosensor could be extended to the detection of other small RNAs including miRNA. The proposed biosensor showed ideal selectivity, specificity, sensitivity, and precision and was expected to become a universal tool for miRNA- specific detection.Author contributions
Jiatong Liu: conceptualization, data curation, formal analysis, writing – original draft. Minzhe Shen: methodology. Jadera Talap: visualization. Xudan Shen: data curation. Zihan Song: visualization. Haihong Hu: visualization. Su Zeng: validation, funding acquisition, supervision. Sheng Cai: methodology, funding acquisition, supervision, project administration.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
We acknowledge financial support from the National Key R&D Project of China (Grant 2019YFC1520500), the National Natural Science Foundation of China (Grant 81673399), and the Natural Science Foundation of Zhejiang Province (LGC21H300004).Notes and references
- V. Ambros, Nature, 2004, 431, 350–355 CrossRef CAS PubMed.
- R. Blanco Dominguez, R. Sanchez Diaz and H. de la Fuente, N. Engl. J. Med., 2021, 384, 2014–2027 CrossRef CAS PubMed.
- A. Kozomara, M. Birgaoanu and S. Griffiths Jones, Nucleic Acids Res., 2019, 47, D155–D162 CrossRef CAS PubMed.
- A. Kozomara and S. Griffiths Jones, Nucleic Acids Res., 2014, 42, D68–D73 CrossRef CAS PubMed.
- Y. Tomimaru, H. Eguchi, H. Nagano, H. Wada, S. Kobayashi, S. Marubashi, M. Tanemura, A. Tomokuni, I. Takemasa, K. Umeshita, T. Kanto, Y. Doki and M. Mori, J. Hepatol., 2012, 56, 167–175 CrossRef CAS PubMed.
- E. van Rooij and E. N. Olson, Nat. Rev. Drug Discovery, 2012, 11, 860–872 CrossRef CAS PubMed.
- H. Ma, Y. Lin, Z. A. Zhao, X. Lu, Y. Yu, X. Zhang, Q. Wang and L. Li, J. Biol. Chem., 2016, 291, 12126–12135 CrossRef CAS PubMed.
- K. Wu, L. Yang, C. Li, C. H. Zhu, X. Wang, Y. Yao and Y. J. Jia, Asian Pac. J. Cancer Prev., 2014, 15, 5583–5586 CrossRef PubMed.
- A. A. Farooqi, M. Z. Qureshi, E. Coskunpinar, S. K. H. Naqvi, I. Yaylim and M. Ismail, Asian Pac. J. Cancer Prev., 2014, 15, 1909–1912 CrossRef PubMed.
- M. Y. Xie, L. J. Hou, J. J. Sun, B. Zeng, Q. Y. Xi, J. Y. Luo, T. Chen and Y. L. Zhang, J. Agric. Food Chem., 2019, 67, 9477–9491 CrossRef CAS PubMed.
- P. Kantharidis, B. Wang, R. M. Carew and H. Y. Lan, Diabetes, 2011, 60, 1832–1837 CrossRef CAS PubMed.
- Y. Zhang and Z. C. Lai, Biochem. Biophys. Res. Commun., 2013, 439, 438–442 CrossRef CAS PubMed.
- C. Xiao and K. Rajewsky, Cell, 2009, 136, 26–36 CrossRef CAS PubMed.
- H. Chen, Y. Liu, S. Feng, Y. Cao, T. Wu and Z. Liu, Biosens. Bioelectron., 2022, 200, 113913 CrossRef CAS PubMed.
- Y. N. Wang, Y. Sun, C. W. Lau and J. Z. Lu, J. Pharm. Anal., 2018, 8, 265–270 CrossRef PubMed.
- J. Alles, T. Fehlmann, U. Fischer, C. Backes, V. Galata, M. Minet, M. Hart, M. Abu Halima, F. A. Graesser, H. P. Lenhof, A. Keller and E. Meese, Nucleic Acids Res., 2019, 47, 3353–3364 CrossRef CAS PubMed.
- T. Kilic, A. Erdem, M. Ozsoz and S. Carrara, Biosens. Bioelectron., 2018, 99, 525–546 CrossRef CAS PubMed.
- H. Baigude and T. M. Rana, Nanomedicine, 2014, 9, 2545–2555 CrossRef CAS PubMed.
- C. F. Chen, D. A. Ridzon, A. J. Broomer, Z. H. Zhou, D. H. Lee, J. T. Nguyen, M. Barbisin, N. L. Xu, V. R. Mahuvakar, M. R. Andersen, K. Q. Lao, K. J. Livak and K. J. Guegler, Nucleic Acids Res., 2005, 33, e179 CrossRef PubMed.
- R. Liu, X. Chen, Y. Du, W. Yao, L. Shen, C. Wang, Z. Hu, R. Zhuang, G. Ning, C. Zhang, Y. Yuan, Z. Li, K. Zen, Y. Ba and C. Y. Zhang, Clin. Chem., 2012, 58, 610–618 CrossRef CAS PubMed.
- P. Mestdagh, N. Hartmann, L. Baeriswy, D. Andreasen, N. Bernard, C. Chen, D. Cheo, P. D'Andrade, M. DeMayo, L. Dennis, S. Derveaux and Y. Feng, Nat. Methods, 2014, 11, 809–815 CrossRef CAS PubMed.
- D. d. Xiong, Y. w. Dang, P. Lin, D. y. Wen, R. q. He, D. z. Luo, Z. b. Feng and G. Chen, J. Transl. Med., 2018, 16, 220 CrossRef CAS PubMed.
- R. Deng, L. Tang, Q. Tian, Y. Wang, L. Lin and J. Li, Angew. Chem., Int. Ed., 2014, 53, 2389–2393 CrossRef CAS PubMed.
- E. M. Harcourt and E. T. Kool, Nucleic Acids Res., 2012, 40, e65 CrossRef CAS PubMed.
- H. Liu, L. Li, L. Duan, X. Wang, Y. Xie, L. Tong, Q. Wang and B. Tang, Anal. Chem., 2013, 85, 7941–7947 CrossRef CAS PubMed.
- J. Ye, M. Xu, X. Tian, S. Cai and S. Zeng, J. Pharm. Anal., 2019, 9, 217–226 CrossRef PubMed.
- R. Duan, X. Zuo, S. Wang, X. Quan, D. Chen, Z. Chen, L. Jiang, C. Fan and F. Xia, J. Am. Chem. Soc., 2013, 135, 4604–4607 CrossRef CAS PubMed.
- K. A. Cissell and S. K. Deo, Anal. Bioanal. Chem., 2009, 394, 1109–1116 CrossRef CAS PubMed.
- J. Y. Kishi, T. E. Schaus, N. Gopalkrishnan, F. Xuan and P. Yin, Nat. Chem., 2018, 10, 155–164 CrossRef CAS PubMed.
- R. Tavakoli Koopaei, F. Javadi-Zarnaghi and H. Mirhendi, Sens. Actuators, B, 2022, 357, 131409 CrossRef CAS PubMed.
- X. Li, L. Liao, B. Jiang, W. Zhou, R. Yuan and Y. Xiang, Anal. Chim. Acta, 2022, 1197, 339521 CrossRef CAS PubMed.
- Z. Xie, S. Chen, W. Zhang, S. Zhao, Z. Zhao, X. Wang, Y. Huang and G. Yi, Biosens. Bioelectron., 2022, 206, 114135 CrossRef CAS PubMed.
- J. Zhang, M. He, C. Nie, M. He, Q. Pan, C. Liu, Y. Hu, T. Chen and X. Chu, Chem. Sci., 2020, 11, 7092–7101 RSC.
- Y. X. Wang, D. X. Wang, J. Wang, B. Liu, A. N. Tang and D. M. Kong, Talanta, 2022, 236, 122846 CrossRef CAS PubMed.
- P. Yin, H. M. T. Choi, C. R. Calvert and N. A. Pierce, Nature, 2008, 451, 318–322 CrossRef CAS PubMed.
- W. W. Liu, X. L. Zhang, L. Zhu, S. Xu, Y. Q. Chai, Z. H. Li and R. Yuan, Anal. Chim. Acta, 2022, 1204, 339663 CrossRef CAS PubMed.
- X. Sun, S. Dong and W. Zhao, Anal. Chim. Acta, 2022, 1205, 339735 CrossRef CAS PubMed.
- P. Miao and Y. G. Tang, ACS Cent. Sci., 2021, 7, 1036–1044 CrossRef CAS PubMed.
- X. Y. Li, X. M. Li, D. D. Li, M. Zhao, H. P. Wu, B. Shen, P. Liu and S. J. Ding, Biosens. Bioelectron., 2020, 168, 112554 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sd00115b |
| This journal is © The Royal Society of Chemistry 2022 |