Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Recent progress on synthetic and protein-based genetically encoded sensors for fluorimetric Cu(I) recognition: binding and reaction-based approaches
Sushil
Kumar
 *a,
Jolly
Kaushal
b,
Tapas
Goswami
*a,
Jolly
Kaushal
b,
Tapas
Goswami
 a,
Pankaj
Kumar
a and
Pramod
Kumar
a,
Pankaj
Kumar
a and
Pramod
Kumar
 *c
*c
aDepartment of Chemistry, University of Petroleum and Energy Studies (UPES), Bidholi, Dehradun-248007, Uttarakhand, India. E-mail: sushil.k@ddn.upes.ac.in
bDepartment of Chemistry, School of Physical Sciences, Doon University, Dehradun-248001, Uttarakhand, India
cDepartment of Chemistry, Mahamana Malviya College Khekra (Baghpat), C.C.S. University Meerut, Uttar Pradesh, India
First published on 11th April 2022
Abstract
Due to the extensive biological significance and applications of copper, the development of different fluorescent probes for Cu ions has been an active area of research over the past 10 years. The present study focuses on the recent progress in the fluorescent-based in vitro and in vivo sensing of Cu(I) ions and understanding copper functions using small molecules and modified protein-based fluorescent probes. With a brief outline of cellular Cu-homeostasis and focus on Cu(I)-responsive probe design, we review a wide range of fluorescent probes for Cu(I) ions and assess their characteristic features in the context of their utility in live cell Cu(I) imaging. In particular, traditional binding-based and chemical reaction-based approaches used to develop fluorimetric probes for Cu(I) ions are highlighted.
1. Introduction
Cation-responsive chemosensors have consistently exhibited potential applications in various research areas including chemical, biological, environmental and medical sciences.1–12 Fluorescence-based cation-recognition methods have dramatically changed the way that different physiological and cellular events are visualized and understood in biological systems. For many years, fluorescent methods have attracted significant attention from chemists and biologists13–17 because of their promising features such as precision, low-cost, versatility, excellent sensitivity and selectivity and real-time detection of analytes. Nevertheless, although many fluorescent sensors have been developed to recognise non-redox active metal ions, the sensing of metals having readily accessible multiple oxidation states, for instance, Fe (II)/Fe(III) or Cu(I)/(II), is still a challenging task. The underlying reason for this is the interfering metal-mediated turn-off fluorescent quenching pathway.16–18Although copper (Cu) is an essential element for life and plays crucial role in a wide range of biological processes,19–22 an alteration or disruption in Cu-homeostasis is linked to several serious neurodegenerative diseases such as Alzheimer's disease,23–25 Menkes diseases and Wilson's disease,26–28 familial amyotrophic lateral sclerosis, bipolar disorder, depression, and anxiety.29–31 Cu is a redox-active element and exists in both reduced (Cu+) and oxidized (Cu2+) forms in all living organisms. The redox features of copper such as reversible Cu+ oxidation and Cu2+ reduction make it a potential candidate for catalysing an extensive range of chemical reactions. Both the 1+ and 2+ states are required as cofactors for the functioning of different vesicular, cytosolic and mitochondrial enzymes such as copper/zinc superoxide dismutase (Cu/Zn-SoD), metallothionein and cytochrome-c-oxidase enzymes in living systems.32,33 It is also evident that a disturbance in the levels of Cu in humans is related to the progression of cancer.34,35 Furthermore, owing to the wide-spread use of copper in agriculture, industrial, and environmental processes, it has become a serious contaminant in water systems.36–38 Therefore, the detection and assessment of Cu-based ions is important to get deep insight into Cu homeostasis and its related diseases.
Over the past few years, a broad range of fluorescent probes based on small organic38–80 and inorganic molecules,16,17,81–94 nano-materials,17,95–97 modified proteins,98–103 metal–organic frameworks (MOFs)104–107 and sol-gels17,108–110 have been developed for the recognition of Cu(I) and Cu(II) ions.
2. Fluorescent probe design
Two common strategies, namely, binding-based and reaction-based strategies, have been employed to construct probes for fluorescence-based Cu recognition experiments. In the binding-based approach, the reversible, non-covalent binding of the target ion induces an emission change in the probe, resulting in the form of an optical response from the fluorophore.48,49,111–114 The probe design is based on a donor-acceptor skeleton combining a cation-binding unit (i.e., a receptor) with a suitable reporter (i.e., a fluorophore or signalling unit). The reporter serves as an optical transducer upon the binding of a cation to the receptor (Scheme 1(A)). A target-selective probe may be designed by considering the balanced interactions between the receptor and the cation of interest. Alternatively, in the reaction-based pathway, the binding of cation first induces a chemical transformation in the probe, which eventually leads to the production of an optical signal due to the formation of a fluorescent product.48,73–80 The selectivity of a particular probe is determined by its specificity for chemical reactions (Scheme 1(B)).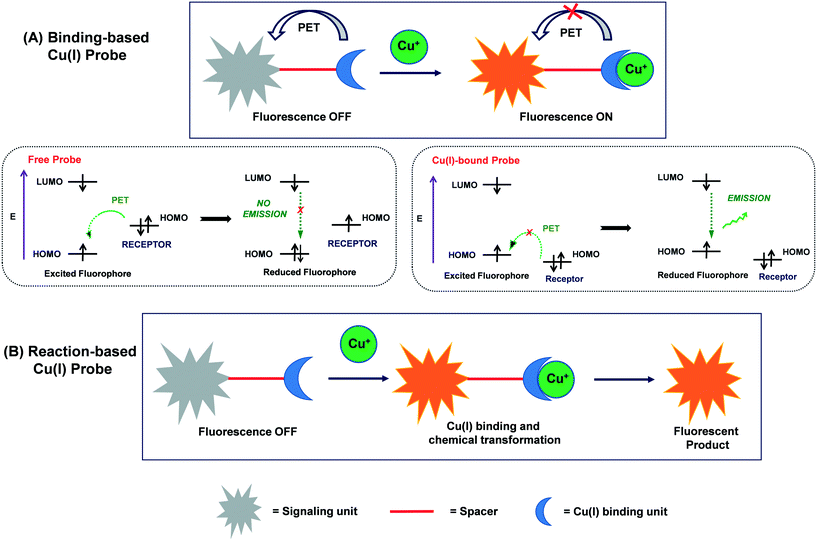 | ||
| Scheme 1 Schematic representation of (A) binding-based and (B) reaction-based approaches for Cu(I)-selective fluorescent probes. | ||
The majority of the Cu(I)-selective fluorescent probes have been developed using the binding-based approach. An important event usually observed in these fluorescent probes is photoinduced electron transfer (PET), which translates Cu(I)-binding reaction into a fluorescent enhancement.48–50 PET is considered to be the most widely accepted mechanism in the design and development of fluorescent Cu(I) probes and biosensors. In this process, intramolecular electron transfer takes place from the electron-rich receptor (i.e., donor) to the fluorophore (i.e., acceptor), resulting in the quenching of fluorescence. However, the Cu(I) interaction with the receptor inhibits PET (or completely suppresses in some cases), which results in the restoration of the fluorescence of the probe. Generally, PET-based Cu(I) probes display a turn-on fluorescence response, which favours Cu(I) imaging with improved signal to noise ratios.
Upon excitation of a fluorophore with light of a suitable wavelength, electron transfer occurs from the highest-occupied molecular orbital (HOMO) to the lowest-unoccupied molecular orbital (LUMO) of the fluorophore. If the HOMO of the appended receptor lies between the HOMO and LUMO of the fluorophore, e− transfer takes place from the HOMO of the receptor to the HOMO of the fluorophore via the PET process. Consequently, one electron is “locked” into the LUMO of the fluorophore, which leads to fluorescence quenching in the probe. The Cu(I)-binding to receptor reduces the energy of the HOMO of the receptor below than that of the HOMO of the fluorophore. The lowering in the energy level of the HOMO of the receptor blocks the PET process, leading to the restoration of fluorescence (Scheme 1(A)).
3. Detection of copper(II) vs. copper(I)
Numerous fluorescent Cu(II)-responsive probes have been established for both in vitro and in vivo applications; however, thus far, only a few probes are known for imaging Cu(I) ions.50–80 This is due to the stability of resultant cation-bound-receptor moiety. The magnitude of the cation–receptor interaction is directly dependent on both the metal ion and coordinating atoms equipped with a receptor. The principle of hard and soft acids and bases (HSAB) may be employed to rationalize these interactions in terms of hardness and softness.115,116 Copper(II), a borderline acid, displays strong interaction with an extensive range of borderline bases such as imidazole, pyrazole and pyridine donors. Moreover, the receptor architecture can also be controlled by considering the geometric aspects of the resultant cation-bound receptor. For instance, the d9 Cu(II) ion, which exhibits Jahn–Teller distortion, may adopt a square planar, square pyramidal or distorted octahedral geometry.In contrast, the soft copper(I) ion prefers to bind to a receptor containing soft bases such as RSR′ and PR3.16–18 Usually, Cu(I) probes are designed with the aim of selective Cu(I) binding to polydentate thioether-based receptors. Considering the HSAB principle, it becomes easy to discriminate soft Cu(I) ions from other biologically important main group (such as Na(I), K(I), Mg(II) and Ca(II)) and transition (such as Cu(II), Fe(II)/(III) and Zn(II)) metal ions, which fall in the range of hard acids to borderline acids. Moreover, with the judicious choice of geometry, size and number of donors in the probe receptors, Cu(I) can be easily recognised over other soft acids such as Pb(II) and Hg(II). Therefore, the majority of established Cu(I) probes contain thioether-based frameworks such as bis{2-[2-(2-ethylthio)ethylthio]ethyl}amine (BETA) or (1,4,7,10-tetrathia-13-aza)-15-crown-5 to coordinate with Cu(I).48,49
After the breakthrough report by Fahrni and group50 on a thioether-rich receptor-based fluorescent probe for intracellular Cu(I) imaging in live cells, many fluorescent Cu(I) probes appended with similar receptors have been reported. This important work, published in 2005, paved the way to utilize thioether-based receptors in the design of Cu(I)-selective probes. However, thus far, only a few Cu(I)-selective probes are suitable for live cell imaging. The reason for this is the inherent stability of aqua-copper(I) and the lipophilic nature of the commonly used thioether-based receptors. Furthermore, several probes require a short-wavelength excitation, which can cause damage to living biological samples.69
To the best of our knowledge, to date, only one review is available on the detection of only Cu(I) ions by turn-on/off-responsive fluorescent sensors, which was published in 2013.49 However, the combined fluorimetric detection of both Cu(I) and Cu(II) ions was reviewed by Chang's group in 2015.48 Since then, several Cu(I)-selective fluorescent sensors have been reported, which demonstrate a response both in vitro and in vivo. Meanwhile, to achieve a bright and high contrast towards Cu(I) ions, many fluorophore architectures are employed to rationally design completely water-soluble probes. However, a compiled study on these Cu(I)-responsive fluorescent probes has not been reviewed thus far in the literature. The main focus of the present study is to outline the recent approaches used for fluorescence-based sensing of Cu(I) with emphasis on their underlying solution chemistry and photophysical pathways. All the noteworthy Cu(I)-responsive fluorescent probes are covered herein. This perspective broadly classifies the probes into small-molecule and genetically encoded proteins, which are subdivided based on their chelating or reaction-based approaches for the detection of Cu(I).
4. Small molecule-based Cu(I) probes
4.1. Binding-based probes
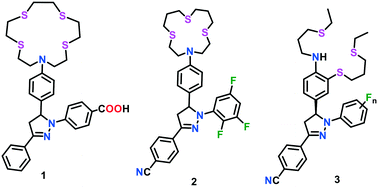
A wide range of binding-based fluorescent probes for Cu(I) ions has been constructed using the selective response of soft S-rich receptors towards the soft Cu(I) ion. To obtain a suitable fluorescent probe design for Cu(I) ions, thioether-based receptors are coupled with a signalling unit or fluorescent reporter whose optical properties can be adjusted through the PET mechanism.48,49 The Cu(I)-selective probes containing BETA or tetrathiaza crown as receptors have two important structural features, as follows: (i) high affinity of electron-rich S donor (soft base) towards Cu(I) ions (soft acid) and (ii) lipophilicity of receptors, which allows the diffusion of the probe across cellular membranes for in vivo imaging.50–56 However, in contrast, thioether-based Cu(I) probes are found to be poorly soluble in water and prone to colloidal aggregation in aqueous medium, which may ultimately lead to alterations in the optical response of the fluorophore.However, although probe 1 was successfully applied for Cu(I) imaging experiments in live cells, the binding of Cu(I) did not result in an efficient quantum yield, which further required the optimization of the ligand architecture. In a process to optimize the fluorescence contrast ratio of probes upon Cu(I) binding, the electron-withdrawing character of the aryl rings of pyrazoline moiety was increased stepwise to form probe type 2 (Fig. 1).51,52 The electronically tuned probes resulted in a 30.0- to 50.0-fold increase in fluorescence response when saturated with Cu(I) ions in methanol. Nonetheless, upon protonation with CF3COOH, an unprecedented increase in fluorescence (greater than 300-fold) was noted in the present probe. A kinetically controlled PET switching mechanism was established to assess the behaviour of these probes in the presence of Cu(I) ions. However, the ligand optimization efforts did not result in much improvement in fluorescence contrast and quantum yield. It was suggested that the weak coordination of Cu(I) to the aryl amine electron donor may be responsible for the incomplete fluorescence recovery and low emission quantum yield.
In continuation of this endeavour, a series of Cu(I)-responsive probes (3) with rigid ligand architectures was constructed by the same group (Fig. 1).53 The probes consisted of an aniline donor-based acyclic NS3 receptor and substituted pyrazoline-based fluorophores. These probes displayed a strong emission upon the addition of Cu(I) ions. The fluorescence was saturated at an equimolar concentration of Cu(I), which indicated the formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex in all cases. Cu(I) binding with the probe induced a conformational change in the receptor, which resulted into adverse steric interactions between the aniline ring and receptor backbone. Further interactions with the solvent molecule afforded the formation of less hindered ternary complexes. Solvent coordination increases the e− donating ability of aniline, which concomitantly led to a more constructive PET event, and thus incomplete fluorescence recovery. Instead of the redox-active metal-induced quenching pathway, the role of ternary complex formation in the presence of solvent molecules was addressed as the major factor in the incomplete recovery of the fluorescence. By switching the behaviour of the PET mechanisms, a tremendous enhancement (>200-fold) in the fluorescence of these probes was successfully achieved. This investigation demonstrated that high-contrast ratios could easily be achieved even in the case of redox-active metals, provided that the metal-mediated quenching is kinetically unfavourable.
1 complex in all cases. Cu(I) binding with the probe induced a conformational change in the receptor, which resulted into adverse steric interactions between the aniline ring and receptor backbone. Further interactions with the solvent molecule afforded the formation of less hindered ternary complexes. Solvent coordination increases the e− donating ability of aniline, which concomitantly led to a more constructive PET event, and thus incomplete fluorescence recovery. Instead of the redox-active metal-induced quenching pathway, the role of ternary complex formation in the presence of solvent molecules was addressed as the major factor in the incomplete recovery of the fluorescence. By switching the behaviour of the PET mechanisms, a tremendous enhancement (>200-fold) in the fluorescence of these probes was successfully achieved. This investigation demonstrated that high-contrast ratios could easily be achieved even in the case of redox-active metals, provided that the metal-mediated quenching is kinetically unfavourable.
The optical properties of the above-mentioned probes were investigated using their apparently transparent homogeneous solutions in aqueous buffer diluted with DMSO (1 mM stock solution). However, the dynamic-light-scattering experiments clearly revealed the formation of colloidal aggregates (average hydrodynamic radius of ∼100 nm), which may lead to striking alterations in the photophysical properties of the probes.
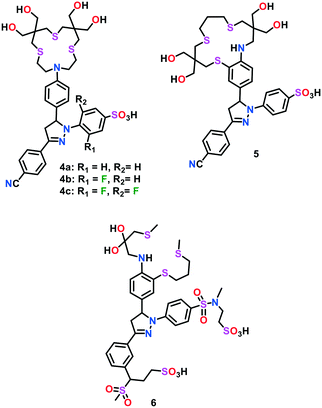
To address the issue of colloidal aggregation, Fahrni and group54 further constructed electronically tuned triarylpyrazoline-based turn-on fluorescent indicators 4a–4c for the fast, reversible and selective detection of Cu(I) ions in aqueous buffer (Fig. 2). Unlike the previously reported Cu(I)-responsive sensors, these probes were found to be directly soluble in pure water up to millimolar (mM) concentrations. The direct solubility of the Cu(I) sensors in water circumvented the possible formation of colloidal aggregates. These probes consisted of a hydroxymethyl-substituted thiocrown receptor appended with triarylpyrazoline-based fluorophores. To further increase the solubility of the probes in water, the pyrazoline and thiocrown units were functionalized with sulfonate and hydroxymethyl groups, respectively. Sulfonation of one aryl ring in the triarylpyrazoline unit of probe 4 resulted in the suppression of colloid aggregation in water. The absence of colloid aggregates was confirmed via dynamic-light-scattering experiments with a 5 μM solution of probe in aqueous buffer. Probes 4a–4c were found to detect Cu(I), which was bound to a metallochaperone Atox1 (Antioxidant Protein 1). The Cu(I) binding affinity of different variants of the probes was investigated via UV-vis and fluorescence spectral studies, and the emission maxima of the probes were observed in the range of 467–508 nm. Upon the addition of Cu(I), a 65-fold increase in fluorescence could easily be achieved via the PET switching mechanism. However, the moderate increase in quantum yield (∼8%) still limited the sensing ability of these probes. In general, PET-based Cu(I)-selective fluorescent probes display an incomplete restoration of their fluorescence signal. Earlier reports on probes having N-aryl thiaza crown receptors suggested that their fluorescence recovery may be compromised by partial Cu–N coordination and/or resulting ternary complexes in the presence of solvent molecules. However, upon saturation with Cu(I), a 65-fold increase in the fluorescence signal of probes 4a–4c is favourable compared to the previously reported Cu(I) probes, but their quantum yield of 0.083 is considerably low.
In an effort to further increase the fluorescence contrast and the quantum yield of these probes, the phenylamine PET donor was integrated into the backbone of the thiaza crown receptor.55 The optimization of the ligand frame was mainly focused on maximizing the PET switching via a synergistic Cu(I)-induced conformational change. Probe 5 displayed an emission maximum at 506 nm and a strong turn-on fluorescent enhancement (ca. 57-fold) could be observed upon saturation with Cu(I). Even with their excellent water solubility in water, it was noticed that probes 4–5 displayed a modest hydrophobic interaction with cellular membranes. This interaction, probably arising from the 3-aryl ring present in the triarylpyrazoline motif, may result in a strong analyte-independent fluorescence response. In this view of ligand architecture, the same group optimized probe 4 and constructed a high-contrast water soluble Cu(I)-selective probe 6.56 The latter probe was devised by replacing the sulfonate group with an N,N′-dialkylsulfonamide group in the pyrazoline reporter and inserting an aniline group into the receptor framework. Compared to 4 and 5, the optimized probe 6 was found to be well-suited for lipid membranes and featured an improved fluorescence signalling response. In the presence of Cu(I) ions, a 180-fold increase in the fluorescence intensity of probe 6 was observed with an appreciable change in quantum yield (∼41%). This probe was found to sense Cu(I) concentrations up to the part-per-trillion (ppt) range.
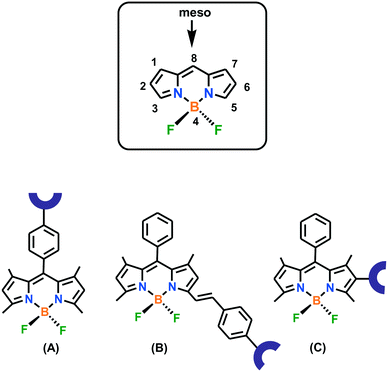
The BODIPY core can easily be functionalized at the boron atom, the pyrrole carbon atoms and the meso carbon atom. The absorption and emission behaviours of BODIPY derivatives are highly affected by a variation in the substituents around the core. Therefore, the photophysical characteristics of these derivatives can easily be tuned on demand. The mechanisms usually involved in BODIPY-based chemosensors include intramolecular charge transfer (ICT), photoinduced electron transfer (PET), Forster resonance energy transfer (FRET), and resonance energy transfer (RET). The BODIPY-based structures commonly used in cation-responsive probe design are shown in Fig. 3. Two important factors are considered when selecting the probe design, as follows: (i) the methyl groups present at positions 1 and 7 hinder the rotation of the aryl ring at position 8, which ultimately results in an enhancement in the emission response and (ii) substitution of different groups at positions 5 or 6 in structure types B and C increases the extended delocalization, which may lead to a color change with the introduction of metal ions. Thousands of BODIPY derivatives with exciting structural variations have been investigated in the past decades. However, in the present review, only Cu(I)-responsive fluorescent probes will be considered.
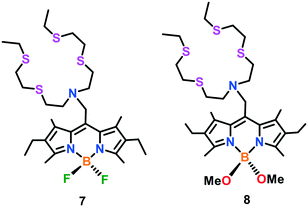
Chang and group57 presented the first BODIPY-based fluorescent probe 7 coupled to a thiaza crown receptor for the selective detection of Cu(I) ions in aqueous medium (HEPES buffer of pH 7.0) (Fig. 4). The presence of the BODIPY fluorophore made probe 7 exhibit emission and excitation in the visible region. The emission maximum of probe 7 was measured at 566 nm (λex 488 nm). Upon the addition of Cu(I) ions, about a 10-fold higher fluorescence response (free probe: Φ = 0.016 and Cu(I)-bound probe: Φ = 0.013) was observed with a slight blue shift in the band at 561 nm. The Job's plot analyses established the formation of 7–Cu(I) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry. Probe 7 was found to be permeable in the cellular membrane, as evidenced from confocal microscopy experiments and successfully used for intracellular Cu(I) detection in live HEK 293T (human embryonic kidney) cells. However, the relatively low quantum (Φ = 0.013) of probe 7 limited its utility to examine the dynamic behaviour of endogenous Cu pools at the basal level. To further improve the turn-on response and quantum efficiency, another fluorescent probe 8 was constructed by replacing the fluoro substituents with methoxy groups.58 Emission spectral analysis of probe 8 in HEPES buffer (20 mM, pH 7.0) exhibited the emission maximum at 560 nm with weak fluorescence (Φ = 0.007). The introduction of Cu(I) ions resulted in a 75-fold increase (Φ = 0.40) in the fluorescence of the probe with a blue-shift to 548 nm. Similar to 7, the 1
1 binding stoichiometry. Probe 7 was found to be permeable in the cellular membrane, as evidenced from confocal microscopy experiments and successfully used for intracellular Cu(I) detection in live HEK 293T (human embryonic kidney) cells. However, the relatively low quantum (Φ = 0.013) of probe 7 limited its utility to examine the dynamic behaviour of endogenous Cu pools at the basal level. To further improve the turn-on response and quantum efficiency, another fluorescent probe 8 was constructed by replacing the fluoro substituents with methoxy groups.58 Emission spectral analysis of probe 8 in HEPES buffer (20 mM, pH 7.0) exhibited the emission maximum at 560 nm with weak fluorescence (Φ = 0.007). The introduction of Cu(I) ions resulted in a 75-fold increase (Φ = 0.40) in the fluorescence of the probe with a blue-shift to 548 nm. Similar to 7, the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 formation of 8–Cu(I) was found to be responsible for the turn-on emission response. This probe could efficiently monitor labile copper pools in living cells at the basal level and copper-depleted conditions. The use of probe 8 in conjunction with XRFM indicated that neuronal cells move considerable Cu pools from their somatic cells bodies to extended outer processes when activated by depolarization. Further imaging experiments exhibited that the redistribution of copper is dependent on Ca(II) release, paving the way to explore cross-talk between Cu(I) and Ca(II) ions.
1 formation of 8–Cu(I) was found to be responsible for the turn-on emission response. This probe could efficiently monitor labile copper pools in living cells at the basal level and copper-depleted conditions. The use of probe 8 in conjunction with XRFM indicated that neuronal cells move considerable Cu pools from their somatic cells bodies to extended outer processes when activated by depolarization. Further imaging experiments exhibited that the redistribution of copper is dependent on Ca(II) release, paving the way to explore cross-talk between Cu(I) and Ca(II) ions.
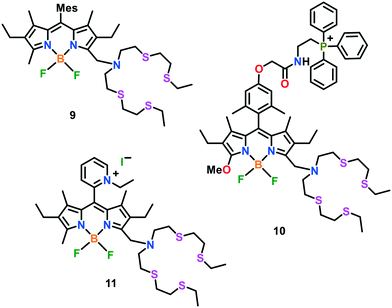
The ratiometric in vitro and in vivo detection of Cu(I) ions has been highly recommended by researchers. This is due to the fact that the intensity ratio of two emission bands may be utilized to verify the probe concentration in a particular medium, which may offer some integral modifications in the environmental parameters (for instance, temperature, pH and polarity of medium) to change the emission intensity of the probe. Meanwhile, a ratiometric BODIPY-based fluorescent probe 9 was reported for imaging Cu(I) ions in live cells (Fig. 5).59 Unlike earlier reported probes, sensor 9 exhibits two emission maxima of nearly equal intensity centred at 505 nm and 570 nm with a quantum yield of 0.002 and 0.003, respectively (λex 488 nm). Upon the addition of Cu(I) ions, the fluorescence response increased by ca. 20-fold with a quantum yield of 0.05 in HEPES buffer (20 mM, pH 7.0). Confocal microscopy experiments revealed that probe 9 was capable of monitoring the changes in the intracellular Cu(I) levels in living cells. Probe 9 was utilized to image the ascorbate-induced intracellular Cu(I) levels in live HEK and C6 rat glioma cells via a ratiometric response.
Fluorescent Cu(I)-responsive probes directed to a specific organelle are gaining increasing attention from biomedical researchers. These organelle-targeting probes may allow the direct, real-time monitoring of copper pools in specific compartments of a live cell. For instance, copper-homeostasis in mitochondria is related to several neurogenerative diseases. Therefore, the development of copper-selective probes specifically targeting mitochondria represents an important tool for biomedical diagnosis. Chang and group60 developed the first mitochondrial-targetable turn-on fluorescent sensor 10 for the imaging of Cu(I) in the mitochondria of living cells. Two of the important characteristics of this probe architecture are as follows: i) fluorescence-based Cu(I) detection using a thioether-rich receptor and ii) mitochondrial-targeting triphenylphosphonium tag to localize the probe in the mitochondria. The UV-vis and emission spectral behaviours of this probe were evaluated in PBS buffer of pH 7.4. Probe 10 featured its emission maximum at 569 nm with a low quantum yield (Φ = 0.009). Upon the addition of 1.0 equiv. of Cu(I) ions, a 10-fold increase in its fluorescence (Φ = 0.05) was observed, with a blue shift in its emission maximum from 569 nm to 558 nm. To gain deep insights into the copper-based homeostasis, a combined imaging and biochemical study was established to monitor mitochondrial Cu(I) pools in the ATP7A fibroblasts of patients with mutations in SCO (synthesis of cytochrome c oxidase).
Giuffrida et al. reported61 another BODIPY-based mitochondria-specific fluorescent probe 11, having a tetrathiaza receptor for live cell Cu(I) imaging. To make it completely soluble in water and specific to mitochondria, a hydrophilic alkyl-pyridinium group was incorporated in the probe (Fig. 5). The UV-vis spectroscopic studies exhibited two bands at 400 nm and 550 nm in HEPES buffer (10 mM, pH 7.2) together with an emission maximum centred at 575 nm. The fluorescence intensity of probe 11 declined upon the addition of Cu(I) in aqueous buffer solution. The quenching in fluorescence was assigned to the photoinduced electron transfer occurring between the fluorophore unit and Cu(I)-bound receptor. The diffusion-ordered spectroscopy (DOSY) experiment clearly revealed the absence of colloidal aggregates in aqueous media. Probe 11 was utilized for Cu(I)-imaging in live cells (neuroblastoma SH-SY5Y cells). Cellular assays revealed that probe 11 could selectively monitor Cu(I) in the cellular environment and found to be non-cytotoxic with long-time incubation. Job's plot and ESI-MS analyses confirmed the binding of Cu(I) to the probe with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 host–guest stoichiometry.
1 host–guest stoichiometry.
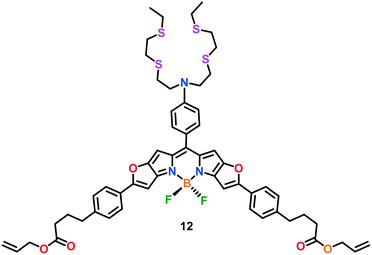
In a recent study, Vilar, Kuimova and co-workers117 developed a new optical BODIPY-based probe 12, which exhibits a turn-on emission response in the presence of Cu(I) ions over other physiologically abundant cations (Fig. 6). Probe 12 displayed an absorbance maximum at 646 nm, with a broad band near 594 nm in methanol. The absorption peak at 646 nm was slightly red-shifted upon the addition of Cu(I) ions to the probe solution. A methanolic solution of 12 was found to be almost non-fluorescent (Φ ∼ 0.032) in the absence of Cu(I) ions. However, upon the introduction of Cu(I) ions, the fluorescence response of 12 was switched-on at 660 nm, with a significant increase in its emission lifetime. The Cu(I) sensing capability of the probe was investigated using neuroblastoma SH-SY5Y cells. The incubation of 12 with these live cells for 24 h exhibited that the probe was non-toxic towards the cellular environment and mainly localised in the lysosome.
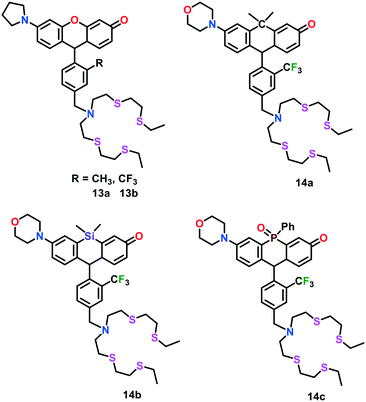
To gain more insights into the spatial and temporal distribution of copper in living systems, Chang and group63 further developed an elegant series of fluorescent Cu(I) probes 14a–14c. These probes were comprised of rhodol dyes substituted with carbon-, silicon- and phosphorous-analogs by replacing the O atom in the classical xanthene moiety. The integration of different non-metallic analogs in the rhodol dyes not only allowed these probes to display rational tuning of their excitation and emission colors from orange to NIR but also resulted in an improvement in their tissue-penetrating ability and minimized sample photodamage. Probes 14a–14c exhibited a turn-on emission response upon binding with Cu(I) in aqueous buffer, with an approximately 5.0-fold, 17.0-fold and 7.0-fold enhancement in fluorescence intensities for 14a, 14b and 14c, respectively. These probes were utilized to monitor labile Cu pools in different live cells.
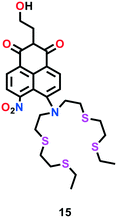
Satriano et al.64 developed a turn-on ratiometric fluorescent probe 15 based on a naphthalimide scaffold for the highly selective sensing of Cu(I) in aqueous solution and neuronal cell lines (Fig. 8). This probe was comprised of a tetrathiaza crown motif, which is well known as a Cu(I) receptor. The Cu(I) sensing ability of probe 15 was investigated using fluorescence titration experiments. A gradual enhancement in the fluorescence intensity of probe 15 was observed in the range of 380–510 nm upon the addition of Cu(I) in HEPES buffer solution. A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry to form complex 15–Cu(I) was suggested by the Job's plot analysis. To assess its practical ability for live cells Cu(I) imaging, probe 15 was successfully applied in neuroblastoma SH-SY5H cells.
1 binding stoichiometry to form complex 15–Cu(I) was suggested by the Job's plot analysis. To assess its practical ability for live cells Cu(I) imaging, probe 15 was successfully applied in neuroblastoma SH-SY5H cells.
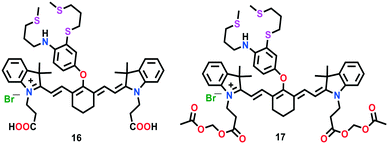
The first-generation NIR-based probe 16 was developed by Chang and group (Fig. 9).65 This probe was constructed by linking an NIR-emitting cyanine dye (cyanine 7) as an optical reporter with a soft sulphur-rich receptor for the selective imaging of Cu(I) ions both in vitro and in vivo. The interesting features of probe 16 such as its emission and excitation in the NIR region make this probe ideal for its utility in thick biological specimens. In the presence of Cu(I) ions, probe 16 displayed a 15-fold increase in the fluorescence intensity of its emission band centred at 790 nm, with an improvement in quantum efficiency (Φ = 0.0042 for 16 and Φ = 0.072 for 16–Cu(I)). A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry for complex 16–Cu(I) was established using Job's plot analysis. Due to the high lipophilicity of lipid bilayers, the carboxyl groups of probe 16 were replaced with acetoxymethyl groups to afford another probe 17. Unlike 16, probe 17 displayed efficient diffusion across cellular membranes. In the cytoplasm, probe 17 could easily be de-esterified to generate 16 by intracellular enzymes. Furthermore, the imaging experiments in live mice cells displayed the excellent ability of probe 17 to report in vivo fluctuations in exchangeable copper(I) pools. In addition, probe 17 successfully visualized the exchangeable Cu levels in a murine model of Wilson's disease. These findings revealed that non-cytotoxic probe 17 is capable of monitoring labile Cu changes in living systems.
1 binding stoichiometry for complex 16–Cu(I) was established using Job's plot analysis. Due to the high lipophilicity of lipid bilayers, the carboxyl groups of probe 16 were replaced with acetoxymethyl groups to afford another probe 17. Unlike 16, probe 17 displayed efficient diffusion across cellular membranes. In the cytoplasm, probe 17 could easily be de-esterified to generate 16 by intracellular enzymes. Furthermore, the imaging experiments in live mice cells displayed the excellent ability of probe 17 to report in vivo fluctuations in exchangeable copper(I) pools. In addition, probe 17 successfully visualized the exchangeable Cu levels in a murine model of Wilson's disease. These findings revealed that non-cytotoxic probe 17 is capable of monitoring labile Cu changes in living systems.
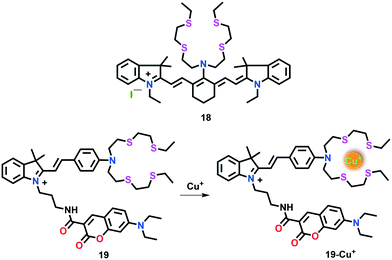
Cao et al.66 developed another first-generation NIR fluorescent probe 18 suitable for the detection of intracellular Cu(I) in live cells (Fig. 10). This probe is based on a tricarbocyanine scaffold and a high affinity Cu(I) BETA receptor. The tricarbocyanine moiety was incorporated in the probe given that its absorption and emission maximum fall in the NIR range. It has been established that the electron-rich S atoms in the BETA receptor quench the fluorescence of the tricarbocyanine dye. However, the fluorescence quenching effect is inhibited upon Cu(I) binding and a turn-on emission response appears. The spectral behaviour of probe 18 was investigated in 25 mM PBS buffer with 10% ethanol (pH 7.0) with and without Cu(I). The absorption band of free probe 18 near 696 nm was red-shifted to 750 nm upon the addition of Cu(I), which was attributed to the coordination of Cu(I) to the N atom of the receptor. The introduction of Cu(I) in 18 was also corroborated with the marked turn-on response in the emission peak at 792 nm (λex = 750 nm). Upon the addition of 1.0 equiv. of Cu(I) ions to 18 (2.5 mM), a nearly 10-fold emission enhancement was observed, which was found to be superior to the probes sensitive only to visible light. Due to its interesting features such as absorption and emission in the NIR region, probe 18 has been successfully employed for Cu(I) imaging in live cells (MG63 cells). The formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 18–Cu(I) complex was suggested with the help of Job's plot analyses.
1 18–Cu(I) complex was suggested with the help of Job's plot analyses.
Shen et al.67 reported a hemicyanine-based ratiometric Cu(I) probe 19 selective for both in vitro and in vivo imaging studies (Fig. 10). Two fluorophore components, indolinium-based hemicyanine and coumarin were linked through an alkyl linker to evade any possible conjugation between them. The indolinium moiety was further attached to a thioether receptor. The incorporation of a coumarin moiety provided a non-responsive emission as an internal standard.
The emission properties of 19 in HEPES buffer (20 mM, pH 7.4) displayed two separate fluorescent bands with emission maxima near 480 nm and 600 nm, which were assigned to the coumarin and indolinium groups, respectively. Upon the addition of Cu(I) ions, the emission peak of the indolinium component at 600 nm decreased; however, a negligible change was observed at 480 nm. The fluorescence intensity ratio of coumarin to indolinium (I600/I480) linearly increased with the progressive addition of Cu(I) (0–1.0 equiv.). Probe 19 was treated with DLD-1 cells to investigate its applications to monitor Cu(I) ions in the cellular environment. Interestingly, this probe displayed specific localization in mitochondria and could be utilized to assess intracellular copper pools.
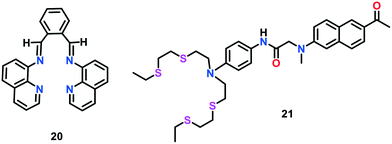
Farhi et al.68 developed a quinoline-based fluorescent and colorimetric sensor 20 with improved selectivity towards Cu(I) ions (Fig. 11). The previously reported Cu(I)-selective probes lack the ability to visually detect Cu(I). The colorimetric Cu(I) sensing by 20 was monitored using UV-vis spectroscopic studies. Upon the addition of Cu(I) to 20 in an acetonitrile: water mixture (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v), a new absorption band at 450 nm appeared with a remarkable increase in peak intensity up to 1.0 equiv. concentration of Cu(I) ions. Moreover, 20 displayed several-fold increase in fluorescence intensity at 530 nm upon the addition of Cu(I) ions together with a bathochromic shift of 35 nm. The Job's plot analyses confirmed the 1
2, v/v), a new absorption band at 450 nm appeared with a remarkable increase in peak intensity up to 1.0 equiv. concentration of Cu(I) ions. Moreover, 20 displayed several-fold increase in fluorescence intensity at 530 nm upon the addition of Cu(I) ions together with a bathochromic shift of 35 nm. The Job's plot analyses confirmed the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry for the formation of complex 20–Cu(I). The weak fluorescence of free 20 was attributed to the flexibility and fast isomerization of the C
1 stoichiometry for the formation of complex 20–Cu(I). The weak fluorescence of free 20 was attributed to the flexibility and fast isomerization of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bond, which may lead to excited state deactivation, inducing the quenching effect. The coordination of Cu(I) with 20 resulted in the isomerisation of the C
N bond, which may lead to excited state deactivation, inducing the quenching effect. The coordination of Cu(I) with 20 resulted in the isomerisation of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bond, leading to an enhancement in fluorescence intensity.
N bond, leading to an enhancement in fluorescence intensity.
In comparison to the conventional one-photon excitation (one-photon microscopy), two-photon microscopy (TPM) has become a more popular tool for the detection of metals in live cells and tissues.118,119 In TPM, two near-infrared photons of low energy are used as the excitation source, which offer a relatively deeper penetration (greater than 500 μM), lower auto-emission and self-absorption of tissues and reduction in photo-damage and photo-bleaching effects. Due to these combined features, TPM results in 3D-imaging of tissues for a long duration. However, the development in this area is hampered due to the lack of two-photon probes for a specific application. Moreover, the previously reported one-photon probes utilized for two-photon microscopy have a lower TP cross-section value (i.e., δTPA < 50 GM), which limits their employment in TPM study. Therefore, many researchers are focused on constructing more efficient TP probes (with a large cross section value) for specific applications and in vivo imaging experiments. In the present study, we only summarized the recent progress in the development of two-photon fluorescent probes for imaging Cu(I) ions.
Lim et al.69 developed the first two-photon fluorescent probe 21 for the selective recognition of Cu(I) ions in live cells and tissues (Fig. 11). Probe 21 consisted of an acyclic NS4 motif as the receptor and a donor–acceptor tethered naphthalene unit as the two-photon excitable PET reporter. Upon the successive addition of Cu(I) to 21 in HEPES buffer (20 mM, pH 7.0), the emission intensity was gradually enhanced; however, a negligible change in its absorption spectrum was observed. The increase in the fluorescence emission is assigned to the blockage of PET event upon Cu(I) coordination. A 4.0-fold increase in emission intensity was observed in the presence of 360 pM free Cu(I). Similar results were obtained for the TP process (excitation wavelength 750 nm) and a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexation between 21 and Cu(I) was demonstrated with the help of Job's plot analyses. Probe 21 was also assessed for the visualization of the distribution of Cu(I) ions in live HeLa cells and tissues at a depth of 90–220 μm with no intervention from other bio-relevant ionic species. In comparison to previously reported BODIPY-based Cu(I) sensors, probe 21 displayed a 13-fold stronger TPEF emission.
1 complexation between 21 and Cu(I) was demonstrated with the help of Job's plot analyses. Probe 21 was also assessed for the visualization of the distribution of Cu(I) ions in live HeLa cells and tissues at a depth of 90–220 μm with no intervention from other bio-relevant ionic species. In comparison to previously reported BODIPY-based Cu(I) sensors, probe 21 displayed a 13-fold stronger TPEF emission.
During the past few years, peptides have also been gaining significant attention as receptors for fluorescent probes due to their efficient cell permeability, appreciable water solubility, bio-compatibility, targeting of specific cell organelles, and effective binding with biologically important cationic species. Moreover, in peptide-based receptors, the binding affinity for metal ions can easily be tuned with an alteration in the sequence of their amino acids.
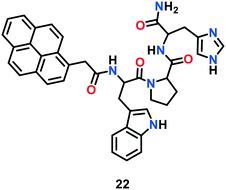
In this case, Mehta et al.70 developed a tripeptide receptor-based ratiometric fluorescent probe, which mimics the binding domain of the metalloprotein (CusF), to monitor Cu(I) ions (Fig. 12). Among the various bio-relevant metals, probe 22 selectively recognised Cu(I) ions via a ratiometric fluorescence response. Probe 22 displayed a marked monomer emission band near 370–410 nm with a tiny emission of excimer in 10 mM HEPES buffer of pH 7.4. A significant decrease in monomer emission at 396 nm and marked increase in excimer emission at 472 nm were observed upon the progressive addition of Cu+ ions to 22.
These findings indicated the dimerization of probe 22 in the presence of Cu+ ions. As calculated from the intensity ratio (I472/I396) at 472 and 396 nm, the fluorescence response of the probe increased up to 130 fold with an increase in Cu(I) concentration. The strong Cu(I)-binding to the peptide receptor is attributed to the presence of imidazole and indole motifs in the long chains of the receptor. Probe 22 exhibited no cytotoxicity towards living A549 cells and was successfully utilized for intracellular Cu(I) imaging via a ratiometric response.
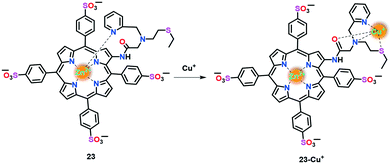
Yi et al.71 reported a Zn(II)-porphyrin-based fluorescent probe 23 for the fast, reversible and selective monitoring of Cu(I) ions in aqueous buffer (Fig. 13). In probe 23, the electron transfer (ET) acceptor pyridine, appended with a thioether moiety, acts as the Cu(I) receptor and the electron transfer (ET) donor Zn(II)–porphyrin unit as the fluorophore. It was speculated that in the absence of Cu(I) ions, pyridine-N is perpendicularly coordinated to Zn(II) to afford a penta-coordinated geometry. This ligation draws Zn(II) slightly out of the porphyrin plane and induces intramolecular ET from the excited fluorophore to the receptor, leading to fluorescence quenching. However Cu(I)-binding to the receptor inhibits the ET process and allows the Zn centre to reside in the porphyrin plane, which results in an enhancement in fluorescence.
The Cu(I) sensing ability of 23 was tested using UV-vis and fluorescence spectroscopic techniques. Upon the addition of Cu(I) to 23 in aqueous buffer (PBS, 20 mM, pH 7.0), a decrease in the Soret band at 430 nm with the concomitant appearance of a new band at 415 nm was observed, which was assigned to the detachment of the pyridyl-N from the Zn centre. Probe 23 exhibited a turn-on fluorescence response at 632 nm in the presence of Cu(I), which is probably due to the inhibition of the intramolecular electron transfer event. A 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry was suggested with the help of Job's analyses. This probe has been utilized for intracellular Cu(I)-imaging applications in HeLa and A549 cells.
1 binding stoichiometry was suggested with the help of Job's analyses. This probe has been utilized for intracellular Cu(I)-imaging applications in HeLa and A549 cells.
The emission response of binding-based Cu probes is an outcome of reversible Cu(I) binding to the probe. Therefore, these probes may be advantageous in monitoring the level of labile Cu in a time-dependent mode. However, the comparative strength of Cu(I) binding and the dynamic equilibrium of Cu(I) between the receptor and bio-ligands/proteins of the probe (which constitute the labile Cu pool) should be carefully characterized and interpreted to address the key roles of the labile Cu pools in the bio-system under investigation (Table 1).
| Probe | Absorbance λmax (nm) | Emission λmax (nm) | Φ | f e | Mechanismc | pH/medium | Ref. |
|---|---|---|---|---|---|---|---|
| a Fluorescence quantum yield for Cu(I)-bound probe. b Enhancement factor in emission intensities between Cu(I)-bound probe and free probe. c Photoinduced electron transfer (PET). Charge transfer (CT). d Two-photon (T-P) absorption. | |||||||
| 1 | 365 | 485 | 0.14 | 4.6 | PET | PIPES/pH 7.2 | 50 |
| 2 | 373 | 464 | 0.048 | 20 | PET | CH3OH (0.1% CH3CN) | 52 |
| 3 | 391–346 | 486–432 | 0.57–0.21 | 34–160 | PET | CH3OH (0.1% CH3CN) | 53 |
| 4a | 396 | 508 | 0.083 | 65 | PET | MOPS/pH 7.2 | 54 |
| 4b | 376 | 498 | 0.033 | 41 | |||
| 4c | 358 | 467 | 0.010 | 9 | |||
| 5 | 388 | 506 | 0.074 | 57 | PET | MOPS/pH 7.2 | 55 |
| 6 | 364 | 455 | 0.41 | 180 | PET | PIPES/pH 7.2 | 56 |
| 7 | 540 | 561 | 0.13 | 10 | PET | 7.0 (HEPES) | 57 |
| 8 | 540 | 548 | 0.40 | 75 | PET | 7.0 (HEPES) | 58 |
| 9 | 548 | 556 | 0.05 | 20 | PET | 7.0 (HEPES) | 59 |
| 10 | 550 | 558 | 0.05 | 10 | CT | 7.4 (PBS) | 60 |
| 11 | 550 | 575 | 0.11 | — | PET | 7.2 (HEPES) | 61 |
| 13a | 529 | 545 | 0.15 | 13 | PET | — | 62 |
| 13b | 534 (910)d | 557 | 0.22 | 40 | |||
| 14a | 580 | 608 | 0.30 | 5 | PET | — | 63 |
| 14b | 616 | 639 | 0.053 | 17 | |||
| 14c | 654 | 680 | 0.0020 | 7 | |||
| 15 | 435 | 470 | 0.088 | 2 | CT | 7.2 (HEPES) (50% CH3CN) | 64 |
| 16 | 760 | 790 | 0.072 | 15 | PET | 7.0 (HEPES) | 65 |
| 18 | 750 | 792 | — | 9.6 | — | 7.0 (PBS with 10% ethanol) | 66 |
| 19 | 430, 550 | 480, 600 | — | — | — | 7.0 (HEPES) | 67 |
| 20 | 450 | 530 | — | — | — | CH3CN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (8 H2O (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) 2) |
68 |
| 21 | 363 (750)d | 482 | 0.13 | 0.4 | PET | 7.0 (HEPES) | 69 |
| 22 | 343 | 472 | 0.0145 | 2 | PET | 7.4 (HEPES, 1% DMF) | 70 |
| 23 | 415 | 623 | 0.169 | 11.8 | PET | 7.0 (PBS) | 71 |
4.2. Reaction-based probes
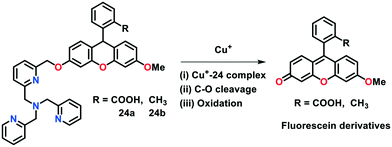
Rather than a traditional lock-and-key binding approach, reaction-based probes, in principle, have been found to be more specific towards analyte sensing in biological systems given that they do not interfere with intracellular chemical processes and generate non-toxic procucts.72 The only challenge in the development of reaction-based probes is identifying the suitable chemical reaction that fulfils the need of chemoselectivity. In this section, we summarize the research progress employed in developing fluorescent Cu(I)-selective probes featuring a specific Cu(I)-catalysed chemical reaction for in vitro and in vivo fluorescence sensing.In an elegant study, Taki et al.73 constructed a reaction-based Cu(I) probe 24 containing a tris-(2-pyridyl)-methyl)amine (TPA) tethered with fluorescein via a benzylic-ether linker (Fig. 14). In the absence of Cu(I), 24 displayed weak fluorescence and was found to be insensitive to oxygen. The introduction of Cu(I) to 24 in HEPES buffer (50 mM, pH 7.2) promoted the cleavage of the C–O bond of benzylic-ether to afford fluorescent product 3′-O-methylfluorescein, with a significant increase (greater than 100-fold) in fluorescence response at 513 nm. The ESI-MS and HPLC analyses revealed that the fluorescent product (i.e., 3′-O-methyl fluorescein; Φ = 0.37) was obtained with a moderate yield of ca. 70% by following a 2 h reaction. Probe 24a was not found to be cell permeable, presumably due to the hydrophilic character of the –COOH group present on the fluorescein moiety. Therefore, the –COOH group of 24a was replaced by a methyl group to obtain cell-permeable probe 24b for potential applications towards Cu(I) imaging in live cells.
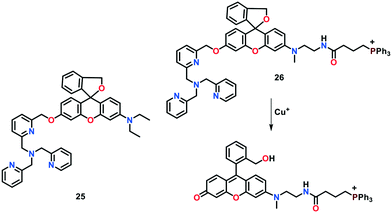
There were two key reasons for not utilizing probe 24a for monitoring the Cu(I) levels in live cells, as follows: i) its cell permeability and ii) the diffusion of its resultant fluorescent product into the cytosol. Thus, to overcome these issues, the same group presented two rhodol-based fluorogenic probes 25 and 26 which contain an aminoethyl function at the xanthene-based scaffold (Fig. 15).74 These probes were found to be capable of being modulated with an organelle-targeting functional group. In the absence of Cu(I) ions, 25 exhibited a negligible emission in HEPES buffer (50 mM) of pH 7.2 due to its spirocyclic structure. However, the addition of Cu(I) to 25 resulted in a 100-fold increase in the fluorescence response at 542 nm. The treatment of 25 with Cu(I) induced the cleavage of the C–O bond of the benzylic-ether and a fluorescent ring-opened form of the cleavage product was obtained. Further analyses by ESI-MS and HPLC studies exhibited the formation of a fluorescent reaction product in a significant yield (about 99%) after a reaction for 3 h. Probe 25 showed detectable fluorescence at Cu+ concentrations up to 1.0 micromolar even under the physiologically reducing environment. Probe 25 was found to be selective for Cu-based ions; however, a slight change in its emission was observed in the presence of Co(II) ions. The possible interference of Co(II) ions with 25 during intracellular Cu(I) imaging studies prompted Taki's group to develop an organelle-specific and copper-selective probe. Probe 26 was found to be mitochondria-targeting and utilized for monitoring mitochondrial Cu(I) pools in live Hela cells.
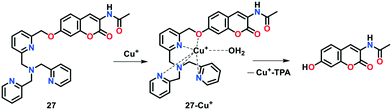
Inspired from the results obtained with indicator 24, a reaction-based coumarin-TPA-based fluorogenic derivative 27 was developed by Yu and group75 for the efficient detection of Cu(I) ions in aqueous buffer solution (Fig. 16). To construct 27, a coumarin-based fluorophore was combined with the tetradentate N4 TPA ligand via a benzylic-ether bond. Probe 27 displayed a violet-colored fluorescence at 410 nm in PBS buffer (25 mM, pH 7.2). Under the physiologically reducing environment, the treatment of Cu(I) with 27 led to a considerable fluorescent enhancement with a significant red shift in emission from 410 nm to 472 nm. This sensing behaviour is attributed to the Cu(I)-catalysed oxidative cleavage of the benzylic ether C–O bond in the presence of O2. In addition, an increase in the emission intensity ratio (i.e., I472/I410) from 0.23 to 7.66 was observed upon the addition of 5 μM Cu(I) ions. The limit of detection for probe 27 was determined to be 2.29 × 10−7 M.
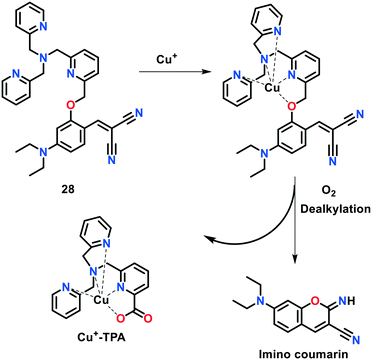
Hu et al.76 constructed another coumarin-TPA-based fluorogenic probe 28 for monitoring Cu(I) ions in aqueous solution and in living systems (Fig. 17). The sensing behaviour of this probe was investigated via the Cu(I)-induced formation of the fluorescent product (imino)coumarin. The fluorescence Cu(I) sensing ability of 28 was investigated in aqueous buffer. The fluorescence intensity of 28 at 480 nm was considerably increased up to ca. 60-fold with the addition of increasing concentrations of Cu(I) ions (0–5 μM) to the probe solution. Hence, the non-fluorescent TPA-based indicator led to the formation of fluorescent (imino)coumarin. Probe 28 exhibited a detection limit of 1.08 × 10−8 M towards Cu(I) ions.
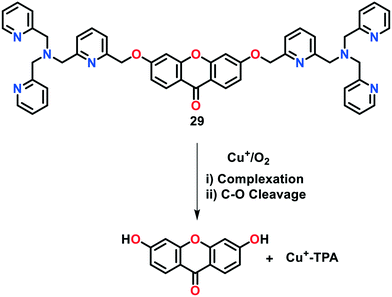
Govindaraju and group77 presented a reaction-based fluorescent probe 29 (XanCu) for the selective monitoring of Cu(I) ions under the physiologically-reducing environment (Fig. 18). In probe 29, xanthone (a blue-fluorescent dye) and tripicolylamine moieties were incorporated to act as reporter and Cu(I)-binding units, respectively. The parent probe without Cu(I) displayed a negligible fluorescence response with an emission maximum at 445 nm. However, the presence of Cu(I) promoted the oxidative cleavage of the C–O bond of benzyl-ether, with the formation of a fluorescent hydroxyxanthone product. This Cu-catalysed reaction resulted in 30-fold increase in the fluorescence response of the probe at 445 nm (quantum yield 0.265). The concentration-dependent studies revealed the fluorescence detection of Cu(I) ions at concentrations of up to submicromolar levels given that this concentration level was sufficient to release the phenolic xanthone (Xan) dye from XanCu. Interestingly, probe 29 was utilized as a two-photon fluorescent indicator for Cu(I) imaging experiments.
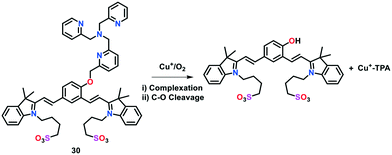
Later, the same group78 constructed a highly water soluble probe 30 for the reaction-based turn-on detection of Cu(I) ions. A very weak fluorescence was displayed by probe 30 without Cu(I) ions (Fig. 19). However, treatment of 30 with Cu(I) in water resulted in a 30-fold increase in fluorescence intensity at 695 nm. This turn-on emission response was assigned to the cleavage of the typical benzylic-ether C–O bond. Under physiological conditions, this probe reacted with sub-micromolar concentrations of Cu(I) in HEPES buffer (50 mM, pH 7.2) to generate a quinone-based cyanine dye, which emitted in the near-infrared region. Furthermore, the ability of 30 to monitor Cu(I) in living systems was investigated by loading it with HEK293T live cells.
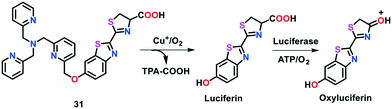
Chang and group79 developed a highly selective and tissue-specific bioluminescent probe 31 (copper-caged luciferin-1) to track the real-time changes in labile copper pools in live cells and animals (Fig. 20). In the presence of Cu(I) ions, probe 31 generated D-luciferin upon a Cu-based oxidative cleavage reaction, subsequently exhibiting a bioluminescent reaction with firefly luciferase. The probe coupled with firefly luciferase reporter was also utilized to monitor the imbalanced levels of Cu in a murine-model of NAFLD. In the case of Cu deficiency or excess, the probe was found to be capable of detecting the physiological changes in the labile Cu(I) levels in mice and living cells. The Cu(I)-induced production of luminescence in 31 was studied in Tris buffer solution (50 nM, pH 7.4). Probe 31 displayed concentration-dependent monitoring towards Cu(I), exhibiting an approximately 80-fold increase in bioluminescence with the addition of 1 equiv. of Cu(I) ions. A high contrast of fluorescence (greater that 80-fold) was achieved with an increase in Cu concentration.
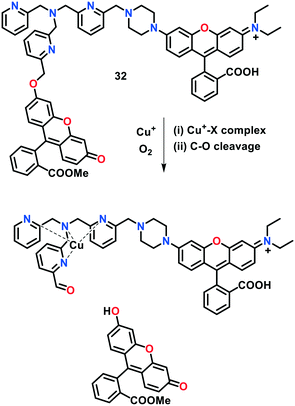
In another report, Chang's group reported a first-generation ratiometric FRET-based fluorescent probe 32 in which a fluorescein unit was connected with rhodamine dye via a TPA linker (Fig. 21).120 The Cu(I)-induced oxidative cleavage occurring between fluorescein (i.e., electron donor) and rhodamine (i.e., electron acceptor) led to a significant decrease in the FRET process. Probe 32 was highly selective towards Cu(I) and facilitated ratiometric measurements, leading to a decrease in the potential interferences raised by varying sample thicknesses, concentration of dye, and intensity of light. This activity-based FRET probe enabled the selective and specific imaging of labile copper pools in live cells by utilizing a typical TPA motif for the Cu(I)-induced oxidative cleavage reaction. Due to the ratiometric response of probe 32, the dynamic changes in the labile copper levels were visualized in living cells with either Cu supplementation or Cu depletion. Moreover, this probe could monitor the obvious decrease in the endogenous labile Cu levels in MEF cells deficient in Cu transporter CTR1. A link between labile Cu pool, glutathione metabolism and oncogenic transformation was established, which paves the way to understand how decreased labile Cu(I) levels may participate into cancer development and progression.
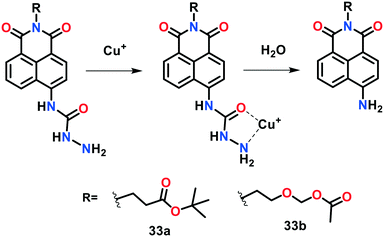
Taking advantage of the strong binding of hydrazide moiety with Cu(I), Park et al.80 developed two reaction-based fluorogenic probes 33a–33b for the ratiometric fluorescent sensing of Cu(I) ions in living cells (Fig. 22). Coordination of CuI or CuII with fluorescent probes 33a and 33b and their concomitant hydrolysis in aqueous medium afforded the formation of the corresponding molecules with a ratiometric change in fluorescence. The formation of the hydrolytic products was confirmed using ESI-MS and 1H NMR spectroscopic studies. The sensing behaviour of 33a and 33b towards different cationic species was examined by monitoring their absorption and emission spectral changes in HEPES buffer (10 mM, pH = 7.4) with 1% DMF. Probe 33a displayed an emission maximum at 480 nm (Φ = 0.014), which was red-shifted to 545 nm (Φ = 0.018) upon the addition of Cu(I) or Cu(II), with an increase in fluorescence. A color change from blue to yellow-green was also observed in the fluorescence of 33a upon the addition of only copper ions over other cations. Similar emission patterns were observed for probe 33a (original probe, λem = 480 nm, Φ = 0.012 and copper bound 33a, λem = 550 nm, Φ = 0.020). The detection limit of probe 33a was determined to be 0.7 μM for Cu(I) ions. These investigations indicated that 33a and 33b are highly responsive ratiometric fluorescent probes for Cu(I) and/or Cu(II) ions. The copper imaging capability of these indicators was also demonstrated in live HeLa cells, where 33a was found to be localized predominantly into the endoplasmic reticulum (ER).
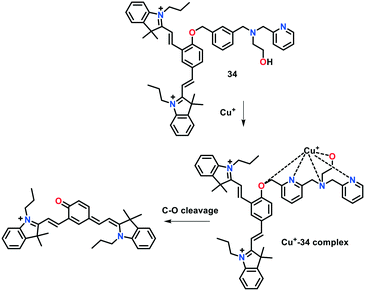
In a recent report, Zhang, Qi and co-workers presented NIR-active fluorescent probe 34 for monitoring of Cu(I) ions (Fig. 23).121 This probe consisted of a cyanine–quinone signalling unit and N3O receptor, displaying absorption and emission maxima near 415 nm and 710 nm (λex 560 nm), respectively, in 25 mM phosphate buffer solution of pH 7.2. The addition of 10 equiv. of Cu(I) ions to the probe solution resulted in a significant redshift (ca. 150 nm) in its absorption spectrum. Interestingly, a 30-fold enhancement was observed in the fluorescence intensity of 34 with a turn-on response at 710 nm upon the introduction of Cu(I) ions. This probe exhibited high selectivity and sensitivity for Cu(I) ions with a detection limit of 9.1 × 10−5 M. The introduction of Cu(I) induced the cleavage of the benzylic ether bond (C–O) of the probe, leading to the release of the cyanine–quinone dye. The bioimaging of Cu(I) in live cells (cultured A549 cells) clearly revealed that this probe is highly biocompatible and can penetrate the cell membrane.
5. Protein-based genetically encoded probes
Live cells may only sense the availability of Cu, not its total concentration in the cytoplasm, and react accordingly. However, although a few small-molecular fluorescent probes have been developed for cellular copper visualization, their incubation requirement with high Cu concentration may disturb the cellular Cu level beyond its physiological amount. Moreover, small-molecular probes may suffer from several issues such as water insolubility, cellular permeability, and cytotoxicity, which limit their practical applications in real biological samples. To overcome these issues, protein-based genetically encoded Cu(I) probes relying on the structural change of the probe domain may be an alternative and appealing approach. During the past few years, several genetically encoded sensors have been reported for the successful in vivo monitoring of metal ions.98–103Based on the previously reported sensors, Wegner et al.98 developed a first-generation protein-based genetically encoded probe (referred to as Amt1-FRET) to detect copper(I) in 2010 (Fig. 24a). Amt1-FRET was constructed by subcloning a Cu binding domain of Amt1 (a Cu(I)-regulatory protein in yeast Candida glarata) between a yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP). Amt1 is a metallothionein-based protein, which consists of eight cysteine groups, creating a Cu(I)-binding region. Excitation of FRET (i.e., fluorescence resonance energy transfer) donor CFP (λex ∼ 433 nm) resulted in the appearance of emission bands near 527 nm and 477 nm for the YFP and CFP motifs, respectively. The FRET response between the protein pair was significantly increased with the introduction of Cu(I), as evidenced by the increase in the emission intensity ratio (I527/I477) from 1.95 to 2.26. The ICP-MS analysis clearly revealed the formation of a tetracopper(I) (Cu4) cluster of Amt1-FRET. The authors demonstrated that Amt1-FRET displays strong binding to Cu(I) inside mammalian CHO-K1 cells (Chinese hamster ovary cells) in a controlled manner.
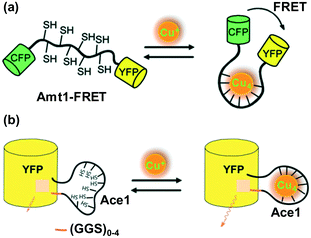 | ||
| Fig. 24 Design of FRET-based genetically encoded Cu(I) reporters. (a) Amt1-FRET and (b) YFP-Ace1 containing GGS linkers of different lengths. Adapted from ref. 98 and 99 and modified, respectively. | ||
Even though Amt1-FRET exhibited excellent Cu(I)-imaging in a biological environment, issues such as its low FRET response (only up to 15%) and rigid non-variable design limit the utility of this probe for further biological applications. Thus, to improve the Cu(I)-binding affinity and FRET response between protein partners, Chen, He and co-workers99 developed a new series of genetically coded fluorescent Cu+ probes, which were designated as YFP-Ace1 (Fig. 24b). A variation in the GGS linker length induced marked changes in the Cu(I)-binding affinity response level of the probes. To construct YFP-Ace1, the Cu(I) binding domain of Ace1 (a Cu(I) regulator in Saccharomyces cerevisiae and an analogue to Amt1) was subcloned between the Y145 and H146 residues of the yellow fluorescent protein, with the aim to place it close to the signalling unit. Upon the addition of Cu(I) to YFP-Ace1, the conformation of the binding unit remarkably altered to form a tetracopper(I) cluster. The conformational change in the binding region led to the tuning of the emissive response of the reporter, which stemmed from the changes occurring in the local environment of the reporter. Excitation with 496 nm light displayed an emission band near 515 nm. Cu(I)-Imaging, both in vitro and in vivo, was investigated using YFP-Ace1 and its variants, which exhibited a relatively more significant ratiometric response with a shorter linker length.
Yan et al.100 constructed a genetically encoded fluorescent protein PMtb-FRET (Mtb = Mycobacterium tuberculosis) as a highly sensitive and selective probe for Cu(I) ions in the presence of other cations (Fig. 25a). Taking advantage of the Cu(I)-binding mediated conformational change of PMtb, this probe was constructed by flanking the Cu(I)-binding domain of Mtb-CDC 1551 (residues 1–162) with the cyan and yellow fluorescent protein pair (CFP and YFP, respectively). The emission spectra of PMtb-FRET with and without Cu(I) were measured by exciting the FRET donor CFP (λex ∼ 433 nm) and the fluorescence intensity of YFP (527 nm) and CFP (477 nm) was recorded. PMtb-FRET affinity to Cu(I) was monitored following the FRET signal change, which exhibited the high sensitivity of PMtb-FRET towards Cu(I). The tight Cu(I)-PMtb-FRET binding indicated the limited availability of Cu(I) ions inside Mtb cells.
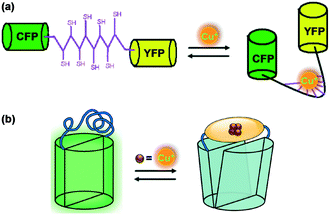 | ||
| Fig. 25 Design of genetically encoded Cu(I) probes. (a) PMtb-FRET based on the FRET method and (b) EGFP-145Amt1 based on engineered structural distortion of a green fluorescent protein (EGFP). Adapted from ref. 100 and 101, respectively. | ||
Liang et al.101 presented a different design strategy to construct a genetically encoded fluorescent sensor for copper(I) imaging in vivo. This approach was based on the engineered structural distortion of a green fluorescent protein (EGFP) and found to be advantageous over previously reported FRET methods. Amt1 was incorporated between two residues (145 and 146) of EGFP to afford the fluorescent Cu(I)-selective probe EGFP-145Amt1 (Fig. 25b). The fluorescence intensity of EGFP-145Amt1 at 512 nm significantly turned down (up to 50% of original fluorescence) upon the addition of 4.0 equiv. of Cu(I) ions. The binding of Cu(I) to Amt1 was found to be reversible with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 stoichiometric ratio. The original fluorescence of the probe could easily be revived in the presence of a Cu(I) chelator, cyanate. Probe EGFP-145Amt1 was applied to mammalian CHO cells to perform Cu(I) imaging experiments in vivo.
4 stoichiometric ratio. The original fluorescence of the probe could easily be revived in the presence of a Cu(I) chelator, cyanate. Probe EGFP-145Amt1 was applied to mammalian CHO cells to perform Cu(I) imaging experiments in vivo.
Koay et al.102 constructed a FRET fluorescent probe to assess the fluctuations observed simultaneously in the intracellular Cu(I) and Zn(II) levels in a cell. Different probe variants were developed by systematic replacement of a few cysteine groups with methionine in previously reported eCALWY Zn(II) probes.103 These probes contain two terminal metal-binding domains, ATOX1 and WD4 (the fourth domain of WDP), and a flexible (GGSGGS)9 peptide linker (Fig. 26). Fluorescent proteins namely cerulean (Cer) and citrine (Cit), served as the donor–acceptor pair flanking the cation binding domains. In particular, the present investigation focused on reengineering the metal-binding site of the original eCALWY-1 probe to maintain its Cu(I) affinity, while simultaneously eliminating its Zn(II) binding property. Taking advantage of the distinguished geometric and coordination preferences of Zn(II) and Cu(I) ions, the effect of cysteine-to-methionine mutations on the sensitivity and affinity of the probe to metals was investigated. Although genetically encoded FRET-based probes display unprecedented Cu(I)-imaging applications in vivo, their fluorescence change is still small. This may be due to the fact that the fluorescent proteins used in these probes are comprised of long tags, which influence the folding of the central probe domain. Moreover, it is not easy to find an appropriate probe domain whose conformational change can well-match the long Forster radius (i.e. ∼5 nm) of the majority of fluorescence proteins. Therefore, sophisticated image processing is in high demand to enhance the image contrast.
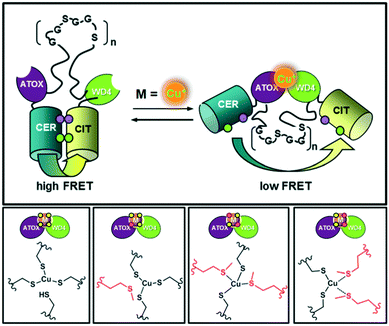 | ||
| Fig. 26 Design of a series of FRET-based fluorescent Cu(I) reporters. Adapted from ref. 102. | ||
Luminescent biological probes containing trivalent lanthanide (Ln3+) ions have also been developed for monitoring Cu(I) ions. Lanthanides exhibit interesting luminescent features such as line-like visible to NIR emission profile, large Stokes shift, long luminescence lifetime and good photostability.122,123 However, the direct excitation of lanthanides is not efficient due to their forbidden f–f transitions (ε < 10 M−1 cm−1), as evidenced by Laporte's rule. The efficient lanthanide luminescence depends on the “antenna effect” in which an organic luminophore (i.e., antenna) acts as a light harvesting device to transfer electronic energy to the lanthanide ion to populate its emissive excited state.
In this case, McClenaghan, Sénèque and co-workers presented a series of lanthanide conjugated cyclic peptides as Cu(I)-responsive probes, named LCC1Tb (Fig. 27).122 These cyclic peptides were inspired by the copper-chaperone CusF, i.e., a Cu-trafficking periplasmic protein of Gram-negative organisms (Fig. 27a). The effect of the amino acid sequence on the Cu(I)-recognition properties of these cyclic peptides was examined in this report. The probes were 18-amino acid peptides comprised of the 16-amino acid Cu(I)-binding loop of CusF, which was closed by a dipeptide turn. In probes LCC1Tb (Fig. 27b), the Cu(I) receptor site was composed of one imidazole ring from histidine, two thioether groups from methionine and one indole ring from tryptophan, which displayed a cation–π type interaction with Cu(I) in CusF. After binding with Cu(I), the resultant Cu-LCC1Tb existed in two forms in equilibrium, i.e., one in which tryptophan forms a cation–π interaction and other without this interaction (Fig. 27c). A Tb3+ complex, for providing a luminescence reporter, was conjugated to the peptide probes, close to tryptophan. The tryptophan residue acts as an antenna for Tb3+ luminescence. The luminescence mechanism depends on the inflection of the antenna affect between the tryptophan amino acid residue and Tb3+ ion in the probe when Cu(I) interacts with tryptophan. Several important factors that govern the cation–π interaction, and thus the behaviour of the probe were highlighted in this study.
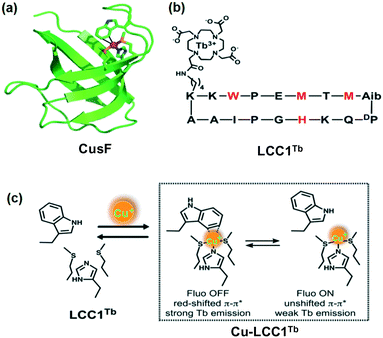 | ||
| Fig. 27 (a) Crystallographic structure of CusF exhibiting the Cu(I) binding site. (b) Amino acid sequence of LCC1Tb, chelating moieties in red. (c) Metal binding equilibrium showing the two forms of the Cu·LCC1Tb complex. Adapted from ref. 122 and modified. | ||
In a recent report, the same group reported another lanthanide-based Cu(I)-responsive probe, i.e., LCCRTbEu (Fig. 28).123 This probe was designed by appending a naphthalene/Eu3+ complex pair to probe LCC1Tb. The new probe LCCRTbEu containing two distinct antenna/lanthanide couples displayed a ratiometric Cu(I) response. The addition of Cu(I) increased the luminescence intensity of Tb3+ and Eu3+ up to 1.35 and 3.15 fold, respectively. The lifetime of the Tb3+ luminescence was also influenced by Cu(I) binding. Hence, LCCRTbEu could be utilized for the ratiometric detection of Cu(I) by monitoring either its spectral variations or luminescence lifetime change. This strategy of using antenna/lanthanide couples looks easy to implement. Different ratiometric systems may be designed and used in many instances, notably in luminescence sensing applications.
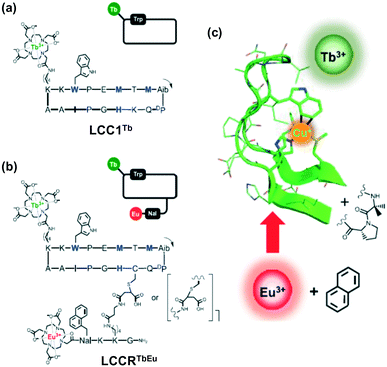 | ||
| Fig. 28 Chemical structure and schematic representation of (a) LCC1Tb and (b) LCCRTbEu, Cu-chelating amino acids in blue and black. The arrow indicates the N-to-C direction within the cyclic peptide. Nal = 3-(1-naphthyl)-L-alanine; Aib = 2-aminoisobutyric acid; DP = D-proline. (c) Cu+ binding site of CusF and design principle showing the positioning of the various components. Adapted from ref. 123. | ||
6. Conclusion and outlook
Herein, the recent progress on small synthetic molecules and genetically encoded proteins as Cu(I)-selective fluorescent probes has been highlighted. As can be seen from the examples described in this study, either a binding- or reaction-based approach is usually employed for fluorimetric Cu(I) detection, whereas some studies adopted a ratiometric detection strategy. Most of the established fluorescent probes rely on the specific attachment of a thioether-based receptor (such as tetrathiaza crown) with Cu(I) ions. These probes have undoubtedly shown huge potential in Cu(I) imaging, which is attributed to the lipophilic character of their receptors containing soft S-donor atoms.The selectivity of the receptor unit for a particular metal ion can easily be achieved by careful ligand design using fundamental coordination chemistry. By considering a few basics of Cu homeostasis (such as S binding and low coordination number), possible strategies can be envisioned to build highly selective and specific Cu(I)-responsive receptors. In this case, the selective ligation of Cu using thioether-based receptors has been extensively exploited in biology and synthetic chemistry. Consequently, thioether ligation is widely implemented to develop Cu-specific receptor motifs for imaging applications. Indeed, the diverse panel of small molecule-based fluorescent Cu(I) probes with variable characteristics (visible light to NIR excitation profiles, hydrophilic nature, signal-to-noise contrast, etc.) has resulted in the identification of important roles of copper as a signalling body in biology. The structural and spectral analyses of cupro-protein binding sites, combined with the study of small-molecular model complexes, has presented the privileged behaviour of the thioether unit through methionine coordination as a key strategy for pH-independent and oxidation-resistant Cu ion binding.
Despite the successful copper recognition of thioether-based fluorescent probes, the structural variation of these receptors is limited to the use of macrocyclic and amino-thioether derivatives. In addition, the utility of many Cu(I) probes in biological media is still limited due to their requirement of an organic solvents (either pure organic or organic-buffer system) for functioning. Thus, the further expansion of the chemical toolbox of molecular sensors would certainly result into new opportunities and challenges to develop new biology of Cu and other essential metals. Consequently, we anticipate that this article will act as a guide to design and develop cation-responsive probes in the future with improved high fluorescence contrast and quantum yields.
Conflicts of interest
There is no conflict to state.Acknowledgements
SK acknowledges Department of Science and Technology (DST), New Delhi for financial assistance as Inspire Faculty Award (IFA CH-213) [DST/INSPIRE/04/2015/002971]. PK thanks DST-SERB, New Delhi for TARE project (TAR/2021/000204).References
- D. Wu, A. C. Sedgwick, T. Gunnlaugsson, E. U. Akkaya, J. Yoon and T. D. James, Chem. Soc. Rev., 2017, 46, 7105 RSC.
- K. P. Carter, A. M. Young and A. E. Palmer, Chem. Rev., 2014, 114, 4564 CrossRef CAS PubMed.
- T. Hirayama, M. Niwa, S. Hirosawa and H. Nagasawa, ACS Sens., 2020, 5(9), 2950 CrossRef CAS PubMed.
- Y. Li, M. Chen, Y. Han, Y. Feng, Z. Zhang and B. Zhang, Chem. Mater., 2020, 32(6), 2532 CrossRef CAS.
- M. L. Zastrow, Z. Huang and S. J. Lippard, ACS Chem. Biol., 2020, 15(2), 396 CrossRef CAS PubMed.
- S. Singha, Y. W. Jun, S. Sarkar and K. H. Ahn, Acc. Chem. Res., 2019, 52(9), 2571 CrossRef CAS PubMed.
- A. Sarkar, S. Chakraborty, S. Lohar, E. Ahmmed, N. C. Saha, S. K. Mandal, K. Dhara and P. Chattopadhyay, Chem. Res. Toxicol., 2019, 32(6), 1144 Search PubMed.
- U. Haldar and H. Lee, ACS Appl. Mater. Interfaces, 2019, 11(14), 13685 CrossRef CAS PubMed.
- M. J. Reimann, D. R. Salmon, J. T. Horton, E. C. Gier and L. R. Jefferies, ACS Omega, 2019, 4(2), 2874 CrossRef CAS PubMed.
- S. Jia, K. M. Ramos-Torres, S. Kolemen, C. M. Ackerman and C. J. Chang, ACS Chem. Biol., 2018, 13(7), 1844 CrossRef CAS PubMed.
- J. F. Zhang, Y. Zhou, J. Yoon and J. S. Kim, Chem. Soc. Rev., 2011, 40, 3416 RSC.
- B. Kaur, N. Kaur and S. Kumar, Coord. Chem. Rev., 2018, 358, 13 CrossRef CAS.
- J. Wu, W. Liu, J. Ge, H. Zhang and P. Wang, Chem. Soc. Rev., 2011, 40, 3483 RSC.
- Q. Zhao and F. Li, Chem. Soc. Rev., 2010, 39, 3007 RSC.
- D. Udhayakumari, S. Naha and S. Velmathi, Anal. Methods, 2017, 9, 552 RSC.
- (a) A. Ramdass, V. Sathish, E. Babu, M. Velayudham, P. Thanasekaran and S. Rajagopal, Coord. Chem. Rev., 2017, 343, 278 CrossRef CAS; (b) A. Ramdass, V. Sathish, M. Velayudham, P. Thanasekaran, S. Umapathy and S. Rajagopal, Sens. Actuators, B, 2017, 240, 1216 CrossRef CAS.
- S. Kumar, S. Singh, A. Kumar and P. Kumar, Dalton Trans., 2021, 50, 2705 RSC.
- C. He, S. Yu, S. Ma, Z. Liu, L. Yao, F. Cheng and P. Liu, Molecules, 2019, 24, 4032 CrossRef CAS PubMed.
- E. L. Que, D. W. Domaille and C. J. Chang, Chem. Rev., 2008, 5, 1517 CrossRef PubMed.
- M. Valko, K. Jomova, C. J. Rhodes, K. Kuča and K. Musílek, Arch. Toxicol., 2016, 90, 1 CrossRef CAS PubMed.
- R. A. Festa and D. J. Thiele, Curr. Biol., 2011, 21, R877 CrossRef CAS PubMed.
- (a) H. Shi, Y. Jiang, Y. Yang, Y. Peng and C. Li, BioMetals, 2021, 34(1), 3–14 CrossRef CAS PubMed; (b) J. Chen, Y. Jiang, H. Shi, Y. Peng, X. Fan and C. Li, Pflugers Arch., 2020, 472(10), 1415 CrossRef CAS PubMed.
- D. C. Brady, M. S. Crowe, M. L. Turski, G. A. Hobbs, X. Yao, A. Chaikuad, S. Knapp and K. Xiao, Nature, 2014, 509, 492 CrossRef CAS PubMed.
- Y. F. Luo, J. Zhang, N. Q. Liu, Y. Luo and B. L. Zhao, Sci. China: Life Sci., 2011, 54, 527 CrossRef CAS PubMed.
- D. Strausak, J. F. B. Mercer, H. H. Dieter, W. Stremmel and G. Multhaup, Brain Res. Bull., 2001, 55, 175 CrossRef CAS PubMed.
- M. Andersson, D. Mattle, O. Sitsel, T. Klymchuk, A. Nielsen, L. B. Møller, S. H. White, P. Nissen and P. Gourdon, Nat. Struct. Mol. Biol., 2014, 21, 43 CrossRef CAS PubMed.
- M. T. Morgan, D. Bourassa, S. Harankhedkar, A. M. McCallum, S. A. Zlatic, J. S. Calvo, G. Meloni, V. Faundez and C. J. Fahrni, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 12167 CrossRef CAS PubMed.
- E. Gaggelli, H. Kozlowski, D. Valensin and G. Valensin, Chem. Rev., 2006, 106, 1995 CrossRef CAS PubMed.
- Y. Sheng, M. Chattopadhyay, J. Whitelegge and S. J. Valentine, Curr. Top. Med. Chem., 2012, 12, 2560–2572 CrossRef CAS PubMed.
- U. C. Müller, T. Deller and M. Korte, Nat. Rev. Neurosci., 2017, 18, 281 CrossRef PubMed.
- N. N. Pavlova and C. B. Thompson, Cell Metab., 2016, 23, 27 CrossRef CAS PubMed.
- X. He, J. Zhang, X. Liu, L. Dong, D. Li, H. Qiu and S. Yin, Sens. Actuators, B, 2014, 192, 29 CrossRef CAS.
- K. R. Bandi, A. K. Singh and A. Upadhyay, Mater. Sci. Eng., C, 2014, 34, 149 CrossRef CAS PubMed.
- G. J. Brewer, Chem. Res. Toxicol., 2010, 23, 319 Search PubMed.
- A. Gupte and R. J. Mumper, Cancer Treat. Rev., 2009, 35, 32 CrossRef CAS PubMed.
- C. Liu, R. Bai and L. Q. San, Water Res., 2008, 42, 1511 CrossRef CAS PubMed.
- L. Kiaune and N. Singhasemanon, Rev. Environ. Contam. Toxicol., 2011, 213, 1 CAS.
- M. Saleem, M. Rafiq, M. Hanif, M. A. Shaheen and S. Y. Seo, J. Fluoresc., 2018, 28, 97 CrossRef PubMed.
- M. Yang, L. Ma, J. Li and L. Kang, RSC Adv., 2019, 9, 16812 RSC.
- (a) W. N. Wu, H. Wu, R. B. Zhong, Y. Wang, Z. H. Xu, X. L. Zhao, Z. Q. Xu and Y. C. Fan, Spectrochim. Acta, Part A, 2019, 212, 121–127 CrossRef CAS PubMed; (b) W. W. Wang, Y. Wang, W. N. Wu, X. L. Zhao, Z. Q. Xu, Z. H. Xu, X. X. Li and Y. C. Fan, Spectrochim. Acta, Part A, 2020, 226, 117592 CrossRef CAS PubMed.
- A. Sil, S. N. Islama and S. K. Patra, Sens. Actuators, B, 2018, 254, 618 CrossRef CAS.
- S. H. Park, N. Kwon, J. H. Lee, J. Yoon and I. Shin, Chem. Soc. Rev., 2020, 49, 143 RSC.
- S. Liu, Y. M. Wang and J. Han, J. Photochem. Photobiol., C, 2017, 32, 78 CrossRef CAS.
- K. Huang, D. Han, X. Li, M. Peng, Q. Qiu and D. Qin, J. Fluoresc., 2019, 29, 727 CrossRef CAS PubMed.
- M. L. Giuffrida, G. T. Sfrazzetto, C. Satriano, S. Zimbone, G. A. Tomaselli, A. Copani and E. Rizzarelli, Inorg. Chem., 2018, 57(5), 2365 CrossRef CAS PubMed.
- Q. Qiu, B. Yu, K. Huang and D. Qin, J. Fluoresc., 2020, 30, 859 CrossRef CAS PubMed.
- K. M. Ramos-Torres, S. Kolemen and C. J. Chang, Isr. J. Chem., 2016, 56, 724 CrossRef CAS PubMed.
- J. A. Cotruvo Jr., A. T. Aron, K. M. Ramos-Torres and C. J. Chang, Chem. Soc. Rev., 2015, 44, 4400 RSC.
- C. J. Fahrni, Curr. Opin. Chem. Biol., 2013, 17, 656 CrossRef CAS PubMed.
- L. Yang, R. McRae, M. M. Henary, R. Patel, B. Lai, S. Vogt and C. J. Fahrni, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 11179 CrossRef CAS PubMed.
- M. Verma, A. F. Chaudhry, M. T. Morgan and C. J. Fahrni, Org. Biomol. Chem., 2010, 8, 363 RSC.
- A. F. Chaudhry, M. Verma, M. T. Morgan, M. M. Henary, N. Siegel, J. M. Hales, J. W. Perry and C. J. Fahrni, J. Am. Chem. Soc., 2010, 132, 737 CrossRef CAS PubMed.
- A. F. Chaudhry, S. Mandal, K. I. Hardcastle and C. J. Fahrni, Chem. Sci., 2011, 2, 1016 RSC.
- M. T. Morgan, P. Bagchi and C. J. Fahrni, J. Am. Chem. Soc., 2011, 133, 15906 CrossRef CAS PubMed.
- M. T. Morgan, P. Bagchi and C. J. Fahrni, Dalton Trans., 2013, 42, 3240 RSC.
- M. T. Morgan, A. M. McCallum and C. J. Fahrni, Chem. Sci., 2016, 7, 1468 RSC.
- L. Zeng, E. W. Miller, A. Pralle, E. Y. Isacoff and C. Chang, J. Am. Chem. Soc., 2006, 128, 10 CrossRef CAS PubMed.
- S. C. Dodani, D. W. Domaille, C. I. Nam, E. W. Miller, L. A. Finney, S. Vogt and C. J. Chang, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 5980 CrossRef CAS PubMed.
- D. W. Domaille, L. Zeng and C. J. Chang, J. Am. Chem. Soc., 2010, 132, 1194–1195 CrossRef CAS PubMed.
- S. C. Dodani, S. C. Leary, P. A. Cobine, D. R. Winge and C. J. Chang, J. Am. Chem. Soc., 2011, 133, 8606 CrossRef CAS PubMed.
- M. L. Giuffrida, E. Rizzarelli, G. A. Tomaselli, C. Satriano and G. T. Sfrazzetto, Chem. Commun., 2014, 50, 9835 RSC.
- S. C. Dodani, A. Firl, J. Chan, C. I. Nam, A. T. Aron, C. S. Onak, K. M. Ramos-Torresa, J. Paek and C. M. Webster, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 16280 CrossRef CAS PubMed.
- S. Jia, K. M. Ramos-Torres, S. Kolemen, C. M. Ackerman and C. J. Chang, ACS Chem. Biol., 2018, 13(7), 1844 CrossRef CAS PubMed.
- C. Satriano, G. T. Sfrazzetto, M. E. Amato, F. P. Ballistreri, A. Copani, M. L. Giuffrida, G. Grasso, A. Pappalardo, E. Rizzarelli, G. A. Tomaselli and R. M. Toscano, Chem. Commun., 2013, 49, 5565 RSC.
- T. Hirayama, G. C. Van de Bittner, L. W. Gray, S. Lutsenko and C. J. Chang, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 2228 CrossRef CAS PubMed.
- X. Cao, W. Lin and W. Wan, Chem. Commun., 2012, 48, 6247 RSC.
- C. Shen, J. L. Kolanowski, C. M. N. Tran, A. Kaur, M. C. Akefeldt, M. S. Rahme, T. W. Hambley and E. J. New, Metallomics, 2016, 8, 915 CrossRef CAS PubMed.
- A. Farhi, F. Firdaus, H. Saeed, A. Mujeeb, M. Shakir and M. Owais, Photochem. Photobiol. Sci., 2019, 18, 3008 CrossRef CAS PubMed.
- C. S. Lim, J. H. Han, C. W. Kim, M. Y. Kang, D. W. Kang and B. R. Cho, Chem. Commun., 2011, 47, 7146 RSC.
- P. K. Mehta, E. T. Oh, H. J. Park and K. H. Lee, Sens. Actuators, B, 2018, 256, 393 CrossRef CAS.
- X. Q. Yi, Y. F. He, Y. S. Cao, W. X. Shen and Y. Y. Lv, ACS Sens., 2019, 4, 856 CrossRef CAS PubMed.
- J. Chan, S. C. Dodani and C. J. Chang, Nat. Chem., 2012, 4, 973 CrossRef CAS PubMed.
- M. Taki, S. Iyoshi, A. Ojida, I. Hamachi and Y. Yamamoto, J. Am. Chem. Soc., 2010, 132, 5938 CrossRef CAS PubMed.
- M. Taki, K. Akaoka, K. Mitsui and Y. Yamamoto, Org. Biomol. Chem., 2014, 12, 4999 RSC.
- K. K. Yu, K. Li, J. T. Hou and X. Q. Yu, Tetrahedron Lett., 2013, 54, 5771 CrossRef CAS.
- Z. Hu, J. Hu, H. Wang, Q. Zhang, M. Zhao, C. Brommesson, Y. Tian, H. Gao, X. Zhang and K. Uvdal, Anal. Chim. Acta, 2016, 933, 189 CrossRef CAS PubMed.
- D. Maity, B. Sarkar, S. Maiti and T. Govindaraju, ChemPlusChem, 2013, 78, 785 CrossRef CAS PubMed.
- D. Maity, A. Raj, D. Karthigeyan, T. K. Kundu and T. Govindaraju, Supramol. Chem., 2015, 27, 589 CrossRef CAS.
- M. C. Heffern, H. M. Park, H. Y. Au-Yeung, G. C. Van de Bittner, C. M. Ackerman, A. Stahl and C. J. Chang, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 14219 CrossRef CAS PubMed.
- S. Y. Park, W. Kim, S. H. Park, J. Han, J. Lee, C. Kang and M. H. Lee, Chem. Commun., 2017, 53, 4457 RSC.
- (a) A. Arora, J. Kaushal, A. Kumar, P. Kumar and S. Kumar, ChemistrySelect, 2019, 4, 6140 CrossRef CAS; (b) P. Kumar and S. Kumar, J. Mol. Struct., 2020, 1202, 127242 CrossRef CAS.
- X. W. Liu, Y. Xiao, S. B. Zhang and J. L. Lu, Inorg. Chem. Commun., 2017, 84, 56 CrossRef CAS.
- M. Li, Q. Liang, M. Zheng, C. Fang, S. Peng and M. Zhao, Dalton Trans., 2013, 42, 13509 RSC.
- Z. Ye, X. An, B. Song, W. Zhang, Z. Dai and J. Yuan, Dalton Trans., 2014, 43, 13055 RSC.
- Y. Zhang, Z. Liu, Y. Zhang, Y. Xu, H. Li, C. Wang, A. Lu and S. Sun, Sens. Actuators, B, 2015, 211, 449 CrossRef CAS.
- Z. B. Zheng, Q. Y. Huang, Y. F. Han, J. Zuo and Y. N. Ma, Sens. Actuators, B, 2017, 253, 203 CrossRef CAS.
- M. Li, S. Sheth, Y. Xu and Q. Song, Microchem. J., 2020, 156, 104848 CrossRef CAS.
- P. Zhang, L. Pei, Y. Chen, W. Xu, Q. Lin, J. Wang, J. Wu, Y. Shen, L. Ji and H. Chao, Chem. – Eur. J., 2013, 19, 15494 CrossRef CAS PubMed.
- F. Cheng, C. He, M. Ren, F. Wang and Y. Yang, Spectrochim. Acta, Part A, 2015, 136, 845 CrossRef CAS PubMed.
- (a) M. Ramachandran and S. Anandan, New J. Chem., 2020, 44, 6186 RSC; (b) M. Ramachandran, S. Anandan and M. Ashokkumar, New J. Chem., 2019, 43, 9832 RSC.
- Z. B. Zheng, S. Y. Kang, Y. Zhao, N. Zhang, X. Yi and K. Z. Wang, Sens. Actuators, B, 2015, 221, 614 CrossRef CAS.
- S. T. Zhang, P. Li, C. Liao, T. Luo, X. Kou and D. Xiao, Spectrochim. Acta, Part A, 2018, 201, 161 CrossRef CAS PubMed.
- Y. Zhang, Z. Liu, K. Yang, Y. Zhang, Y. Xu, H. Li, C. Wang, A. Lu and S. Sun, Sci. Rep., 2015, 5, 8172 CrossRef CAS PubMed.
- S. Ji, M. Chen, G. Gan, H. Li and W. Li, Spectrochim. Acta, Part A, 2012, 88, 124 CrossRef CAS PubMed.
- K. M. Santos, A. R. Santos and I. M. Raimundo Jr., Sens. Actuators, B, 2018, 255, 2367 CrossRef CAS.
- F. Chen, F. Xiao, W. Zhang, C. Lin and Y. Wu, ACS Appl. Mater. Interfaces, 2018, 10, 26964 CrossRef CAS PubMed.
- Y. Shi, Q. Liu, W. Yuan, M. Xue, W. Feng and F. Li, ACS Appl. Mater. Interfaces, 2019, 11, 430 CrossRef CAS PubMed.
- S. V. Wegner, H. Arslan, M. Sunbul, J. Yin and C. He, J. Am. Chem. Soc., 2010, 132, 2567 CrossRef CAS PubMed.
- J. Liu, J. Karpus, S. V. Wegner, P. R. Chen and C. He, J. Am. Chem. Soc., 2013, 135, 3144 CrossRef CAS PubMed.
- X. Yan, X. Li, S. S. Lv and D. C. He, Dalton Trans., 2012, 41, 727 RSC.
- J. Liang, M. Qin, R. Xu, X. Gao, Y. Shen, Q. Xu, Y. Cao and W. Wang, Chem. Commun., 2012, 48, 3890 RSC.
- M. S. Koay, B. M. G. Janssen and M. Merkx, Dalton Trans., 2013, 42, 3230 RSC.
- J. L. Vinkenborg, T. J. Nicolson, E. A. Bellomo, M. S. Koay, G. A. Rutter and M. Merkx, Nat. Methods, 2009, 6, 737–740 CrossRef CAS PubMed.
- Y. Shu, J. N. Hao, D. Niu and Y. Li, J. Mater. Chem. C, 2020, 8, 8635 RSC.
- T. T. Zheng, J. Zhao, Z. W. Fang, M. T. Li, C. Y. Sun, X. Li, X. L. Wang and Z. M. Su, Dalton Trans., 2017, 46, 2456 RSC.
- L. Li, S. Shen, R. Lin, Y. Bai and H. Liu, Chem. Commun., 2017, 53, 9986–9989 RSC.
- M. Pamei and A. Puzari, Nano-Struct. Nano-Objects, 2019, 19, 100364 CrossRef CAS.
- T. L. Mako, J. M. Racicot and M. Levine, Chem. Rev., 2019, 119, 322 CrossRef CAS PubMed.
- J. Liu, Y. Q. Fan, S. S. Song, G. F. Gong, J. Wang, X. W. Guan, H. Yao, Y. M. Zhang, T. B. Wei and Q. Lin, ACS Sustainable Chem. Eng., 2019, 7(14), 11999 CAS.
- H. Fang, G. Cai, Y. Hu and J. Zhang, Chem. Commun., 2018, 54, 3045 RSC.
- S. Pal, N. Chatterjee and P. K. Bharadwaj, RSC Adv., 2014, 4, 26585 RSC.
- L. He, B. Dong, Y. Liu and W. Lin, Chem. Soc. Rev., 2016, 45, 6449 RSC.
- J. K. H. Wong, M. H. Todd and P. J. Rutledge, Molecules, 2017, 22, 200 CrossRef PubMed.
- J. S. Kim and D. T. Quang, Chem. Rev., 2007, 107, 3780 CrossRef CAS PubMed.
- R. G. Pearson, J. Am. Chem. Soc., 1963, 85, 3533 CrossRef CAS.
- K. L. Haas and K. J. Franz, Chem. Rev., 2009, 109, 4921 CrossRef CAS PubMed.
- M. Priessner, P. A. Summers, B. W. Lewis, M. Sastre, L. Ying, M. K. Kuimova and R. Vilar, Angew. Chem., Int. Ed., 2021, 60, 23148 CrossRef CAS PubMed.
- H. M. Kim and B. R. Cho, Chem. – Asian J., 2011, 6, 58 CrossRef CAS PubMed.
- F. Helmchen and W. Denk, Nat. Methods, 2005, 2, 932 CrossRef CAS PubMed.
- C. Y.-S. Chung, J. M. Posimo, S. Lee, T. Tsang, J. M. Davis, D. C. Brady and C. J. Chang, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 18285 CrossRef PubMed.
- Y. Liu, T. Kang, Q. He, Y. Hu, Z. Zuo, Z. Cao, B. Ke, W. Zhang and Q. Qi, RSC Adv., 2021, 11, 14824 RSC.
- A. Roux, M. Isaac, V. Chabert, S. A. Denisov, N. D. McClenaghan and O. Sénèque, Org. Biomol. Chem., 2018, 16, 5626 RSC.
- C. Cepeda, S. A. Denisov, D. Boturyn, N. D. McClenaghan and O. Sénèque, Inorg. Chem., 2021, 60, 17426 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |