Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Electroanalytical overview: screen-printed electrochemical sensing platforms for the detection of vital cardiac, cancer and inflammatory biomarkers
Robert D.
Crapnell
 ,
Alejandro
Garcia-Miranda Ferrari
,
Alejandro
Garcia-Miranda Ferrari
 ,
Nina C.
Dempsey
,
Nina C.
Dempsey
 and
Craig E.
Banks
and
Craig E.
Banks
 *
*
Faculty of Science and Engineering, Manchester Metropolitan University, Chester Street, M1 5GD, UK. E-mail: c.banks@mmu.ac.uk; Tel: +44 (0)1612471196
First published on 3rd March 2022
Abstract
Biomarkers play an important and irrefutable role in the screening, diagnosis, monitoring and treatment of a wide variety of human diseases. As these biomarkers inevitably feature more and more prominently in the patient care pathway, there is a growing need for technologies that can provide rapid, accurate and sensitive test results at low cost. In this review we showcase, discuss, evaluate, and explain some recent advances in screen-printed electrodes (SPEs) and the modification strategies used for the electrochemical biosensing of some of the important, established biomarkers related to 1) cardiac injury, 2) cancer diagnostics and 3) acute inflammatory conditions, three areas of medicine currently associated with significant healthcare costs. Electroanalytical biosensors are proven to be an attractive alternative to benchtop conventional testing techniques, saving space, whilst allowing enhanced portability, a reduction in testing costs and test turnaround times. Electrochemical-based point-of-care (POC) testing technologies are still in the early stages of commercial, and hence clinical, uptake. Due to the design flexibility, low-cost and reliability of SPEs we expect to see a significant acceleration in the development of SPE-based electrochemical approaches to POC in these areas of medicine. Rapid, simultaneous detection of multiple important analytes in a single test at the point of patient's care will undoubtedly be the driver for uptake into clinical settings; their potential for impact is discussed herein.
Introduction to screen-printed electrodes
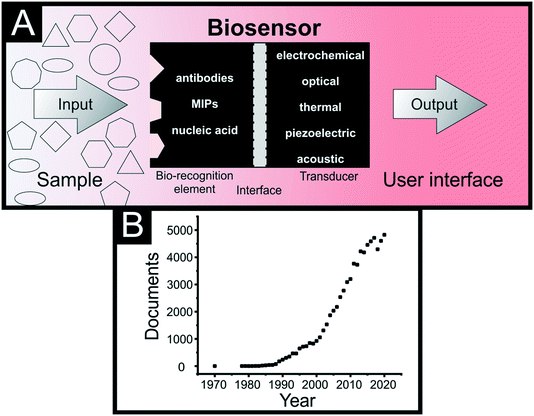
Screen-printed electrodes (SPEs) are the result of decades of component miniaturization applied to electrochemistry. They offer an economical yet highly reproducible, disposable and robust platform as an alternative against the classic electrochemistry setup based on three-external solid electrodes (such as glassy carbon working, platinum counter and Ag/AgCl reference electrodes) and a glass volumetric cell. SPEs are fabricated through depositing a combination of various layers onto a flat substrate that comprises the entire three-electrode basic configuration often used in electroanalysis (working, counter and reference electrodes respectively; WE, CE and RE) in a miniaturised chip of a few cm2.Large scale manufacturing methods such as roll-to-roll, pad-printing and screen-printing have been applied to electrode manufacturing since the 1990s, providing great versatility and precision in the manufacturing process, offering great ink, substrate and precision compatibility for the final electrode.1,2 The use of industrial coating and printing manufacturing such as screen-printing and roll-to-roll processes for graphitic-solvent mix deposition has been used to replace less reproducible and more time-consuming electrode manufacture and modification techniques such as drop-casting, dip-coating, spin-coating, pad-printing and spray coating.3,4 The now extended use of miniaturised electrode/sensor systems, such as SPEs, pushes the boundaries of classic benchtop lab-based test kits towards the on-site, point-of-care (POC) solutions for forensic, environmental and healthcare monitoring to name a few. As shown in Fig. 1A the fabrication of SPEs involves complex parameters such as the used substrate, ink, electrode design and electrode pre-treatment. Fig. 1B includes a scheme of the ink deposition process that eventually becomes a SPE, showing that the ink composition can be bulk-modified and is a complex mixture of binders, solvents and can also have modifiers. Fig. 1C depicts how the successive deposition of ink layers with a defined design/drawing creates a SPE.
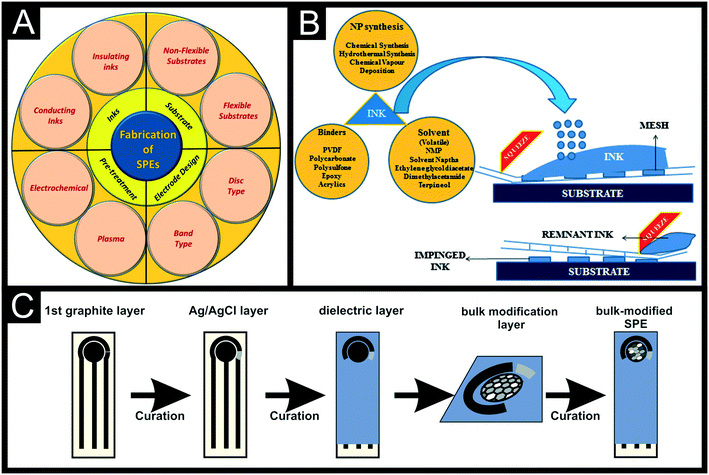 | ||
| Fig. 1 Overall scheme of screen-printing parameters (A) and process of depositing ink through the screen mesh by the squeegee motion (B). Reproduced from ref. 1 with permission from Springer, copyright 2021. (C) Schematic representation of the bulk-manufacture of modified-SPEs with a defined design. | ||
Screen-printed electrodes are now widely used throughout the academic literature and have transitioned into commercial products due to their wide range of advantageous properties, in addition to their well-stated low-cost, in comparison to commercial external electrodes:
1) Combination of all three electrodes on one device – instead of requiring three separate external electrodes as in a traditional electrochemical set-up, all of the working, counter and reference electrode can be printed onto the same substrate, see Fig. 1C.
2) Miniaturisation of the electrochemical platform – as seen in Fig. 1, the size of the required set-up and solution volume is massively reduced through the use of macro SPEs in comparison to traditional commercial electrodes. This can be taken even further through printing of SP microelectrodes, further reducing the size of the required system.5
3) Flexibility of the electrode shape and size – there is a huge range of electrode shapes and sizes seen in the literature. Many research groups produce their own bespoke SPEs through simply changing the stencil design. This can be used to change the size of the electrodes, length of connections, amount of electrodes on one sheet etc.2 This can be seen in the literature through the development of recessed microelectrode arrays,6 back-to-back SPEs7 and microbands8,9 among other examples.
4) Wide ranging options of electrode materials – there is a wide range of base inks that can be used as the electrode surface. The choice of this material will typically come down to the desired application of the electrode. For example, if a thiol self-assembled monolayer was desired a gold SPE could be produced. SPEs based on graphite,10 graphene,11 gold,12 platinum,13 silver14 and copper15 have been reported in the literature to name a few.
5) Wide array of printable substrates – screen printing can be applied to a wide range of substrates depending on the final desired application including paper, polymers, textiles, wood, ceramics and metal to name a few. In recent years, paper based sensors have become increasingly popular due to environmental reasons.16,17
6) Mass production capabilities – machinery is readily available for the production of thousands of electrodes per day. This will obviously depend on the resources available for purchasing the equipment.18
7) High reproducibility, sensitivity and accuracy – when the production methods and materials are optimised (considerations are outlined below) it is possible to produce highly reproducible electrodes with excellent inter- and intra-batch reproducibility.19 These have been shown to achieve excellent sensitivities when used in biosensing applications.20,21
8) Ability to bulk-modify inks for bespoke production – bespoke electrode configurations can be achieved through mixing active materials into standard inks before the printing process. This ensures a consistent presence of the active material with excellent adhesion to the electrode which can be an issue with drop-casting for example. Some interesting examples of this can be seen for both electroanalytical sensing22–24 and energy applications.25,26
There are of course drawbacks to the use of SPEs, including batch to batch variation and poor benchmarking standards for some commercially acquired SPEs. These variations will greatly affect development of electrochemical sensing platforms in research labs and therefore we recommend either finding a consistent and trusted supplier, benchmarking in-house each batch when purchased, collaborating with other research groups who produce SPEs in-house or even starting to produce your own. The use of an internal standard during measurements might help alleviate such issues.27 It has recently been shown how the connection length of graphitic carbon SPEs can have a significant effect on the electrochemical performance of the electrode.28 It has therefore been recommended that all reports in literature should ideally characterise the connection resistance of their SPEs to allow true comparisons to be made across literature reports. When performed to a high standard with consistent methods, screen-printing offers a way to produce thousands of electrodes a day at low-cost, with excellent electrochemical performance and with high intra- and inter-batch reproducibility.
There are important factors to consider when manufacturing screen-printed electrodes (SPEs):
1) What is the general architecture of the printer? Screen mesh, substrate, squeegee specifications and printing parameters play an important role in the final performance of the device. The mesh defines the area to be printed and allows the ink to be deposited in a specific pattern and thickness. The squeegee will push the ink through the empty holes of the mesh, facilitating the ink deposition in the defined pattern.
2) What mesh to use? When screen-printing, the desired pattern is negatively imprinted onto a mesh that allows the pass of the ink in that particular, desired way. A mesh has both porous (empty) and non-porous (blocked) holes that allowed the ink to be deposited onto the substrate. Meshes are often manufactured with nylon, polyester or stainless steel. The mesh is usually built on a metal or wooden frame able to hold the pressure from the mesh and printing tension. For further information on how to choose the right mesh for screen-printing for electrochemical applications, please read Foster et al.29
3) What squeegee to use? The squeegee pushes the ink through the mesh at high pressure, and sometimes speed, creating a controlled flow of ink upon the substrate. In order to avoid damaging the screen stainless steel is often avoided, therefore softer materials such as polyurethane are used, which is reported to possess extreme long-lasting capabilities, achieving up to 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 prints before visible damage occurs. The softness of the squeegee allows to have an increased contact with the screen, being the softness one the more efficient grade to use. Also, the width of the squeegee is needed to be at least 10 mm larger than the screen drawing to spread homogeneously the pressure.
000 prints before visible damage occurs. The softness of the squeegee allows to have an increased contact with the screen, being the softness one the more efficient grade to use. Also, the width of the squeegee is needed to be at least 10 mm larger than the screen drawing to spread homogeneously the pressure.
4) What is the ink composition? Electrochemical sensors often use thixotropic inks, meaning that their viscosity decreases when there is a shear rate increase, which decreases the spread or bleed of the ink, obtaining a better definition/precision in the final print.30 For electrochemical purposes, the use of conductive inks is required, often achieved by using graphitic active materials which are then mixed in the binder matrix. The chosen conductive/active material is needed to allow electron tunnelling and optimal electrical conductivity throughout the printed circuit. Regarding the binder selection, these can be divided depending on their solubility (water-, solvent-based etc.), sourcing (natural or synthetic), molecular weight (MW) (solution (low MW) or emulsion resins (high MW) etc.). Solvent selection tailors the printability properties of the final ink in terms of the viscosity and rheology. Literature reports strongly suggest the use of inert solvent-free of non-volatile impurities with moderate boiling points for screen-printing applications. Due to the electrical conductivity requirements for electrochemical applications, the selection of solvents take a paramount role in the curing process, where changes in the rheology can have a substantial effect on the final cured printed circuit. Finally, the overall ink performance can be tuned by using additives, that when added in the right amounts, can help improving the homogeneity, conductivity and viscosity of the ink. These additives are often waxes, plasticisers and surfactants.31 It is because of this that the reader should pay careful attention to the printability, electrical and electrochemical properties reported by manufacturers, when purchasing commercial SPE platforms if the exact ink composition is not described within the vendor's datasheet.
5) What electrode geometry to use? SPE platforms offer the freedom of exhibiting a wide range of electrode sizes that can be bulked manufactured. SPE dimensions range from micro-electrode configuration at the μm scale to the cm or larger. Other than less ink usage, micro-electrodes are reported to exhibit an enhanced electrochemical response than those of macro-size. As an example, Damiati et al.32 reported a ×100 improvement in LOD when using graphitic carbon SPE (C-SPE) when sensing human chorionic gonadotropin (hCG) from 100 ppb for a 4 mm SPE to 1 ppb LOD for a 400 μm working electrode respectively. The main reason for this electrochemical enhancement is the mass transport rate and diffusion profile, that changes from planar (at a macro-electrode) to radial (at a micro-electrode) as reported by Compton et al.,33 depicted in Fig. 2. Fig. 2 depicts the changes in diffusion profiles, that when using multiple single micro-electrodes in parallel (micro-array) have been reported to exhibit lower LODs and greater sensitivities in comparison with their equivalent macro-electrodes.34 It is also important to note that, the ability of manufacturing multiple electrode configurations allow their use for simultaneous analyte determination, where each electrode can be tailored or modified differently, as reported multiple times within the literature towards for example, immuno-sensor applications.35–37 Added to the size manipulation and multi-electrode configurations, screen-printing allows designing the shape of the electrodes depending on the desired application, being circle, bands, arrays, interdigitated, back-to-back etc. as examples of some of the many reported electrode shapes.6–8,38–40
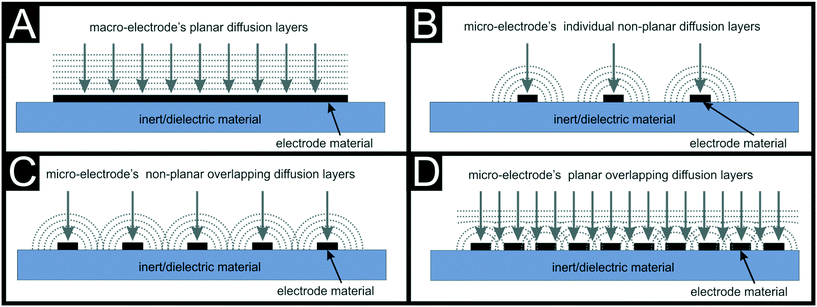 | ||
| Fig. 2 Shows the planar diffusion at macro-electrodes (A), the radial non-planar diffusion at individual micro-electrodes (B), the radial overlapping of diffusion layers at micro-electrodes (C) and the planar overlapping of diffusion layers at micro-electrodes arrays (D). Reproduced from ref. 1 with permission from Royal Society of Chemistry, copyright 2020. | ||
As a summary, screen-printing offers a linking bridge to transition from small batches to large-scale electrode manufacturing and is therefore suited for both research and commercial solutions, however there are some important challenges that still need addressing. In order to move from centralised labs to POC sensing devices, some these challenges include the transition to flexible substrates and electrodes for wearable applications, in conjunction with other bottom-up fabrication methods, to bring the cost of the final sensor down. Mass-production remains the paramount goal of printed electrodes, therefore studies where bulk fabrication/modification of electrodes should be prioritised. Although we describe herein some of the complicated parameters that take part when choosing a SPE platform, this is a guide for non-expert experimentalists and therefore we suggest the reader to address their most deep inquiries to some of the more concrete references included herein.
In this perspective we have highlighted some key parameters that experimentalists should carefully consider when designing or choosing their screen-printed sensing platforms for useful electroanalytical investigations. It is important to also note that often these parameters are taken with little thought or consideration, and sometimes that same literature fails to properly explain to the unexperienced researcher the reason behind choosing certain screen-printed platforms for their experiments. Consequently, herein we aim to highlight the various methodologies reported for the modification of SPEs toward the incorporation of recognition elements for detecting vital biomarkers.
Recognition elements and their integration into SPE biomarker platforms
Recognition elements are a vital part of the development of electrochemical sensor platforms, especially in cases where the target is itself not electro-active. In the context of this work, the term recognition element will cover both non-bio and biorecognition elements. We note that not all of the sensor platforms discussed herein are strictly biosensors, as the IUPAC definition for such devices specifically requires a biorecognition element.41Fig. 3A shows a classic biosensor configuration diagram, where the biosensor is made by a bio-recognition element for a specific target in a specific sample matrix, a transducer then converts the signal from the chemical/physical contact to an electrical/electronic signal output that will be processed in the processor/computer unit. The purpose of recognition elements in electrochemical sensing platforms is to provide specificity toward the target, through a strong and selective affinity. The reports of “biosensor” in the literature have accelerated in the last few decades as shown in Fig. 3B, especially since the beginning of the 21st century with the latest advances in computer powering and miniaturisation. Among those, there are several recognition elements used throughout the literature, ranging from naturally occurring to completely synthetic. The main ones that will be seen throughout this work include antibodies, aptamers, enzymes, nucleic acids and molecularly imprinted polymers (MIPs). Antibodies remain the most widely used recognition element. They are 3D protein structures that have unique recognition patterns, allowing for high specificity and binding affinity towards their specific target. This structure is commonly depicted as the distorted “Y” shape; whereby the two arms are known as the Fab region and the “tail” is known as the Fc region.42 It is the Fab region that is relied upon for the specific binding toward antigens, and the majority of cartoons/schematics depicting antibody binding to electrodes has the Fc region bound to the surface with the Fab region conveniently orientated perpendicularly. We note that this is a common trend throughout sensing literature and not solely a problem for antibody schematics. Unless the sensor developer has included specific mechanisms to orientate the recognition element, it is suggested authors and reviewers take care to include more realistic depictions of electrode surfaces. Antibody based electrochemical sensors tend to operate through either amperometric or impedimetric approaches, whereby the binding phenomena is observed through either increases or reductions in the measured current or resistance. Although still the most utilised, there has recently been a shift away from using antibodies due to issues with their reproducibility, batch-to-batch variations, long-term stability and use of animals in production.Enzymes represent the other most often used biorecognition element throughout the literature and gain their specificity from non-covalent recognition patterns within their 3D structure.43 Once integrated into devices they help with the detection of targets through biocatalytic effects, whereby the target analyte is typically captured and converted into an electrochemically measurable product. This platform most commonly utilises amperometric approaches for the detection of these products. These systems, although simple and specific can struggle from poor working lifetimes, requirements for optimum pH/temperature tuning and costly purification strategies.44 Nucleic acids rely on the complementary binding of DNA to achieve their high specificity. Through the identification of a target DNA sequence, a complimentary fragment can be artificially manufactured to provide the desired recognition element. These geno-sensors function through recognising when hybridisation occurs and therefore have a very limited range of targets, however they do present improved stability over antibodies.
More recently, aptamers have increased in popularity. They are short oligonucleotides comprising of nucleobases that have been synthesized without the need for animals or cell cultures.45 Aptamers are more thermally stable than the other biorecognition elements mentioned, and provide the ability to design bespoke recognition elements for the user's desired targets. In this regard, they are similar to MIPs, which are also chemically, thermally and mechanically stable, are re-usable and are low-cost.46 MIPs are fully synthetic recognition elements, whereby a polymerisation process is initiated around the target molecule, effectively freezing it in place. Once removed, cavities specific to the size, shape and functionalities of the target molecule are left within the polymer backbone. There is a plethora of different synthesis strategies to create MIPs, such as UV-polymerisation,47 thermal polymerisation48 and electropolymerisation49 to name a few. There has been a significant increase in the amount of research on MIPs in recent years and significant improvements in performance have been made, however they typically still suffer from a time-consuming production process, template leakage, complex fabrication processes and relatively poor synergy with electrochemical platforms. In addition to those mentioned above, there are other recognition elements available such as phages50 and affibodies51 for example. We direct you to some excellent reviews discussing recognition elements in more detail.44,52
The choice of recognition element is one of the most important parameters when developing a biosensor and this depends on the target analyte and desired characteristics of the sensing platform (cost, lifetime, sensitivity etc.). The recognition element should have not only high affinity towards the target molecule, but should exhibit stability too.53Table 1 compares the advantages and limitations of recognition elements in chemical sensors and biosensors, with their respective advantages and limitations.53 As shown in Fig. 4, the integration of these recognition elements can take on many forms depending on the chosen bioreceptor elements. Pérez-Fernández et al. recently summarised the main immobilisation methods of bio-receptors when applied to screen-printed electrodes.54 Adsorption, covalent bonding, electrochemical deposition and electrochemical polymerization are the most common surface modifications used with SPEs.3,55 Adsorption relies on the creation of non-covalent bonds between the modified and the electrode's surface. Electrochemical modifications are based on passing voltage, current or charge through the electrolyte and working electrode to deposit stable films onto the electrode's surface. One of the main advantages of SPE manufacture is the bulk modification of the inks with the desired materials. This provides a convenient mass-producible approach that improves the manual modification of electrodes, increases the cycling stability and improves the homogeneity of the newly-created modification. For more details on SPE modifications, please read García-Miranda Ferrari et al.1 The reliable attachment of biorecognition elements to the solid support of a sensor platform is one of the key parameters that underpins the design of a biosensor.56 Covalent coupling is commonplace throughout the literature with various chemical processes available, depending on the system (substrate functionalities, pH, temperature, coverage required). We highlight one of the most popular cross-linkers 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) for bio-chemical conjugations due to its efficient and powerful ability to conjugate primary amines to carboxylic acids for peptide and protein crosslinking, thereby immobilising them onto the working electrode's surface. For its application with higher stability and efficiency, it is often used in conjunction with N-hydroxysuccinimide (NHS; or its water soluble analogue sulfo-NHS). When used together, EDC/NHS allow for a two-step coupling of proteins without altering the second protein's carboxylic groups. The authors note that it is important to consider the mechanism behind this coupling chemistry when applying it as the EDC activates the carboxylic acid groups present, which can then couple to any amine groups present. Both one and two-step methods are possible, where the two-step method is recommended for the immobilisation onto substrates where the biorecognition element contains both carboxylic acid and amine groups. This allows for activation of the carboxylic acid group on a substrate first, eliminating the possible activation and reaction of the biorecognition element with others around it. We direct the reader toward review papers discussing the different immobilisation techniques, emphasising the importance of biorecognition element orientation to the end performance of the biosensor.56–59
| Recognition element | Sensor designation | Advantages | Limitations | |
|---|---|---|---|---|
| Classical | Enzymes | Enzymatic biosensor | Specificity | Purification is costly and time consuming |
| Simple procedure and apparatus | Poor stability | |||
| Efficient only at optimum pH and temperature | ||||
| Antibodies | Immunosensor | High affinity | Limited target (protein) | |
| Specificity | Laborious production | |||
| Production requires use of animals | ||||
| Poor stability | ||||
| Nucleic acids | Geno-sensor | Stability | Limited target (complementary nucleic acid) | |
| Whole cells | Whole-cell biosensor | Low-cost preparation | Expensive macromolecule isolation costs | |
| Reduced purification requirements | Limited detection capability | |||
| Short useable lifetime | ||||
| Recent | Phages | Phage biosensor | Specificity and sensitivity | Optimisation of the phage size |
| Stability | Expression on phage's surface | |||
| Discerning live vs. dead bacteria | ||||
| Phage multiplication inhibition | ||||
| Antiviral bacterial immunity system | ||||
| Presence of specific phage inhibition genes | ||||
| Aptamers (DNA, RNA or peptides) | Aptasensor (DNA or RNA sensor) | Easy to modify | Target's molecular weight (MW) is proportional to the binding affinity of the aptamer | |
| Possibility to design structure | Limited functional groups in small molecules | |||
| Possibility to denaturalize and to rehybridize | Chemisorption of molecules to conductive surface | |||
| Possibility to distinguish targets with different functional groups | Quality of semiconductor material | |||
| Thermally stable | Improvement of signal-to-noise ratio | |||
| Peptide sensor | ||||
| In vitro synthesis | Need of a systematic aptamer discovery process for biosensor | |||
| Molecularly-imprinted polymers (MIPs) | MIP sensor | High thermal, chemical, and mechanical tolerance | Complex fabrication methodology | |
| Reusability | Time-consuming process | |||
| Low cost | Incompatibility with aqueous media | |||
| Leakage of template molecules | ||||
| Affibodies | Affibody sensor | Lack of disulfide bonds that enable intracellular applications | Expensive | |
| Long shelf-life | ||||
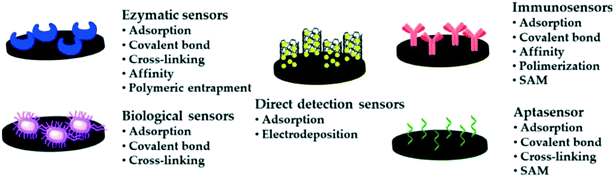 | ||
| Fig. 4 Schematic depiction of the main immobilisation methods for bioreceptors on screen-printed electrodes. Reproduced from ref. 54 with permission from MDPI, copyright 2020. | ||
Recent technological advances in the detection of vital biomarkers using SPE based platforms
Cardiac biomarkers
Cardiovascular disease (CVD) is the number one cause of deaths worldwide, responsible for approximately 18 million deaths annually, according to the World Health Organisation (WHO).60 CVD imposes huge burdens on healthcare services around the globe; In the United Kingdom (UK) alone, approximately £7 billion is spent by the National Health Service (NHS) annually on treating CVD.61 It is estimated that four out of every five deaths related to CVD are the result of acute myocardial infarction ((AMI), commonly referred to as a heart attack) and strokes.60 AMI occurs when blood flow to the heart via the coronary artery is blocked, causing ischaemia and myocyte necrosis (cell death).62 Rapid diagnosis post onset of chest pain is vital for effective treatment and for improving patient outcome.63 The Joint European Society of Cardiology/American College of Cardiology Committee (ESC/ACC) have emphasised the importance of utilising cardiac biomarkers in the diagnosis of AMI. Cardiac biomarkers (CBs) are produced as a result of pathological processes in the cardiovascular system. There are a vast number of CBs encompassing enzymes, hormones, and proteins, each with their own set of characteristics.In more recent updates to the (ESC/ACC) guidelines, particular importance is placed on the use of cardiac troponins (cTn), recognising them as the gold standard in AMI diagnosis.64–66 As such, development of electrochemical sensors aimed at rapidly detecting both cardiac troponin I (cTnI) and T (cTnT) at point-of-care has dominated the literature. These cTn have typically taken the place of the previously used cardiac biomarkers creatinine kinase-MB (CK-MB) and myoglobin, given their superior sensitivity, but yet, there are still a plethora of examples targeting CK-MB and myoglobin due to their reduced comparative cost. A number of commercial analysers for cTn are now available, for example the TnI-Ultra assay (ADVIA Centaur XP immunoanalyzer, Siemens Healthcare Diagnostics) and the cTnT assay (Elecsys TnT-hs, Roche Diagnostics). The cTnI assay can achieve detection in plasma as low as 0.006 ng mL−1 and spanning a range of 0.006–50 ng mL−1; whereas, the cTnT assay has a limit of detection (LOD) of 0.005 ng mL−1 and can detect its presence up to 50 ng mL−1. These lab-based methodologies have improved significantly, with the Roche Troponin T assay able to produce results in a single hour.67 But there is still a huge drive for portable, reliable and low-cost devices. The major limitation with using cTn is their low sensitivity within the first few hours, with circulating concentrations spiking between 6–12 hours post-AMI.68 This, can lead to delayed diagnosis and treatment, increasing morbidity and mortality. As such, there is a desire within the healthcare community to combine the characteristics of multiple biomarkers, providing greater amounts of data which may help to improve patient outcomes. Amongst many emerging biomarkers for AMI are B-type natriuretic peptide (BNP, NT-pro-BNP), atrial natriuretic peptide (ANP), soluble ST2 (sST2), copeptin and heart-fatty acid binding protein (h-FABP). These markers have the potential to be added to the detection of cTn to help vastly improve diagnostic capabilities in healthcare, dramatically improving patient outcome. For example, h-FABP can be detected within 2–3 h of AMI compared to the 3–6 h for cTn,69,70 and its rapid clearance (within 24 hours post-cardiac event), means it is useful to identify repeated cardiac events.
The literature on the development of SPE platforms for the detection of cardiac biomarkers, Table 2, is predominantly based around cTn (to be expected as they remain the gold standard) and myoglobin; the latter is expected due to the significant savings financially when choosing to work with this marker. One of the first platforms reported for the detection of cTnI using a carbon based SPE (C-SPE) was by Jagadeesan and co-workers,71 who used filter paper as the base substrate due to its hydrophilicity and biocompatibility. They first deposited a silver paste onto the substrate to aid with connections, followed by the carbon paste, which was left to dry for 20–30 min. The carbon electrode was modified further with the conductive polymer polyaniline (PANI) through electrodeposition, which provided the appropriate amine functionalisation. This was followed by EDC/NHS coupling of the cTnI specific monoclonal antibodies and further electrode blocking via bovine serum albumin (BSA, 1 wt%). We note that BSA is an extremely popular electrode blocking agent found regularly throughout the literature, however there are various other options such as blocking buffers (e.g. SuperBlock™), milk and casein.72,73 The sensor produced by Jagadeesan utilised cyclic voltammetry for detection, achieving a linear range of 1–100 ng mL−1. This sensor platform although reported to be reproducible, will suffer in terms of reproducibility and extended linear range due to the random orientation of the immobilised antibodies. Additionally, this work although a good foundation study did not test the functionality of their sensor in a real sample medium such as serum or plasma, which we would expect for future work published in the area, ideally alongside ELISA validation. It is this comparison and validation alongside industry and clinical gold standards that will increase the confidence and acceptance of technology by professionals and consumers not in the field.
| Biomarker | Electrode & Modification | Recognition element | Electrochemical technique | Linear range | Limit of detection | Real sample matrix | Ref. |
|---|---|---|---|---|---|---|---|
| Key: cTnT: cardiac troponin T; Abs: antibodies; C-SPE: carbon screen-printed electrode; Au-SPE: gold screen-printed electrode; CA: chronoamperometry; CV: cyclic voltammetry; DPV: differential pulse voltammetry; SWV: square-wave voltammetry; EIS: electrochemical impedance spectroscopy; SWASV: square wave adsorptive stripping voltammetry; MIP: molecularly imprinted polymer; GQD: TTCA: 5,2′:5′2′′-terthiophene-3′-carboxylic acid; NTH: nanotetrahedron; graphene quantum dots; PLL: poly-L-lysine; BP: black phosphorous; PANI: polyaniline; MoS2NF: MoS2 nanoflowes; rGO: reduced graphene oxide; CdS NCs: CdS semiconducting nanocrystals; PPy: polypyrrole; MWCNT: multi-walled carbon nanotubes; MB: methylene blue; Ph: phenol; PVC: polyvinyl chloride; AAM: acrylamide; PAP: polyaminophenol; o-PD: o-phenylenediamine; NPs: nanoparticles; PEI: polyethyleneimine; propargyl: propargylacetic acid; Str-MS: streotavidin-microsphere; CA: cellulose acetate; MOF: metal–organic framework. | |||||||
| cTnI | C-SPE/Au-NPs/TTCA | Aptamer | CA | 1–100 pM | 1 pM | Human serum | 83 |
| cTnI | C-SPE/Fe3O4@UiO-66/Cu@Au | Aptamer | DPV | 0.05–100 ng mL−1 | 16 pg mL−1 | Human serum | 82 |
| cTnI | C-SPE/PANI | Abs | CV | 1–100 ng mL−1 | N/A | N/A | 71 |
| cTnI | C-SPE/AuNPs | Abs | EIS | 0.2–12.5 ng mL−1 | 0.2 ng mL−1 | N/A | 77 |
| cTnI | C-SPE/MOF/PANI | Abs | EIS | 1–400 ng mL−1 | 0.8 ng mL−1 | Mouse serum | 81 |
| cTnI | C-SPE/MoS2/CA | Aptamer | EIS | 10 fM–1 nM | 10 fM | Human serum | 75 |
| cTnI | G-SPE/GO | Abs | SWV | 1–100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ng mL−1 000 ng mL−1 |
0.38 ng mL−1 | Serum | 79 |
| cTnI | C-SPE/MoS2NF | Aptamer | EIS | 10 fM–1 nM | 10 fM | Human serum | 76 |
| cTnI | C-SPE/GO/ aminotrimesic acid | Abs | EIS | 0.1–100 ng mL−1 | 0.08 ng mL−1 | Mouse serum | 80 |
| cTnT | C-SPE | Abs | CV | 0–700 ng mL−1 | 0.15 ng mL−1 | N/A | 84 |
| cTnT | C-SPE/rGO | PANI–MIP | DPV | 0.02–0.09 ng mL−1 | 0.008 ng mL−1 | Blood serum | 89 |
| cTnT | C-SPE/rGO | PPy–MIP | DPV | 0.01–0.1 ng mL−1 | 0.006 ng mL−1 | Blood serum | 90 |
| cTnT | C-SPE/MWCNT/MB | PANI–MIP | DPV | 0.1–8.0 pg mL−1 | 0.04 pg mL−1 | Human plasma | 87 |
| cTnT | C-SPE/MWCNT | Abs | DPV | 2.5–500 pg mL−1 | 35 pg mL−1 | Human serum | 86 |
| cTnT | C-SPE/Str-MS | Abs | Amperometry | 0.1–10 ng mL−1 | 0.2 ng mL−1 | Human serum | 85 |
| cTnT | C-SPE/CdS NCs | Abs | SWASV | 5–1000 ng L−1 | 2 ng L−1 | Human serum | 78 |
| Myoglobin | C-SPE/GQD | Abs | EIS | 0.01–100 ng mL−1 | 0.01 ng mL−1 | Human serum | 96 |
| Myoglobin | C-SPE/rGO/CNT | Aptamer | CV | 1–4000 ng mL−1 | 0.34 ng mL−1 | N/A | 95 |
| Myoglobin | C-SPE/PLL-BP | Aptamer | CV | 1 pg mL−1–16 μg mL−1 | 0.524 pg mL−1 | Serum | 95 |
| Myoglobin | Au-SPE | Ph–MIP | SWV | 0.01 ng mL−1–100 μg mL−1 | 2.1 pg mL−1 | Artificial serum | 93 |
| Myoglobin | Au-SPE/PVC | AAM–MIP | EIS | 0.852–4.26 μg mL−1 | 2.25 μg mL−1 | Artificial serum | 91 |
| Myoglobin | Au-SPE | PAP–MIP | SWV | 2.22–16 μg mL−1 | 0.8 μg mL−1 | Artificial serum | 94 |
| Myoglobin | C-SPE | o-PD–MIP | DPV | 1 nM–1 μM | 9 ng mL−1 | Human plasma | 92 |
| NT-proBNP | Au-SPE | Abs | EIS | 0.1–5 ng mL−1 | 0.1 ng mL−1 | Porcine plasma | 97 |
| BNP-32 | Au-SPE/rGO/PEI/propargyl | DPV | 1 pg mL−1–1 μg mL−1 | 0.9 pg mL−1 | Human serum | 98 | |
| cTnI | Aptamer | 1 pg mL−1–10 ng mL−1 | 1 pg mL−1 | ||||
It is commonplace in electrochemical biosensor development to try and improve the sensor performance through the incorporation of nanomaterials and nanoparticles on the electrode surface.74 This can be seen for the detection of cTnI with reports incorporating MoS2,75,76 gold nanoparticles (AuNPs),77 quantum dots78 and graphene oxide.79,80 This last example by Boonkaew et al.79 utilised identical modification strategies to produce a sensing platform for cTnI, and two inflammatory biomarkers procalcitonin (PCT) and C-reactive protein (CRP). They used a graphene modified SPE (G-SPE), drop-casted with graphene oxide as their electrode base, Fig. 5A. They anodised the graphene oxide through the application of +1.4 V for 30 s to produce a surface functionalised with carboxyl groups. This allowed for the two-step EDC/NHS coupling, whereby the surface carboxyl groups were functionalised prior to the droplet of monoclonal antibodies being added. Unreacted carboxyl groups were then reduced through the application of −1.4 V for 30 s followed by electrode blocking with BSA. Detection of the biomarkers was achieved through a reduction in observed square-wave signal for [Fe(CN)6]3−/4− upon binding of the analyte to the immobilised antibody. Through this, cTnI detection was achieved with a linear response of 0.001–250 ng mL−1 with a detection limit of 0.16 pg mL−1. This work was tested against the interferents glycine, creatinine, L-cysteine, homocysteine, albumin, haemoglobin and myoglobin, showing no significant deviation in results. Additionally, it was shown that detection was possible in human serum samples with good recoveries between 98.73–103.0%. An alternative material used due to their large surface area are metal organic frameworks (MOFs). Although they possess insulating properties, when used on the electrode surface they are commonly used in conjunction with electrochemically active nanomaterials or another conducting material. Gupta et al.81 utilised PANI to provide conductivity and functionality to the MOF in their biosensor that achieved a LOD of 0.8 ng mL−1 using electrochemical impedance spectroscopy (EIS). Alternatively, Sun et al.82 utilised a MOF as a nanocarrier in their system, rather than attaching it directly to the SPE surface. They immobilised DNA nanotetrahedrons (DNA NTH) with aptamers onto gold SPEs (Au-SPE) and blocked the remaining surface through the formation of a self-assembled monolayer (SAM) using 6-mercapto-1-hexanol. Aptamer labelled MOFs with Cu@Au nanoparticles can then join with bound cTnI analyte, which can catalyze the oxidation of hydroquinone. This was used in conjunction with differential pulse voltammetry (DPV) to produce a linear range of 0.05–100 ng mL−1, a LOD of 16 pg mL−1 and achieve recoveries between 96.2–102% in human serum samples. These sandwich style assays are commonplace throughout the literature and typically function through having one recognition element bound to the electrode surface to capture the analyte, then a second tagged detection antibody is introduced to bind to the analyte and produce a signal. This technology has been used for SPE electrochemical platforms for cardiac biomarkers, for example Jo and co-workers utilising aptamers for the detection of cTnI83 and Dempsey et al.84 reporting this concept using Abs for cTnT.
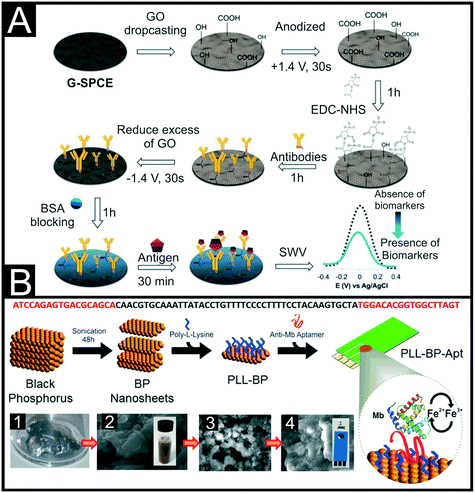 | ||
| Fig. 5 (A) Step-by-step schematic of the preparation of an electrochemical sensor for cTnI through the immobilisation of antibodies onto a modified graphene SPE. Reproduced from ref. 79 with permission from Elsevier, copyright 2021. (B) Schematic representation of the workflow for the liquid-phase exfoliation of black phosphorous (BP) nanosheets and their surface modification for bio-interface development. Panels show depict: (1) bulk BP prior to liquid-phase exfoliation, (2) SEM image of exfoliated few layer BP from aqueous 1% w/v Triton X-100 solution and a photograph of the stable colloid, (3) SEM image of few-layer BP with poly-L-lysine (PLL), (4) SEM image of few layer BP functionalised with PLL and myoglobin aptamer and a photograph of the SPE used throughout. The DNA sequence of the aptamer is shown above. Reproduced from ref. 95 with permission from American Chemical Society, copyright 2016. | ||
One of the first reported SPE based electrochemical biosensors for cTnT utilised a sandwich-type assay, using cTnT Abs attached to streptavidin microspheres to capture the antigen and Abs modified with horseradish peroxidase as the detection Ab.85 In this work they produced the SPE by mixing together epoxy resin silver (40 wt%), hardener epoxy silver (45 wt%), graphite (10 wt%) and tetratcyanoquinodimethane (TCNQ, 5 wt%), in which the TCNQ acts as a mediator, followed by printing onto a polyethylene terephthalate wafer substrate. The in-house production of SPEs allowed for the customisation of the electrode dimensions, where the authors defined their working electrode as a 0.15 cm2 disk. In this work, the use of the streptavidin microspheres is stated to offer 8.5 times increase in the analytical sensitivity of the system, producing a linear range between 0.1–10 ng mL−1, a LOD of 0.2 ng mL−1 and a 95% recovery in undiluted human serum. As seen above for cTnI, there are examples of biosensors for cTnT that utilise nanomaterials such as carbon nanotubes (CNTs) to boost the performance of biosensors. This can be for systems using EDC/NHS coupling to Abs,86 where the CNTs are amine functionalised, or to enhance the analytical signal obtained from molecularly imprinted polymers (MIPs).87 Surprisingly there were limited examples of cTnI MIPs on SPE electrochemical platforms, although we point out recent work using nano-MIPs on SPEs alongside a thermal detection methodology.88 In contrast, there have been more reports for cTnT, with two reports of electrodeposition of the MIPs onto reduced graphene oxide (rGO) modified SPEs, one using PANI89 and one using polypyrrole (PPy).90 Recently, Phonklam et al.87 reported a PANI MIP based sensor for the detection of cTnT. In this work they modified a C-SPE with multi-walled carbon nanotubes (MWCNTs) followed by electrodeposition of polymethylene blue (PMB). The PMB acted as the redox probe, on which the PANI MIP layer was electrodeposited. Upon binding of the cTnT, there was an observed reduction in the DPV signal with a linear range of 0.1–8.0 pg mL−1, LOD of 0.04 pg mL−1 and recoveries between 91–112% in diluted human blood plasma samples. There are an even greater proportion of reported MIP-based sensors for the detection of myoglobin. This makes sense as optimization of MIP composition, synthesis parameters, template removal and detection methodology require a significant number of sensors to be made. Therefore, using a cheaper reagent and cheaper electrode platform to provide evidence in support of a system is logical.
Various MIP platforms for myoglobin have been reported, one using ammonium persulfate catalyzed acrylamide MIPs91 but the majority using the inherent synergy between SPEs and electropolymerisation.46 These platforms are all very similar with the MIP formed directly onto the SPE surface through electropolymerisation, with no added nanomaterials to improve performance. They explore utilising poly(o-phenylenediamine) as a conductive polymer,92 as well as polyphenol93 and poly(o-aminophenol)94 as non-conductive options. Careful consideration must be made when designing these platforms as conductive MIPs can synergise well with the detection methodology, but non-conductive MIPs can provide useful self-limiting growth to aid reproducibility.
An interesting alternative approach to myoglobin sensing has been reported by Kumar et al.,95 where black phosphorous (BP) nanosheets were functionalised with aptamers, Fig. 5B. BP nanosheets were produced through sonication in a Triton X-100 surfactant solution and collected through centrifugation. The BP nanosheets were functionalised first with poly-L-lysine (PLL) before being drop-cast onto the SPE surface, before the aptamer recognition element was drop-cast on top. The modification with PLL altered the zeta potential of the surface from −23.4 mV to +13.5 mV, which significantly aided the strong electrostatic interactions between the positive NH3+ groups on the PLL and the negatively charged backbone of the DNA on the aptamer. This system managed to produce a response over a wide range between 1 pg mL−1–16 μg mL−1, produced a detection limit of 0.52 pg mL−1 and was successfully applied to measurements in serum samples. An alternative nanostructure explored toward improving the performance of electrochemical cardiac biosensors is graphene quantum dots (GQDs). They are zero dimensional nanostructures with large surface areas, low synthesis cost, excellent aqueous solubility and the co-presence of carboxyl and hydroxyl groups at their edges allowing for conjugation to biomolecules.74 Tuteja and co-workers96 produced their GQDs using waste graphite electrodes from spent alkaline batteries, coating them onto a C-SPE surface through bulk electrolysis at a constant potential of +0.7 V (vs. Ag|AgCl) for 10 min. These were then activated using EDC/NHS coupling and incubated with antibodies specific for myoglobin, followed by standard blocking with BSA (0.5 mg mL−1). This platform achieved a linear response from 0.01–100 ng mL−1 using EIS with a detection limit of 0.01 ng mL−1 and produced recoveries between 95–105% in human serum samples.
We note that the vast majority of the literature on cardiac biomarker detection using SPEs has been focussed on the three proteins mentioned above, leaving plenty of scope for researchers to introduce novel biosensors for other biomarkers, or to combine multiple markers into an array detection system. There has been reports for the detection of NT-proBNP,97 as well as BN-32.98 This latter example from Grabowska et al.98 focussed on developing a generic approach for multi-analyte sensing using aptamers as a recognition element. They modified Au-SPE surfaces with rGO/polyethyleneimine films. Following this they covalently grafted propargylacetic acid onto which azide terminated aptamers were grafted with click chemistry. With this amount of modification, it is not clear why Au-SPEs were used instead of cheaper C-SPE counterparts. Even so, this sensor achieved a linear response in serum from 1 pg ml−1–1 μg mL−1 for cTnI and from 1 pg mL−1–10 ng mL−1 for BNP-32. This is a good example of where the field can progress to, highlighting the detection of multiple markers to offer improved validation and confidence in results. This will encourage the commercialisation of these sensing platforms in the future. We now turn our attention to the progress in the development of SPE based electrochemical platforms for the detection of cancer biomarkers (Fig. 6).
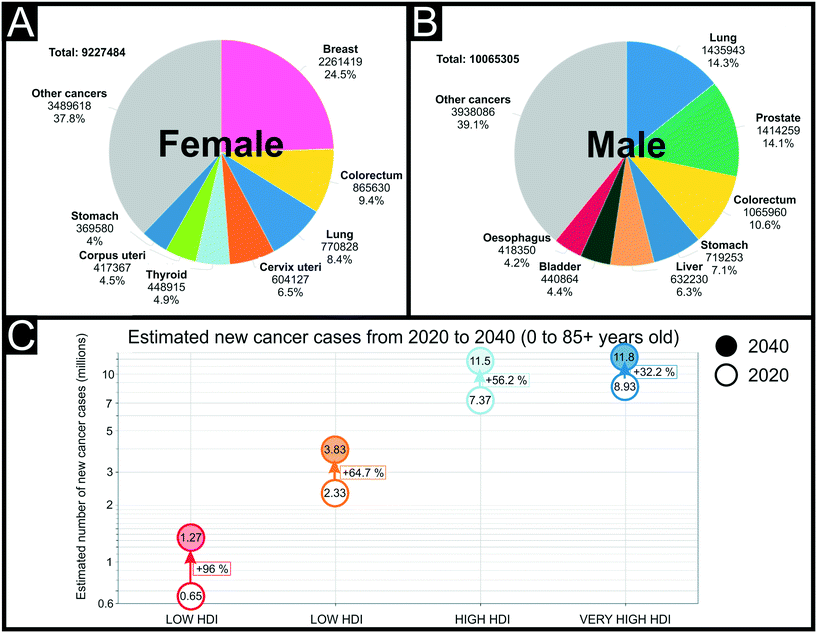 | ||
| Fig. 6 Estimated number of new cancer cases in 2020, worldwide, for females (A) and males (B) worldwide at all ages. Inequity in health care between higher and lower human development index (HDI) countries (C). Reproduced from ref. 184 with permission from UICC, copyright 2020. | ||
Cancer biomarkers
Biomarkers play an important role in the screening, diagnosis, monitoring and treatment of a wide range of different cancer types, and the number of emerging biomarkers being validated and entering routine clinical use is continuing to grow. From a screening and diagnostics perspective, analysis of cancer biomarkers can prevent the need for more invasive, risky and costly practices.99–101 From a monitoring perspective, they allow for easy and frequent repeated testing and have been shown to be extremely useful for detection of minimal residual disease and risk of relapse.102,103 The advent of ‘personalised medicine’ in which a patient's treatment is tailored towards their specific genetic or molecular profile has certainly been a key driver of the push for biomarker validation. Indeed, there are numerous examples of cancer biomarkers that are used to quantify the residual risk faced by patients and identify those groups of patients likely to benefit from costly, novel treatment approaches.104–106 The growing importance of biomarker analysis in the cancer setting is being met with a desire for cheaper and more rapid testing technologies.Cancer biomarkers encompass genes, gene products, DNA, RNA, proteins, enzymes, hormones and/or specific cells,107 and as different cancers vary so significantly in terms of their aetiology and pathogenesis, so too do their associated specific biomarkers. SPE-based electrochemical approaches to the analysis of cancer biomarkers typically focus on the detection of proteins, a summary of these SPE approaches can be found in Table 3. Direct comparisons between different testing systems proves difficult as the appropriate clinical ranges for different markers varies widely, meaning that one system can work excellently for one marker but would not necessarily translate to the analysis of others. Human epidermal growth factor receptor-2 (HER2) has certainly received the most attention in terms of SPE-based approaches to biomarker detection. A member of the HER family of receptors, HER2 plays a central role in the pathogenesis of several human cancers, but most notably is an important predictor and prognostic marker for breast cancer.108 Indeed, HER2 is overexpressed in 15–30% of invasive breast cancers and is associated with shorter disease-free survival and overall survival.109 Perhaps more importantly though, HER2 expression is a sole predictive marker for treatment benefits from HER2-targeting agents such as trastuzumab, lapatinib, and pertuzumab, and as such, testing HER2 status is an essential step in the treatment of breast cancer.110
| Biomarker | Electrode & Modification | Recognition element | Electrochemical technique | Linear range | Limit of detection | Sample testing matrix | Ref. |
|---|---|---|---|---|---|---|---|
| Key: PSA: prostate specific antigen; CA 15-3: cancer antigen 15-3; C-SPE: carbon screen-printed electrode; Au-SPE: gold screen-printed electrode; 2AP: 2-aminophenol; DPV: differential pulse voltammetry; U mL−1: units per millilitre; Ph: phenol; HER2: human epidermal growth factor receptor 2; AGR2: anterior gradient-2; GA: glutaraldehyde; Abs: antibodies; CA 19-9: cancer antigen 19-9; EIS: electrochemical impedance spectroscopy; Ag-SPE: silver screen-printed electrode; CNOs: carbon nano-onions; PEI: polyethyleneimine; GO: graphene oxide; SWV: square-wave voltammetry; CEA: carcinogenic embryonic antigen; PPy: polypyrrole; MIP: molecularly imprinted polymer; CV: cyclic voltammetry; NPs: nanoparticles; SAM: self-assembled monolayer; Gr-SPE: graphene screen-printed electrode; PANI: polyaniline; MALAT1: metastasis-associated lung adenocarcinoma transcript 1; AuNCs: gold nanocages; MWCNT: multi-walled carbon nanotubes; RNA: ribonucleic acid; PIK3CA: phosphatidylinositol-4,5-biphosphate 3-kinase catalytic subunit alpha; CRISPR: clustered regularly interspaced short palindromic repeats; MBs: magnetic beads; PLL: poly-L-lysine; LSV: linear sweep voltammetry; Nbs: nanobodies. | |||||||
| HER2 | Au-SPE | Ph–MIP | DPV | 10–70 ng L−1 | 1.6 ng L−1 | Human serum | 114 |
| HER2 | Au-SPE/SAM | Aptamer | EIS | 1 pg mL−1–100 ng mL−1 | 148.1 pg mL−1 | Human serum | 115 |
| HER2 | C-SPE | Affibody | DPV | 0–20 ng mL−1 | 1.8 ng mL−1 | Diluted serum | 126 |
| HER2 | C-SPE/AuNPs/CeO2 | Abs | CV | 1–500 pg mL−1, 5–20 ng mL−1 | 34.9 pg mL−1 | Human serum | 122 |
| HER2 | C-SPE/MBs | Abs | CV | 0–15 ng ml−1 | 6 ng mL−1 | Human serum | 128 |
| HER2 | C-SPE | Abs | SWV | 1–100 ng mL−1 | 0.28 ng mL−1 | Human serum | 130 |
| HER2 | C-SPE/AuNPs | Aptamer | DPV | 0.0001–100 ng mL−1 | 0.001 ng mL−1 | N/A | 116 |
| HER2 | C-SPE/PLL | Aptamer | DPV | 10–60 ng mL−1 | 3 ng mL−1 | Human serum | 113 |
| HER2 | C-SPE | Abs | CV | 5–20, 20–200 ng mL−1 | 4 ng mL−1 | Human serum | 125 |
| HER2 | C-SPE/MBs | Abs | LSV | 5–50, 50–100 ng mL−1 | 2.8 ng mL−1 | Human serum | 129 |
| HER2 | C-SPE/AuNPs | Abs | LSV | 15–100 ng mL−1 | 4.4 ng mL−1 | Human serum | 185 |
| HER2 | C-SPE/AuNPs | Abs | LSV | 0–50 ng mL−1 | 2.9 ng mL−1 | N/A | 118 |
| CA 15-3 | 0–70 U mL−1 | 5.0 U mL−1 | |||||
| HER2 | C-SPE/AuNPs | Abs | EIS | 0.01–100 ng mL−1 | 0.01 ng mL−1 | Human serum | 120 |
| HER2 | C-SPE | Abs | DPV | 10–150 ng mL−1 | 2.1 ng mL−1 | Human serum | 131 |
| HER2 | C-SPE/MBs | Abs | DPV | 0.5–50 ng mL−1 | 0.29 ng mL−1 | Human serum | 186 |
| HER2 | C-SPE | Nbs | Amperometric | 1–200 μg mL−1 | 4.4 μg mL−1 | Cell lysates | 127 |
| HER2 | C-SPE/AuNPs | Affibody | EIS | 0–40 ng mL−1 | 6.0 ng mL−1 | Human serum | 119 |
| HER2 | C-SPE /MWCNT/AuNPs | Abs | LSV | 7.5–50 ng mL−1 | 0.16 ng mL−1 | Human serum | 124 |
| CA 15-3 | Au-SPE | 2AP–MIP | DPV | 5–50 U mL−1 | 1.5 U mL−1 | Human serum | 114 |
| PSA | C-SPE | Abs | Amperometric | N/A | 0.25 ng mL−1 | N/A | 137 |
| PSA | C-SPE/AuNPs | Aptamer | DPV | 1 pg mL−1–200 ng mL−1 | 0.077 pg mL−1 | Human serum | 138 |
| PSA | C-SPE/AuNP | Abs | Amperometry | 0.225–5 pg mL−1 | 0.1 pg mL−1 | Human serum | 139 |
| CEA | AgxO-SPE | PPy–MIP | CV | 1.25–125 pg mL−1 | 1.25 pg mL−1 | Urine | 135 |
| CA 19-9 | Ag-SPE/CNOs/PEI/GO | Abs | EIS | 0.3–100 U mL−1 | 0.12 U mL−1 | Cell lysates | 40 |
| CA 125 | Gr-SPE/PANI | Abs | EIS | 0.92 pg μL−1–15.2 ng mL−1 | 0.92 pg mL−1 | N/A | 147 |
| MALAT1 | C-SPE/MWCNT/AuNCs | RNA | DPV | 10−7–10−14 M | 42.8 fM | Human serum | 141 |
| PIK3CA exon mutation 9 | GO-SPE/CRISPR | RNA | EIS | 0–20 nM | 0.65 nM | Human blood | 187 |
| AGR2 | Au-SPE/GA | Abs | EIS | 0.01–10 fg mL−1 | 0.093 fg mL−1 | N/A | 136 |
SPE-based testing approaches have, to a lesser degree, focused on the detection of another important breast cancer biomarker, cancer antigen 15-3 (CA 15-3). Although this antigen is not specific to breast cancer and can be detected in other cancer types, testing for this marker in the serum of breast cancer patients has been adopted by the NHS to aid in determining response to therapy in patients with metastatic breast cancer, and for the early detection of recurrence in patients with previously treated stage II and stage III breast cancer.111,112
As seen in the above section on cardiac markers there are various strategies used alongside SPEs to achieve relevant detection of the biomarker HER2, such as using aptamers113 and MIPs. In terms of MIPs, Pacheco and co-workers described a simple system electro-synthesising polyphenol directly onto the surface of an Au-SPE in the presence of the HER2 template.114 The template was removed using a mix of water, SDS (0.5%) and acetic acid (0.5%), leaving cavities through which the [Fe(CN)6]3−/4− redox couple could be monitored by DPV. Upon rebinding of the HER2, these pores were blocked giving detection between 10–70 ng mL−1 and a detection limit of 1.6 ng mL−1. Alternatively, Ferreira et al.115 recently reported and aptasensor, where they utilised a ternary SAM architecture made from a mix of mixed DNA, 1-hexanethiol and hexandithiol (HDT). The HDT provides a horizontal configuration, bridging gaps between surface irregularities and helping to reduce the adsorption of non-specific proteins. As such, using EIS the sensor achieved a wide linear range of 1 pg mL−1–100 ng mL−1, a LOD of 172 pg mL−1 and appropriate detection in undiluted human serum. Instead of utilising an full Au-SPE, it is common practice to utilise Au nanoparticles (NPs) on a standard C-SPE,116 providing the beneficial properties of nanoparticles and the potential for bioconjugation using simple thiols.117 Marques and co-workers utilised the AuNPs on a C-SPE surface in conjunction with a sandwich style assay. They deposited the AuNPs on the carbon surface through electrodeposition, applying a constant current of −100 μA for 240 s, followed by the application of +0.1 V for 120 s to produce nanoparticles of 32 ± 10 nm in size. The antibodies were deposited through adsorption followed by surface blocking with β-casein to finish the biorecognition system. After incubation with a sample, the system was incubated with a biotinylated antibodies and their detection solution for LSV. They reported similar work for the dual detection of HER2 and CA 15-3, where the sandwich assay is incubated with a solution of Streptomyces avidinii conjugated to alkaline phosphatase, followed by a solution of 3-indoxyl phosphate disodium salt and AgNO3.118 The same system was used for both markers, simply changing the antibodies used, producing a linear range of 0–50 ng mL−1 and 0–70 U mL−1, and LODs of 2.9 ng mL−1 and 5.0 U mL−1 for HER2 and CA 15-3 respectively. Although showing good results in buffered solutions, this system was not tested in real samples which raises questions about the application. Similarly, Ravalli et al.119 utilised electrodeposition of AuNPs onto C-SPEs, but through cyclic voltammetry in the range −0.2–+1.6 V at 0.1 V s−1. They then immobilised a C-terminal cysteine affibody to the gold surface, followed by blocking the remaining gold surface with a blocking thiol solution and the bare carbon surface with non-fat milk powder (1% w/v). Affibodies are engineered versions of one of the five stable three-α-helix bundle domains from the immunoglobulin Fc-binding region. They can be grown in prokaryotes rather than the animal system required for antibody synthesis and can include specific labels such as cysteine groups to help couple the affibody to surfaces. Using this system in conjunction with EIS, the authors report a linear detection range for HER2 between 0–40 ng mL−1, a LOD of 6 ng mL−1 and successfully applied the system in human serum. An alternative method for producing AuNPs was utilised by Sharma and co-workers for the detection of HER2;120 whereby colloidal AuNPs were formed through citrate reduction of chloroauric acid utilising a modified Turkevich method.121 The SPE surface was then modified with cysteamine hydrochloride to produce a surface functionalised with thiol groups, allowing the citrate-coated AuNPs to form highly stable Au–S bonds when incubated. This surface was then further modified with amine groups for antibody attachment, allowing for the sensitive detection of HER2 using EIS. This produced a sensor capable of detecting HER2 in the range of 0.01–100 ng mL−1, with a LOD of 0.01 ng mL−1 and good performance in foetal calf serum. They report the detection in this medium in only 15 min, using a 3 μL droplet of sample, indicating the possibility for this technology to be tested further toward real applications. Another alternative methodology is presented by Hartati et al.122 who proposed a sensor utilising a polyethylene glycol (PEG) nano-structured cerium oxide (CeO2) modified SPE. Cerium oxide has good electrical conductivity, chemical inertness, a large surface area and good biocompatibility.123 They CeO2 was functionalised with PEG, followed by attachment of antibodies specific for HER2. The bioconjugate was coupled to the electrode surface through EDC coupling and the uncovered surface blocked with BSA. The biosensor managed to achieve detection of HER2 using CV and [Fe(CN)6]3−/4− between 1–500 pg mL−1, with a LOD of 34.9 pg mL−1 and achieving good recovery between 101–103% when tested in human serum. Alternatively, Freitas et al.124 used AuNPs in conjunction with MWCNTs to produce a sandwich assay platform for HER2. They studied the effect of utilising the nanomaterials individually and then in unison, finding that an improved performance was found. We note it is important to validate the reasoning behind using multiple nanomaterials rather than just using them for the sake of the next hot topic.
As seen above, sandwich assay style platforms have been used regularly;125 for example Ilkhani et al.126 utilised affibodies and Patris and co-workers reported the use of Nanobodies®, which are the smallest known single entity with full antigen binding properties.127 They used EDC/NHS coupling to functionalise the SPE surface with the capture nanobody and then achieved detection using a nanobody tagged with hydrogen peroxidase. Another popular biological tag for the detection antibodies has been streptavidin-conjugated alkaline phosphatase.128 Freitas and co-workers also used this as their detection antibody, with their capture antibody immobilized onto carboxyl-modified magnetic beads (MBs), Fig. 7A.129 The MBs allow for a high loading of Abs, using EDC/NHS, which can bind to the target analyte in solution. After set periods of incubation, a magnet can be placed under the system to attract the MBs to the surface, bringing the detection Abs with them. Using this method they achieved a LOD of 2.8 ng mL−1 and was applied successfully to HER+ breast cancer cell lines. The reliance on two biological components, an antibody and enzyme, can bring with it problems in terms of storage, stability and lifetime of the sensor platform. One method to overcome these issues that has been seen regularly with regards to electrochemical biosensors is to incorporate antibody modified quantum dots (QDs) as the detection system. Ahmad and co-workers utilised PbS quantum dots, using 1,1′-carbonyldiimidazole to bioconjugate them to the appropriate antibodies.130 SWV was used to detect the presence of the PbS QDs when attached to the surface bound target giving a linear range of 1–100 ng mL−1 and a LOD of 0.28 ng mL−1. This work showed good recoveries in human serum but required dilution of 1000 times to remove any matrix effects. Alternatively, Freitas and co-workers have utilised CdSe@ZnS quantum dots for the detection of HER2. They report two methodologies, one similar to the above, with capture antibodies immobilized on the surface of the working electrode through covalent linkages.131 The other uses MBs tagged with capture antibodies on the surface. They noticed an increase in electrochemical signal using micro-sized MBs that formed an organised layer on top of the electrode surface. In both systems differential pulse anodic stripping voltammetry was used to detect the Cd2+ ions released from the QDs. The former produced a linear range of 1–150 ng mL−1 with a LOD of 2.1 ng mL−1; compared to the MBs platform that achieved a linear range of 0.5–50 ng mL−1 and a LOD of 0.29 ng mL−1. This provides evidence toward the MBs helping with the sensor platform sensitivity, but this comes at a cost elsewhere.
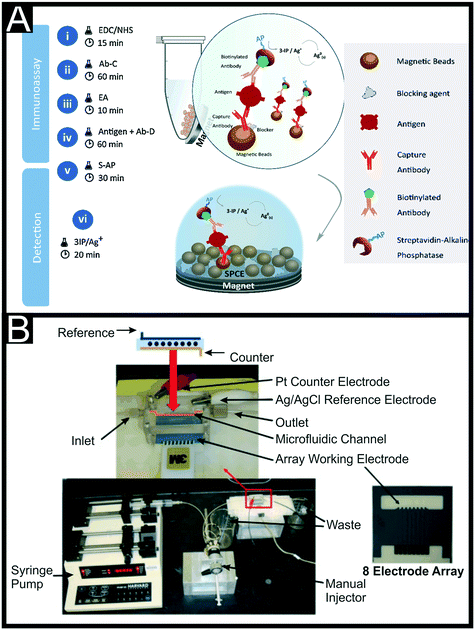 | ||
| Fig. 7 (A) Schematic representation of the sandwich-immunoassay produced using carboxyl-modified magnetic beads as the platform for the capture antibody and a streptavidin–alkaline phosphatase modified detection antibody. Reproduced from ref. 129 with permission from Elsevier, copyright 2020. (B) Microfluidic array device showing 8-electrode C-SPE with microfluidic flow channel, manual injector and syringe pump. Reproduced from ref. 139 with permission from Elsevier, copyright 2011. | ||
Trau et al. reported the use of a hydrophilic polyacrylic acid brush-modified (pAA) indium tin oxide substrate as an antifouling substrate to minimise the non-specific serum protein adsorption when working in human serum.132 They reported that when applied to sandwich immunoassays, their method would increase the sensitivity of the biosensors when compared to CNTs and graphene-based amplification strategies. They applied it to the detection of epithelial cell adhesion molecule (EpCAM) antigen achieving femtomolar-level detection, although could be potentially applied to a large number of immunosensors that are based on similar sandwich assays.
In terms of alternative markers for other cancers very similar strategies of biosensor development to the ones highlighted for HER2 have been used. For example, there have been reports for the determination of miRNA-155 (a specific microRNA (non-coding RNA) shown to have diagnostic value in several cancer types including prostate, lung, squamous cell carcinoma and, most notably breast),133 arginase-2 (AGR2) (an enzyme, which when overexpressed in many cancer types is responsible for the destruction of L-arginine and hence creates a localised immunosuppressive state)134 using an Au-SPE modified with their biorecognition elements through Au–S bonds135,136 and for prostate specific antigen (PSA) (one of the most well established cancer biomarkers for the diagnosis, treatment and follow-up of prostate cancer cases) using a simple antibody sandwich assay on C-SPEs,137 aptamers deposited on AuNPs138 and detection systems combining these two methods.139,140 This latter example is reported for PSA initially, then followed up for the detection of both PSA and interleukin-6 (IL-6), a commonly used biomarker for many conditions that will be discussed in more depth in the latter section. Chikkaveeraiah and co-workers used an 8 C-SPE microelectrode array, with a microfluidic channel directing the flow of solution from a pump and injector across the electrode surfaces, Fig. 7B. They produced glutathione modified AuNPs, immobilizing these onto the C-SPE surface through utilising a layer of poly(diallydimethylammonium chloride) (PDDA) to create a favourable environment for electrostatic interactions. Capture antibodies were immobilised onto these AuNPs through EDC/NHS coupling, with the rest of the electrode surface being blocked by a solution of BSA (2%). To increase the signal generation from the detection part of the sandwich assay, the immobilised large amounts of secondary antibodies and horseradish peroxidase (HRP) labels onto 1 μm tosylated superparamagnetic MBs, estimating approximately 9 × 104 molecules of Abs and 2.4 × 105 HRP labels per MB. They achieved a linear range of 0.225–5 pg mL−1 and a LOD of 0.1 pg mL−1 for PSA and validated their results in human serum achieving a LOD of 0.23 pg mL−1 in this medium. An interesting comparison between AuNPs and Au nanocages (AuNCs) has been presented in recent work by Chen et al.141 for the detection of lung cancer marker MALAT1. They use a combination of AuNCs and MWCNTs to provide enhanced electrochemical signals compared to the materials on their own and in comparison to the use of AuNPs. The AuNCs (40–50 nm in size) are also used to bind the S′-thiolated RNA biorecognition element through the strong Au–S linkages. This system utilised DPV for its detection and achieved a linear range between 10−7–10−14 M, with a very low LOD of 42.8 fM. They also achieved excellent results when measuring in 6 human serum samples, achieving recoveries between 94–102%. Koo et al.142 reported, following on from previous work,143 the use of bare commercial Au-SPEs for an amplification-free miRNA assay based on elevated affinity interaction, were a poly(A) extension on the 3′ ends of magnetically isolated miRNA targets enhanced and facilitated the high adsorption efficiency onto the Au-SPE. They reported its use towards the electrochemical quantification of miR-107 in human cell lines and clinical urine at 10 fM. Shiddiky et al. reported the use of gold-loaded nanoporous iron oxide nanocubes (Au@NPFe2O3NC) towards the detection of p53 (ref. 144 and 145) antigens and miR-107.146 Their p53 detection method applies a two-step strategy where i) a magnetic capture and isolation of autoantibodies uses p53/Au@NPFe2O3NC as dispersible nanocapture agents in serum samples and ii) a subsequent detection of those autoantibodies via a peroxidase-catalysed reaction on commercial Au-SPEs or a naked-eyed colorimetric detection in Eppendorf tubes. Their method demonstrated a good sensitivity and reproducibility for p53 autoantibodies in serum, reporting a LOD of 0.02 U mL−1. They successfully applied it to a small cohort of clinical colorectal samples too.
A final examples have utilised graphene based SPEs (G-SPEs) for their electrode surface.147 Gazze et al.147 modified a G-SPE with PANI through electropolymerisation between −0.1 V and +1.2 V. Antibodies specific for their target of cancer antigen 125 (CA-125) (a biomarker routinely used as part of the diagnostic pathway in ovarian cancer)148 were then attached to the polymer through EDC/NHS coupling and the remaining free surface blocked with BSA (0.5 mg mL−1). Using EIS, they achieved detection in a linear range between 0.00096–15.3 ng μL−1 and a LOD of 0.923 pg μL−1. This work does not report testing in real sample matrices, which we would advise for anyone looking to disseminate work in this field.
As reported above, screen-printed electrochemical sensing platforms have the potential to offer affordable, quick detection of cancer biomarker levels. This suggests that with further work and validation, these sensors could be useful in the monitoring of cancer biomarker levels, improving treatment. Due to the rapid nature of electroanalytical platforms, they offer the possibility of providing clinicians with real-time data on a patient's condition, which could allow faster treatment. This is vital in the area of sepsis, where a delay in treatment of an hour can lead to an increase in mortality of up to 7%.149 Screen-printed electrochemical sensors to help in the diagnosis of this condition is where our focus turns next.
Biomarkers of inflammation
Inflammation is the immune system's response to harmful stimuli such as pathogens, damaged cells or toxic compounds. It acts to remove or neutralise these stimuli and initiate the healing process. Various pathogenic factors, such as infection or tissue injury can induce inflammation by causing tissue damage. Assessment of the degree of inflammation is an extremely important part of the diagnostic workup of numerous diseases including autoimmune diseases, infections, inflammatory disorders such as inflammatory bowel disease, and most notably, sepsis. Indeed, rapid assessment of a patient's inflammatory profile is vital in Intensive Care Units, to help aid identification of patient deterioration and requirement for organ support. Sepsis is defined as a life-threatening organ dysfunction caused by a dysregulated host response to infection.150,151 Sepsis has been recognised as a global health priority by the United Nations World Health Assembly, since more than 6 million deaths and 19 million sepsis related cases occur annually.152 It is the over-exaggerated (hyperinflammatory) immune response that causes the widespread tissue and organ damage in the patient, rather than the infectious pathogen itself. Rapid assessment of the inflammatory status of a patient with sepsis is important for accurate diagnosis and timely intervention. Indeed, it has been reported that there is a 7.6% decrease in patient survival rate for every hour that sepsis is not diagnosed and treatment not initiated.149C-Reactive protein (CRP) and procalcitonin (PCT) are well-established markers of inflammation that are frequently analysed in hospital laboratories using sophisticated biochemistry analysers. As acute phase proteins produced primarily by the liver, CRP and PCT are elevated in response to pro-inflammatory cytokines (particularly Interleukin-6, (IL-6), IL-1β and Tumour Necrosis Factor-α (TNF-α)) that are themselves produced early in the inflammatory response to harmful stimuli.153 The turn-around time for CRP and PCT tests (approximately 4 to 6 hours), and the lack of testing facilities in low-resource settings has led to a flurry of research into rapid testing methodologies for these biomarkers. The plethora of data evidencing the clinical utility of CRP and PCT testing in a variety of acute inflammatory settings154–158 including sepsis, provides justification for their suitability as subjects of POC technology. Lactate analysis in the setting of inflammation is also extremely useful. Although primarily a marker of metabolic derangement, lactate clearance is associated with biomarkers of inflammation, organ dysfunction and mortality in severe sepsis and septic shock,159 and forms a key part of sepsis management and risk stratification in current international guidelines from both NICE and the surviving sepsis campaign.160 Hence, the ability for rapid and repeated analysis of lactate is warranted and has led to development of technologies for POC lactate sensing. For advancements in electrochemical lactate sensing and their transition to critical care we direct the reader towards a recent review.161
Recent developments in advanced electrochemical, immunological or magnetic sensors provide rapid highly sensitive and selective detection of PCT and CRP which might offer an alternative solution to current testing methods. There have been several reports for the detection of CRP alone primarily following similar trends to that seen in previous sections, namely the use of MIPs162 or nanomaterials and nanoparticles with a view to improve electrode performance and EDC/NHS or glutaraldehyde coupling to immobilise recognition elements.163,164 These include modifying a C-SPE surface with rGO and 1-pyrenebutryic acid,165 electropolymerisation of PANI onto a G-SPE166 and coating with chitosan.167 There are many examples that utilise the deposition of AuNPs onto C-SPE surfaces, followed by L-cysteine and then EDC coupling of an antibody. Boonkaew and co-workers used this set-up alongside EIS to produce an origami paper-based immunoassay producing a LOD of 15 ng mL−1.168 This system was tested using diluted serum samples, which is common place for CRP as the cut-off limit is expected at 2 mg mL−1. Jampasa et al.165 utilised this methodology for the immobilisation of their capture antibodies onto a G-SPE in their sandwich assay, Fig. 8A, depositing the AuNPs through electrodeposition of potassium tetrachloroaurate(III) at −0.5 V for 100 s. They modified their secondary detection antibody with an anthraquinone label through EDC/NHS coupling of the carboxyl group on 4-(anthraquinone-2-oxy)butyric acid to the amine group present on the anti-CRP. Using optimised DPV parameters they achieved a linear range between 0.01–150 μg mL−1 and a detection limit of 1.5 ng mL−1 in buffered solutions with a good % RSD of lower than 5%. This sensing platform was validated in diluted human serum samples achieving a paired t-test result at a 95% confidence level, with a reported error no larger than 0.5% against standard methods. Other sandwich assays reported for the detection of CRP include the use of a bismuth-citrate modified SPE alongside a biotinylated detection antibody with streptavidin modified PbQDs169 and the use of MBs.170 In this latter example, Molinero-Fernández et al.170 reported a dual-magneto-immunosensor for the simultaneous detection of CRP and PCT, utilising amperometric measurements performed at −0.2 V (versus an Ag wire pseudo-reference). This followed on from previous work looking at just PCT.171 They used a commercial streptavidin-functionalised MB solution, further modifying them with biotinylated antibodies specific for either CRP or PCT. For the detection antibody they utilised HRP modified antibodies, which produced the required amperometric signal when incubated in the hydrogen peroxide solution. Using this method, they achieved linear ranges of 0.01–5 μg mL−1 and 0.25–100 ng mL−1, LODs of 8 ng mL−1 and 0.09 ng mL−1 and 8% inter-assay precision for CRP and PCT detection respectively. It can be seen from inspection of Table 4 that there are far fewer examples of PCT detection or dual-detection platforms and we suggest this is an area of considerable interest for future research. In regards to other markers, IL-6 has been one of the most reported due to its link to a wide variety of conditions including sepsis, cardiac disease172 and COVID-19.173 Indeed, IL-6 appears as a central driver of COVID-19-associated inflammation and this has led to the use of the IL-6 receptor inhibitor, Tocilizumab, for the treatment of critically-ill COVID-19 patients.174,175 Its appearance in the blood at an earlier stage of inflammation than CRP and PCT also makes it an attractive analyte, and we expect significantly more reports of low-cost screen-printed sensors to be published in this area in the coming years. Recently an example has been reported utilising a thermal read-out platform,176 but we will focus on electrochemical methodologies. One of the first reports of an IL-6 screen-printed electrochemical platform was that of modifying the C-SPE surface with HRP tagged antibodies through the use of glutaraldehyde, which has been seen multiple times throughout the literature.177 Tertiş and co-workers described a platform depositing PPy onto an C-SPE surface, followed by AuNPs for aptamer binding through the formation of Au–S bonds.178 They produce a sensor for IL-6 with a wide linear range (1–15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pg mL−1), however they do not fully discuss why the PPy is integral to the performance of the sensor platform. Data presented shows the charge transfer resistance is significantly reduced with only the AuNPs present on the C-SPE surface, with these also providing the binding point for bio-conjugation. In any case, they report a low LOD of 0.33 pg mL−1 and show the successful determination of IL-6 in a diluted serum sample. There have also been reports of IL-6 detection in conjunction with other biomarkers of inflammation. Shi et al.179 utilised polymer@metal core–shell nanocomposites for the detection of IL-6 and matrix metallopeptidase-9 (a marker for ischemic strokes). They immobilised capture antibodies onto a C-SPE surface previously modified with graphene nanorods (used to improve the sensitivity of the sensor). Polystyrene (PS) and polydopamine (PDA) were used to create nanocomposites with specific antibodies and different metals for each of the targets PS@PDA–Ag for the metallopeptidase and PS@PDA–Cd for IL-6. This allowed them to monitor SWV peaks at −0.7 V for the oxidation of Cd when IL-6 was present and +0.1 V for Ag. Through this they report a linear range for IL-6 of 10−3 – 103 ng mL−1, a LOD of 0.1 pg mL−1 and successful validation of the sensor in human serum against a commercial ELISA. An interesting application that reports both the detection of IL-6 and another pro-inflammatory cytokine, tumour-necrosis factor-α (TNF-α), is the muscle-on-a-chip with in situ monitoring reported by Ortega and co-workers, Fig. 8B.180 TNF-α has additionally been shown to have links to sepsis,181 COVID-19 (ref. 182) as well as heart failure,183 and as such we expect significantly more reports to be published in the coming years. The immunoassay reported by Ortega uses amperometric measurements of the HRP at an applied potential of −0.2 V vs. Ag wire. They functionalised their Au-SPEs through the formation of a SAM, followed by EDC/NHS coupling of the capture antibody and free surface blocking through BSA (1%). The functionalised Au-SPEs were placed in the system connected with a peristaltic pump, which flowed the tissue solution over the working electrode surface. After binding occurs, the electrodes were removed, placed in a static cell and the secondary antibodies were added. Performing this method, they achieved LODs of 8 and 2 ng mL−1 for IL-6 and TNF-α, respectively and compared these to commercial ELISAs. They continued to show that the detection of these biomarkers was possible in 3D muscle microtissues mimicking exercise and allowed for real-time monitoring of the markers.
000 pg mL−1), however they do not fully discuss why the PPy is integral to the performance of the sensor platform. Data presented shows the charge transfer resistance is significantly reduced with only the AuNPs present on the C-SPE surface, with these also providing the binding point for bio-conjugation. In any case, they report a low LOD of 0.33 pg mL−1 and show the successful determination of IL-6 in a diluted serum sample. There have also been reports of IL-6 detection in conjunction with other biomarkers of inflammation. Shi et al.179 utilised polymer@metal core–shell nanocomposites for the detection of IL-6 and matrix metallopeptidase-9 (a marker for ischemic strokes). They immobilised capture antibodies onto a C-SPE surface previously modified with graphene nanorods (used to improve the sensitivity of the sensor). Polystyrene (PS) and polydopamine (PDA) were used to create nanocomposites with specific antibodies and different metals for each of the targets PS@PDA–Ag for the metallopeptidase and PS@PDA–Cd for IL-6. This allowed them to monitor SWV peaks at −0.7 V for the oxidation of Cd when IL-6 was present and +0.1 V for Ag. Through this they report a linear range for IL-6 of 10−3 – 103 ng mL−1, a LOD of 0.1 pg mL−1 and successful validation of the sensor in human serum against a commercial ELISA. An interesting application that reports both the detection of IL-6 and another pro-inflammatory cytokine, tumour-necrosis factor-α (TNF-α), is the muscle-on-a-chip with in situ monitoring reported by Ortega and co-workers, Fig. 8B.180 TNF-α has additionally been shown to have links to sepsis,181 COVID-19 (ref. 182) as well as heart failure,183 and as such we expect significantly more reports to be published in the coming years. The immunoassay reported by Ortega uses amperometric measurements of the HRP at an applied potential of −0.2 V vs. Ag wire. They functionalised their Au-SPEs through the formation of a SAM, followed by EDC/NHS coupling of the capture antibody and free surface blocking through BSA (1%). The functionalised Au-SPEs were placed in the system connected with a peristaltic pump, which flowed the tissue solution over the working electrode surface. After binding occurs, the electrodes were removed, placed in a static cell and the secondary antibodies were added. Performing this method, they achieved LODs of 8 and 2 ng mL−1 for IL-6 and TNF-α, respectively and compared these to commercial ELISAs. They continued to show that the detection of these biomarkers was possible in 3D muscle microtissues mimicking exercise and allowed for real-time monitoring of the markers.
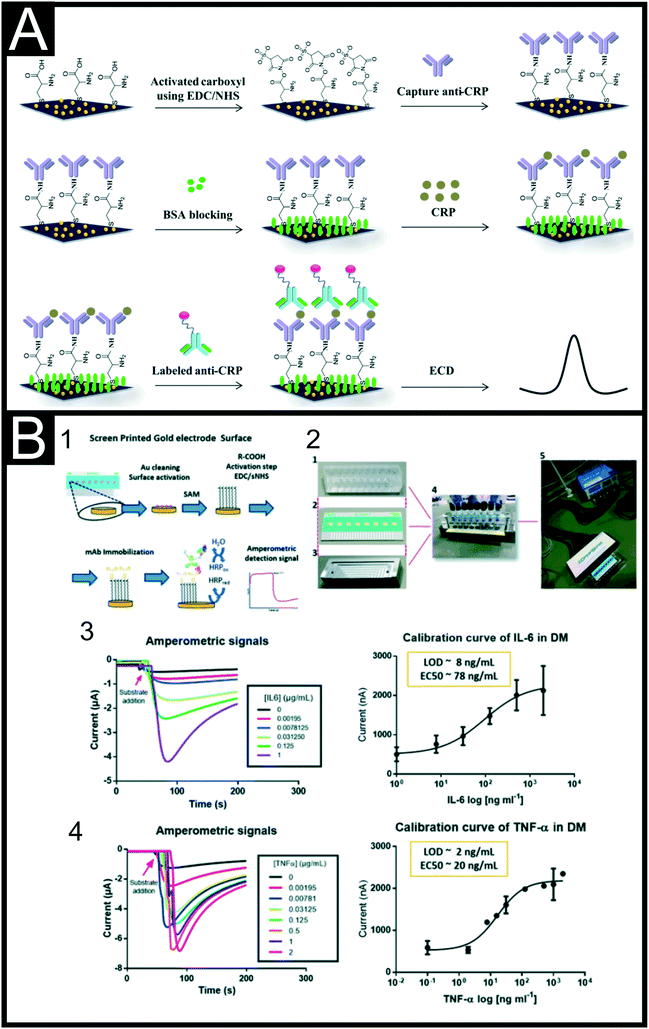 | ||
| Fig. 8 (A) Schematic illustration of the preparation of a CRP biosensor using AuNPs, L-cysteine, EDC/NHS coupling and a sandwich assay. Reproduced from ref. 165 with permission from Elsevier, copyright 2018. (B) On-site multiplex sensing system for IL-6 and TNF-α, highlighting (1) the fabrication of the sandwich assay; (2) photographs of the physical set-up; (3) and (4) plots of the amperometric signals obtained for IL-6 and TNF-α respectively in conjunction with their calibration curves. Reproduced from ref. 180 with permission from Royal Society of Chemistry, copyright 2019. | ||
| Biomarker | Electrode & Modification | Recognition element | Electrochemical technique | Linear range | Limit of detection | Sample testing matrix | Ref. |
|---|---|---|---|---|---|---|---|
| Key: CRP: c reactive protein; PCT: procalcitonin; C-SPE: carbon screen-printed electrode; G-SPE: graphene screen-printed electrode; MWCNT: multi-walled carbon nanotube; ADEP: 2-acrylamidoethyldihydrogen phosphate; DMAA: N-(4-dimethylaminophenyl)-acrylamide; EIS: electrochemical impedance spectroscopy; PEDOT: poly(3,4-ethlenedioxythiophene); 4AP: 4-aminophenol; PEBT: poly(eriochrome black T); ASV: adsorptive stripping voltammetry; G: graphene; MBs: magnetic beads; SWV: square-wave voltammetry; IL: interleukin; PPy: polypyrrole; GNR: graphene nanoribbons; HRP: horseradish peroxidase; Py-MMPs: pyrrole coated magnetite magnetic microparticles; PANI: polyaniline. | |||||||
| CRP | C-SPE/MWCNT | AEDP/DMAA–MIP | DPV | 0.18–8.51 μg mL−1 | 0.04 μg mL−1 | Human blood serum | 162 |
| CRP | G-SPE/AuNPs | Abs | DPV | 0.01–150 μg mL−1 | 1.5 ng mL−1 | Human serum | 165 |
| CRP | C-SPE/bi-citrate | Abs | ASV | 0.2–100 ng mL−1 | 0.05 ng mL−1 | Human serum | 169 |
| CRP | C-SPE/G/AuNP | Abs | EIS | 0.05–100 μg mL−1 | 15 ng mL−1 | Human serum | 168 |
| CRP | C-SPE/rGO | Abs | EIS | 0.01–10 μg mL−1 | N/A | N/A | 188 |
| CRP | C-SPE/MWCNT/ProteinA | HARP-Abs | Amperometry | N/A | 0.5 ng mL−1 | N/A | 189 |
| CRP | C-SPE/AuNPs | Abs | Amperometry | 0.047–23.6 μg mL−1 | 17 ng mL−1 | Human serum | 163 |
| CRP | G-SPE/PANI | Abs | EIS | 0.25–2 μg mL−1 | 0.5 μg mL−1 | Fetal bovine serum | 166 |
| CRP | C-SPE | Phosphocholine | EIS | 0.005–500 mg L−1 | 0.001 mg L−1 | Human blood | 167 |
| CRP | C-SPE/GQDs | Abs | Amperometry | 0.5–10 ng mL−1 | 36 pg mL−1 | Ringer lactate solution | 164 |
| CRP | G-SPE/GO | Abs | SWV | 0.01–100 μg mL−1 | 0.38 ng mL−1 | Serum | 79 |
| PCT | 0.5 pg mL−1–250 ng mL−1 | 0.27 pg mL−1 | |||||
| CRP | C-SPE/MBs | Abs | Amperometry | 0.01–5 μg mL−1 | 0.008 μg mL−1 | Human plasma | 170 |
| PCT | 0.25–100 ng mL−1 | 0.09 ng mL−1 | |||||
| PCT | C-SPE/MBs | Abs | Amperometry | 0.5–1000 ng mL−1 | 0.1 ng mL−1 | Human plasma and serum | 171 |
| IL-6 | C-SPE | PPy–MIP | EIS | 0.02–20 ng mL−1 | 0.02 pg mL−1 | Serum | 190 |
| IL-6 | C-SPE/GNR | Abs | SWV | 0.001–1000 ng mL−1 | 0.1 pg mL−1 | Human serum | 179 |
| IL-6 | C-SPE/nanoAu | HRP-Abs | DPV | 5–100 pg mL−1 | 1 pg mL−1 | Human serum | 177 |
| IL-6 | C-SPE/PPy/AuNPs | Aptamer | EIS | 1 pg mL−1–15 μg mL−1 | 0.33 pg mL−1 | Human serum | 178 |
| IL-6 | C-SPE/AuNP | Abs | Amperometry | 0.3–20 pg mL−1 | 0.25 pg mL−1 | Human serum | 139 |
| IL-6 | Au-SPE | Abs | Amperometry | N/A | 8 ng mL−1 | Differentiation medium | 180 |
| TNF-α | 2 ng mL−1 | ||||||
| TNF-α | Au-SPE/Py-MMPs | HRP-Abs | Amperometry | 1–15 pg mL−1 | 0.3 pg mL−1 | Artificial saliva | 183 |
| IL-1β | C-SPE/PEDOT/4AP | PEBT–MIP | EIS | 0.06–600 nM | 1.5 pM | Artificial serum | 191 |
This review has highlighted that there are a significant amount of work published towards the detection of vital biomarkers for cardiac disease, cancer and inflammation using screen-printed platforms. Many of these systems use similar methodologies for sensor creation. We note a large proportion of work still utilises antibodies and although reporting good performance, we would expect certain amounts of reliability due to antibody orientation. Many papers neglect to discuss the orientation of their recognition elements, which is a key component to producing a reliable and sensitive sensing platform.
Unpublished research:
In recent years the progress in electroanalysis and electrochemical devices has shown a trend in miniaturisation, low-cost and disposability of sensors. As a response, the use of screen-printed electrodes (SPEs) towards biosensing applications and point-of-care (POC) platforms has become more popular with time. SPE-based biosensors provide an alternative analytical tool to conventional lab-based techniques, allowing the reduction of analysis time and their difficulty of use. In addition, the integration of advanced novel materials and nanotechnology with electroanalysis, has transformed the POC applications like with the glucose sensor. The ability of bulk-manufacturing modified SPEs will help translating novel research advances from the lab into the healthcare industry and clinical applications. Cardiac injury, cancer diagnostics and acute inflammatory conditions are currently associated with significant healthcare costs, and although there are a large number of academic studies and manuscripts reporting their excellent performance towards their vital analyte of choice, there is a need of increasing the general efforts onto developing mass-producible and affordable bottom-up biosensors solutions for clinical environments. It is likely that screen-printed electrodes can help overcoming these challenges, however further research on wearable devices, in situ healthcare applications and the ability of simultaneous multi-analyte detection and their application to clinical settings is needed.
Conclusions and future outlook
In this perspective we have reported, discussed and reviewed some of the most recent advances in biosensing vital biomarkers for cardiac disease, cancer and inflammation. It can be observed that electrochemical biosensors are becoming a promising alternative to benchtop conventional techniques, offering advantages in terms of reduction of test turnarounds and costs in POC diagnosis. Although the use of electrochemical biosensors as POC solutions towards vital biomarker testing is still in its early stages, we believe that, electrochemical sensors have the potential to replace, or certainly complement traditional lab-based methods of analyte testing and thereby improve test turnaround times and ultimately patient care. However, as development of these SPE-based technologies for different biomarkers continues, thought should be given to the settings in which POC analysis is a fundamental factor; the identification of an AMI in patients with chest pain requires rapid test turnaround times, as does the identification of systemic inflammation in a deteriorating patient on the Intensive Care Unit. However, the assessment of CA 15-3, for example, during a patient's cancer treatment to evaluate the response to therapy could justifiably be performed remotely.Furthermore, there are still some technological and commercial challenges that need addressing before clinical application of these technologies can be widely applied:
a) Miniaturisation of biosensors: current literature focuses on two main goals around electrochemical POCs: miniaturization of the electrodes, connections, readouts etc., which is mainly supported by recent technological and computer powering advances, and the improvement of the biosensing itself, which is being addressed by the use of novel nanomaterials and molecules for a specific application.
b) Sensitivity of biosensors: the focus of experimentalists is not only on developing new detection setup/platforms, but also on targeting the discovery of new materials that can overcome sensitivity and selectivity problems that some signal amplification methods have.
c) Stability of biosensors: the signal of biosensors is often criticised due to poor reproducibility without accurately controlled conditions. Extra efforts in mass-producible modification and manufacturing methods for biosensing applications need improvements, such as reducing batch-to-batch variability, introduction of internal standards and their sensitivity to real samples with complex matrices.
d) Commercialisation of biosensors: large-scale electrode manufacture and modification methods need to be prioritised in order to be able to translate literature reports to commercial products. Enzymes, antibodies, aptamers etc. are often characterised as reliable and robust, however some of them exhibit poor chemical stability, short shelf-life and high cost. Also, it is important to note the importance of using sustainable materials when developing biosensors with 2D-nanomaterials. This could not only lead to lowering the cost and decrease the sourcing and manufacture challenges, but also to reduce the humanitarian and environmental impact of certain mining activities requited to obtain certain materials. It could also provide recyclable and circular materials, and the systems to support them. This will be paramount to the new generation of rapid biosensors built on advanced materials.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
A. G. M. F. would like to acknowledge Innovate UK for funding his Knowledge Transfer Partnership (KTP Reference: 11606).References
- A. García-Miranda Ferrari, P. Carrington, S. J. Rowley-Neale and C. E. Banks, Environ. Sci.: Water Res. Technol., 2020, 6, 2676–2690 RSC.
- J. P. Metters, R. Kadara and C. Banks, Analyst, 2011, 136, 1067–1076 RSC.
- A. J. Bard, J. Chem. Educ., 1983, 60, 302 CrossRef CAS.
- A. García-Miranda Ferrari, S. J. Rowley-Neale and C. E. Banks, Talanta Open, 2021, 3, 100032 CrossRef.
- R. O. Kadara, N. Jenkinson and C. E. Banks, Electrochem. Commun., 2009, 11, 1377–1380 CrossRef CAS.
- R. O. Kadara, N. Jenkinson and C. E. Banks, Sens. Actuators, B, 2009, 142, 342–346 CrossRef CAS.
- J. P. Metters, E. P. Randviir and C. E. Banks, Analyst, 2014, 139, 5339–5349 RSC.
- J. P. Metters, R. O. Kadara and C. E. Banks, Sens. Actuators, B, 2012, 169, 136–143 CrossRef CAS.
- J. P. Metters, R. O. Kadara and C. E. Banks, Analyst, 2013, 138, 2516–2521 RSC.
- P. M. Hallam, D. K. Kampouris, R. O. Kadara and C. E. Banks, Analyst, 2010, 135, 1947–1952 RSC.
- E. P. Randviir, D. A. Brownson, J. P. Metters, R. O. Kadara and C. E. Banks, Phys. Chem. Chem. Phys., 2014, 16, 4598–4611 RSC.
- J. P. Metters, R. O. Kadara and C. E. Banks, Analyst, 2012, 137, 896–902 RSC.
- J. P. Metters, F. Tan, R. O. Kadara and C. E. Banks, Anal. Methods, 2012, 4, 1272–1277 RSC.
- D. Martín-Yerga, A. Pérez-Junquera, M. B. González-García, J. V. Perales-Rondon, A. Heras, A. Colina, D. Hernández-Santos and P. Fanjul-Bolado, Electrochim. Acta, 2018, 264, 183–190 CrossRef.
- N. A. Choudhry, D. K. Kampouris, R. O. Kadara, N. Jenkinson and C. E. Banks, Anal. Methods, 2009, 1, 183–187 RSC.
- P. Yáñez-Sedeño, S. Campuzano and J. M. Pingarrón, Biosensors, 2020, 10, 76 CrossRef.
- E. Costa-Rama and M. T. Fernández-Abedul, Biosensors, 2021, 11, 51 CrossRef CAS.
- C. W. Foster, R. O. Kadara and C. E. Banks, in Screen-printing electrochemical architectures, Springer, 2016, pp. 13–23 Search PubMed.
- W. Vastarella, B. Lanza, A. Masci and R. Pilloton, in Sensors And Microsystems, World Scientific, 2005, pp. 19–24 Search PubMed.
- F. Arduini, L. Micheli, D. Moscone, G. Palleschi, S. Piermarini, F. Ricci and G. Volpe, TrAC, Trends Anal. Chem., 2016, 79, 114–126 CrossRef CAS.
- J. P. Hart, A. Crew, E. Crouch, K. C. Honeychurch and R. M. Pemberton, Anal. Lett., 2004, 37, 789–830 CrossRef CAS.
- C. W. Foster, J. Pillay, J. P. Metters and C. E. Banks, Sensors, 2014, 14, 21905–21922 CrossRef.
- M. P. O'Halloran, M. Pravda and G. G. Guilbault, Talanta, 2001, 55, 605–611 CrossRef.
- S. J. Rowley-Neale, D. A. Brownson, G. Smith and C. E. Banks, Biosensors, 2020, 10, 27 CrossRef CAS.
- J. P. Hughes, S. Rowley-Neale and C. Banks, RSC Adv., 2021, 11, 8073–8079 RSC.
- S. J. Rowley-Neale, C. W. Foster, G. C. Smith, D. A. Brownson and C. E. Banks, Sustainable Energy Fuels, 2017, 1, 74–83 RSC.
- D. A. Brownson, P. J. Kelly and C. E. Banks, RSC Adv., 2015, 5, 37281–37286 RSC.
- M. J. Whittingham, N. J. Hurst, R. D. Crapnell, A. Garcia-Miranda Ferrari, E. Blanco, T. J. Davies and C. E. Banks, Anal. Chem., 2021, 93, 16481–16488 CrossRef CAS.
- C. Banks, C. Foster and R. Kadara, Fundamentals of screen-printing electrochemical architectures, 2016, DOI:10.1007/978-3-319-25193-6.
- H.-W. Lin, C.-P. Chang, W.-H. Hwu and M.-D. Ger, J. Mater. Process. Technol., 2008, 197, 284–291 CrossRef CAS.
- E. Jewell, B. Philip and P. Greenwood, Biosensors, 2016, 6, 30 CrossRef.
- S. Damiati, C. Haslam, S. Søpstad, M. Peacock, T. Whitley, P. Davey and S. A. Awan, IEEE Access, 2019, 7, 94048–94058 Search PubMed.
- R. G. Compton and C. E. Banks, Understanding Voltammetry, Imperial College Press, 2nd edn, 2010 Search PubMed.
- T. J. Davies, S. Ward-Jones, C. E. Banks, J. del Campo, R. Mas, F. X. Muñoz and R. G. Compton, J. Electroanal. Chem., 2005, 585, 51–62 CrossRef CAS.
- D. Ortiz-Aguayo, M. Bonet-San-Emeterio and M. del Valle, Sensors, 2019, 19, 3286 CrossRef CAS.
- V. Ruiz-Valdepeñas Montiel, E. Povedano, S. Benedé, L. Mata, P. Galán-Malo, M. Gamella, A. J. Reviejo, S. Campuzano and J. M. Pingarrón, Anal. Chem., 2019, 91, 11266–11274 CrossRef.
- E. C. Rama and A. Costa-García, Electroanalysis, 2016, 28, 1700–1715 CrossRef CAS.
- D. H. Craston, C. P. Jones, D. E. Williams and N. El Murr, Talanta, 1991, 38, 17–26 CrossRef CAS.
- D. Li, C. Batchelor-McAuley, L. Chen and R. G. Compton, ACS Sens., 2019, 4, 2250–2266 CrossRef CAS.
- G. Ibáñez-Redín, R. H. Furuta, D. Wilson, F. M. Shimizu, E. M. Materon, L. M. R. B. Arantes, M. E. Melendez, A. L. Carvalho, R. M. Reis and M. N. Chaur, Mater. Sci. Eng., C, 2019, 99, 1502–1508 CrossRef.
- D. R. Thevenot, K. Toth, R. A. Durst and G. S. Wilson, Pure Appl. Chem., 1999, 71, 2333–2348 CrossRef CAS.
- D. R. Davies and S. Chacko, Acc. Chem. Res., 1993, 26, 421–427 CrossRef CAS.
- S. Oue, A. Okamoto, T. Yano and H. Kagamiyama, J. Biol. Chem., 1999, 274, 2344–2349 CrossRef CAS.
- C. I. Justino, A. C. Freitas, R. Pereira, A. C. Duarte and T. A. R. Santos, TrAC, Trends Anal. Chem., 2015, 68, 2–17 CrossRef CAS.
- K.-M. Song, S. Lee and C. Ban, Sensors, 2012, 12, 612–631 CrossRef PubMed.
- R. D. Crapnell, N. C. Dempsey-Hibbert, M. Peeters, A. Tridente and C. E. Banks, Talanta Open, 2020, 100018 CrossRef.
- O. Jamieson, T. C. Soares, B. A. de Faria, A. Hudson, F. Mecozzi, S. J. Rowley-Neale, C. E. Banks, J. Gruber, K. Novakovic and M. Peeters, Chemosensors, 2020, 8, 5 CrossRef CAS.
- K. Betlem, I. Mahmood, R. Seixas, I. Sadiki, R. Raimbault, C. Foster, R. Crapnell, S. Tedesco, C. Banks and J. Gruber, Chem. Eng. J., 2019, 359, 505–517 CrossRef CAS.
- R. D. Crapnell, A. Hudson, C. W. Foster, K. Eersels, B. v. Grinsven, T. J. Cleij, C. E. Banks and M. Peeters, Sensors, 2019, 19, 1204 CrossRef CAS PubMed.
- G. Ertürk and R. Lood, Sens. Actuators, B, 2018, 258, 535–543 CrossRef.
- M. Thaler and P. B. Luppa, Anal. Bioanal. Chem., 2019, 411, 7623–7635 CrossRef CAS.
- M. A. Morales and J. M. Halpern, Bioconjugate Chem., 2018, 29, 3231–3239 CrossRef CAS.
- C. I. L. Justino, A. C. Freitas, R. Pereira, A. C. Duarte and T. A. P. Rocha Santos, TrAC, Trends Anal. Chem., 2015, 68, 2–17 CrossRef CAS.
- B. Pérez-Fernández, A. Costa-García and A. d. l. E. Muñiz, Biosensors, 2020, 10, 32 CrossRef.
- Y. Lu, X. Liang, C. Niyungeko, J. Zhou, J. Xu and G. Tian, Talanta, 2018, 178, 324–338 CrossRef CAS.
- N. G. Welch, J. A. Scoble, B. W. Muir and P. J. Pigram, Biointerphases, 2017, 12, 02D301 CrossRef.
- R. M. Bednar, T. W. Golbek, K. M. Kean, W. J. Brown, S. Jana, J. E. Baio, P. A. Karplus and R. A. Mehl, ACS Appl. Mater. Interfaces, 2019, 11, 36391–36398 CrossRef CAS.
- Y. Liu and J. Yu, Microchim. Acta, 2016, 183, 1–19 CrossRef CAS.
- S. V. Rao, K. W. Anderson and L. G. Bachas, Microchim. Acta, 1998, 128, 127–143 CrossRef CAS.
- W. H. Organisation, Cardiovascular diseases, https://www.who.int/health-topics/cardiovascular-diseases#tab=tab_1, (accessed 19/08/2021, 2021) Search PubMed.
- NHS Blog by M. Kearney, entitled The Long Term Plan is a game changer, 2019, available at: https://www.longtermplan.nhs.uk/blog/the-long-term-plan-is-a-game-changer/.
- M. Chiong, Z. Wang, Z. Pedrozo, D. Cao, R. Troncoso, M. Ibacache, A. Criollo, A. Nemchenko, J. Hill and S. Lavandero, Cell Death Dis., 2011, 2, e244–e244 CrossRef CAS.
- T. Keller, T. Zeller, D. Peetz, S. Tzikas, A. Roth, E. Czyz, C. Bickel, S. Baldus, A. Warnholtz and M. Fröhlich, N. Engl. J. Med., 2009, 361, 868–877 CrossRef CAS.
- K. Thygesen, J. S. Alpert, A. S. Jaffe, B. R. Chaitman, J. J. Bax, D. A. Morrow and H. D. White, J. Am. Coll. Cardiol., 2018, 72, 2231–2264 CrossRef.
- J. J. Bax, H. Baumgartner, C. Ceconi, V. Dean, C. D. Uk, R. Fagard, C. Funck-Brentano, D. Hasdai, A. Hoes and P. Kirchhof, J. Am. Coll. Cardiol., 2012, 60(16), 1581–1598 CrossRef PubMed.
- E. Antman, J.-P. Bassand, W. Klein, M. Ohman, J. L. Lopez Sendon, L. Rydén, M. Simoons and M. Tendera, J. Am. Coll. Cardiol., 2000, 36, 959–969 CrossRef.
- R. Body, G. Burrows, S. Carley, L. Cullen, M. Than, A. S. Jaffe and P. S. Lewis, Clin. Chem., 2015, 61, 983–989 CrossRef CAS.
- T. Reichlin, W. Hochholzer, S. Bassetti, S. Steuer, C. Stelzig, S. Hartwiger, S. Biedert, N. Schaub, C. Buerge and M. Potocki, N. Engl. J. Med., 2009, 361, 858–867 CrossRef CAS.
- R. D. Crapnell, F. Canfarotta, J. Czulak, R. Johnson, K. Betlem, F. Mecozzi, M. P. Down, K. Eersels, B. van Grinsven and T. J. Cleij, ACS Sens., 2019, 4, 2838–2845 CrossRef CAS PubMed.
- S. Banu, S. Tanveer and C. N. Manjunath, Saudi J. Biol. Sci., 2015, 22, 56–61 CrossRef CAS.
- K. K. Jagadeesan, S. Kumar and G. Sumana, Electrochem. Commun., 2012, 20, 71–74 CrossRef CAS.
- J. E. Contreras-Naranjo and O. Aguilar, Biosensors, 2019, 9, 15 CrossRef CAS.
- W. Shen, S. Li, M.-K. Park, Z. Zhang, Z. Cheng, V. A. Petrenko and B. A. Chin, J. Electrochem. Soc., 2012, 159, B818 CrossRef CAS.
- R. D. Crapnell and C. E. Banks, Microchim. Acta, 2021, 188, 1–23 CrossRef.
- K. Gobalu, M. Vasudevan, S. C. Gopinath, V. Perumal and M. Ovinis, Cellulose, 2021, 28, 5761–5774 CrossRef CAS.
- M. Vasudevan, M. J. Tai, V. Perumal, S. C. Gopinath, S. S. Murthe, M. Ovinis, N. M. Mohamed and N. Joshi, Biotechnol. Appl. Biochem., 2021, 68, 1386–1395 CAS.
- V. Bhalla, S. Carrara, P. Sharma, Y. Nangia and C. R. Suri, Sens. Actuators, B, 2012, 161, 761–768 CrossRef CAS.
- A. Pourali, J. Barar, M. R. Rashidi, G. Pavon-Djavid and Y. Omidi, Microchem. J., 2021, 165, 106107 CrossRef CAS.
- S. Boonkaew, I. Jang, E. Noviana, W. Siangproh, O. Chailapakul and C. S. Henry, Sens. Actuators, B, 2021, 330, 129336 CrossRef CAS.
- A. Gupta, S. Kumar Sharma, A. L. Sharma and A. Deep, Phys. Status Solidi A, 2021, 218, 2000700 CrossRef CAS.
- A. Gupta, S. K. Sharma, V. Pachauri, S. Ingebrandt, S. Singh, A. L. Sharma and A. Deep, RSC Adv., 2021, 11, 2167–2174 RSC.
- D. Sun, Z. Luo, J. Lu, S. Zhang, T. Che, Z. Chen and L. Zhang, Biosens. Bioelectron., 2019, 134, 49–56 CrossRef CAS PubMed.
- H. Jo, J. Her, H. Lee, Y.-B. Shim and C. Ban, Talanta, 2017, 165, 442–448 CrossRef CAS.
- E. Dempsey and D. Rathod, IEEE Sens. J., 2018, 18, 1828–1834 CAS.
- B. V. Silva, I. T. Cavalcanti, A. B. Mattos, P. Moura, T. S. Maria Del Pilar and R. F. Dutra, Biosens. Bioelectron., 2010, 26, 1062–1067 CrossRef CAS PubMed.
- B. V. Silva, I. T. Cavalcanti, M. M. Silva and R. F. Dutra, Talanta, 2013, 117, 431–437 CrossRef CAS PubMed.
- K. Phonklam, R. Wannapob, W. Sriwimol, P. Thavarungkul and T. Phairatana, Sens. Actuators, B, 2020, 308, 127630 CrossRef CAS.
- J. McClements, P. M. Seumo Tchekwagep, A. L. Vilela Strapazon, F. Canfarotta, A. Thomson, J. Czulak, R. E. Johnson, K. Novakovic, P. Losada-Pérez and A. Zaman, ACS Appl. Mater. Interfaces, 2021, 13(24), 27868–27879 CrossRef CAS.
- M. Karimi, M. Rabiee, M. Tahriri, R. Salarian and L. Tayebi, Synth. Met., 2019, 256, 116136 CrossRef CAS.
- B. V. Silva, B. A. Rodríguez, G. F. Sales, T. S. Maria Del Pilar and R. F. Dutra, Biosens. Bioelectron., 2016, 77, 978–985 CrossRef CAS.
- F. T. Moreira, R. A. Dutra, J. P. Noronha and M. G. F. Sales, Electrochim. Acta, 2013, 107, 481–487 CrossRef CAS.
- V. V. Shumyantseva, T. V. Bulko, L. V. Sigolaeva, A. V. Kuzikov and A. I. Archakov, Biosens. Bioelectron., 2016, 86, 330–336 CrossRef CAS PubMed.
- J. Ribeiro, C. Pereira, A. Silva and M. G. F. Sales, Anal. Chim. Acta, 2017, 981, 41–52 CrossRef CAS.
- F. T. Moreira, S. Sharma, R. A. Dutra, J. P. Noronha, A. E. Cass and M. G. F. Sales, Sens. Actuators, B, 2014, 196, 123–132 CrossRef CAS.
- V. Kumar, J. R. Brent, M. Shorie, H. Kaur, G. Chadha, A. G. Thomas, E. A. Lewis, A. P. Rooney, L. Nguyen and X. L. Zhong, ACS Appl. Mater. Interfaces, 2016, 8, 22860–22868 CrossRef CAS.
- S. K. Tuteja, R. Chen, M. Kukkar, C. K. Song, R. Mutreja, S. Singh, A. K. Paul, H. Lee, K.-H. Kim and A. Deep, Biosens. Bioelectron., 2016, 86, 548–556 CrossRef CAS PubMed.
- P. D. Sinawang, D. Harpaz, L. Fajs, R. C. S. Seet, A. I. Y. Tok and R. S. Marks, EuroBiotech J., 2017, 1, 165–176 CrossRef.
- I. Grabowska, N. Sharma, A. Vasilescu, M. Iancu, G. Badea, R. Boukherroub, S. Ogale and S. Szunerits, ACS Omega, 2018, 3, 12010–12018 CrossRef CAS.
- R. S. Bresalier, W. M. Grady, S. D. Markowitz, H. J. Nielsen, S. K. Batra and P. D. Lampe, Cancer Epidemiol. Biomark. Prev., 2020, 29, 2431–2440 CrossRef CAS.
- J. Li, X. Guan, Z. Fan, L.-M. Ching, Y. Li, X. Wang, W.-M. Cao and D.-X. Liu, Cancers, 2020, 12, 2767 CrossRef CAS.
- M. Matuszczak, J. A. Schalken and M. Salagierski, Cancers, 2021, 13, 3373 CrossRef CAS.
- S. Guo, B. Yang, H. Liu, Y. Li, S. Li, L. Ma, J. Liu and W. Guo, J. Cancer Res. Ther., 2017, 13, 689 CrossRef CAS.
- P. Sardarabadi, A. A. Kojabad, D. Jafari and C.-H. Liu, Biosensors, 2021, 11, 394 CrossRef CAS.
- S. Kopetz, A. Grothey, R. Yaeger, E. Van Cutsem, J. Desai, T. Yoshino, H. Wasan, F. Ciardiello, F. Loupakis and Y. S. Hong, N. Engl. J. Med., 2019, 381, 1632–1643 CrossRef CAS PubMed.
- A. M. Poma, R. Bruno, I. Pietrini, G. Alì, G. Pasquini, A. Proietti, E. Vasile, S. Cappelli, A. Chella and G. Fontanini, Cancers, 2021, 13, 3828 CrossRef CAS.
- S. Siena, M. Di Bartolomeo, K. Raghav, T. Masuishi, F. Loupakis, H. Kawakami, K. Yamaguchi, T. Nishina, M. Fakih and E. Elez, Lancet Oncol., 2021, 22, 779–789 CrossRef CAS.
- Y. Chen, L. Sun, X. Qiao, Y. Zhang, Y. Li and F. Ma, Anal. Chim. Acta, 2020, 1103, 67–74 CrossRef CAS.
- J. Carlsson, H. Nordgren, J. Sjöström, K. Wester, K. Villman, N. O. Bengtsson, B. Ostenstad, H. Lundqvist and C. Blomqvist, Br. J. Cancer, 2004, 90, 2344–2348 CrossRef CAS.
- D. J. Slamon, G. M. Clark, S. G. Wong, W. J. Levin, A. Ullrich and W. L. McGuire, Science, 1987, 235, 177–182 CrossRef CAS.
- S. Ahn, J. W. Woo, K. Lee and S. Y. Park, J. Pathol. Transl. Med., 2020, 54, 34 CrossRef.
- NHS, Cancer antigen 15-3 (CA 15-3), https://www.nwlpathology.nhs.uk/tests-database/cancer-antigen-15-3-ca-15-3/, (accessed 27/10/2021, 2021) Search PubMed.
- NHS, CA 15–3 (Tumour marker), https://www.gloshospitals.nhs.uk/our-services/services-we-offer/pathology/tests-and-investigations/ca-15-3-tumour-marker/, (accessed 27th October 2021, 2021) Search PubMed.
- G. Bezerra, C. Córdula, D. Campos, G. Nascimento, N. Oliveira, M. A. Seabra, V. Visani, S. Lucas, I. Lopes and J. Santos, Anal. Bioanal. Chem., 2019, 411, 6667–6676 CrossRef CAS.
- J. G. Pacheco, P. Rebelo, M. Freitas, H. P. Nouws and C. Delerue-Matos, Sens. Actuators, B, 2018, 273, 1008–1014 CrossRef CAS.
- D. C. Ferreira, M. R. Batistuti, B. B. Junior and M. Mulato, Bioelectrochemistry, 2021, 137, 107586 CrossRef CAS PubMed.
- T. Harahsheh, Y. F. Makableh, I. Rawashdeh and M. Al-Fandi, Biomed. Microdevices, 2021, 23, 1–11 CrossRef.
- R. Crapnell and C. E. Banks, Sens. Diagn., 2022, 1, 71–86 RSC.
- R. C. Marques, E. Costa-Rama, S. Viswanathan, H. P. Nouws, A. Costa-García, C. Delerue-Matos and M. B. González-García, Sens. Actuators, B, 2018, 255, 918–925 CrossRef CAS.
- A. Ravalli, C. G. da Rocha, H. Yamanaka and G. Marrazza, Bioelectrochemistry, 2015, 106, 268–275 CrossRef CAS PubMed.
- S. Sharma, J. Zapatero-Rodríguez, R. Saxena, R. O'Kennedy and S. Srivastava, Biosens. Bioelectron., 2018, 106, 78–85 CrossRef CAS PubMed.
- S. Sharma, N. Gupta and S. Srivastava, Biosens. Bioelectron., 2012, 37, 30–37 CrossRef CAS.
- Y. W. Hartati, L. K. Letelay, S. Gaffar, S. Wyantuti and H. H. Bahti, Sens. Bio-Sens. Res., 2020, 27, 100316 CrossRef.
- F. Charbgoo, M. B. Ahmad and M. Darroudi, Int. J. Nanomed., 2017, 12, 1401 CrossRef CAS PubMed.
- M. Freitas, H. P. Nouws and C. Delerue-Matos, Electroanalysis, 2019, 31, 121–128 CrossRef CAS.
- S. D. Tallapragada, K. Layek, R. Mukherjee, K. K. Mistry and M. Ghosh, Bioelectrochemistry, 2017, 118, 25–30 CrossRef CAS PubMed.
- H. Ilkhani, A. Ravalli and G. Marrazza, Chemosensors, 2016, 4, 23 CrossRef.
- S. Patris, P. De Pauw, M. Vandeput, J. Huet, P. Van Antwerpen, S. Muyldermans and J.-M. Kauffmann, Talanta, 2014, 130, 164–170 CrossRef CAS.
- Q. Al-Khafaji, M. Harris, S. Tombelli, S. Laschi, A. Turner, M. Mascini and G. Marrazza, Electroanalysis, 2012, 24, 735–742 CrossRef CAS.
- M. Freitas, H. P. Nouws, E. Keating and C. Delerue-Matos, Sens. Actuators, B, 2020, 308, 127667 CrossRef CAS.
- S. A. A. Ahmad, M. S. Zaini and M. A. Kamarudin, J. Pharm. Biomed. Anal., 2019, 174, 608–617 CrossRef PubMed.
- M. Freitas, M. M. Neves, H. P. Nouws and C. Delerue-Matos, Talanta, 2020, 208, 120430 CrossRef PubMed.
- M. J. A. Shiddiky, P. H. Kithva, S. Rauf and M. Trau, Chem. Commun., 2012, 48, 6411–6413 RSC.
- Y. Hou, J. Wang, X. Wang, S. Shi, W. Wang and Z. Chen, Medicine, 2016, 95(2), e2450 CrossRef CAS.
- T. M. Grzywa, A. Sosnowska, P. Matryba, Z. Rydzynska, M. Jasinski, D. Nowis and J. Golab, Front. Immunol., 2020, 11, 938 CrossRef CAS.
- A. R. Cardoso, F. T. Moreira, R. Fernandes and M. G. F. Sales, Biosens. Bioelectron., 2016, 80, 621–630 CrossRef CAS PubMed.
- W. Białobrzeska, K. Dziąbowska, M. Lisowska, M. A. Mohtar, P. Muller, B. Vojtesek, R. Krejcir, R. O'Neill, T. R. Hupp and N. Malinowska, Biosensors, 2021, 11, 184 CrossRef PubMed.
- P. Sarkar, P. S. Pal, D. Ghosh, S. J. Setford and I. E. Tothill, Int. J. Pharm., 2002, 238, 1–9 CrossRef CAS PubMed.
- S. Hassani, A. S. Maghsoudi, M. R. Akmal, S. R. Rahmani, P. Sarihi, M. R. Ganjali, P. Norouzi and M. Abdollahi, J. Pharm. Pharm. Sci., 2020, 23, 243–258 CAS.
- B. V. Chikkaveeraiah, V. Mani, V. Patel, J. S. Gutkind and J. F. Rusling, Biosens. Bioelectron., 2011, 26, 4477–4483 CrossRef CAS PubMed.
- V. Mani, B. V. Chikkaveeraiah, V. Patel, J. S. Gutkind and J. F. Rusling, ACS Nano, 2009, 3, 585–594 CrossRef CAS.
- M. Chen, D. Wu, S. Tu, C. Yang, D. Chen and Y. Xu, Sci. Rep., 2021, 11, 1–11 CrossRef CAS PubMed.
- K. M. Koo, L. G. Carrascosa, M. J. A. Shiddiky and M. Trau, Anal. Chem., 2016, 88, 6781–6788 CrossRef CAS PubMed.
- K. M. Koo, L. G. Carrascosa, M. J. Shiddiky and M. Trau, Anal. Chem., 2016, 88, 2000–2005 CrossRef CAS PubMed.
- S. Yadav, M. K. Masud, M. N. Islam, V. Gopalan, A. K.-y. Lam, S. Tanaka, N.-T. Nguyen, M. S. A. Hossain, C. Li, M. Y. Yamauchi and M. J. A. Shiddiky, Nanoscale, 2017, 9, 8805–8814 RSC.
- M. K. Masud, S. Yadav, M. N. Islam, N.-T. Nguyen, C. Salomon, R. Kline, H. R. Alamri, Z. A. Alothman, Y. Yamauchi and M. S. A. Hossain, Anal. Chem., 2017, 89, 11005–11013 CrossRef CAS.
- M. N. Islam, M. K. Masud, N. T. Nguyen, V. Gopalan, H. R. Alamri, Z. A. Alothman, M. S. A. Hossain, Y. Yamauchi, A. K. Lamd and M. J. A. Shiddiky, Biosens. Bioelectron., 2018, 101, 275–281 CrossRef CAS PubMed.
- A. Gazze, R. Ademefun, R. S. Conlan and S. R. Teixeira, J. Interdiscip. Nanomed., 2018, 3, 82–88 CrossRef CAS.
- NICE, Ovarian Cancer, (accessed 27th October 2021, 2021) Search PubMed.
- A. Kumar, D. Roberts, K. E. Wood, B. Light, J. E. Parrillo, S. Sharma, R. Suppes, D. Feinstein, S. Zanotti and L. Taiberg, Crit. Care Med., 2006, 34, 1589–1596 CrossRef PubMed.
- M. Singer, C. S. Deutschman, C. W. Seymour, M. Shankar-Hari, D. Annane, M. Bauer, R. Bellomo, G. R. Bernard, J.-D. Chiche and C. M. Coopersmith, JAMA, J. Am. Med. Assoc., 2016, 315, 801–810 CrossRef CAS.
- A. Pant, I. Mackraj and T. Govender, J. Biomed. Sci., 2021, 28, 6 CrossRef.
- C. Fleischmann, A. Scherag, N. K. Adhikari, C. S. Hartog, T. Tsaganos, P. Schlattmann, D. C. Angus and K. Reinhart, Am. J. Respir. Crit. Care Med., 2016, 193, 259–272 CrossRef CAS PubMed.
- D. G. Remick, G. Bolgos, S. Copeland and J. Siddiqui, Infect. Immun., 2005, 73, 2751–2757 CrossRef CAS PubMed.
- P. Chen, G. Zhou, J. Lin, L. Li, Z. Zeng, M. Chen and S. Zhang, Front. Med., 2020, 7, 123 CrossRef PubMed.
- E. de Jong, J. A. van Oers, A. Beishuizen, P. Vos, W. J. Vermeijden, L. E. Haas, B. G. Loef, T. Dormans, G. C. van Melsen and Y. C. Kluiters, Lancet Infect. Dis., 2016, 16, 819–827 CrossRef CAS.
- J. D. Faix, Crit. Rev. Clin. Lab. Sci., 2013, 50, 23–36 CrossRef CAS PubMed.
- N. Jones, A. Tridente and N. C. Dempsey-Hibbert, Platelets, 2021, 32, 941–949 CrossRef CAS PubMed.
- S. G. Vishalashi, P. Gupta and P. K. Verma, Indian J. Crit. Care Med., 2021, 25, 507 CrossRef CAS PubMed.
- H. B. Nguyen, M. Loomba, J. J. Yang, G. Jacobsen, K. Shah, R. M. Otero, A. Suarez, H. Parekh, A. Jaehne and E. P. Rivers, J. Inflammation, 2010, 7, 1–11 CrossRef.
- M. M. Levy, L. E. Evans and A. Rhodes, Intensive Care Med., 2018, 44, 925–928 CrossRef CAS PubMed.
- R. D. Crapnell, A. Tridente, C. E. Banks and N. C. Dempsey-Hibbert, Sensors, 2021, 21, 879 CrossRef CAS PubMed.
- D. Kumar and B. B. Prasad, Sens. Actuators, B, 2012, 171, 1141–1150 CrossRef.
- M. Thangamuthu, C. Santschi and O. J. F. Martin, Biosensors, 2018, 8, 34 CrossRef PubMed.
- M. Lakshmanakumar, N. Nesakumar, S. Sethuraman, U. M. Krishnan and J. B. B. Rayappan, ACS Omega, 2021, 6, 32528–32536 CrossRef CAS PubMed.
- S. Jampasa, W. Siangproh, R. Laocharoensuk, T. Vilaivan and O. Chailapakul, Talanta, 2018, 183, 311–319 CrossRef CAS PubMed.
- A. Baradoke, R. Hein, X. Li and J. J. Davis, Anal. Chem., 2020, 92, 3508–3511 CrossRef CAS PubMed.
- Y. Boonyasit, O. Chailapakul and W. Laiwattanapaisal, Biosens. Bioelectron., 2019, 130, 389–396 CrossRef CAS PubMed.
- S. Boonkaew, S. Chaiyo, S. Jampasa, S. Rengpipat, W. Siangproh and O. Chailapakul, Microchim. Acta, 2019, 186, 153 CrossRef.
- C. Kokkinos, M. Prodromidis, A. Economou, P. Petrou and S. Kakabakos, Anal. Chim. Acta, 2015, 886, 29–36 CrossRef CAS.
- A. g. Molinero-Fernández, M. Moreno-Guzman, L. Arruza, M. A. n. López and A. Escarpa, ACS Sens., 2019, 4, 2117–2123 CrossRef PubMed.
- Á. Molinero-Fernández, M. Moreno-Guzmán, M. Á. López and A. Escarpa, Biosensors, 2020, 10, 66 CrossRef PubMed.
- T. Hou, B. C. Tieu, S. Ray, A. Recinos III, R. Cui, R. G. Tilton and A. R. Brasier, Curr. Cardiol. Rev., 2008, 4, 179 CrossRef CAS PubMed.
- T. L. Hedrick, B. P. Murray, R. S. Hagan and J. R. Mock, Am. J. Respir. Cell Mol. Biol., 2020, 63, 541–543 CrossRef CAS PubMed.
- N. Biran, A. Ip, J. Ahn, R. C. Go, S. Wang, S. Mathura, B. A. Sinclaire, U. Bednarz, M. Marafelias and E. Hansen, Lancet Rheumatol., 2020, 2, e603–e612 CrossRef.
- J. H. Stone, M. J. Frigault, N. J. Serling-Boyd, A. D. Fernandes, L. Harvey, A. S. Foulkes, N. K. Horick, B. C. Healy, R. Shah and A. M. Bensaci, N. Engl. J. Med., 2020, 383, 2333–2344 CrossRef CAS.
- R. D. Crapnell, W. Jesadabundit, A. García-Miranda Ferrari, N. C. Dempsey-Hibbert, M. Peeters, A. Tridente, O. Chailapakul and C. E. Banks, Anal. Chem., 2021, 93, 5931–5938 CrossRef CAS PubMed.
- K. Z. Liang and W. J. Mu, Anal. Chim. Acta, 2006, 580, 128–135 CrossRef CAS PubMed.
- M. Tertiş, B. Ciui, M. Suciu, R. Săndulescu and C. Cristea, Electrochim. Acta, 2017, 258, 1208–1218 CrossRef.
- J.-J. Shi, T.-T. He, F. Jiang, E. Abdel-Halim and J.-J. Zhu, Biosens. Bioelectron., 2014, 55, 51–56 CrossRef CAS PubMed.
- M. A. Ortega, X. Fernández-Garibay, A. G. Castaño, F. De Chiara, A. Hernández-Albors, J. Balaguer-Trias and J. Ramón-Azcón, Lab Chip, 2019, 19, 2568–2580 RSC.
- H. Chaudhry, J. Zhou, Y. Zhong, M. M. Ali, F. McGuire, P. S. Nagarkatti and M. Nagarkatti, In Vivo, 2013, 27, 669–684 CAS.
- H. Han, Q. Ma, C. Li, R. Liu, L. Zhao, W. Wang, P. Zhang, X. Liu, G. Gao and F. Liu, Emerging Microbes Infect., 2020, 9, 1123–1130 CrossRef CAS.
- L. Barhoumi, F. G. Bellagambi, F. M. Vivaldi, A. Baraket, Y. Clément, N. Zine, M. Ben Ali, A. Elaissari and A. Errachid, Sensors, 2019, 19, 692 CrossRef PubMed.
- U. F. I. C. C. (UICC), GLOBOCAN 2020: New Global Cancer Data, https://www.uicc.org/news/globocan-2020-new-global-cancer-data, (accessed 31/08/21) Search PubMed.
- R. C. Marques, S. Viswanathan, H. P. Nouws, C. Delerue-Matos and M. B. González-García, Talanta, 2014, 129, 594–599 CrossRef CAS PubMed.
- M. Freitas, H. P. Nouws, E. Keating, V. C. Fernandes and C. Delerue-Matos, Microchim. Acta, 2020, 187, 1–10 CrossRef PubMed.
- Z. O. Uygun, L. Yeniay and F. G. R. N. Sağın, Anal. Chim. Acta, 2020, 1121, 35–41 CrossRef CAS PubMed.
- S. Singal and R. K. Kotnala, Appl. Biochem. Biotechnol., 2017, 183, 672–683 CrossRef PubMed.
- M. Buch and J. Rishpon, Electroynalysis, 2008, 20, 2592–2594 CrossRef CAS.
- M. D. L. Gonçalves, L. A. Truta, M. G. F. Sales and F. T. Moreira, Anal. Lett., 2021, 1–13 Search PubMed.
- A. R. Cardoso, M. de Sá and M. G. F. Sales, Bioelectrochemistry, 2019, 130, 107287 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |