Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Rapid detection of major Gram-positive pathogens in ocular specimens using a novel fluorescent vancomycin-based probe†
Richa
Sharma
 a,
Harinee
Rajagopalan
b,
Maxime
Klausen
a,
Harinee
Rajagopalan
b,
Maxime
Klausen
 a,
Mani Vimalin
Jeyalatha
b,
Muhammed
Üçüncü
a,
Mani Vimalin
Jeyalatha
b,
Muhammed
Üçüncü
 ac,
Seshasailam
Venkateswaran
a,
Appakkudal R.
Anand
*b and
Mark
Bradley
ac,
Seshasailam
Venkateswaran
a,
Appakkudal R.
Anand
*b and
Mark
Bradley
 *a
*a
aEaStCHEM, School of Chemistry, University of Edinburgh, King's Buildings, Edinburgh, EH9 3FJ, UK. E-mail: Mark.Bradley@ed.ac.uk
bL & T Microbiology Research Centre, Medical Research Foundation, Sankara Nethralaya, Chennai, India. E-mail: aranand@gmail.com
cDepartment of Analytical Chemistry, Faculty of Pharmacy, Izmir Katip Celebi University, Izmir, Turkey
First published on 30th June 2022
Abstract
Bacterial infections of the eye are significant causes of morbidity that can lead to permanent visual loss without rapid and adequate treatment, with Gram-positive bacteria causing the majority of ocular infections. Here we report a novel probe, based on the reductive amination of vancomycin with a 4-nitrobenzoxadiazole (NBD)-based aldehyde, that rapidly and specifically detects Gram positive infections from ocular samples.
Introduction
Bacterial infections of the eye are significant causes of morbidity that can lead to permanent visual loss without rapid and adequate treatment.1,2 Aetiological studies have shown that Gram-positive bacteria cause the majority of ocular infections.1 Accurate information of the Gram status of the bacterial aetiology is critical to tailor appropriate antimicrobial therapy. Current rapid diagnosis of bacterial infection relies on observing the Gram-stained specimens by direct microscopy, which can provide information on the presence and type of bacteria. However, Gram staining generates a high rate of false negative results especially in ocular specimens where the sample volume is minimal. In order to increase sensitivity of detection of bacteria, isolation of the bacterium by microbial culture followed by identification is a commonly adopted procedure. This is considered the gold standard for laboratory diagnosis, although it may take many days for growth, delaying treatment. Hence, there is an urgent need to develop alternate rapid and accurate labelling techniques to detect and identify bacteria in clinical specimens3 and a few studies have demonstrated the application of Gram-selective optical probes with clinical specimens.Several strategies have been used to develop Gram-specific fluorescent probes,4–7 including the development of positively charged dyes8 or aggregation-induced emission systems,9–12 or the direct covalent conjugation of known fluorophores to pathogen-binding ligands.13–16 In this exploratory study, we designed and evaluated a fluorescent probe for the rapid detection of Gram-positive bacteria in ocular specimens in comparison with the conventional Gram stain.17,18 The fluorescent probe developed (a vancomycin–NBD conjugate here called Van-Green) discriminated Gram-positive bacteria from other bacteria with excellent selectivity and vitally did not require any modification to the readily available microscope systems widely used in the laboratory. The optical molecular imaging agent used here, Van-Green, was designed with a solvato-fluorogenic dye16 conjugated to the clinically approved glycopeptide antibiotic vancomycin. The design of the probe imitates the structure of oritavancin, a semi-synthetic vancomycin derivative in which the vancosamine moiety (i.e. the amino sugar) was modified with a bis-aryl group by reductive amination.19,20 This modification dramatically improves biological activity21,22 even on vancomycin-resistant strains of Gram-positive bacteria.20,23 In the present design, a benzaldehyde derivative of the 4-nitrobenzoxadiazole (NBD) fluorophore (a bis-aryl type structure) was prepared and conjugated to vancomycin so as to take advantage of the same enhancement of binding effect. NBD, a well-described “environmentally-sensitive” green fluorophore,24 emits in hydrophobic environments, and is non-fluorescent in aqueous solutions thus giving high signal-to-noise ratios without the need to wash-off excess of probe from the sample. In addition, NBD is a robust, oxygen insensitive and easily accessible dye, making the synthesis of this new probe quick and cost-efficient compared to other custom-made dyes.
Results and discussion
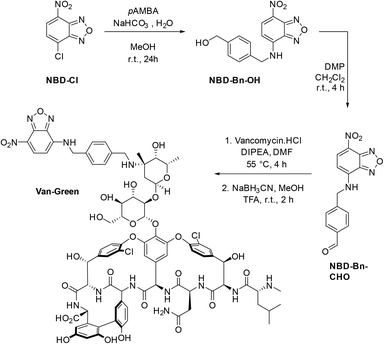
The benzaldehyde-modified NBD analogue (NBD-Bn-CHO) was synthesized in two steps starting from the commercially available dye precursor 4-chloro-7-nitrobenzofurazan (NBD-Cl). Nucleophilic aromatic substitution with p-(aminomethyl)benzylalcohol was followed by Dess–Martin periodinane (DMP) oxidation to afford the key aldehyde intermediate in a good, two-step, yield of 57%. The fluorophore was subsequently conjugated to the amino-sugar moiety of vancomycin using reductive amination, as previously described,19 to give Van-Green after purification by reverse phase HPLC. This transformation was selective and orthogonal to all the other vancomycin functional groups, and gave access to the final probe in only 3 steps and on a 100 mg scale (Scheme 1).Van-Green was fully characterized chemically (HPLC, HRMS and FTIR, Fig. S4 and S5†) and photophysically. In accordance with the optical properties of other NBD probes,24 Van-Green had an excitation maximum around 480 nm and emitted in the green (≈540 nm). In addition, the fluorescence intensity of the imaging agent proved to be sensitive to changes in the polarity of the environment, as demonstrated by the alteration of its brightness in PBS solutions containing different percentages of DMSO (Fig. 1). By determining ε and Φf values for each PBS/DMSO mixture (Table S1†), we found a progressive, 22-fold, increase in brightness (εΦf) in pure DMSO compared to aqueous buffers with negligible variations in excitation and emission wavelengths. This solvato-fluorogenic character is the key to labelling, selectively, pathogen membranes/cell walls without the need for washing steps,16 as the low fluorescence in aqueous media reduces background signal and “lights-up” the bacteria.
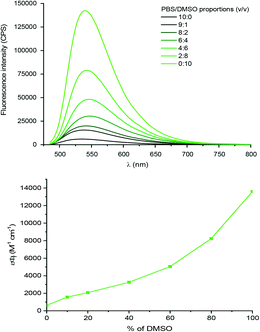 | ||
| Fig. 1 Top panel: Evolution of the fluorescence intensity and lower panel: brightness (εΦf) of solutions of Van-Green (5 μM) with increasing percentages of DMSO in PBS upon excitation at 475 nm. | ||
Biological validation
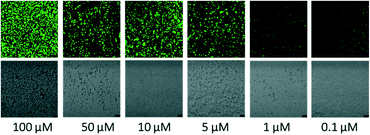
The probe showed good labelling of bacteria down to 0.1 μM for the target microbe (Fig. 2), which is lower than the reported minimum inhibitory concentration (MIC) of the parent vancomycin.22 Intense fluorescence was observed for 5–100 μM probe, showing that the vancomycin ligand has strong affinity for the cell wall of Gram positive bacteria and the dye does not affect its binding.
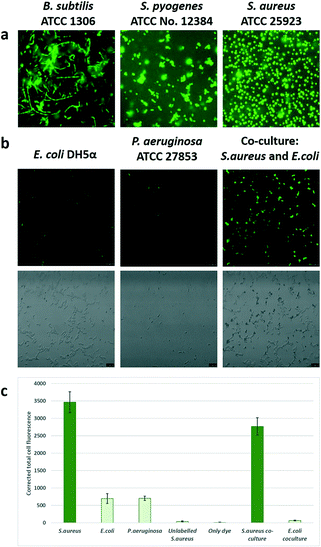
However, the intensity of staining of Gram positive bacteria differed, with S. aureus showing uniform bright fluorescence, while B. cereus showed irregular staining. Time for detection was <20 minutes similar to the time taken for Gram staining. When a mixture of Gram positive and Gram negative bacteria were stained, the probe specifically stained the Gram positive bacteria and Gram negative bacteria remained unstained (see Fig. 3 and S6†).
| S. no. | Sample type | Gram stain results | Identified culture | Van-Green results |
|---|---|---|---|---|
| 1 | Pus from lid abscess | GP cocci | Staphylococcus aureus (GP) | +++ |
| 2 | Pus from lid abscess | GP cocci | Staphylococcus aureus (GP) | +++ |
| 3 | Eviscerated material | GP cocci | Staphylococcus aureus (GP) | +++ |
| 4 | Canalicular pus | Filamentous GP bacilli | Actinomyces spp (GP) | +++ |
| 5 | Corneal scraping | GP cocci | Streptococcus pneumoniae (GP) | +++ |
| 6 | Canalicular pus | Filamentous GP bacilli | Actinomyces spp (GP) | +++ |
| Klebsiella oxytoca (GN) | ||||
| 7 | Corneal scraping | GN bacilli | Pseudomonas aeruginosa (GN) | − |
| 8 | Corneal scraping | GP bacilli | Corynebacterium amycolatum (GP) | +++ |
| Corynebacterium urealyticum (GP) | ||||
| 9 | Scleral scraping | GP cocci | Staphylococcus aureus (GP) | +++ |
| 10 | Eviscerated material | GN cocco bacilli | Neisseria subflava (GN) | + |
| 11 | Eviscerated material | GN bacilli | Pseudomonas aeruginosa (GN) | − |
| 12 | Pus from orbit | GP bacilli | Bacillus cereus (GP) | ++ |
| 13 | Corneal scraping | GN bacilli | Pseudomonas aeruginosa (GN) | − |
| 14 | Eviscerated material | GP cocci | Streptococcus pneumoniae (GP) | ++ |
| 15 | Aqueous humor | Gram variable cocci | Streptococcus pyogenes (GP) | +++ |
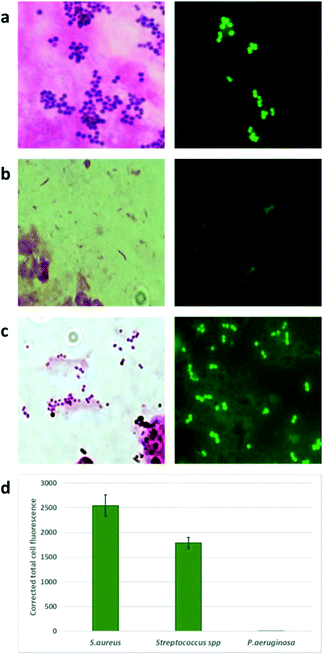
The brightness of the staining was characterized as +++, ++, + and −. As shown in Table 1, all specimens that showed Gram positive (GP) bacteria by the traditional Gram stain and culture were also positive using the NBD (Van-Green) probe. Gram negative (GN) bacteria showed little or no staining.
There was complete concordance (100%) in specimens with Gram positive infections, namely, Staphylococcus, Streptococcus, Actinomycetes spp and Corynebacterium spp. In one sample where Bacillus cereus was seen and grown, the probe did stain the bacteria, but the staining was not uniform. Two specimens grew P. aeruginosa and two grew Neisseria subflava and Klebsiella oxytoca respectively, which did not take up the NBD stain indicating its selectivity for Gram-positive bacteria. Representive microscopy images are shown in Fig. 4 and S7.†
Experimental section
Materials
Clinical specimens: included corneal scrapings (4), eviscerated material (4), canalicular pus (2), pus from eyelids (2), pus from the orbit (1), scleral scrapings (1) and aqueous humor (1).
Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc, 1
EtOAc, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v) (orange solid, 464 mg, yield 60%).
2, v/v) (orange solid, 464 mg, yield 60%).
1H NMR (500 MHz, DMF-d7) δ ppm: 8.51 (d, J = 8.9 Hz, 1H), 7.49 (d, J = 8.1 Hz, 2H), 7.38 (d, J = 8.2 Hz, 2H), 6.49 (d, J = 8.9 Hz, 1H), 4.86 (s, 2H), 4.61 (s, 2H). 13C NMR (126 MHz, DMF-d7) δ ppm: 145.2, 144.9, 144.3, 142.1, 137.1, 135.5, 127.3, 126.6, 121.6, 99.6, 63.1, 46.7. HRMS (ESI)+ calcd for C14H13N4O4 [M + H]+m/z 301.09313 found m/z 301.09360. MS (ES)+m/z 301.1 [M + H]+. HPLC tR 3.9 min.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EtOAc, 1
EtOAc, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v) (orange solid, 284 mg, yield 95%).
2, v/v) (orange solid, 284 mg, yield 95%).
1H NMR (500 MHz, DMF-d7) δ ppm: 10.09 (s, 1H), 9.92 (s, 1H), 8.55 (d, J = 8.8 Hz, 1H), 7.97 (d, J = 8.2 Hz, 2H), 7.78 (d, J = 8.1 Hz, 2H), 6.51 (d, J = 8.7 Hz, 1H), 5.04 (s, 2H). 13C NMR (125 MHz, DMF-d7) δ ppm: 192.6, 145.4, 145.2, 144.7, 144.5, 137.6, 136.3, 130.1, 128.4, 122.7, 100.1, 46.9. MS (ES)+m/z 299.1 [M + H]+. HRMS (ESI)+ calcd for C14H11N4O4 [M + H]+m/z 299.07748 found m/z 299.07710. HPLC tR 4.5 min. FT-IR νmax (cm−1): 3227 (NH), 3099, 3033 (CH aryl), 2953, 2918 (CH alkyl), 2850 (CH benzaldehyde), 1672 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1555 (NO).
O), 1555 (NO).
HRMS (ESI)+ calcd for C80H86Cl2N13O27 [M + H]+m/z 1730.51277 found m/z 1730.51290.
HPLC tR(Prep) 7.97 min. tR(analytical) 5.36 min.
FT-IR νmax (cm−1): 3280 (br, OH, NH), 3097 (CH aryl), 2984 (CH alkyl), 1668 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1587 (NH), 1505 (NO).
O), 1587 (NH), 1505 (NO).
Optimization of probe concentration. μ-Slide fifteen-well glass bottom confocal chambers (Ibidi) were coated with poly-D-lysine (0.1 mg mL−1 in PBS) for 20 minutes at 37 °C, after which the wells were washed with PBS. Forty μL of the S. aureus ATCC 29213 liquid culture (concentrations were adjusted to 0.5 × 109 cfu mL−1) were added to the coated wells and incubated for 1 hour. The wells were washed with PBS and probe of concentrations 100, 50, 10, 5, 1 and 0.1 μM were added (30 μL). After an incubation period of 1 hour, the wells were washed to remove unbound probe, the slide was sealed and imaged on a Leica SP5 confocal microscope (excitation laser: 476 nm, 10%, HyD detector: 510–590 nm with gain 100, brightfield PMT trans-detector with 250 gain).
Specificity of the probe. In order to ascertain if the probe labelled across Gram-positive strains, a range of microorganisms, namely, S. aureus ATCC 25923, Streptococcus pyogenes (ATCC no. 12384), Bacillus cereus (ATCC 13061) were tested. To check the specificity, Gram negative Escherichia coli DH5α, Escherichia coli (ATCC no. 25922) and Pseudomonas aeruginosa (ATCC 27853) were used. The slides were observed under FITC filters (Leica SP5 confocal and a Nikon Eclipse Ni microscopes). A mixture of S. aureus (Gram positive bacteria) and E. coli (Gram negative bacteria) were prepared by adding equal volumes of saline suspensions of both strains, followed by labelling with the Van-Green probe.
For quantification images were analyzed by Image J (Fiji, National Institutes of Health). For analysis, an average intensity of 25 labelled bacteria distributed over the area of interest were analysed (an average of 10 dark spots was taken as the background). The corrected intensity was calculated by subtracting the product of the mean area and the mean intensity of the background from the integrated density of the labelled cells. In order to determine if Van-Green was able to permeabilise the bacteria we performed live-dead staining experiments (LIVE/DEAD BacLight Bacterial Viability Kit, ThermoFisher Scientific) and viewed the bacteria under a fluorescent microscope using FITC filters.
Determination of antibacterial activity of the probe. The MIC of Van-Green and the parent compound vancomycin was determined using a broth microdilution method against standard strains, namely Staphylococcus aureus (ATCC no. 25923) and Escherichia coli (ATCC no. 25922) according to the CLSI guidelines. Briefly, bacteria were cultured in Muller Hinton broth (MHB) (Himedia) at 37 °C overnight, after which the cell density was standardized to match 0.5 McFarland standard (1.5 × 108 CFU mL−1). The compounds were serially diluted two-fold across the wells, with concentrations ranging from 256 μM to 0.25 μM plated in duplicate. The mid-log phase cultures of bacteria were diluted to the final concentration of 5 × 105 CFU mL−1 after which 100 μL of bacterial suspension was added to 100 μL of medium containing various concentrations of the compounds in a 96-well plate. Controls included growth control and a broth control. All the plates were covered and incubated at 37 °C for 18–24 h with the MIC defined as the lowest compound concentration at which no bacterial growth was visible.
Conventional microbiological investigations. The ocular specimens were processed immediately after receipt at the laboratory for conventional microbiological investigations. Smears were made for Gram staining using a 10% KOH/calcofluor white wet mount. The specimens were inoculated into the following solid media (5% sheep blood agar (BA), Brucella blood agar (BBA), chocolate agar (CA), MacConkey agar (MAC) and Sabouraud's dextrose agar (SDA)) and liquid media (brain heart infusion broth (BHIB) and thioglycolate broth). Once bacteria or fungi were isolated, the isolates were identified by standard procedures including colony morphology and biochemical tests. When significant pathogens were grown, fresh isolates were identified using GN/GP/NH cards from the VITEK 2®system (BioMérieux, France).
Labelling by Van-Green. A total of 15 Gram stain positive ocular samples were used for the study. 5 μM was the standardized concentration of the probe used for analysis of the clinical specimens. A total of 20 μL of 5 μM Van-Green was added directly to the methanol fixed slides. A coverslip was mounted on the smear and fluorescent imaging was done immediately. Images were captured using a camera fixed to the fluorescent microscope. The results were compared with the Gram stain and conventional culture results.
Conclusions
Conjugation of the fluorophore NBD to the antibacterial agent vancomycin gave a fluorescent probe that was able to efficiently label Gram positive clinical pathogens with excellent sensitivity and specificity. The probe showed excellent Gram specificity, and did not label Gram negative pathogens. This green-emitting probe constitutes a reliable, convenient and versatile diagnostic tool. As the “green channel” is the most common fluorescent wavelength found in standard microscope systems, with other wavelengths often limited, the optical properties of Van-Green make it usable on standard equipment, across different laboratories and facilities. Additionally, the solvato-fluorogenicity of the NBD dye switches “on” the emission specifically in the hydrophobic cell membrane of the target pathogen, without causing significant background signal in the usual aqueous environment of clinical samples. The probe therefore showed bright fluorescence, with negligible background, for multiple clinical ocular specimens under practical, wash-free, conditions. This easily synthesized, selective fluorescent probe allows the rapid and accurate diagnosis of Gram-positive microbes that dominate bacterial eye infections, thereby enabling early follow-up treatment.Author contributions
Richa Sharma: data curation, formal analysis, investigation, methodology, validation, visualization, writing – original draft, writing – review & editing. Harinee Rajagopalan: formal analysis, investigation, validation, visualization. Maxime Klausen: data curation, formal analysis, investigation, methodology, visualization, writing – original draft, writing – review & editing. Mani Vimalin Jeyalatha: formal analysis, investigation, validation, visualization. Muhammed Üçüncü: data curation, formal analysis, investigation, methodology, validation, visualization, writing – review & editing. Seshasailam Venkateswaran: conceptualization, supervision, visualization, writing – review & editing. Appakkudal R. Anand: data curation, formal analysis, investigation, methodology, validation, visualization, writing – original draft, writing – review & editing. Mark Bradley: conceptualization, funding acquisition, methodology, project administration, resources, supervision, visualization, writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by Engineering and Physical Sciences Research Council (EPSRC, United Kingdom) (grant number EP/K03197X and EP/R005257/1). We thank the Royal Society (United Kingdom) and the Science and Engineering Research Board (SERB, India) (grant number NIF/R1/192688) for a Newton International Fellowship.Notes and references
- M. Teweldemedhin, H. Gebreyesus and A. H. Atsbaha, et al., Bacterial profile of ocular infections: a systematic review, BMC Ophthalmol., 2017, 17, 212 CrossRef PubMed.
- D. S. J. Ting, C. S. Ho, R. Deshmukh, D. G. Said and H. S. Dua, Eye, 2021, 35, 1084–1101 CrossRef PubMed.
- M. van Oosten, M. Hahn, L. M. Crane, R. G. Pleijhuis, K. P. Francis, J. M. van Dijl and G. M. van Dam, FEMS Microbiol. Rev., 2015, 39, 892–916 CrossRef CAS PubMed.
- L. Miao, W. Liu, Q. Qiao, X. Li and Z. Xu, J. Pharm. Anal., 2020, 10, 444–451 CrossRef PubMed.
- M. R. L. Stone, M. S. Butler, W. Phetsang, M. A. Cooper and M. A. T. Blaskovich, Trends Biotechnol., 2018, 36, 523–536 CrossRef CAS PubMed.
- A. Baibek, M. Üçüncü, E. A. Blackburn, M. Bradley and A. Lilienkampf, Pept. Sci., 2021, 113, e24167 CAS.
- H.-Y. Kwon, X. Liu, E. G. Choi, J. Y. Lee, S.-Y. Choi, J.-Y. Kim, L. Wang, S.-J. Park, B. Kim, Y.-A. Lee, J.-J. Kim, N. Y. Kang and Y.-T. Chang, Angew. Chem., Int. Ed., 2019, 58, 8426–8431 CrossRef CAS PubMed.
- S. Kang, Y. Jung, J. Jung, K.-H. Park and D. Kim, Dyes Pigm., 2021, 185, 108939 CrossRef CAS.
- X. Liu, Z. Yang, W. Xu, Y. Chu, J. Yang, Y. Yan, Y. Hu, Y. Wang and J. Hua, J. Mater. Chem. C, 2019, 7, 12509–12517 RSC.
- P. Bao, C. Li, H. Ou, S. Ji, Y. Chen, J. Gao, X. Yue, J. Shen and D. Ding, Biomater. Sci., 2021, 9, 437–442 RSC.
- S. M. Sayed, K.-F. Xu, H.-R. Jia, F.-F. Yin, L. Ma, X. Zhang, A. Khan, Q. Ma, F.-G. Wu and X. Lu, Anal. Chim. Acta, 2021, 1146, 41–52 CrossRef CAS PubMed.
- T. Gao, X. Cao, J. Dong, Y. Liu, W. Lv, C. Li, X. Feng and W. Zeng, Dyes Pigm., 2017, 143, 436–443 CrossRef CAS.
- M. van Oosten, T. Schäfer, J. A. C. Gazendam, K. Ohlsen, E. Tsompanidou, M. C. de Goffau, H. J. M. Harmsen, L. M. A. Crane, E. Lim, K. P. Francis, L. Cheung, M. Olive, V. Ntziachristos, J. M. van Dijl and G. M. van Dam, Nat. Commun., 2013, 4, 2584 CrossRef PubMed.
- A. R. Akram, S. V. Chankeshwara, E. Scholefield, T. Aslam, N. McDonald, A. Megia-Fernandez, A. Marshall, B. Mills, N. Avlonitis, T. H. Craven, A. M. Smyth, D. S. Collie, C. Gray, N. Hirani, A. T. Hill, J. R. Govan, T. Walsh, C. Haslett, M. Bradley and K. Dhaliwal, Sci. Transl. Med., 2018, 10, eaal0033 CrossRef PubMed.
- A. Baibek, M. Üçüncü, B. Short, G. Ramage, A. Lilienkampf and M. Bradley, Chem. Commun., 2021, 57, 1899–1902 RSC.
- A. Megia-Fernandez, M. Klausen, B. Mills, G. E. Brown, H. McEwan, N. Finlayson, K. Dhaliwal and M. Bradley, Chemosensors, 2021, 9, 117 CrossRef CAS.
- A. Labbé, C. Khammari, B. Dupas, E. Gabison, E. Brasnu, M. Labetoulle and C. Baudouin, Ocul. Surf., 2009, 7, 41–52 CrossRef.
- Y. E. Wang, T. C. Tepelus, L. A. Vickers, E. Baghdasaryan, W. Gui, P. Huang, J. A. Irvine, S. Sadda, H. Y. Hsu and O. L. Lee, Int. Ophthalmol., 2019, 39, 2865–2874 CrossRef PubMed.
- R. D. Cooper, N. J. Snyder, M. J. Zweifel, M. A. Staszak, S. C. Wilkie, T. I. Nicas, D. L. Mullen, T. F. Butler, M. J. Rodriguez, B. E. Huff and R. C. Thompson, J. Antibiot., 1996, 49, 575–581 CrossRef CAS PubMed.
- D. Guan, F. Chen, L. Xiong, F. Tang, Y. Faridoon, N. Qiu, L. Zhang, J. Gong, L. Lan Li and W. Huang, J. Med. Chem., 2018, 61, 286–304 CrossRef CAS PubMed.
- A. Okano, A. Nakayama, A. W. Schammel and D. L. Boger, J. Am. Chem. Soc., 2014, 136(39), 13522–13525 CrossRef CAS PubMed.
- S. J. Kim, K. S. E. Tanaka, E. Dietrich, A. Rafai Far and J. Schaefer, Biochemistry, 2013, 52, 3405–3414 CrossRef CAS PubMed.
- S. N. Leonard and M. J. Rybak, Pharmacotherapy, 2008, 28, 458–468 CrossRef CAS PubMed.
- A. S. Klymchenko and Y. Mely, in Progress in Molecular Biology and Translational Science, ed. M. C. Morris, Academic Press, 2013, vol. 113, pp. 35–58 Search PubMed.
- D. Guan, F. Chen, L. Xiong, F. Tang, F. Faridoon, Y. Qiu, N. Zhang, L. Gong, J. Li, L. Lan and W. Huang, J. Med. Chem., 2017, 61, 286–304 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sd00061j |
| This journal is © The Royal Society of Chemistry 2022 |