Open Access Article
Open Access ArticleUltrasensitive and label-free detection of prognostic and diagnostic biomarkers of sepsis on a AgNP-laden black phosphorous-based SERS platform†
Anirban
Kundu
 ,
Renu
Rani
,
Anas
Ahmad
,
Renu
Rani
,
Anas
Ahmad
 ,
Ajay
Kumar
,
Mamta
Raturi
,
Tanya
Gupta
,
Rehan
Khan
,
Ajay
Kumar
,
Mamta
Raturi
,
Tanya
Gupta
,
Rehan
Khan
 * and
Kiran Shankar
Hazra
* and
Kiran Shankar
Hazra
 *
*
Institute of Nano Science and Technology, Knowledge City, Sector 81, Mohali, Punjab 140306, India. E-mail: rehankhan@inst.ac.in; kiran@inst.ac.in
First published on 16th March 2022
Abstract
Rapid and early detection of sepsis biomarkers is essential and an immediate need to prevent significant casualties of ICU patients caused by an uncontrolled infection in the patient's body. A highly sensitive and selective surface-enhanced Raman spectroscopy (SERS) based detection protocol has been established to detect the sepsis biomarkers. For this purpose, uniquely designed SERS substrate on black phosphorus (BP) flake are fabricated by a one-step and straightforward process where uniform silver nanoparticles (AgNPs) are synthesized over the BP surface, which in turn enhance the stability of BP flakes. Both the prognostic [interleukin-3 (IL-3)] and diagnostic [procalcitonin (PCT)] biomarkers are detected up to a limit of detection (LOD) as low as 1000 fM and 100 fM, respectively, where the SERS enhancement factor (EF) is obtained as high as in the order of ∼1014. A comprehensive study of Raman spectra has been carried out to explore the origin of observed Raman peaks of IL-3 and PCT, arising from various amino acids, which has never been reported earlier. The unknown signature Raman peaks for such biomarkers are identified at low energy regions, occurring due to structural variation of the two different protein chains even though both consist of the same possible amino acids. The findings of signature Raman modes of sepsis biomarkers help distinguish the Raman peaks of sepsis biomarkers in spiked clinical samples even in the simultaneous presence of both the biomarkers.
Introduction
Sepsis affects about 30 million humans globally and is the leading cause of death of ICU patients worldwide due to its complicated detection techniques and the absence of prompt treatment.1,2 Sepsis is a severe inflammatory reaction to infectious agents, and readily advances to a sustained immunosuppressive condition. Thus, it has become the reason for significant mortality rates.3 Sepsis requires immediate medical treatment to avoid casualties within hours of the disease onset due to its rapid growth of infection in the body. This leads to the condition of organ failure, termed as septic shock.4 Various pathogens, such as bacteria, fungi or viruses, can cause sepsis, but most of the sepsis cases are caused by bacterial infections.5 Sepsis is also observed as a common complication directly caused by the new coronavirus (SARS-CoV2) first reported in late 2019.6,7 The optimized sepsis management requires the rapid, early and sensitive detection of various diagnostic and prognostic sepsis biomarkers, which can be diagnosed on a single platform. However, such kinds of detection and diagnostic techniques are still rarely accessible and create significant lacunae in the timely management of the pathophysiological consequences of sepsis.8,9 Rapid and accurate detection has also become significant as it helps in the well-timed administration of appropriate antimicrobial agents that potentially obviate the attainment of critical septic stages, and can thus prevent fatalities.10Techniques, like enzyme-linked immunosorbent assay (ELISA), polymerase chain reaction (PCR), fluorescence in situ hybridization (FISH), microarray hybridization and proteomics spectrometry phage assays,11–14 are available for the detection of sepsis. However, they have limitations due to their selectivity and longer process time in the typical range of 12 hours to few days.15 Since the affected patients have a rapid progression of underlying infection, the level of most of the biomarkers shoots up during the standard testing duration, which disables the identification of the real-time infection stage. In recent years, serum biomarkers, such as C-reactive protein (CRP), tumor necrosis factor-alpha (TNF-α), interleukin-6 (IL-6), and Fc-gamma receptor-1 (FcγR1) (also called CD64), have been explored to identify the progress and prediction of the sepsis.10,13,16–18 However, the desired selectivity and sensitivity of these techniques for early detection are not adequate. The absence of a specific treatment and the complicated pathogenesis of sepsis make it necessary for the early detection of disease biomarkers, so that supportive medications can be implemented at an early stage when it is easily manageable.
Among several biomarkers of sepsis, lipopolysaccharide (LPS) has proven to be an essential mediator in the pathogenesis of Gram-negative bacterial sepsis, which is associated with several other clinical manifestations, like hypertension, blood clotting, diarrhea, pyrogenic reactions, and infections of Gram-negative bacteria such as meningococcus.19–21 There is no single ideal biomarker of sepsis present that helps to identify critically ill patients who need immediate medical attention with possible diagnosis and treatment. The category of different sepsis biomarkers and their detection techniques are mentioned in Table 1, which gives an idea about the detection process and required time. Recently, Weber et al. identified interleukin-3 (IL-3), a cytokine, as a prognostic biomarker or independent predictor of septic shock and death.22 Furthermore, Min et al. showed that when using a hybrid magneto-electrochemical sensor, the limit of detection (LOD) for IL-3 can be achieved up to <10 pg ml−1, which produces the test results within 1 h.23 Along with the prognostic biomarker IL-3, the diagnostic biomarker procalcitonin (PCT) has a vital role in determining the severity of sepsis at different stages. The role of PCT for determining the stage of sepsis has already been proven and explained in several reports.24–26
| Phase | Category | Markers | Type | Method | Process time |
|---|---|---|---|---|---|
| Hyper-inflammatory | Cytokines | TNF-α, IL-1β, IL-6 | Prognostic | Immunoassay61 | 2–5 days |
| Chemokines | IL-8, IL-10 | Diagnostic | ELISA62 | 2–5 days | |
| Protein | CRP, procalcitonin (PCT) | Diagnostic | ELISA17 | 2–5 days | |
| Complementary peptides | C3b, C5a | Diagnostic | ELISA63 | 2–5 days | |
| Antigens on neutrophils and monocytes | CD64, CD11b, CD14 | Diagnostic | Flow cytometry; immunoassay64 | 3–6 hours; 2–5 days | |
| Detection of microbes | RNA | Diagnostic | PCR65 | 2–5 days | |
| Immuno-suppressive | Surface antigens | ↓MHC II, ↓CD28, ↑CD152 (CTLA) IL-10 | Diagnostic | ELISA66 | 2–5 days |
| Organ dysfunction | Proteins | ↑lactate, IL-6, PCT, fibrinolysis (↑coagulation factors) | Diagnostic | Immunoassays, DIC (disseminated intravascular coagulation) score for fibrinolysis17 | 2–5 days; over hours or days |
In recent times, 2D layered material-based SERS substrates have emerged as a potential tool for the identification of various biomarkers present in blood or serum. This is due to their advantages of having improved sensitivity, reusability, enhanced stability, stronger surface adsorption capability, and feasible scalability.27–29 Reports show that the graphene and MoS2-based SERS platform has application in bio-sensing for the sensitive detection of diseases, like bacterial infections,30 malaria,31 diabetes,32 cardiovascular,33 and brain tumors.34 However, the SERS signal enhancement of such systems mostly relies on charge transfer between the 2D van der Waals (vdW) nano-sheet/flake and the analyte, rather than the electromagnetic field enhancement.35 Very recently, layered black phosphorus (BP) has emerged as one of the most suitable 2D materials for SERS applications, as it has intrinsic in-plane ferroelectricity unlike other 2D members, which is favorable for SERS signal enhancement. The laser-induced nano-structuring of the BP flake can create arrays of hotspots, which has emerged as a metal-free SERS substrate.36 Along with the pristine layered BP flake and its nanostructures, the BP-metal nanoparticle heterostructures have also emerged as a potential platform that can be used for the detection of biomolecules, where the intrinsic biocompatibility of layered BP can be an added advantage.36–38 In recent times, the heterostructures of the layered BP flake and metal nanoparticles have proved to be a stable and potential platform for the development of bio-sensors.38–42 Although the SERS-based detection is precise to the target analyte and capable of ultra-low detection limits,43,67–69 very little effort has been made so far on the detection of sepsis biomarkers due to the fact that the signature Raman modes of these biomarkers are still unexplored. Although a recent report by Cao et al.44 demonstrated the SERS-based diagnosis for sepsis biomarkers of the C-reactive protein (CRP), procalcitonin (PCT), the limit of detection (LOD) was still only achieved in the range of hundred pM, where the sensitivity may not reach the required level. In such labeled detection technique, no signature Raman modes of the sepsis biomarkers have been identified, which is essential and a primary need for the SERS-based detection technique. Thus, the absence of any label-free detection of prognostic and diagnostic biomarkers of sepsis on a single platform with minimum processing time is a vital necessity to ease the timely detection and management of septic patients.
Here, the ultrasensitive detection of prognostic and diagnostic biomarkers of sepsis is demonstrated using a uniquely designed BP-based SERS substrate. The possible signature Raman modes of the IL-3 and PCT protein chains are distinguished by the Raman signals arising from the amino acid building blocks of IL-3 and PCT. In situ growth of silver nanoparticles (AgNPs) over the BP flake is demonstrated, where BP itself acts as a precursor to achieving the uniform distribution of the AgNPs over the BP flake surface. The as-grown AgNPs were used as hotspots for local electric field enhancement in order to obtain enhanced Raman signal of the biomarkers, i.e., LPS, IL-3, and PCT, where the detection limit was investigated to as low as ∼10 fM.
Experimental details
Materials used
Single crystal of black phosphorus (BP) from Manchester Nanomaterials (MN), UK; lipopolysaccharide (LPS) and silver nitrate (AgNO3) were purchased from Sigma Aldrich, and used without further modification. Human IL3/IL-3 protein (IL-3) and human procalcitonin/CALCA (PCT) were purchased from Sino Biological Inc. Purified human serum IgA was purchased from MP Biomedicals (Catalogue No. 0855979). No study was performed directly on humans and no blood or serum was isolated from any human subject.Preparation of the AgNP@BP SERS platform
Pristine BP flake was mechanically exfoliated from a bulk BP single crystal on top of a silicon substrate using the standard micromechanical exfoliation technique via the scotch-tape method. Few layered-BP flakes were identified in an optical microscope and characterized by a WITec alpha 300R Raman spectrometer, followed by AFM measurement using Bruker Multimode 8 atomic force microscope (AFM) in tapping mode. The as-exfoliated BP flake supported by the silicon substrate was immersed in the AgNO3 precursor solution with varying concentrations and kept at ∼90 °C for ∼5 min. The as-fabricated AgNP@BP flake was characterized by Raman spectroscopic measurement and AFM techniques. All of the Raman spectra were recorded in a back-scattering configuration with a Nd:YAG 532 nm laser line using a 100× objective, where the laser power was kept at ∼2 mW to avoid the possibilities of chemical alteration of the molecules by laser heating. The SERS EF was calculated by considering the Raman spectra of the PBS buffer solution on the AgNP@BP flake as reference spectra.TEM sample preparation
Liquid exfoliation of a tiny piece of single-crystal BP was performed by sonication in NMP for 12 h. The exfoliated BP flakes were immediately used for TEM sample preparation by drop-cast on a Cu-grid with 300 mesh, followed by optical microscope identification and Raman measurement. The Cu-grid with the layered BP flake was used for TEM analysis. The same was further used for the deposition of AgNPs as described earlier. The same grid was also further used for TEM analysis of the AgNP@BP flake. The TEM micrographs were recorded using a JEOL JEM2100 transmission electron microscope with 200 kV accelerating voltage.Preparation of IL-3 and the PCT solution in PBS
To prepare different concentrations of the biomarkers, the IL-3 and PCT were diluted in phosphate-buffered saline (PBS) solution at pH 7.4.Preparation of IL-3 and the PCT solution in sterile human serum
Different concentrations of interleukin-3 (IL-3) and procalcitonin (PCT) biomarkers were prepared as spiked samples using sterile human serum. Briefly, the known amounts of IL-3 and PCT were dissolved in sterile PBS solution at pH 7.4 to prepare the final concentrations of the spiked samples in the fM range (10 fM–100 nM). The samples were cooled at 4 °C and then gently stirred over vortexing to obtain a homogenous solution, where a range of six to seven spiked samples were selected for biomarker detection.Results and discussion
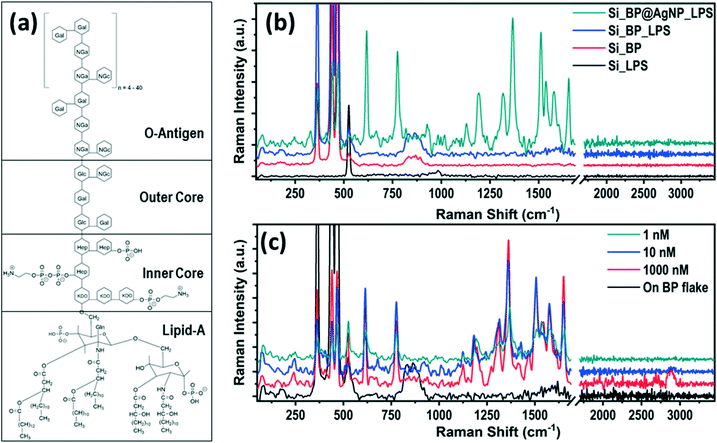
The BP flakes were mechanically exfoliated on a Si substrate, using the standard scotch tape technique and immediately immersed in an aqueous AgNO3 precursor solution for 5 min at 90 °C to avoid any oxidation. Lone pair electrons of phosphorus atoms, evenly distributed all over the BP surface, interacted with the AgNO3 precursor solution to adsorb Ag+ ions on its surface by reducing Ag+ to Ag0.45 Due to such spontaneous adsorption of the Ag+ ions on the BP flake by passivating the lone pair electrons, the BP flake surface became stable even under ambient conditions (room temperature ∼25 °C and relative humidity ~60%) as compared to the pristine one, which surpassed one of the significant limitations of BP for its practical application.46,47 The detailed synthesis protocol and characterization of the AgNP-decorated BP flake are described in the ESI† (Fig. S1–S4). The stability of the AgNP-decorated BP (AgNP@BP) flake was examined by AFM (Fig. S5†), where no signs of physical degradation were observed as the thickness, and surface roughness of the flake remained unchanged for up to 72 h under ambient conditions (room temperature ∼25 °C and relative humidity ∼60%).The SERS capability of the designed AgNP@BP substrate for the detection of LPS was investigated, where 10 μL solution of LPS with concentrations varying from 1000 nM to 1 nM were drop-casted onto the SERS substrate to conduct Raman spectroscopic measurements. A low laser power of ∼2 mW was used to avoid the possibilities of chemical alteration of the molecules by laser heating. LPS is a bacterial endotoxin present in the outer cell membrane of Gram-negative bacteria. It is thought to be a key mediator for the pathogenesis of septic shock.17 The LPS structure can be divided into three distinct parts, i.e., hydrophobic lipid-A, hydrophilic polysaccharide chain, and the repeating units of the bacterial serotype specific O-antigen oligosaccharide (hydrophilic),21 as shown in Fig. 1a. Fig. 1b compares the Raman spectra of LPS (concentration ∼1 μM) on different substrate configurations (i.e., bare Si, pristine BP on Si, and AgNP@BP on Si), concluding that the signature peaks of LPS arises only on the AgNP@BP SERS substrate due to signal enhancement. In contrast, the peaks at 362, 442, and 465 cm−1 originated from the BP flake, which corresponds to the out-of-plane vibration, in-plane armchair and zigzag vibration, respectively, and the peak at 521 cm−1 denotes the first order Si Raman mode. The enhancement in the Raman signal is attributed to the increase in the local electric field by the plasmonic effect of the AgNPs decorated on the BP flake. The uniform size and spatial distribution of the nanoparticles with higher density are the key factors for superior SERS enhancement. Here, the electromagnetic enhancement is dominated over the chemical enhancement for contribution towards a higher SERS enhancement factor. Table ST2, in the ESI,† summarizes the observed Raman peaks from LPS and identifies their origins from the specific vibration modes, which has unexplored so far. The signature Raman peaks of LPS are observed at 1131, 1366, and 1655 cm−1, which originated from the presence of lipids48 in the LPS structure. A Raman peak at 1316 cm−1 confirms the presence of aliphatic molecules, whereas the 1579 cm−1 peak originates from graphitic carbon. However, the only Raman peak in the high energy range at ∼2900 cm−1 originated from the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and CH3 stretching modes, which confirms the presence of sugar, lipid and a polyethylene group in the structure.49
O and CH3 stretching modes, which confirms the presence of sugar, lipid and a polyethylene group in the structure.49
The SERS signal of LPS for different concentrations (1000–1 nM) are shown in Fig. 1c, where the Raman intensity decreases with the decrease in the LPS concentration. The enhancement in the Raman signal of LPS is not prominent beyond ∼1 nM concentration, which is the limit of detection (LOD) of the AgNP@BP SERS substrate for LPS as an analyte. The SERS enhancement factor (EF) for LPS was calculated using the two most intense Raman peaks at 1366 cm−1 and 1655 cm−1. The enhancement factor was calculated for different LPS concentrations, which is provided in Table ST4 of the ESI.† The enhancement factor of the reported SERS substrate was calculated from the average area covered by the particles (hotspot area) and the possible number of molecules present at the hotspot vicinity (additional details are provided in the ESI† Fig. S7). The average value of the SERS enhancement factor was found to be surprisingly high, EF ∼2.9 × 1014, for LPS under 532 nm laser excitation.
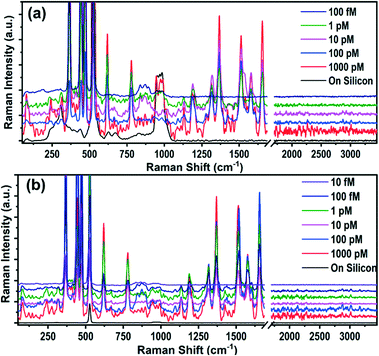
Although an elevated LPS with LOD of 1 nM could be a good prima facie evidence, it cannot be a confirmatory biomarker for the diagnosis of sepsis due to the lack of specificity towards sepsis. Hence, it requires the sensitive detection of more specific biomarkers such as IL-3 (prognostic) and PCT (diagnostic). The Raman spectra of IL-3 and PCT are depicted in Fig. 2a and b, where it is evident that the intense Raman spectra of the biomarker appear only on the AgNP@BP SERS substrate and not on bare Si substrates. In order to identify the detection limit, Raman signals were recorded with varying concentrations from 1000 pM to 10 fM for both IL-3 and PCT. However, the spectra of the biomarkers were observable only up to 1 pM for IL-3 and 100 fM for PCT, which are the LODs of these two biomarkers on the AgNP@BP SERS substrate.
Clinically, it has been proven that the level of biomarkers present in the infected human body is in the pg mL−1 range (∼pM concentration), suggesting that the reported LODs on the AgNP@BP SERS substrate can provide three-digit accuracy. This is sensitive enough and more than sufficient for the early detection of the elevated IL-3 and PCT levels in the blood caused by sepsis.23 The enhancement factor (EF) of the SERS substrate was calculated using the three intense Raman peaks of IL-3 and PCT, appearing at 620, 1366, and 1655 cm−1, where the EF values corresponding to different concentrations of the biomarkers were calculated and are listed in Tables ST5 and ST6.† The obtained average EF values calculated for IL-3 and PCT were as high as ∼2.3 × 1014 and ∼4.5 × 1014, respectively. Table ST7† summarizes the average EF and LODs for all three sepsis biomarkers (LPS, IL-3, and PCT) detected using the AgNP@BP SERS platform.
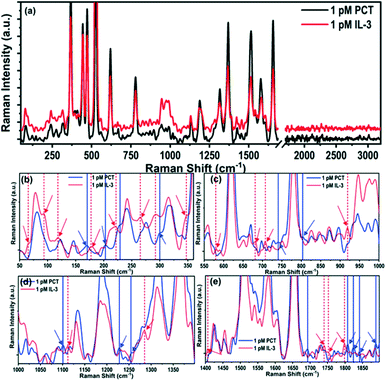
To the best of our knowledge, this is the first scientific report on the Raman spectra of IL-3 and PCT. Hence, it requires the identification of the vibrational modes corresponding to the observed Raman peaks. The prominent characteristic Raman vibrational peaks of IL-3 and PCT corresponding to 1 pM concentration are compared in Fig. 3a, whereas all of the observed Raman modes are summarized in Table 2. The phonon band assignment of the Raman peaks, and the identification of the signature peaks of the IL-3 and PCT protein structure are challenging tasks. Both biomarkers contain the same building blocks of amino acids with multiple variations in sequence. Hence, they end up interfering with the signature peaks, originating from the structural anomalies.
| Wavenumber (cm−1) | Band assignment | Reported band position (cm−1) | Amino acida | |
|---|---|---|---|---|
| PCT | IL-3 | |||
| a A: alanine; R: arginine; N: asparagine; D: aspartic acid; C: cysteine; Q: glutamine; E: glutamic acid; G: glycine; H: histidine; I: isoleucine; L: leucine; K: lysine; M: methionine; F: phenylalanine; P: proline; S: serine; T: threonine; W: tryptophan; Y: tyrosine; V: valine. | ||||
| 81.89 | 79.11 | |||
| — | 111 | |||
| 122.52 | — | |||
| — | 130.11 | |||
| 144.97 | 144.39 | |||
| 169.53 | 170.63 | Lattice stretching | 166 | W |
| — | 192.36 | C–O torsional | 185 | V |
| 202.76 | 206.7 | Torsional CH2–CH | 200–210 | C |
| — | 269.06 | Torsional | 262 | M |
| 275.78 | — | |||
| 300.11 | 304.8 | CCC beading | 300 | S |
| — | 406.54 | Bending N–C–C | 400 | AY |
| 444.39 | 445.36 | Torsional NH3 | 450 | TP |
| 476.5 | 476.34 | Torsional NH2 | 475–480 | AQ |
| Rocking COO | 493 | T | ||
| — | 516.53 | Rocking COO | 515 | S |
| — | 535.36 | Rocking of COO | 530–535 | ARI |
| COO rocking | 544 | H | ||
| 559.72 | — | Wagging COO | 565 | T |
| Bending COO | 564 | Q | ||
| COO rocking | 556 | I | ||
| 620.22 | 618.57 | Bending of COO | 611 | SF |
| Amide bending | 632 | Q | ||
| — | 647.27 | Bending of COO | 655–660 | AR |
| Symmetric C–S stretching | 645 | MQ | ||
| 669.2 | 666.59 | COO scissoring | 684 | H |
| — | 726.87 | Torsional COH | 703 | T |
| C–S–C stretching | 721 | M | ||
| — | 744.11 | CH2 rocking | 740–775 | CM |
| Condensed ring system | 757 | W | ||
| C–H twisting | 749 | I | ||
| 779.81 | 779.51 | OOP of COO | 776 | T |
| COO deformation | 782 | V | ||
| CNH stretching | 784 | R | ||
| — | 794.06 | COO bending | 804 | V |
| — | 840.01 | C–C stretching | 848 | HRNFPI |
| Wagging of H at imidazole ring | 854 | H | ||
| COO OOP | 838 | L | ||
| Ring breathing vibration | 854 | Y | ||
| — | 872.54 | Stretching C–C–N | 878 | T |
| C–C stretching | 877 | ME | ||
| N–H stretching | 875 | W | ||
| — | 886.03 | |||
| 896.39 | 904.63 | NH2 bending | 896 | P |
| NH2 bending | 908 | R | ||
| 930.37 | — | Stretching C–C | 930 | TLSD |
| C–C asymmetric bending | 922 | I | ||
| 972.56 | — | C–C stretching | 900–970 | VH |
| CH2 rocking | 970 | S | ||
| 991.76 | 986.61 | S–H in-plane bending | 990 | C |
| 1016.7 | 1013.1 | C–N stretching | 1010 | S |
| Ring breathing | 1003 | FW | ||
| CC stretching | 1008 | Q | ||
| 1033.54 | — | OH bending | 1032 | R |
| C–N stretching | 1033 | LKN | ||
| In-plane bending | 1032 | F | ||
| 1096.63 | 1098.19 | Symmetric stretching CN | 1091 | HRLMQI |
| C–O stretching | 1090 | S | ||
| Rocking NH3 | 1101 | N | ||
| 1134.67 | 1130.92 | NH3 asymmetric rocking | 1140 | CLSKQE |
| Rocking NH3 | 1113 | H | ||
| Stretching asymmetrical C–C–N | 1114 | T | ||
| CH3 rocking | 1125–1200 | V | ||
| Ring deformation of tyrosine | 1128 | Y | ||
| 1187.54 | 1182.04 | NH3 rocking | 1175 | L |
| C–H stretching | 1175 | M | ||
| NH3 rocking | 1188 | L | ||
| — | 1194.24 | CH2 twisting | 1200 | C |
| Rocking NH3 | 1202 | T | ||
| CO stretching | 1192 | R | ||
| 1279.15 | 1288.21 | CH2 twisting | 1265 | M |
| CH2 bending wagging | 1261 | Q | ||
| 1316.53 | 1317.14 | COO stretching | 1321 | K |
| C–H stretching | 1311 | ED | ||
| 1360.42 | COO symmetric stretching | 1350 | C | |
| C–H twisting | 1360 | K | ||
| Stretching CH | 1358 | NQI | ||
| Bending CH | 1349 | TL | ||
| Pyrrole ring stretching | 1359 | W | ||
| 1371.01 | 1368.3 | OH bending | 1378 | P |
| 1435.9 | 1423.3 | Stretching asymmetric C–O–O | 1420 | TH |
| Stretching CH2 | 1425 | NQED | ||
| Pyrrole ring stretching | 1426 | W | ||
| — | 1477.49 | Bending CH3 | 1465 | T |
| CH3 bending | 1400–1460 | VRI | ||
| CH stretching (asym) | 1465 | M | ||
| 1513.74 | 1513.3 | Asymmetric stretching COO | 1506 | H |
| 1543.33 | 1551.32 | Benzene ring stretching | 1557 | W |
| 1577.19 | 1577.99 | OH bending | 1544 | R |
| CC stretching | 1586 | F | ||
| 1603.9 | — | NH3 bending | 1609 | K |
| CC ring stretching | 1604 | FY | ||
| — | 1650.98 | Stretching CO | 1640 | N |
| 1654.72 | 1655.69 | Stretching CO | 1692 | D |
The amino acid sequence of IL-3 is as follows:50,51
MSRLPVLLLLQLLVRPGLQAPMTQTTPLKTSWVNCSNMIDEIITHLKQPPLPLLDFNNLNGEDQDILMENNLRRPNLEAFNRAVKSLQNASAIESILKNLLPCLPLATAAPTRHPIHIKDGDWNEFRRKLTFYLKTLENAQAQQTTLSLAIF
Whereas, the amino acid sequence of PCT is:52,53
APFRSALESSPADPATLSEDEARLLLAALVQDYVQMKASELEQEQEREGSSLDSPRSKRCGNLSTCMLGTYTQDFNKFHTFPQTAIGVGAPGKKRDMSSD LERDHRPHVS MPQNAN
The origin of the Raman peaks (Fig. 3a) of IL-3 and PCT were identified and are listed in Table 2 by comparing the signals with the previously reported Raman spectra of amino acids.54–56 For instance, the intense Raman peaks that are common for both IL-3 and PCT are observed at 620, 779, 1130, 1187, 1316, 1370, 1513, 1577, and 1655 cm−1, and are the primary characteristics of the amino acids. Common Raman peaks of low intensity are observed for both IL-3 and PCT at ∼896, 991, 1096, 1279, 1423, and 1551 cm−1, originating from the amino acids. The observed Raman peak positions of the biomarkers may have perturbations in the peak positions. They may not match exactly with the reported peak positions of the isolated amino acids, as they exist within a long chain of the protein biomarker. In the low energy region, prominent Raman peaks are also present in matching positions (PCT: 81 cm−1, 144 cm−1, and 169 cm−1; IL-3: 79 cm−1, 144 cm−1, and 170 cm−1), originating from the structural vibrations of the short-range identical amino acid sequence.
To identify the Raman signature modes of IL-3 and PCT, the spectra (Fig. 3a) were compared in four different ranges, i.e., 50–360 cm−1 (Fig. 3b), 550–1000 cm−1 (Fig. 3c), 1000–1400 cm−1 (Fig. 3d) and 1400–1900 cm−1 (Fig. 3e). The Raman signature peaks of IL-3 and PCT are marked in Fig. 3b–e with dotted and solid vertical lines, respectively, whereas, all the signature Raman modes are tabulated in Table ST8.† For instance, a few distinguished low frequency (<350 cm−1) Raman peaks are observed for PCT, such as 122, 170, 229, 203, and 299 cm−1, which are not observed in IL-3. Similarly, the Raman peaks at 65, 92, 118, 177, 221, 265, 292, 347 cm−1 are observed in IL-3 but not in the Raman spectra of PCT. Such low energy Raman peaks may originate from the complete structural vibration, and can be considered as the signature Raman modes of IL-3 and PCT. Apart from the low energy Raman modes, a few distinguished high energy (>500 cm−1) Raman peaks are also observed such as 579, 681, 702, 919, 1111, 1285, 1406, 1739, 1752 and 1798 cm−1 for IL-3 and 740, 803, 1100, 1228, 1254, 1692, 1807, 1824, 1841 and 1888 cm−1 for PCT, which helps to identify the biomarkers precisely. The high energy signature Raman modes of the biomarkers may have originated owing to the long or short-range structural deformation of the long-chain proteins.
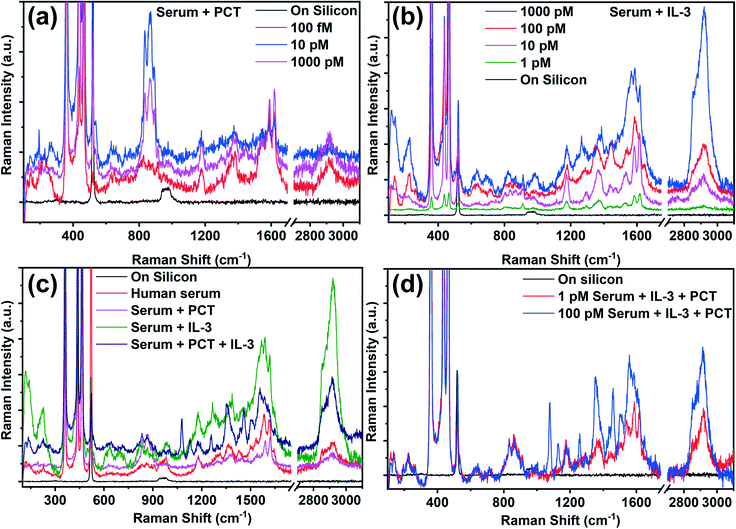
In order to confirm the SERS capability of the developed AgNP@BP substrate for the detection of sepsis biomarkers (PCT and IL-3) in spiked clinical samples, solutions of the biomarkers with varying concentrations were prepared in sterile human serum to mimic the real serum sample.57–60 The as-prepared spiked biomarker samples were drop-casted onto the fabricated SERS substrate, and kept for vacuum drying for two hours. The Raman spectra were recorded using a very low power (<2 mW) laser irradiation to avoid any kind of possible degradation of the molecular structure. The Raman spectra of the IL-3 and PCT in the spiked clinical samples are shown in Fig. 4a and b, where the prominent Raman spectral features were observed in solutions with concentrations as low as 1 pM and 100 fM for IL-3 and PCT, respectively. Such low LOD of both biomarkers obtained in the spiked clinical samples are similar to the obtained LOD of the pure biomarkers (Fig. 2a and b), which establishes the detection capability of the developed AgNP@BP SERS substrate for spiked clinical samples. The identification of biomarkers in clinical samples through SERS is one of the major limitations for the diagnosis of sepsis due to the presence of indistinguishable Raman peaks from several other biomolecules present in the sample. However, the established signature Raman modes of the sepsis biomarkers (Table ST8†) help us to identify and distinguish the presence of biomarkers in the spiked clinical samples. The calculated SERS EF was obtained on the order of ∼1014 as well, i.e., similar to that of the pure biomarker sample. The in-depth analysis of the Raman spectra of the spiked clinical samples was performed by rigorous Lorentzian fitting to identify all of the possible Raman peaks of the biomarkers. The obtained Raman peaks of PCT and IL-3 in the spiked clinical samples are listed in Table ST9,† which matches well with the Raman peaks observed for the pure biomarkers. By comparing the Raman spectra of the spiked clinical samples, it was observed that a strong peak at ∼2900 cm−1 is present in the case of IL-3, while the same is weak for PCT. Thus, the presence of the Raman peak at ∼2900 cm−1 significantly denoted the presence of IL-3 in the spiked clinical sample. Such strong peak can be considered as the signature Raman peak of IL-3. To distinguish the Raman modes of the biomarkers from the sterile human serum, Fig. 4c compares the Raman spectra of IL-3, PCT, and serum along with the spiked clinical sample, where both the biomarkers are present simultaneously. As shown in Fig. 4a and b, the Raman peak at ∼2900 cm−1 has a strong intensity in the case of the IL-3 and mixed (IL-3 + PCT) samples. This further confirmed the Raman peak at ∼2900 cm−1 as a signature mode of IL-3, in addition to the previously identified signature Raman modes. Here, it is worthy to note that the Raman peak at ∼2900 cm−1 is not observed with nearly as strong intensity for the pure biomarkers, although a small peak is observed in that range for IL-3 (Fig. 3b). The possible Raman peaks of the sterile human serum and the corresponding origin of such Raman modes were obtained by analyzing the Raman spectra of sterile human serum (Fig. 4c). The results are listed in Table ST9,† where it was observed that most of the Raman peaks appear in the range of 850–1700 cm−1. The origin of such high energy Raman modes was mainly due to the presence of several proteins and their amino acid building blocks, as mentioned in Table ST10.† Surprisingly, in the case of sterile human serum, no such low energy Raman peak is observed. This becomes an added advantage for the identification of biomarkers in spiked clinical samples, as most of the signature Raman modes of the biomarkers lie in the low energy range (<400 cm−1). Although it can be concluded that the identification of sepsis biomarkers in a spiked clinical sample is possible, the detection of the biomarkers in the presence of each other, i.e., in a mixed state is crucial. To separate the peaks of the biomarkers in the spiked clinical sample, where both of the biomarkers are present, the Raman spectra of the biomarkers in the mixed state for different concentrations were recorded (Fig. 4d). The Raman spectra of the mixed biomarkers suggests that the presence of the strong Raman peak at ∼2900 cm−1 confirms the presence of IL-3. Meanwhile, the low frequency Raman modes help to identify the distinct biomarkers from each other. All of the possible Raman peaks of the mixed biomarkers in the clinical samples were analyzed and are listed in Table ST10,† where the origin of each Raman peak was assigned to sterile human serum, IL-3 and PCT according to their signature Raman modes. For instance, the Raman modes observed at 114, 135, 194, 221, and 268 cm−1 solely originated from IL-3. These peaks were not observed for PCT nor for sterile human serum, and matched well with the previously identified signature Raman modes of IL-3 (Table ST9†). In a similar fashion, the Raman modes observed at 122, 228 cm−1 were observed in the case of PCT only, which are also the signature Raman modes of PCT. Along with the low frequency Raman modes, several other high frequency signature Raman modes of the biomarkers were identified, as mentioned in Table ST10.† Such prominent identification of the distinct Raman peaks for both the biomarkers in the spiked clinical samples, having LOD values as low as 100 fM (for PCT) and 1 pM (for IL-3), confirms the specificity and sensitivity of the developed SERS protocol for the detection of sepsis biomarkers. Thus, an in-depth analysis of the IL-3 and PCT Raman spectra confirmed that the developed SERS substrate may have great potential for immediate application for the detection of ultralow amounts of sepsis biomarkers. This may predict the possibility of septic shock in the amateur stage to prevent deaths caused in septic shock. Such ultrasensitive and stable AgNP@BP flake may also be useful for the development of layered BP based sensors, actuators, and optoelectronic devices.
Conclusions
In summary, the ultrasensitive detection of prognostic and diagnostic biomarkers of sepsis has been established using the AgNP@BP SERS platform. A one-step protocol has been optimized for developing a highly stable BP-based SERS substrate having a uniform distribution of AgNPs. The developed AgNP@BP SERS substrate demonstrates the ultrasensitive detection of sepsis biomarkers, i.e., LPS, IL-3, and PCT, where the limit of detection (LOD) value was as low as 1 nM, 1 pM and 100 fM with an enhancement factor as high as on the order of ∼1014 for LPS, IL-3 and PCT, respectively. The detection of sepsis biomarkers was demonstrated for the spiked clinical samples as well. All of the possible signature Raman modes of IL-3 and PCT were identified and reported for the first time. Thus, for the detection of prognostic and diagnostic biomarkers of sepsis using the same sensing platform, the developed AgNP@BP SERS substrate may pave a path towards the rapid and early detection of sepsis biomarkers. The main advantage of Raman-based diagnostics over state-of-art fluorescence diagnostics is its signal selectivity to molecular structure. Hence, it is considered as a fingerprint technique and provides flexibility for the real-time monitoring of the disease condition. Hence, the immediate proposition is to establish a Raman library, identifying the signature peaks of various biomarkers. This is the key in distinguishing various biomarkers having molecular structures of close similarities, and would allow for greater speed, accuracy, online monitoring, and multiplexity during diagnosis.Author contributions
Anirban Kundu: main conceptualization, methodology, experiment perform, data analysis, original manuscript writing, reviewing & editing; Renu Rani: data curation, reviewing; Anas Ahmad: biomarkers solution preparation, reviewing; Ajay Kumar: writing; Mamta Raturi: writing; Tanya Gupta: data curation; Rehan Khan: conceptualization, writing – reviewing and editing, Kiran Shankar Hazra: supervision, conceptualization, writing – reviewing and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
A. K., R. R., A. A., and M. R. would like to acknowledge the Department of Science and Technology, Govt. of India, for financial support through a fellowship. K. S. H. would like to acknowledge the core funding of INST, Mohali, and DST-SERB Project EEQ/2017/000497 for supporting the project. R. K. would like to acknowledge DST-SERB Project Grant No. CRG/2019/004018 for supporting the project. The authors would also like to acknowledge Dr. Samrat Mukhopadhyay from the Indian Institute of Science Education and Research, Mohali for his kind support for the Raman spectroscopy measurement of the spiked clinical samples.References
- R. D. Schachter and S. Finfer, Recognizing Sepsis as a Global Health Priority — A WHO Resolution, 2017, pp. 414–417 Search PubMed.
- C. Fleischmann, et al., Assessment of global incidence and mortality of hospital-treated sepsis current estimates and limitations, Am. J. Respir. Crit. Care Med., 2016, 193, 259–272 CrossRef CAS PubMed.
- X. Hou, et al., Vitamin lipid nanoparticles enable adoptive macrophage transfer for the treatment of multidrug-resistant bacterial sepsis, Nat. Nanotechnol., 2020, 15(1), 41–46 CrossRef CAS PubMed.
- R. S. Hotchkiss, L. L. Moldawer, S. M. Opal, K. Reinhart and J. Vincent, Sepsis and septic shock, Nat. Rev. Dis. Primers, 2016, 2(1), 1–21 Search PubMed.
- G. L. Lin, J. P. McGinley, S. B. Drysdale and A. J. Pollard, Epidemiology and Immune Pathogenesis of Viral Sepsis, Front. Immunol., 2018, 9, 2147 CrossRef.
- N. Chen, et al., Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study, Lancet, 2020, 395, 507–513 CrossRef CAS.
- F. Zhou, et al., Articles Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study, Lancet, 2020, 6736, 1–9 Search PubMed.
- X. Y. Shao, et al., Rapid and Sensitive Lateral Flow Immunoassay Method for Procalcitonin (PCT) Based on time-resolved immunochromatography, Sensors, 2017, 17(3), 480 CrossRef PubMed.
- A. Belushkin, et al., Rapid and Digital Detection of Inflammatory Biomarkers Enabled by a Novel Portable Nanoplasmonic Imager, Small, 2020, 16(3), 1906108 CrossRef CAS PubMed.
- S. Kumar, S. Tripathy, A. Jyoti and S. G. Singh, Recent advances in biosensors for diagnosis and detection of sepsis: A comprehensive review, Biosens. Bioelectron., 2019, 124, 205–215 CrossRef PubMed.
- A. H. Nguyen, Y. Shin and S. J. Sim, Development of SERS substrate using phage-based magnetic template for triplex assay in sepsis diagnosis, Biosens. Bioelectron., 2016, 85, 522–528 CrossRef CAS PubMed.
- M. M. Guido, et al., In vitro diagnosis of sepsis: a review, Pathol. Lab. Med. Int., 2016, 8, 1–14 Search PubMed.
- S. Riedel and K. C. Carroll, Laboratory Detection of Sepsis Biomarkers and Molecular Approaches, Clin. Lab. Med., 2013, 33, 413–437 CrossRef PubMed.
- G. Polat, R. A. Ugan, E. Cadirci and Z. Halici, Sepsis and Septic Shock: Current Treatment Strategies and New Approaches, Eurasian J. Med., 2017, 49(1), 53–58 CrossRef CAS.
- S. Haydar, M. Spanier, P. Weems, S. Wood and T. Strout, Comparison of QSOFA score and SIRS criteria as screening mechanisms for emergency department sepsis, Am. J. Emerg. Med., 2017, 35, 1730–1733 CrossRef PubMed.
- J. D. Faix, Biomarkers of sepsis, Infect. Chemother., 2013, 46(1), 23–36 Search PubMed.
- S. Y. Cho and J. H. Choi, Biomarkers of Sepsis, Infect. Chemother., 2014, 46, 1–12 CrossRef CAS PubMed.
- J. Min, et al., Integrated Biosensor for Rapid and Point-of-Care Sepsis Diagnosis, ACS Nano, 2018, 12, 3378–3384 CrossRef CAS.
- P. A. Mosier-Boss, Review on SERS of bacteria, Biosensors, 2017, 7, 51 CrossRef.
- P. Xie, et al., Highly sensitive detection of lipopolysaccharides using an aptasensor based on hybridization chain reaction, Sci. Rep., 2016, 6(29524), 1–8 Search PubMed.
- C. Erridge, E. Bennett-Guerrero and I. R. Poxton, Structure and function of lipopolysaccharides, Microbes Infect., 2002, 4, 837–851 CrossRef CAS PubMed.
- G. F. Weber, et al., Interleukin-3 amplifies acute inflammation and is a potential therapeutic target in sepsis, Science, 2015, 347(6227), 1260–1265 CrossRef CAS PubMed.
- J. Min, et al., Integrated Biosensor for Rapid and Point-of- Care Sepsis Diagnosis, ACS Nano, 2018, 12(4), 3378–3384 CrossRef CAS.
- A. L. Vijayan, et al., Procalcitonin: a promising diagnostic marker for sepsis and antibiotic therapy, J. Intensive Care, 2017, 5(1), 1–7 CrossRef.
- S. E. J. Weitkamp, Procalcitonin versus C-reactive protein : review of kinetics and performance for diagnosis of neonatal sepsis, J. Perinatol., 2019, 39(7), 893–903 CrossRef PubMed.
- A. Hakeem, A. Mohsen and B. A. Kamel, Predictive values for procalcitonin in the diagnosis of neonatal sepsis, Electron. Physician, 2015, 7(4), 1190–1195 Search PubMed.
- Q. Cai, et al., Boron Nitride Nanosheets Improve Sensitivity and Reusability of Surface Enhanced Raman Spectroscopy, Angew. Chem., 2016, 128(29), 8545–8549 CrossRef.
- X. Song, et al., Plasmon-Free Surface-Enhanced Raman Spectroscopy Using Metallic 2D Materials, ACS Nano, 2019, 13(7), 8312–8319 CrossRef CAS PubMed.
- J. Li, W. Zhang, H. Lei and B. Li, Ag nanowire / nanoparticle-decorated MoS 2 monolayers for surface-enhanced Raman scattering applications, Nano Res., 2018, 11(4), 2181–2189 CrossRef CAS.
- Q. Guo, et al., Black Phosphorus Mid-Infrared Photodetectors with High Gain, Nano Lett., 2016, 16(7), 4648–4655 CrossRef CAS PubMed.
- K. Chen, C. Yuen, Y. Aniweh, P. Preiser and Q. Liu, Towards ultrasensitive malaria diagnosis using surface enhanced Raman spectroscopy, Sci. Rep., 2016, 6, 20177 CrossRef CAS PubMed.
- J. T. Walsh, D. Ph, M. R. Glucksberg and D. Ph, Progress Toward an In Vivo Surface-Enhanced Raman Spectroscopy Glucose Sensor, Diabetes Technol. Ther., 2008, 10(4), 257–265 CrossRef.
- F. Mos, T. Takamura and A. Sandhu, Laser Power Dependent Optical Properties of MoS2, J. Nanosci. Nanotechnol., 2015, 15(9), 6843–6846 CrossRef PubMed.
- Y. Yang, L. Chen, M. Ji and H. Hospital, Stimulated Raman scattering microscopy for rapid brain tumor histology, J. Innovative Opt. Health Sci., 2017, 10, 1–12 Search PubMed.
- M. Xia, A Review on Applications of Two-Dimensional Materials in Surface-Enhanced Raman Spectroscopy, Int. J. Spectrosc., 2018, 4861472 Search PubMed.
- A. Kundu, R. Rani and K. S. Hazra, Controlled nanofabrication of metal-free SERS substrate on few layered black phosphorus by low power focused laser irradiation, Nanoscale, 2019, 11, 16245–16252 RSC.
- X. Ge, Z. Xia and S. Guo, Recent Advances on Black Phosphorus for Biomedicine and Biosensing, Adv. Funct. Mater., 2019, 29, 1900318 CrossRef.
- Z. Liu, et al., A two-dimensional fingerprint nanoprobe based on black phosphorus for bio-SERS analysis and chemo-photothermal therapy, Nanoscale, 2018, 10, 18795–18804 RSC.
- M. Liang, et al., Silver-Laden Black Phosphorus Nanosheets for an Efficient In Vivo Antimicrobial Application, Small, 2020, 16, 1905938 CrossRef CAS PubMed.
- J. Ouyang, et al., Two dimensional semiconductors for ultrasound-mediated cancer therapy: The case of black phosphorus nanosheets, Chem. Commun., 2018, 54, 2874–2877 RSC.
- W. Lei, et al., Bandgap- and Local Field-Dependent Photoactivity of Ag/Black Phosphorus Nanohybrids, ACS Catal., 2016, 6, 8009–8020 CrossRef CAS.
- X. Fan, et al., Amperometric sensor for dopamine based on surface-graphenization pencil graphite electrode prepared by in-situ electrochemical delamination, Microchim. Acta, 2019, 186, 324 CrossRef PubMed.
- R. V. Devi, M. Doble and R. S. Verma, Biosensors and Bioelectronics Nanomaterials for early detection of cancer biomarker with special emphasis on gold nanoparticles in immunoassays / sensors, Biosens. Bioelectron., 2015, 68, 688–698 CrossRef PubMed.
- X. Cao, Z. Wang, L. Bi and J. Zheng, Label-Free Detection of Human Serum Using Surface-Enhanced Raman Spectroscopy Based on Highly Branched Gold Nanoparticle Substrates for Discrimination of Non-Small Cell Lung Cancer, J. Chem., 2018, 2018, 9012645 Search PubMed.
- H. Huang, et al., Black phosphorus: a two-dimensional reductant for in situ nanofabrication, npj 2D Mater. Appl., 2017, 1, 1–8 CrossRef.
- F. Alsaffar, et al., Raman Sensitive Degradation and Etching Dynamics of Exfoliated Black Phosphorus, Sci. Rep., 2017, 7, 44540 CrossRef CAS PubMed.
- T. Zhang, et al., Degradation Chemistry and Stabilization of Exfoliated Few-Layer Black Phosphorus in Water, J. Am. Chem. Soc., 2018, 140, 7561–7567 CrossRef CAS PubMed.
- J. V. Trueblood, et al., Heterogeneous Chemistry of Lipopolysaccharides with Gas-Phase Nitric Acid : Reactive Sites and Reaction Pathways, J. Phys. Chem. A, 2016, 120, 6444–6450 CrossRef CAS PubMed.
- C. N. Kotanen, L. Martinez, R. Alvarez and J. W. Simecek, Sensing and Bio-Sensing Research Surface enhanced Raman scattering spectroscopy for detection and identification of microbial pathogens isolated from human serum, Sens. Bio-Sens. Res., 2016, 8, 20–26 CrossRef.
- Y. Feng, B. K. Klein and C. A. McWherter, Three-dimensional solution structure and backbone dynamics of a variant of human interleukin-3, J. Mol. Biol., 1996, 259, 524–541 CrossRef CAS PubMed.
- D. L. Urdal, et al., Molecular Characterization of Colony- Stimulating Factors and Their Receptors: Human Interleukin-3, Ann. N. Y. Acad. Sci., 1989, 554, 167–176 CrossRef CAS PubMed.
- J. A. M. Hubbard, et al., Solution structures of calcitonin-gene-related-peptide analogues of calcitonin-gene-related peptide and amylin, Biochem. J., 1991, 275, 785–788 CrossRef CAS PubMed.
- J. B. Petermann, W. Born, J. Y. Chang and J. A. Fischer, Identification in the human central nervous system, pituitary, and thyroid of a novel calcitonin gene-related peptide, and partial amino acid sequence in the spinal cord, J. Biol. Chem., 1987, 262, 542–545 CrossRef CAS.
- Y. Wang, M. Salehi, M. Schütz, K. Rudi and S. Schlücker, Microspectroscopic SERS detection of interleukin-6 with rationally designed gold/silver nanoshells, Analyst, 2013, 138, 1764–1771 RSC.
- G. Zhu, X. Zhu, Q. Fan and X. Wan, Raman spectra of amino acids and their aqueous solutions, Spectrochim. Acta, Part A, 2011, 78, 1187–1195 CrossRef PubMed.
- J. M. Benevides, S. A. Overman and G. J. Thomas Jr, Raman Spectroscopy of Proteins, Curr. Protoc. Protein Sci., 2004, 33, 17–18 Search PubMed.
- A. H. Nguyen, Y. Shin and S. J. Sim, Development of SERS substrate using phage-based magnetic template for triplex assay in sepsis diagnosis, Biosens. Bioelectron., 2016, 85, 522–528 CrossRef CAS PubMed.
- L. Tang and J. Casas, Quantification of cardiac biomarkers using label-free and multiplexed gold nanorod bioprobes for myocardial infarction diagnosis, Biosens. Bioelectron., 2014, 61, 70–75 CrossRef CAS PubMed.
- B. Zhang, A. W. Morales, R. Peterson, L. Tang and J. Y. Ye, Label-free detection of cardiac troponin I with a photonic crystal biosensor, Biosens. Bioelectron., 2014, 58, 107–113 CrossRef CAS PubMed.
- R. Sharma, et al., Label-free electrochemical impedance biosensor to detect human interleukin-8 in serum with sub-pg/ml sensitivity, Biosens. Bioelectron., 2016, 80, 607–613 CrossRef CAS PubMed.
- H. Chaudhry, J. Zhou, Y. Zhong, M. M. Ali, F. McGuire, P. S. Nagarkatti and M. Nagarkatti, Role of Cytokines as a Double-edged Sword in Sepsis, In Vivo, 2013, 27, 669–684 CAS.
- M. L. Steinhauser, C. M. Hogaboam, A. Matsukawa, N. W. Lukacs and R. M. Strieter, Chemokine C10 Promotes Disease Resolution and Survival in an Experimental Model of Bacterial Sepsis, Infect. Immun., 2000, 68, 6108–6114 CrossRef CAS PubMed.
- J. Charchaflieh, J. Rushbrook, S. Worah and M. Zhang, Activated Complement Factors as Disease Markers for Sepsis, Dis. Markers, 2015, 2015, 382463 Search PubMed.
- D. D. Danikas, M. Karakantza, G. L. Theodorou, G. C. Sakellaropoulos and C. A. Gogos, Prognostic value of phagocytic activity of neutrophils and monocytes in sepsis. Correlation to CD64 and CD14 antigen expression, Clin. Exp. Immunol., 2008, 154, 87–97 CrossRef CAS PubMed.
- K. Reinhart, M. Bauer, N. C. Riedemann and C. S. Hartog, New Approaches to Sepsis: Molecular Diagnostics and Biomarkers, Clin. Microbiol. Rev., 2012, 25, 609–634 CrossRef CAS PubMed.
- S. Newton, et al. , NIH Public Access, 2008, 5, 375–383 Search PubMed.
- H. Lim, et al., Synthesis of Uniformly Sized Mesoporous Silver Films and Their SERS Application, J. Phys. Chem. C, 2020, 124, 23730–23737 CrossRef CAS.
- H. Lim, et al., A mesopore-stimulated electromagnetic nearfield: electrochemical synthesis of mesoporous copper films by micelle self-assembly, J. Mater. Chem. A, 2020, 8, 21016–21025 RSC.
- G. Kwon, et al., Nanoporous cellulose paper-based SERS platform for multiplex detection of hazardous pesticides, Cellulose, 2019, 26, 4935–4944 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d1sd00057h |
| This journal is © The Royal Society of Chemistry 2022 |