Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A novel, proof-of-concept electrochemical impedimetric biosensor based on extracellular matrix protein–adhesin interaction†
Juan
Leva-Bueno
 a,
Ina
Meuskens
a,
Ina
Meuskens
 b,
Dirk
Linke
b,
Dirk
Linke
 c,
Paul A.
Millner
c,
Paul A.
Millner
 d and
Sally A.
Peyman
d and
Sally A.
Peyman
 *ef
*ef
aSchool of Biomedical Sciences, Faculty of Biological Sciences, University of Leeds, UK. E-mail: bsjlev@leeds.ac.uk; Tel: +44 (0) 1133433343
bDepartment of Biosciences, Section for Genetics and Evolutionary Biology, University of Oslo, Oslo, Norway. E-mail: ina.meuskens@ibv.uio.no
cDepartment of Biosciences, Section for Genetics and Evolutionary Biology, University of Oslo, Oslo, Norway. E-mail: dirk.linke@ibv.uio.no; Tel: +47 22857654
dSchool of Biomedical Sciences, Faculty of Biological Sciences, University of Leeds, UK. E-mail: P.A.Millner@leeds.ac.uk; Tel: Phone: +44 (0) 1133433149
eMolecular and Nanoscale Physics Group, School of Physics and Astronomy, University of Leeds, UK. E-mail: S.Peyman@leeds.ac.uk; Tel: +44 (0) 1133433747
fLeeds Institute for Medical Research, St James' Teaching Hospital, Wellcome Trust Brenner Building, Leeds, LS9 7TF, UK
First published on 8th July 2022
Abstract
This work presents a novel perspective for electrochemical biosensors based on the detection of the interaction between extracellular matrix proteins and bacterial adhesins. Two types of impedimetric biosensors using collagen as a bioreceptor were investigated for their ability to detect pathogen adhesion using a well-characterised system of collagen-binding to recombinant E. coli expressing adhesin. First, a collagen–polymer-matrix-based biosensor was shown to detect whole bacteria in buffer media over the range 8 × 105 cfu to 8 × 107 cfu in a 10 μL sample. Second, a step-by-step full optimised biosensor based on direct collagen attachment showed bacterial detection over the range of 8 × 104 cfu to 8 × 107 cfu in a 10 μL sample. Electrochemical impedance spectroscopy and cyclic voltammetry were employed to assess the binding, which was corroborated by fluorescence binding assays. Up to now, a wide range of bioreceptors have been used in biosensor fabrication, including antibodies, oligonucleotides, phages and molecularly imprinted polymers. In this study, we show that extracellular matrix proteins can also be used for whole pathogen recognition by binding to adhesins.
Introduction
Bacterial infections pose an increasing health risk for humans as antibiotic resistance is on the rise.1 As antibiotic resistance in bacteria is increasingly common, selection of an effective antibiotic is crucial and therefore accurate, fast diagnostic tools are of utmost importance.2,3 Biosensors have been developed and shown to be competitive when compared with the current techniques such as ELISA, PCR, SERS and others.4 However, these techniques are time-consuming and expensive and require specialised personnel to carry out measurements. The advantages offered by biosensors are the miniaturisation of the devices and potential for mass-production, providing lower cost with potentially higher specificity and operation by relatively unskilled users.5 This is particularly the case for electrochemical biosensors, where the use of screen-printed electrodes (SPE) has allowed miniaturisation via modern microelectronics without needing classic bulky electrodes.6 Impedimetric biosensors are label-free and the working principle consists mainly on surface immobilisation of a bioreceptor, which directly reports on impedance resulting from pathogen binding.7,8 Due to their sensitivity, biosensors can detect the presence of pathogens in a few minutes, depending on the type of analysis and technique, which is crucial for some post-diagnostic medical interventions.The most common biosensor configuration comprises a bioreceptor molecule attached to a transducer surface. When the bioreceptor–analyte binding occurs, there is a change in the transducer surface which can be monitored by electrochemical, optical or mechanical means. The choice of bioreceptor component is crucial for the biosensor architecture and fabrication route and strongly depends on the affinity and specificity for the target analyte. Many different types of analytes can be detected, from smallest biomolecules to larger entities such as bacteria and viruses. There are different types of bioreceptors employed for pathogen detection. The use of antibodies as bioreceptors relies on the antibody–antigen interaction, and they are the gold standard of bioreceptors for pathogen detection.9 There are many reported immunosensors for pathogen detection. A summary of them can be found in recent reviews either for general10 biosensors or electrochemical sensors.11
Other bioreceptor molecules for pathogen detection are oligonucleotides, DNA or RNA which have been widely used and many examples can be found in the review by Wu et al., 2019.12 Other bioreceptors include proteins, such as lectins;13 phage such as those employed for specific bacterial detection using T4 bacteriophages;14 and sol–gel bacterial imprinted films which have been used for impedimetric detection of E. coli by imprinting the bacteria in organosilica films.15 The concept of using extracellular matrix (ECM) proteins for pathogen detection offers a cheaper option than antibodies and allows detection of groups of pathogens that express adhesins. ECM proteins for pathogen detection in electrochemical biosensors have not been studied extensively although they have been used as bioreceptors for toxin detection.16 In the present article, an ECM protein, collagen, is presented as a bioreceptor for electrochemical pathogen biosensors that target pathogen borne adhesins.
During an infection, interaction between the pathogenic bacterium and the host cell surface is the first and most crucial step.17 For adhesion, bacteria express a multitude of surface proteins able to adhere to specific cell surface factors or, more generally, the extracellular matrix.18 Some of these adhesins are specific for adhesion to only one host cell factor while other adhesins can bind several host cell factors.17,19 One of these adhesins is YadA, the major adhesin A of Yersinia enterocolitica. YadA belongs to the group of autotransporter adhesins (type V secretion systems)20–22 and is a trimeric autotransporter adhesin23 that is presented on the bacterial surface. It has been shown to bind to a variety of extracellular matrix proteins, including collagen, fibronectin and laminin.24 In this article we show a proof-of-concept electrochemical biosensor based on ECM protein–adhesin interaction built upon miniaturised disposable screen-printed gold electrodes (SGPEs). In this research we concentrate on the interaction between YadA and collagen,20,25,26 using collagen as a novel receptor for detection of YadA-expressing bacteria (Fig. 1a).
The characterisation of the biosensors was carried out by cyclic voltammetry (CV) and electrochemical impendence spectroscopy (EIS) and binding monitored by EIS, which is supported by fluorescence assay.
Experimental
Materials
Screen-printed gold electrodes (BVT-AC1.W1.RS.Dw2) from BVT technologies comprised gold working and counter electrodes and an Ag/AgCl reference electrode in a 3-electrode chip. Palmsens4 potentiostat, galvanostat and frequency response analyser (FRA) and PSTrace (5.8) software were from Palmsens BV. A channel multiplexer (MUX8-R2) and an electrode adapter from Palmsens was employed to perform all replicate experiments simultaneously. Na2HPO4 and Na3PO4; octopamine hydrochloride; tryptone, agar and yeast used to make the microbial culture and broth; calf skin collagen; goat HRP-conjugated anti-rabbit; BSA, casein from powder from bovine milk and TRIS were all purchased from Sigma Aldrich. NaCl was purchased from Fisher Chemical. K3Fe(CN)6 and K4Fe(CN)6·3H2O were purchased from VWR. Rabbit anti-collagen type I from bovine and human was purchased from 2BScientific. 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), enhanced chemiluminescence Pierce (ECL) reagent and sulfo-NHS (N-hydroxysulfosuccinimide) and 1 M MES buffer pH 7 were purchased from Thermo Fisher Scientific. Chemiluminescence images were analysed by G:BOX Gel Imaging System (Syngene Ltd.; Cambridge, UK).Cloning —O:8 FL
For the pASK-Iba4c based YadA full length constructs, Gibson cloning was performed. In short: YadA O:8 was PCR-amplified using Q5 polymerase from the plasmid used in ref. 27 using the following primers: 5′-CGACAAAAATCTAGATAACGAGGGCAAAAAatgactaaagattttaagatcagtgt-3′ and 5′-GCCATTTTTCACTTCACAGGTCAAGCTTAGttaccactcgatattaaatgatgca-3′. These primers have overlaps to pASK-Iba4c for later Gibson annealing. pASK-Iba4c was linearized in a PCR reaction using Q5 polymerase (New England Biolabs) and the following primers: 5′-CTAAGCTTGACCTGTGAAGT-3′ and 5′-TTTTTGCCCTCGTTATCTAGATT-3′. Both the plasmid and the insert were PCR purified and DpnI (New England Biolabs) digested for 30 min at 37 °C in Cutsmart buffer (New England Biolabs). DpnI was afterwards inactivated by incubation at 80 °C for 10 min. The Gibson assembly was performed with a self-made Gibson mix for 20 min at 50 °C with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 concentration ratio of plasmid to insert. The assembly mix was transformed into TSS competent E. coli Top10 and incubated in SOC medium for 1 h at 37 °C. The transformed E. coli were plated on LB agar plates with 12.5 μg mL−1 chloramphenicol. From this plate, two colonies were chosen, and 5 mL overnight cultures were prepared with plasmids isolated the next day. After Sanger sequencing, the correct plasmid was transformed into fluorescent E. coli Top10 (1) and used for further experiments.
5 concentration ratio of plasmid to insert. The assembly mix was transformed into TSS competent E. coli Top10 and incubated in SOC medium for 1 h at 37 °C. The transformed E. coli were plated on LB agar plates with 12.5 μg mL−1 chloramphenicol. From this plate, two colonies were chosen, and 5 mL overnight cultures were prepared with plasmids isolated the next day. After Sanger sequencing, the correct plasmid was transformed into fluorescent E. coli Top10 (1) and used for further experiments.
Growth of E coli and induction of YadA expression
E. coli Top10 pARA_sfGFP (ref. 27) were transformed with vector pIBA4c_YadA_FL. The transformants were then streaked out on a plate supplemented with 25 μg mL−1 chloramphenicol and a single colony was picked for further experiments. Overnight cultures were grown in 5 mL LB medium supplemented with 25 μg mL−1 chloramphenicol. The next morning a subculture was prepared in LB medium supplemented with 25 μg mL−1 chloramphenicol and grown at 37 °C in a shaking incubator to an OD600 of 0.5. After reaching the required OD600, YadA surface expression was induced by addition of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10.000 anhydrotetracycline hydrochloride (Abcam, ab145350) (AHTC, 2 mg mL−1 stock). Expression was allowed for another 2 h at 37 °C in the shaking incubator. In case of the negative controls, AHTC was omitted. Proper YadA expression was determined by visual inspection of the bacterial aggregation behaviour indicative of YadA surface presentation.28 After expression, the OD600 was measured again and cfu calculated. The bacteria were diluted to the required cfu in PBS pH 7.4 and immediately used for measurements.
10.000 anhydrotetracycline hydrochloride (Abcam, ab145350) (AHTC, 2 mg mL−1 stock). Expression was allowed for another 2 h at 37 °C in the shaking incubator. In case of the negative controls, AHTC was omitted. Proper YadA expression was determined by visual inspection of the bacterial aggregation behaviour indicative of YadA surface presentation.28 After expression, the OD600 was measured again and cfu calculated. The bacteria were diluted to the required cfu in PBS pH 7.4 and immediately used for measurements.
Preparation of collagen solution
Collagen type I bovine was purchased from Thermo Fisher Scientific (A1064401). Collagen was resuspended to a final stock concentration of 1 mg mL−1 in 10 mM acetic acid.Fluorescence-based detection of YadA-expressing E. coli
The bacteria were grown as described above. To also induce sfGFP expression from the genome, 0.02% w/v arabinose was added at every step of the growth process. Wells of a 96-well clear plate were coated with 100 μL of a 10 μg mL−1 collagen solution. As negative controls, PBS was added to the wells. The plates were incubated at RT for 1 h. After that, the solutions were discarded, and the wells were washed three times with 200 μL PBS and afterwards blocked with 3% w/v BSA in PBS for 1 h at RT. After YadA expression was allowed for 3 h, the OD600 was set to 1.0 for both the uninduced and the induced culture and serial 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilutions were performed in 3% w/v BSA in PBS. 100 μL of each dilution was loaded into the plate. Binding of bacteria was allowed for 1 h at RT under static conditions. After that the wells were washed 3 times with PBS and fluorescence was measured. The measurements were performed in a Synergy H1 plate reader with excitation at 455 nm and an emission 533 nm at gain setting of 100.
10 dilutions were performed in 3% w/v BSA in PBS. 100 μL of each dilution was loaded into the plate. Binding of bacteria was allowed for 1 h at RT under static conditions. After that the wells were washed 3 times with PBS and fluorescence was measured. The measurements were performed in a Synergy H1 plate reader with excitation at 455 nm and an emission 533 nm at gain setting of 100.
Collagen–POc matrix-based biosensor fabrication
SPGEs (BVT-AC1.W1.RS.Dw2) from BVT technologies were employed for biosensor fabrication. Prior to electrochemical measurements, the electrodes were pre-treated in 97% v/v ethanol for 30 min, rinsed with deionised water and dried with N2. Bovine collagen type I was attached onto the electrode surface as a bioreceptor using a protocol provided by ELISHA Systems Ltd: 25 μL of octopamine in 10 mM PB pH 7.2 was mixed with a solution of 200 μg mL−1 collagen at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and kept in ice, yielding a final concentration of 2.5 mM octopamine and 100 μg mL−1. The final solution was spread across the working electrode and electropolymerised for 2 cycles at a scan rate of 100 mV s−1 from +0.0 V to +1.6 V, to entrap the collagen. Finally, once the biosensor was fully constructed and optimised, baseline EIS scans were recorded at 0 V over a frequency range of 5 kHz to 0.1 Hz, with a modulation voltage of +10 mV recording three measurements to ensure signal stabilisation. Then, 10 μL of E. coli from stock solutions containing 8 × 102 cfu to 8 × 107 cfu was applied to the working electrodes which were incubated for 30 min. The electrodes were then washed with PBS prior to EIS and CV. This procedure was also carried out for the uninduced YadA E. coli control. The blank was conducted with the same procedure as per the analyte and the control but substituting PBS buffer for the bacterial sample (Scheme 1).
1 and kept in ice, yielding a final concentration of 2.5 mM octopamine and 100 μg mL−1. The final solution was spread across the working electrode and electropolymerised for 2 cycles at a scan rate of 100 mV s−1 from +0.0 V to +1.6 V, to entrap the collagen. Finally, once the biosensor was fully constructed and optimised, baseline EIS scans were recorded at 0 V over a frequency range of 5 kHz to 0.1 Hz, with a modulation voltage of +10 mV recording three measurements to ensure signal stabilisation. Then, 10 μL of E. coli from stock solutions containing 8 × 102 cfu to 8 × 107 cfu was applied to the working electrodes which were incubated for 30 min. The electrodes were then washed with PBS prior to EIS and CV. This procedure was also carried out for the uninduced YadA E. coli control. The blank was conducted with the same procedure as per the analyte and the control but substituting PBS buffer for the bacterial sample (Scheme 1).
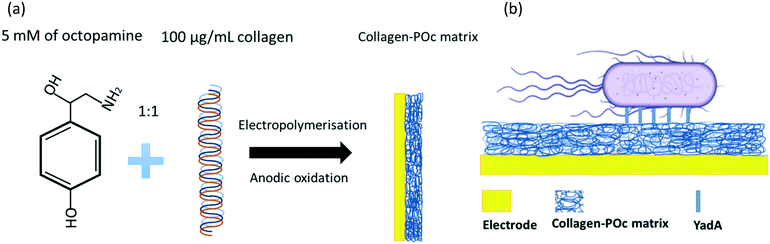 | ||
| Scheme 1 (a) Schematic representation of step-by-step fabrication of collagen–POc matrix biosensor and (b) final biosensor architecture. | ||
Collagen direct attachment biosensor fabrication
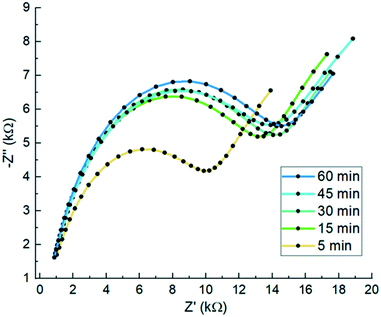
SPGEs (BVT-AC1.W1.RS.Dw2) from BVT technologies were employed for biosensor fabrication. Prior to electrochemical measurements, the electrodes were pre-treated in 97% v/v ethanol for 30 min, rinsed with deionised water and dried with N2. For the collagen direct attachment biosensor, a range of different POc concentrations (1, 2.5, 5, 10, 25, 100, and 250 mM) of octopamine were dissolved in 10 mM PB pH 7.2 and spread across the working electrode and electropolymerised for 2 cycles at a scan rate of 100 mV s−1 from +0.0 V to +1.6 V. Then, different collagen concentrations (5, 10, 50, 100, 500, and 1000 μg mL−1) were dissolved in 50 mM MES pH 5.5. To the last, EDC and sulfo-NHS were added to the collagen solution to a final concentration of 2 mM EDC and 6 mM sulfo-NHS and immediately spread over the working electrode to react with the amine reactive groups from the POc. After 2 h of incubation, the electrodes were washed 3 times with 50 mM MES pH 5.5 and a final wash with TRIS buffer in order to block any carboxyl reactive groups. Following biosensor optimisation, samples of E. coli induced for YadA expression of 8 × 106 cfu in 10 μL sample were tested for different incubation times (5, 15, 30, 45, and 60 min) at RT. For the best optimised biosensor, blocking agents to avoid non-specific binding were assessed. BSA at 1.5 mg mL−1 or 1.5 mg mL−1 casein in 10 mM PBS were incubated for 1 h over the electrodes at RT to block non-specific binding sites on the biosensor. After that, electrodes were washed with PBS buffer plus 0.05% (v/v) Tween-20. Then, fully constructed biosensors blocked either with 1.5 mg mL−1 BSA and 1.5 mg mL−1 casein or non-blocked were tested with 10 μL of sample containing 8 × 106 cfu for both induced and uninduced E. coli systems. Finally, once the biosensor was fully constructed and optimised, baseline EIS scans were recorded at 0 V over a frequency range of 5 kHz to 0.1 Hz, with a modulation voltage of +10 mV recording three measurements to ensure signal stabilisation. Then, 10 μL of E. coli from stock solutions containing 8 × 102 cfu to 8 × 107 cfu was applied to the working electrodes which were incubated for 15 min. The electrodes were then washed with PBS prior to EIS and CV. This procedure was also carried out for the uninduced YadA E. coli control. The blank was conducted with the same procedure as per the analyte and the control but substituting PBS buffer for the bacterial sample (Scheme 2).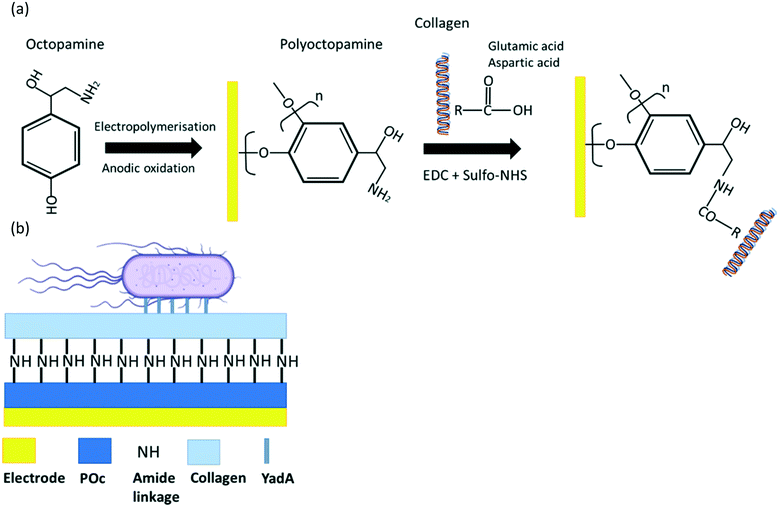 | ||
| Scheme 2 (a) Schematic representation of step-by-step fabrication of collagen direct attachment biosensor and (b) final biosensor architecture. | ||
Chemiluminescence analysis
Midland blotting technique29 was performed to validate the presence of collagen in the collagen direct attachment biosensor. On-sensor chemiluminescence measurement started with incubation of 1 mg mL−1 rabbit anti-collagen type I from bovine and human over the electrode surface. After 1 h incubating in a moist chamber at RT, a washing step with PBS for 5 min was carried out. After that, the electrodes were then incubated with (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 in PBS) goat anti-rabbit HRP secondary antibodies for 1 h at RT in a moist chamber. Then the electrodes were washed for 3 times with PBS, for 5 min each time. The second wash with PBS included 0.1% (v/v) Tween-20 to remove non-specific bound material. Finally, ECL reagent was added onto the electrodes to detect the chemiluminescence after 1 min via a G:BOX gel Imaging System (Syngene Ltd.; Cambridge, UK).
1000 in PBS) goat anti-rabbit HRP secondary antibodies for 1 h at RT in a moist chamber. Then the electrodes were washed for 3 times with PBS, for 5 min each time. The second wash with PBS included 0.1% (v/v) Tween-20 to remove non-specific bound material. Finally, ECL reagent was added onto the electrodes to detect the chemiluminescence after 1 min via a G:BOX gel Imaging System (Syngene Ltd.; Cambridge, UK).
Electrochemical characterisation and data treatment
The electrodes were subjected to EIS and CV characterisation before and after each step of biosensor construction.EIS was performed by spreading the electrodes with 10 mM [Fe(CN)6]3−/4− in 10 mM PBS, pH 7.2, coupled to a potentiostat. EIS was recorded at 0 V over a frequency range of 5 kHz to 0.1 Hz, with a modulation voltage of +10 mV. Three EIS measurements were taken for signal stabilisation at any step of collagen–POc matrix biosensor. Two EIS measurements were taken for signal stabilisation at any functionalisation step in the collagen direct attachment biosensor and 3 EIS measurements before testing the analyte. All EIS analyses were performed in a three-cell system using a Palmsens4 potentiostat, galvanostat and FRA (Palmsens B.V., The Netherlands). PSTrace (5.8) was used for CV and EIS recording. EIS electrochemical circuit fit was assessed with Metrohm Autolab Nova 2.1.4. Data was plotted and analysed in statistical software OriginPro (2019b) and all experiments were replicated with n ≥ 6.
Frequency response from EIS measurements can be observed in a Nyquist plot which was fit to the Randles' equivalent circuit, from which the charge transfer resistance Rct was obtained. Charge-transfer resistance (Rct) was normalised and expressed in Rct (%) with the eqn (1):
| Change in Rct (%) = (Rct analyte − Rct zero)/Rct zero·100 | (1) |
For CV analysis, the potential was swept between −0.6 V to +0.6 V following for 10 scans at a scan rate of 100 mV s−1.
Results & discussion
Collagen-matrix biosensor
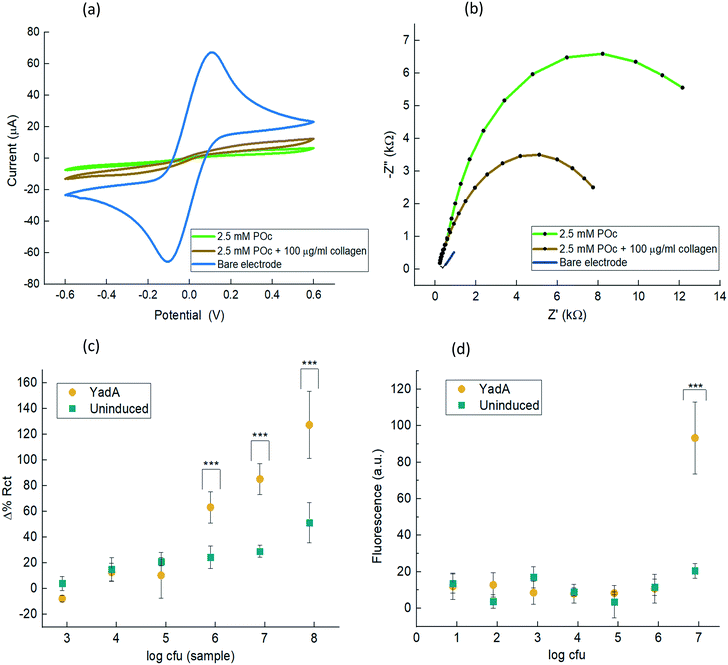
From the voltammograms, the bare electrode showed the two peaks of oxidation and reduction current at clear oxidative and reductive potentials, thus indicating proper electron flow through the metallic surface and presenting a clearly reversible redox reaction. On the other hand, the CV profile shown for POc indicates either oxidative and reductive peak currents of +5.32 μA and −4.17 μA, at +0.3 V and −0.20 V respectively. This shows that once POc is electropolymerised, the redox pair cannot adequately reach the surface to perform exchange of electrons indicating the presence of a non-conducting surface. The collagen–POc matrix voltammogram shows that the observed electrochemical properties are in line with the composition of the interface: the collagen-POc composite presents a more open nanostructure than POc by itself. Consequently, it is easier for the electron carriers to reach the electrode surface and thus presents more conductive properties. The values of the currents achieved were +9.61 μA and −9.93 μA for the oxidative and reductive peaks. This behaviour can be also seen from the electropolymerisation profiles in which less current is observed for the collagen-POc matrix in the oxidative peak compared to only POc (Fig. S1a and b†) (Fig. 1a).
Further assessment through EIS (Fig. 1b) was carried out and the results obtained were in line with what was observed with the CV. The curve obtained for POc was comparatively large when compared to the other systems, as is expected given the insulating effect of the non-conducting polymer. On the other hand, the collagen–POc matrix showed a substantial decrease in impedance compared to POc alone.
The strategy to assess the selectivity of the biosensor consisted of analysing the same range of bacterial samples but for E. coli uninduced for YadA expression. Comparison between the analyte and the control was assessed through changes in Rct, which is represented as Δ% Rct after normalisation, (Fig. 1c). To obtain the Rct values, each Nyquist plot was fitted into a Randle's equivalent circuit model. The bacterial binding of YadA-expressing E. coli appeared significantly different (p ≤ 0.001) from 8 × 105 cfu to 8 × 107 cfu. For this range of cfu, the response to YadA-expressing E. coli was clearly larger compared to their respective uninduced E. coli samples as shown in Fig. 1c. The lower cfu detected that were different from the uninduced control is found at 8 × 105 cfu with a Δ% Rct of 63.20 ± 12.18 for induced and 24.20 ± 8.78 for uninduced samples. Lower cfu samples showed similar values of Δ% Rct between analyte and uninduced samples. In comparison, the fluorescence assay (Fig. 1d) showed a detection limit of 8 × 106 cfu (p ≤ 0.001). More dilute samples did not show significant differences between bacteria expressing YadA and uninduced bacteria and can thus be explained by non-specific binding to the well material. The difference in LOD between the two detection techniques was due to the fact that EIS provided more sensitivity and linear detection compared to the fluorescence assay, which provided a semiquantitative detection.
Collagen direct attachment biosensor
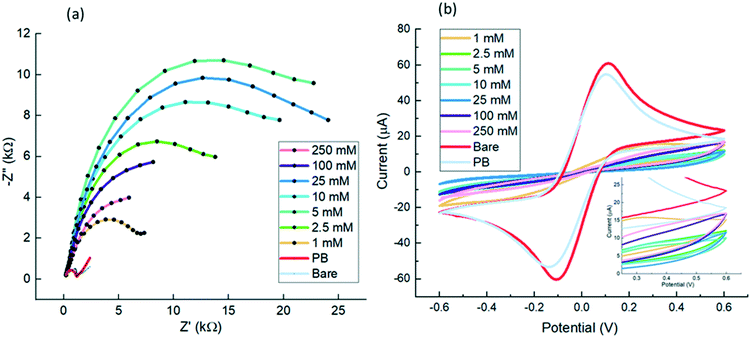
First, 1 mM to 250 mM octopamine in 10 mM PB pH 7.2 was assessed. Also 10 mM PB pH 7.2 alone was valuated to observe any potential component to take into consideration from the buffer. In addition, electrochemical characterisation of the bare electrode was also carried for the different POc concentrations, electropolymerisation was successfully achieved as observed from the oxidation peaks on the first cycle and the absence of peak in the second, indicating an irreversible oxidation in all cases (Fig. S2a and b†). Once each electrode was passivated with the corresponding POc concentration, EIS measurements were performed to evaluate impedance. As previously commented, the Nyquist semicircle diameter increased or decreased in accordance to how insulating or conductive the surface was. As observed in Fig. 2a the Nyquist plots diameter increased with polymer concentration until 5 mM POc. Then, for higher concentrations, the Nyquist semicircle size started decreasing us indicating a more conductive surface at higher concentrations. Therefore, EIS results suggest that the more efficient surface coating was at 5 mM POc.
In addition, CV was performed to corroborate the results obtained by EIS (Fig. 2b). Similarly, 5 mM POc appeared to be one of the most insulating with higher concentrations showed more conductive profiles, which is in line with the EIS analysis. Both EIS and CV results are in accordance with oxidation current observed in the electropolymerisation profiles (Fig. S2†).
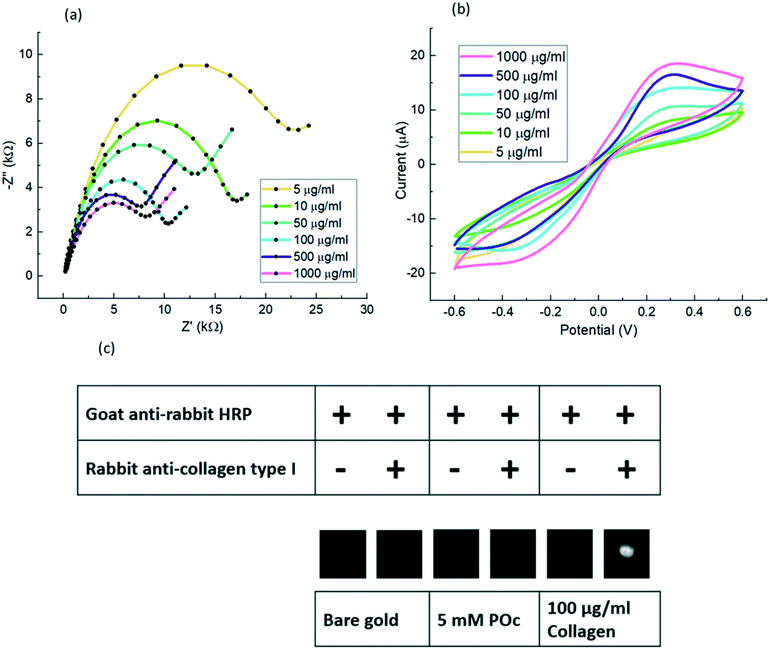 | ||
| Fig. 4 Electrochemical characterisation of a different range of collagen concentrations and validation of collagen direct attachment to POc through on-sensor blotting: direct immobilisation of 5–1000 μg mL−1 of collagen to 5 mM POc via EDC/sulfo-NHS were tested and assessed by EIS and CV and finally validated with on-sensor blotting chemiluminescence. (a), All Nyquist profiles derived from EIS measurements were in 10 mM [Fe(CN)6]3−/4− in 10 mM PBS, pH 7.2. EIS was recorded at 0 V over a frequency range of +5 kHz to +0.1 Hz, with a modulation voltage of +10 mV; (b), CVs were cycled from −0.6 V to +0.6 at a scan rate of 100 mV s−1 in 10 mM [Fe(CN)6]3−/4− in 10 mM PBS, pH 7.2; (c), Midland blotting29 was carried out to validate the collagen binding to POc. The sample was tested with 100 μg mL−1 collagen. Samples were prepared by adding primary rabbit anti-collagen type I, then addition of secondary goat anti-rabbit HRP antibodies as detailed in section Chemiluminescence analysis. Control samples comprised a bare gold surface, coated with 5 mM POc, or omission of primary antibodies as negative controls. | ||
To validate the collagen direct attachment to POc through EDC/sulfo-NHS, one more experiment following the Midland blotting29 technique was carried out as detailed in section Biosensor fabrication. As shown in Fig. 4c the presence of attached collagen was determined by on-sensor chemiluminescence in which two control surfaces were tested.
As observed in Fig. 6, BSA and casein blocked surfaces showed bacterial binding (54.89 ± 12.93 and 41.31 ± 15.71 respectively) as substantial as the non-blocked surface (61.61 ± 7.80). However, the blocked biosensor variability was substantially larger than the unblocked one. When control samples were tested with uninduced bacteria, it was observed that the BSA and casein blocked surfaces showed less non-specific binding, 11.45 ± 10.25 and 9.87 ± 13.31 respectively, compared to the non-blocked surface, 26.12 ± 5.95. Nonetheless, the variability of the control samples were substantially larger than the samples without blocking agent and therefore a blocking step was not deemed essential for this work.
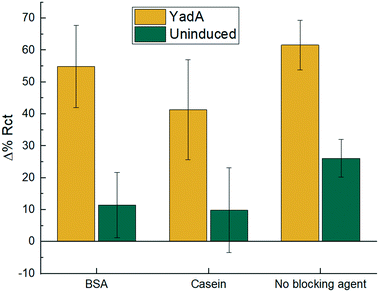 | ||
| Fig. 6 Electrochemical binding measurements for blocked and non-blocked biosensors. 1.5 mg mL−1 BSA, 1.5 mg mL−1 casein and non-blocked biosensor were tested with 8 × 106 cfu in 10 μL sample for E. coli expressing YadA or uninduced E. coli. Incubation was for 15 min and Δ% Rct is calculated with eqn (1). Data are mean ± SD (n = 6). | ||
From the bar chart (Fig. 7), it can be observed that the two lowest bacterial concentrations of 8 × 102 and 8 × 103 cfu were not detected. From 8 × 104 cfu with Δ% Rct 38.98 ± 3.04 for YadA induced and 15.20 ± 4.95 for uninduced, to larger cfu samples, there was a differentiated analyte response proportional to concentration. Sensors challenged with YadA induced bacteria (Fig. 7) showed a steady increase in % ΔRct. However, higher bacterial concentrations gave greater variability. The trend shown in the bar graphs appear to show a plateau in response for uninduced samples. The biosensors also tested bacteria over the polymer in absence of collagen. No significant binding was observed (Fig. S3†).
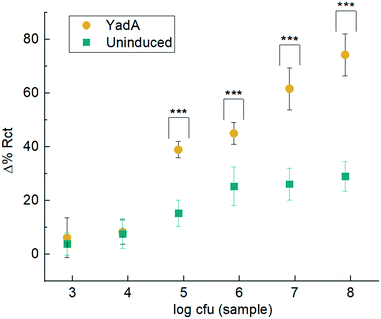 | ||
| Fig. 7 Electrochemical binding measurements. Δ% Rct before and after analyte incubation with uninduced E. coli or E. coli expressing YadA for a range of bacteria from 8 × 102 to 8 × 107 cfu in 10 μL. Incubation was for 15 min and Δ% Rct is calculated in accordance with eqn (1). Data are mean ± SD (n ≥ 6). Statistical analysis was performed using ANOVA and data with p < 0.001 are indicated by ***. | ||
Conclusions
In this study, we present collagen as a bioreceptor for a proof-of-concept impedance-based biosensor for rapid bacterial measurement. The first system, based on a non-optimised polymer-collagen matrix bioreceptor, can detect 8 × 105 cfu to 8 × 107 cfu in 10 μL sample of YadA expressing E. coli. The second system, based on an optimised direct attachment of the collagen, can detect 8 × 104 cfu to 8 × 107 cfu in 10 μL sample of YadA expressing E. coli. The aim of this research was to demonstrate that the ECM protein–adhesin interaction can be utilized for detection of whole pathogens using an electrochemical biosensor. In addition, ECM proteins utilised as bioreceptors could offer pan-specificity, i.e. the biosensor could detect a group of organisms rather than a specific organism. This first-step determination could be a useful tool for clinicians who need to perform a rapid initial ‘sample in, answer out’ diagnostic test in order to begin treatment, before the lengthier investigation of identifying exact species. In contrast, antibody-based biosensors may be too specific for a certain pathogen and may miss the detection of other species. A balance of the specificity of these models will provide more accurate diagnostic results for clinicians.Analogous to YadA used as a model surface adhesin in this study, all pathogenic bacteria have adhesins targeting host cell structures.17,18 As adhesins generally show specificities for different targets, this could in the future be harnessed for diagnostic purposes as previously discussed. A potential device could use panels of biosensors coated with different ECM molecules and host surface structures to identify pathogenic bacteria based on their adhesion patterns. Other common electrochemical biosensors, for instance, can detect E. coli at a concentration of 1.4 × 103 cfu mL−1 in 25 μL sample32 or other bacteria species such as S. typhimurium at a concentration of 103 cfu mL−1 in 1 mL,33 or S. pyogenes detecting 100 cells in 10 μL.8 While this method will likely not replace more accurate methods like mass spectrometry, a biosensor-based approach could function as an initial, rapid point- of-care device for what type of infection is present. In contrast to other methods, biosensor-based methods do not require growth of the pathogen on plates.34,35 While slightly less accurate, biosensors coated with ECM molecules rather than antibodies or enzymes would be less expensive in comparison.
Author contributions
J. Leva-Bueno: investigation, writing – original draft, visualisation. I. Meuskens: validation, writing – original draft, visualisation. D. Linke: conceptualisation, writing – review & editing, supervision. P. A. Millner: conceptualisation, writing – review & editing, supervision, project administration. S. A. Peyman: conceptualisation, writing – review & editing, supervision, project administration.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Funding: this project is funded by the European Union's Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie grant agreement No. 765042. D. Linke also receives funding from the Center for Digital Life, grant 294605 (Research Council of Norway). The data repository for this publication is available at https://doi.org/10.5518/1165.Notes and references
- H. W. Boucher, et al., Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America, Clin. Infect. Dis., 2009, 48, 1–12 CrossRef PubMed.
- M. Gajdács and F. Albericio, Antibiotic resistance: from the bench to patients, Antibiotics, 2019, 8, 8–11 CrossRef PubMed.
- A. Cassini, et al., Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis, Lancet Infect. Dis., 2019, 19, 56–66 CrossRef PubMed.
- C. K. Joung, et al., Ultra-sensitive detection of pathogenic microorganism using surface-engineered impedimetric immunosensor, Sens. Actuators, B, 2012, 161, 824–831 CrossRef CAS.
- J. Leva-Bueno, S. A. Peyman and P. A. Millner, A review on impedimetric immunosensors for pathogen and biomarker detection, Med. Microbiol. Immunol., 2020, 209, 343–362 CrossRef CAS PubMed.
- Z. Taleat, A. Khoshroo and M. Mazloum-Ardakani, Screen-printed electrodes for biosensing: A review (2008-2013), Microchim. Acta, 2014, 181, 865–891 CrossRef CAS.
- M. Xu, R. Wang and Y. Li, Electrochemical biosensors for rapid detection of Escherichia coli O157:H7, Talanta, 2017, 162, 511–522 CrossRef CAS PubMed.
- A. Ahmed, J. V. Rushworth, J. D. Wright and P. A. Millner, Novel impedimetric immunosensor for detection of pathogenic bacteria streptococcus pyogenes in human saliva, Anal. Chem., 2013, 85, 12118–12125 CrossRef CAS PubMed.
- J. R. Birch and A. J. Racher, Antibody production, Adv. Drug Delivery Rev., 2006, 58, 671–685 CrossRef CAS PubMed.
- Y. Chen, et al., Recent advances in rapid pathogen detection method based on biosensors, Eur. J. Clin. Microbiol. Infect. Dis., 2018, 37, 1021–1037 CrossRef CAS PubMed.
- E. Cesewski and B. N. Johnson, Electrochemical biosensors for pathogen detection, Biosens. Bioelectron., 2020, 159, 112214 CrossRef CAS PubMed.
- Q. Wu, Y. Zhang, Q. Yang, N. Yuan and W. Zhang, Review of electrochemical DNA biosensors for detecting food borne pathogens, Sensors, 2019, 19(22), 4916 CrossRef CAS PubMed.
- F. Xi, J. Gao, J. Wang and Z. Wang, Discrimination and detection of bacteria with a label-free impedimetric biosensor based on self-assembled lectin monolayer, J. Electroanal. Chem., 2011, 656, 252–257 CrossRef CAS.
- A. Shabani, et al., Bacteriophage-modified microarrays for the direct impedimetric detection of bacteria, Anal. Chem., 2008, 80, 9475–9482 CrossRef CAS PubMed.
- H. Jafari, et al., Entrapment of uropathogenic E. coli cells into ultra-thin sol-gel matrices on gold thin films: A low cost alternative for impedimetric bacteria sensing, Biosens. Bioelectron., 2019, 124–125, 161–166 CrossRef CAS PubMed.
- S. Xia, et al., Development of a simple and convenient cell-based electrochemical biosensor for evaluating the individual and combined toxicity of DON, ZEN, and AFB1, Biosens. Bioelectron., 2017, 97, 345–351 CrossRef CAS PubMed.
- I. Meuskens, A. Saragliadis, J. C. Leo and D. Linke, Type V secretion systems: An overview of passenger domain functions, Front. Microbiol., 2019, 10, 1–19 CrossRef PubMed.
- B. Westerlund and T. K. Korhonen, Bacterial proteins binding to the mammalian extracellular matrix, Mol. Microbiol., 1993, 9, 687–694 CrossRef CAS PubMed.
- J. Pizarro-Cerdá and P. Cossart, Bacterial adhesion and entry into host cells, Cell, 2006, 124, 715–727 CrossRef PubMed.
- J. C. Leo, H. Elovaara, B. Brodsky, M. Skurnik and A. Goldman, The Yersinia adhesin YadA binds to a collagenous triple-helical conformation but without sequence specificity, Protein Eng., Des. Sel., 2008, 21, 475–484 CrossRef CAS PubMed.
- M. Mühlenkamp, P. Oberhettinger, J. C. Leo, D. Linke and M. S. Schütz, Yersinia adhesin A (YadA) - Beauty & beast, Int. J. Med. Microbiol., 2015, 305, 252–258 CrossRef PubMed.
- H. Nummelin, Y. El Tahir, P. Ollikka, M. Skurnik and A. Goldman, Expression, purification and crystallization of a collagen-binding fragment of Yersinia adhesin YadA, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2002, 58, 1042–1044 CrossRef PubMed.
- D. Linke, T. Riess, I. B. Autenrieth, A. Lupas and V. A. J. Kempf, Trimeric autotransporter adhesins: variable structure, common function, Trends Microbiol., 2006, 14, 264–270 CrossRef CAS PubMed.
- Y. El Tahir and M. Skurnik, YadA, the multifaceted Yersinia adhesin, Int. J. Med. Microbiol., 2001, 291, 209–218 CrossRef CAS PubMed.
- J. C. Leo, et al., First analysis of a bacterial collagen-binding protein with collagen toolkits: Promiscuous binding of YadA to collagens may explain how YadA interferes with host processes, Infect. Immun., 2010, 78, 3226–3236 CrossRef CAS PubMed.
- H. Nummelin, et al., The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel β-roll, EMBO J., 2004, 23, 701–711 CrossRef CAS PubMed.
- A. Saragliadis and D. Linke, Assay development for the discovery of small-molecule inhibitors of YadA adhesion to collagen, Cell Surf., 2019, 5, 100025 CrossRef CAS PubMed.
- U. Grosskinsky, et al., A conserved glycine residue of trimeric autotransporter domains plays a key role in Yersinia adhesin A autotransport, J. Bacteriol., 2007, 189, 9011–9019 CrossRef CAS PubMed.
- J. V. Rushworth, A. Ahmed and P. A. Millner, Midland Blotting: A Rapid, Semi-Quantitative Method for Biosensor Surface Characterization, J. Biosens. Bioelectron., 2013, 4, 146 Search PubMed.
- S. H. Shamsuddin, et al., Reagentless Affimer- and antibody-based impedimetric biosensors for CEA-detection using a novel non-conducting polymer, Biosens. Bioelectron., 2021, 178, 113013 CrossRef CAS PubMed.
- G. T. Hermanson, Bioconjugate Techniques, Elsevier, 1996 Search PubMed.
- R. Wang, et al., A label-free impedance immunosensor using screen-printed interdigitated electrodes and magnetic nanobeads for the detection of E. coli O157:H7, Biosensors, 2015, 5, 791–803 CrossRef CAS PubMed.
- Z. Farka, et al., Rapid Immunosensing of Salmonella Typhimurium Using Electrochemical Impedance Spectroscopy: the Effect of Sample Treatment, Electroanalysis, 2016, 28, 1803–1809 CrossRef CAS.
- A. Croxatto, G. Prod'hom and G. Greub, Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology, FEMS Microbiol. Rev., 2012, 36, 380–407 CrossRef CAS PubMed.
- B. W. Senior, Media and tests to simplify the recognition and identification of members of the Proteeae, J. Med. Microbiol., 1997, 46, 39–44 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sd00075j |
| This journal is © The Royal Society of Chemistry 2022 |