Open Access Article
Open Access ArticleTurn-on fluorescent sensors for Cu-rich amyloid β peptide aggregates†
Yiran
Huang
 ,
Liang
Sun
,
Liang
Sun
 and
Liviu M.
Mirica
and
Liviu M.
Mirica
 *
*
Department of Chemistry, University of Illinois at Urbana-Champaign, 600 S. Mathews Avenue, Urbana, IL 61801, USA. E-mail: mirica@illinois.edu
First published on 9th May 2022
Abstract
Protein misfolding and metal dishomeostasis are two key pathological factors of Alzheimer's disease. Previous studies have shown that Cu-mediated amyloid β (Aβ) peptide aggregation leads to the formation of neurotoxic Aβ oligomers. Herein, we report a series of picolinic acid-based Cu-activatable sensors, which can be used for the fluorescence imaging of Cu-rich Aβ aggregates.
The formation of extracellular amyloid plaques containing the amyloid β (Aβ) peptide is one of the pathological hallmarks of the brains of Alzheimer's disease (AD) patients.1,2 Remarkably high concentrations (up to 20–50 μM) of Cu and Zn have been found within these amyloid plaques,3,4 and several studies have explored the interactions of metals with monomeric Aβ peptides and their correlation with amyloid plaques and reactive oxygen species formation.5–11 These studies show that Cu can slow down the aggregation of Aβ42 and reduce fibrilization to a large extent, and it is considered that Cu(II) ions can stabilize the Aβ42 oligomer species.12–14 In this regard, novel molecules that can modulate the interaction of Cu ions with the soluble Aβ42 species and alleviate their neurotoxicity may serve as novel therapeutic agents for AD.15–20
In addition to the development of new therapeutic agents, it is also highly important to detect the Cu-containing Aβ species in AD. A number of fluorescent sensors have been developed to probe biological copper fluxes.21–25 These reporters can achieve high selectivity and signal-to-noise responses for Cu ion imaging, from cellular to tissue to whole animal settings.21,26–28 However, only a few of these probes were utilized in detecting labile copper pools in AD,29,30 even though countless studies have shown the copper homeostasis is dramatically disrupted. In this regard, to understand and probe the Cu-mediated Aβ aggregation process, it is significantly crucial to develop Cu–Aβ specific probes. Herein, we synthesized a series of Cu-based activatable sensors aimed to potentially detect the Cu–Aβ species in vitro and ex vivo. For the molecular design, we drew inspiration from Chan et al. who have reported the use of a 2-picolinic ester fragment that readily hydrolysed in the presence of Cu(II) but not with other divalent metal ions.25 By linking the picolinic ester moiety with the Aβ-binding molecular fragments, Cu(II) ions can rapidly catalyze the hydrolysis of the ester bond to generate the highly fluorescent Aβ binding molecules in vitro (Fig. 1). Interestingly, the Cu-responsive sensors can also promptly react with Cu–Aβ oligomers and fibrils, resulting in a significant fluorescence turn-on, and indicating that the probes are able to detect Cu–Aβ species in vitro. To confirm the Aβ binding specificity of the probes, brain sections from transgenic 5xFAD mice were stained with the developed sensors. As expected, when the brain sections were only stained with the compounds, the fluorescence images show that the sensor has a poor ability to detect amyloid plaques, given the low fluorescence intensity. However, upon the addition Cu(II) ions to the medium, followed by exhaustive washing of the free unbound Cu ions, the fluorescence images clearly indicate that the Cu-responsive sensors are activated by the Aβ-bound Cu(II) ions and release highly fluorescent amyloid binding fluorophores, which can specifically label the native amyloid plaques in the 5xFAD brain sections.
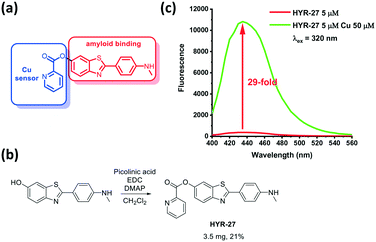 | ||
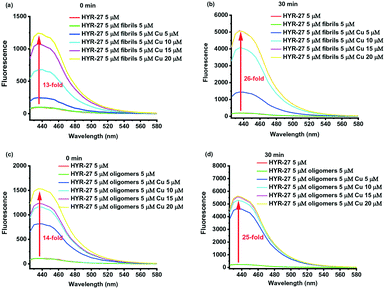
| Fig. 1 a) Design and b) synthesis of Cu-based activatable sensor HYR-27; c) fluorescence turn-on effect of HYR-27 in the presence of Cu(II); [HYR-27] = 5 μM, [Cu] = 50 μM, pH 7.4 PBS. | ||
To detect Cu(II) in various Aβ species, the developed fluorescence probes such as HYR-27 include two components: a Cu(II)-responsive 2-picolinic ester group that chelates Cu(II) ions and selectively activates the ester bond for hydrolysis (Fig. 1),25,31 and a widely-used Aβ-binding molecular fragment derived from Pittsburgh compound B (PiB), which has high fluorescence intensity and can strongly interact with the Aβ fibrils.
To evaluate the copper-responsive activity of the probe, we firstly performed the Cu turn-on assay in PBS, using physiologically relevant Cu concentrations typically employed in in vitro Aβ-binding studies.12–19 With the attachment of the picolinic ester moiety, the fluorescence intensity of the probe is dramatically quenched. However, when HYR-27 was treated with Cu(II) ions, the fluorescence intensity significantly increased ∼29 fold (Fig. 1 and S4†), indicating that the compound can coordinate to Cu(II) to facilitate the hydrolysis reaction and enhance the fluorescence intensity.
Then, we explored the Cu-activatable ability of HYR-27 in the presence of various Aβ species. When the compound was treated with Aβ fibrils only, no fluorescence enhancement was observed after 30 min incubation. However, when Cu–Aβ fibrils were added to the solution, the fluorescence intensity dramatically increased, likely due to the rapid Cu(II)-catalysed hydrolysis reaction (Fig. 2). Addition of excess Cu(II) ions to the solution led to a further increase in the turn-on effect (∼26 fold) to a fluorescence intensity similar to the one observed in the presence of only Cu(II) ions (Fig. 1). Furthermore, since Cu(II) ions were shown to stabilize the formation of Aβ oligomers, which are the most neurotoxic species in vivo, it is also crucial to also investigate if the developed probe can also detect Cu–Aβ oligomers. As a result, we first treated the compound with Aβ oligomers only. Similar to the Aβ fibril studies, no obvious turn-on effect was observed after 30 min incubation. Excitingly, when Cu(II) was added to the solution, the fluorescence intensity significantly increased, indicating that the probe can compete with the Aβ oligomers to chelate Cu(II) and facilitate the hydrolysis reaction to release the highly fluorescent PiB probe (Fig. 2). Importantly, the Cu-mediated hydrolysis of the picolinic ester fragment is slower in the presence of the Aβ aggregates (Fig. S4†), confirming that the Cu ions are indeed bound to the Aβ aggregates in these in vitro studies, and thus mimicking the in vivo conditions. Overall, the Cu-dependent fluorescence turn-on studies with or without various Aβ species clearly demonstrate that the developed probes can be efficiently activated by Cu(II), Cu(II)–Aβ fibrils, and Cu(II)–Aβ oligomers, exhibiting appreciable fluorescence turn-on in vitro.
To further explore the Aβ-binding affinity and Cu-activatable activity of HYR-27, we also performed fluorescence staining studies on brain sections from 11 month-old transgenic 5xFAD mice.17 Firstly, the brain sections were only stained with HYR-27, and the low fluorescence intensity images show that the compound cannot efficiently detect amyloid plaques (Fig. 3), which is consistent with the in vitro studies. To mimic the Cu-rich environment, we first treated the brain sections with 50 μM Cu(II), followed by extensive washings with the buffer to remove any unbound Cu ions. Excitingly, performing the brain section staining following these treatment steps clearly shows that the fluorescence intensity significantly increased after the incubation with Cu(II), indicating that HYR-27 was rapidly hydrolysed to form PiB, which can strongly bind to the native amyloid plaques in the presence of Aβ-bound Cu(II) ions.
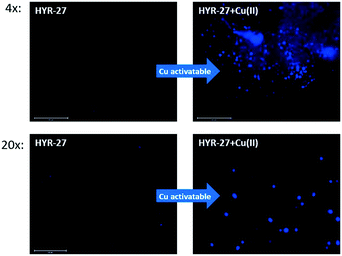 | ||
| Fig. 3 Fluorescence microscopy images of 5xFAD mice brain sections incubated with compounds HYR-27 (left panel) and HYR-27 + Cu (right panel). [HYR-27] = 5 μM, [Cu] = 50 μM. Scale bar (20×): 125 μm. | ||
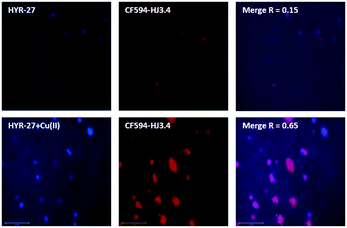
To further confirm the Aβ binding specificity, the brain sections were first stained with HYR-27 or HYR-27 + Cu(II), and sequentially immunostained with CF594-labeled HJ3.4 antibodies, which can bind to all Aβ species.17 The fluorescence images show that while HYR-27 does not show any colocalization with the HJ3.4 antibodies, in the presence of Cu(II), the hydrolysed product of HYR-27 shows a significantly increased fluorescence intensity that is colocalized with the HJ3.4 antibodies (Fig. 4), showing that HYR-27 has a high Cu-responsive ability that could be utilized to detect the Cu–Aβ amyloid plaques present in the 5xFAD brain sections.
Since the chelation of Cu(II) can facilitate the hydrolysis of the picolinic ester bond, a strong Cu(II)-binding chelator, Me2HTACN, was introduced to the HYR-27 fragment, to generate HYR-27-TACN (Fig. 5). However, when Cu(II) was added to HYR-27-TACN, the fluorescence intensity decreased significantly, suggesting that in this case, Cu(II) ions bind strongly to the TACN group and cause paramagnetic quenching of the fluorescence intensity of HYR-27-TACN, instead of catalysing the hydrolysis reaction of the picolinic ester group. Thus, these results suggest that the use of strong Cu chelators may not be optimal for the design of Cu(II)-activatable fluorescent sensors, and a weaker Cu(II)-binding picolinic ester moiety may suffice to promote the hydrolysis of the ester bond.
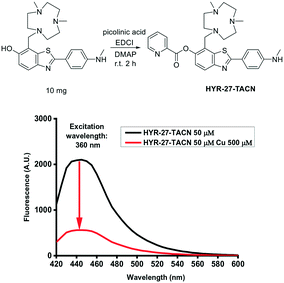 | ||
| Fig. 5 Cu(II) fluorescence turn-on effects of HYR-27-TACN. [HYR-27-TACN] = 50 μM, [Cu] = 500 μM in PBS (pH = 7.4). | ||
After achieving promising results in which HYR-27 can act as a Cu(II)-activatable fluorescent sensor, a series of curcumin derivatives with high Aβ-binding affinity and near-infrared (NIR) emission properties were functionalized with the Cu(II)-responsive picolinic ester fragment (Scheme 1 and Fig. S1–S3†). The syntheses of the curcumin precursors YL-1, YL-3, and YL-5 were carried out by following previously reported procedures,32 and the final Cu(II)-responsive curcumin derivatives YL-2, YL-4, and YL-6 were synthesized via an EDC-mediated coupling reaction between 2-picolinic acid and the curcumin precursors.31
To confirm the Cu-activatable ability of the developed curcumin derivatives, fluorescence turn-on experiments were performed. Since the solubility of the compounds is poor in PBS, methanol was selected as the solvent to perform the Cu(II) turn-on assays. Notably, in the presence of Cu(II), the curcumin derivatives YL-2, YL-4, and YL-6 exhibit a significant fluorescence turn-on signal, proving that the developed curcumin-based compounds can be activated by Cu(II) ions (Fig. 6a and b, respectively).
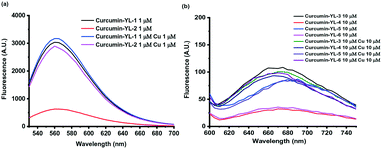 | ||
| Fig. 6 Evaluation of Cu(II) fluorescence turn-on effects for a) YL-1 and YL-2, and b) YL-3–YL-6. [YL-X] = 1 or 10 μM, [Cu] =1 or 10 μM in MeOH. | ||
To investigate the Cu-activatable activity of the curcumin derivatives towards amyloid plaques, fluorescence staining studies of 5xFAD brain sections were performed with YL-4, since YL-4 exhibited the highest solubility in aqueous media. Upon staining of the brain sections with YL-4 only, the fluorescence images show that YL-4 can stain the amyloid plaques on the brain sections (Fig. 7). However, the fluorescence intensity of the YL-4-stained amyloid plaques is low, and it seems that YL-4 can only stain the core regions of the amyloid plaques. By comparison, upon addition of Cu(II) to the staining solution of the 5xFAD brain sections, the fluorescence intensity of the compound-stained amyloid plaques significantly increases, indicating that YL-4 is efficiently activated by Cu(II) to generate the more highly fluorescent curcumin derivative YL-3. Interestingly, upon incubation with YL-4 and Cu(II), the resulting YL-3 compound seems to also stain the periphery of the amyloid plaques where the Aβ oligomers and smaller Aβ aggregates are usually found, suggesting that these compounds have the potential to detect Cu–Aβ oligomers ex vivo. Overall, these brain section staining studies clearly demonstrate that YL-4 can be utilized as a Cu-responsive sensor for the detection of Cu-rich Aβ aggregates of various sizes.
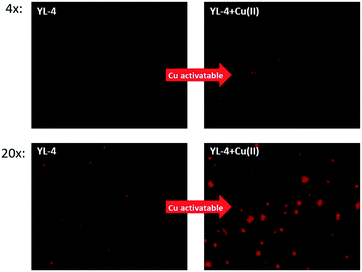 | ||
| Fig. 7 Fluorescence microscopy images of 5xFAD mouse brain sections incubated with YL-4 (left panels) and YL-4 + Cu(II) (right panels). [YL-4] = 50 μM, [Cu] = 50 μM. Scale bar (20×): 125 μm. | ||
In conclusion, we rationally designed and synthesized a series of Cu-activatable sensors to detect Cu–Aβ species in vitro and ex vivo. With the introduction of the picolinate ester moiety attached to an Aβ-binding fragment, these developed fluorescent sensors can chelate Cu(II) to facilitate the hydrolysis of the ester bond and release the highly fluorescent Aβ-binding molecule in vitro. Furthermore, the Cu-responsive sensors can also react with Cu–Aβ oligomers and fibrils, resulting in a significant fluorescence turn-on, and indicating that the probes are able to detect Cu–Aβ species in vitro. To confirm the Aβ-binding specificity of the probes, transgenic AD mouse brain section imaging studies were also performed. As expected, if the brain sections were only stained with the compounds, the fluorescence images show that the sensor has a poor ability to detect amyloid plaques, exhibiting a low fluorescence intensity. However, in the presence of Cu(II) ions, the resulting fluorescence images clearly demonstrate that the sensors are activated by the Cu(II) ions and generate highly fluorescent amyloid-binding fluorophores, which can specifically label the amyloid plaques in the 5xFAD mouse brain sections. Overall, the developed Cu-activatable sensors can be utilized to detect Cu–Aβ species, both in vitro and ex vivo.
Conflicts of interest
There are no conflicts to declare.References
- 2021 Alzheimer's disease facts and figures, Alzheimer's Dementia, 2021, 17, 327–406 Search PubMed.
- S. Tiwari, V. Atluri, A. Kaushik, A. Yndart and M. Nair, Int. J. Nanomed., 2019, 14, 5541–5554 CrossRef CAS PubMed.
- K. P. Kepp, Coord. Chem. Rev., 2017, 351, 127–159 CrossRef CAS.
- K. P. Kepp, Chem. Rev., 2012, 112, 5193–5239 CrossRef CAS PubMed.
- C. J. Maynard, A. I. Bush, C. L. Masters, R. Cappai and Q. X. Li, Int. J. Exp. Pathol., 2005, 86, 147–159 CrossRef CAS PubMed.
- M. A. Smith, G. Perry, P. L. Richey, L. M. Sayre, V. E. Anderson, M. F. Beal and N. Kowall, Nature, 1996, 382, 120–121 CrossRef CAS PubMed.
- D. P. Smith, G. D. Ciccotosto, D. J. Tew, M. T. Fodero-Tavoletti, T. Johanssen, C. L. Masters, K. J. Barnham and R. Cappai, Biochemistry, 2007, 46, 2881–2891 CrossRef CAS PubMed.
- B. Alies, C. Bijani, S. Sayen, E. Guillon, P. Faller and C. Hureau, Inorg. Chem., 2012, 51, 12988–13000 CrossRef CAS PubMed.
- J. T. Pedersen, J. Ostergaard, N. Rozlosnik, B. Gammelgaard and N. H. Heegaard, J. Biol. Chem., 2011, 286, 26952–26963 CrossRef CAS PubMed.
- S. Bagheri, R. Squitti, T. Haertle, M. Siotto and A. A. Saboury, Front. Aging Neurosci., 2017, 9, 446 CrossRef PubMed.
- G. z. Eskici and P. H. Axelsen, Biochemistry, 2012, 51, 6289–6311 CrossRef CAS PubMed.
- A. K. Sharma, S. T. Pavlova, J. Kim, D. Finkelstein, N. J. Hawco, N. P. Rath, J. Kim and L. M. Mirica, J. Am. Chem. Soc., 2012, 134, 6625–6636 CrossRef CAS PubMed.
- Y. Zhang, D. L. Rempel, J. Zhang, A. K. Sharma, L. M. Mirica and M. L. Gross, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 14604–14609 CrossRef CAS PubMed.
- A. K. Sharma, S. T. Pavlova, J. Kim, J. Kim and L. M. Mirica, Metallomics, 2013, 5, 1529–1536 CrossRef CAS PubMed.
- A. K. Sharma, J. Kim, J. T. Prior, N. J. Hawco, N. P. Rath, J. Kim and L. M. Mirica, Inorg. Chem., 2014, 53, 11367–11376 CrossRef CAS PubMed.
- A. K. Sharma, J. W. Schultz, J. T. Prior, N. P. Rath and L. M. Mirica, Inorg. Chem., 2017, 56, 13801–13814 CrossRef CAS PubMed.
- H.-J. Cho, A. K. Sharma, Y. Zhang, M. L. Gross and L. M. Mirica, ACS Chem. Neurosci., 2020, 11, 1471–1481 CrossRef CAS PubMed.
- C. Esmieu, D. Guettas, A. Conte-Daban, L. Sabater, P. Faller and C. Hureau, Inorg. Chem., 2019, 58, 13509–13527 CrossRef CAS PubMed.
- C. Hureau, J. Biol. Inorg. Chem., 2017, 22, S230–S230 Search PubMed.
- H. J. Yu, W. Zhao, Y. Zhou, G. J. Cheng, M. Sun, L. Wang, L. Yu, S. H. Liang and C. Z. Ran, Anal. Chim. Acta, 2020, 1097, 144–152 CrossRef CAS PubMed.
- T. Hirayama, G. C. Van de Bittner, L. W. Gray, S. Lutsenko and C. J. Chang, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 2228–2233 CrossRef CAS PubMed.
- S. Lee, C. Y. S. Chung, P. Liu, L. Craciun, Y. Nishikawa, K. J. Bruemmer, I. Hamachi, K. Saijo, E. W. Miller and C. J. Chang, J. Am. Chem. Soc., 2020, 142, 14993–15003 CrossRef CAS PubMed.
- K. J. Bruemmer, S. W. M. Crossley and C. J. Chang, Angew. Chem., Int. Ed., 2020, 59, 13734–13762 CrossRef CAS PubMed.
- D. A. Iovan, S. Jia and C. J. Chang, Inorg. Chem., 2019, 58, 13546–13560 CrossRef CAS PubMed.
- H. Li, P. Zhang, L. P. Smaga, R. A. Hoffman and J. Chan, J. Am. Chem. Soc., 2015, 137, 15628–15631 CrossRef CAS PubMed.
- M. Saleem, M. Rafiq, M. Hanif, M. A. Shaheen and S. Y. Seo, J. Fluoresc., 2018, 28, 97–165 CrossRef PubMed.
- L. Zeng, E. W. Miller, A. Pralle, E. Y. Isacoff and C. J. Chang, J. Am. Chem. Soc., 2006, 128, 10–11 CrossRef CAS PubMed.
- D. W. Domaille, L. Zeng and C. J. Chang, J. Am. Chem. Soc., 2010, 132, 1194–1195 CrossRef CAS PubMed.
- B. Muthuraj, S. Layek, S. N. Balaji, V. Trivedi and P. K. Iyer, ACS Chem. Neurosci., 2015, 6, 1880–1891 CrossRef CAS PubMed.
- Y. Y. Yu, P. Wang, X. D. Zhu, Q. W. Peng, Y. Zhou, T. X. Yin, Y. X. Liang and X. X. Yin, Analyst, 2018, 143, 323–331 RSC.
- See ESI.†.
- K. Liu, J. M. Chen, J. Chojnacki and S. J. Zhang, Tetrahedron Lett., 2013, 54, 2070–2073 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, spectroscopic data, and copies of NMR spectra. See DOI: https://doi.org/10.1039/d2sd00028h |
| This journal is © The Royal Society of Chemistry 2022 |