Open Access Article
Open Access ArticleSelective and rapid water transportation across a self-assembled peptide-diol channel via the formation of a dual water array†
Debashis
Mondal
 a,
Bhupendra R.
Dandekar
a,
Bhupendra R.
Dandekar
 b,
Manzoor
Ahmad
b,
Manzoor
Ahmad
 a,
Abhishek
Mondal
a,
Abhishek
Mondal
 a,
Jagannath
Mondal
a,
Jagannath
Mondal
 *b and
Pinaki
Talukdar
*b and
Pinaki
Talukdar
 *a
*a
aDepartment of Chemistry, Indian Institute of Science Education and Research Pune, Dr Homi Bhabha Road, Pashan, Pune 411008, Maharashtra, India. E-mail: ptalukdar@iiserpune.ac.in
bCenter for Interdisciplinary Sciences, Tata Institute of Fundamental Research, Hyderabad 500046, Telangana, India. E-mail: jmondal@tifrh.res.in
First published on 20th July 2022
Abstract
Achieving superfast water transport by using synthetically designed molecular artifacts, which exclude salts and protons, is a challenging task in separation science today, as it requires the concomitant presence of a proper water-binding site and necessary selectivity filter for transporting water. Here, we demonstrate the water channel behavior of two configurationally different peptide diol isomers that mimic the natural water channel system, i.e., aquaporins. The solid-state morphology studies showed the formation of a self-assembled aggregated structure, and X-ray crystal structure analysis confirmed the formation of a nanotubular assembly that comprises two distinct water channels. The water permeabilities of all six compounds were evaluated and are found to transport water by excluding salts and protons with a water permeability rate of 5.05 × 108 water molecules per s per channel, which is around one order of magnitude less than the water permeability rate of aquaporins. MD simulation studies showed that the system forms a stable water channel inside the bilayer membrane under ambient conditions, with a 2 × 8 layered assembly, and efficiently transports water molecules by forming two distinct water arrays within the channel.
Introduction
Even though 71% of the Earth's surface is covered with water,1 only a tiny percentage is available as freshwater.2–4 In consequence, more than one-third of the world's population faces severe freshwater scarcity that limits access to safe water for drinking and maintaining basic hygiene.5–8 To solve these issues, scientists have devised various water purification techniques, and in 90% of these cases, the reverse osmosis (RO) method is applied for the desalination of seawater.9–11 In this regard, reverse osmosis membranes or, more precisely, artificial water channels (AWCs), which have the ability to transport water selectively by excluding the salts from the seawater, have an essential role.12 However, one disadvantage of this technique is that the desalination process is highly energy-consuming.13,14 Therefore, an extremely sophisticated membrane material is required, which can complete the process by consuming less energy and making the process economical and available to all.15,16In nature, aquaporin-based protein channels are available, which are known to transport water across lipid bilayer membranes with high permeability and permselectivity.17–20 However, the lack of stability and compatibility of aquaporins outside the cellular environment delimit its widespread use for water purification applications.21 Therefore, the development of biomimetic artificial water channels (AWCs) is fundamentally essential for studying their water transport behavior and can be useful for medical and water purification purposes.12,22
Over the past decade, various AWCs based on unimolecular and supramolecular systems have been reported to achieve superfast water transport across the bilayer membranes.23–30 Some of them have even shown ultrafast water transport rates. For example, carbon nanotube porins (CNTPs) transport water across the bilayer membrane with a transport rate 6 times31 that of AQP1, a natural water channel protein.32,33 However, it also transports monovalent cations such as Na+, K+, and protons.34 Nevertheless, the exclusion of salts and protons by the channel forming substrate is an indispensable criterion for water purification application.35 In this regard, Barboiu, Hou, and their co-workers made tremendous efforts to develop synthetic channels for selective water transport, and later, the studies were further extended by Kumar, Zeng, and their co-workers. The reports from Barboiu and co-workers demonstrated the I-quartet and the hydroxyl-based water channels for selective and fast water permeation across the bilayer membrane.36–38 Contemporarily, Hou and co-workers reported the hydrazide-appended pillar[5]arene derivatives which showed proton excluding water transport behavior.25 Later, Kumar and co-workers reported peptide-appended hybrid[4]arene (PAH[4]) based water transport systems that transport water (water permeability > 109 H2O per s per channel) with a comparable transport rate of AQPs while excluding salts.39 The studies were further extended in recent reports by Zeng and co-workers, where they have reported several foldamer-based water channels that have shown excellent water permeability rates, which exclude salts and protons during the water transport process.40–42
As mentioned earlier, most artificial water channels have shown water permeability slightly less or close to that of aquaporins, but very few have demonstrated good water-salt permselectivity.43 The lack of a water selectivity mechanism (such as that of aquaporins) in most of the synthetic channels makes them less selective for water over the ions.37 Therefore, the proper structural design of the channel, which can interact with the water molecules selectively and softly, can show excellent water transport behavior with high salt rejection. Moreover, the absence of intermolecular hydrogen bonding interactions between two water molecules in the same water array can restrict the Grotthuss mechanism for proton transport, which is essential for showing good water channel behavior.44–46 However, obtaining permselectivity while achieving superfast permeability is still an exciting and challenging task in water purification science.
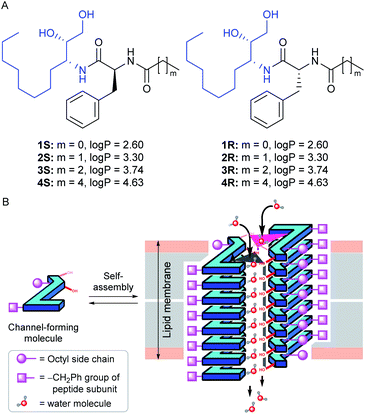
Herein, we report the peptide backbone-based 1,2-diol systems 1S–4S and 1R–4R (Fig. 1A) that form a dimeric rosette assembly and allow water transport across bilayer membranes by forming two distinct water channels. The diol group is used in our previous report to develop an anion-selective self-assembled ion channel,47 where the functional group acts as a hydrogen bond donor to interact with the anion. In contrast, it is also found in the literature that the hydroxyl groups (from the diol unit) have the ability to form the self-assembled water channel while participating either as a hydrogen bond donor or acceptor site (via either O–H⋯OH2 or O⋯H–OH interactions).37
In their proposed design, eight aggregated stacking units come closer to form a cylindrical channel through which the water transport phenomenon was observed. Here, in our design, we attached one alkyl group connected peptide unit with the diol group, which is expected to provide an additional hydrogen bond acceptor site to interact with the water molecules through C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–OH interaction. It is anticipated that this additional hydrogen bonding interaction site will support the formation of a water channel structure utilizing a minimum number of aggregated stacking units compared to the previous report. Overall, the peptide-diol molecules were designed to have multiple hydrogen bond donor and acceptor sites such as O–H, N–H, C
O⋯H–OH interaction. It is anticipated that this additional hydrogen bonding interaction site will support the formation of a water channel structure utilizing a minimum number of aggregated stacking units compared to the previous report. Overall, the peptide-diol molecules were designed to have multiple hydrogen bond donor and acceptor sites such as O–H, N–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, etc., for efficient noncovalent interactions. We expected that these systems would self-aggregate into a stacked sheet by a parallel one-dimensional array of monomers utilizing the intermolecular C
O, etc., for efficient noncovalent interactions. We expected that these systems would self-aggregate into a stacked sheet by a parallel one-dimensional array of monomers utilizing the intermolecular C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–N bonding interactions. Two such one-dimensional sheets would bundle up via intermolecular hydrogen bonding interaction through hydroxyl groups (from the diol subunit) and carbonyl groups (from the peptide subunit) with the water molecules to form a water channel (Fig. 1B). The chirality of the phenylalanine α-carbon was varied for generating two different series. The octyl and alkyl chains were introduced onto the monomers to tune the membrane permeability,43 which was estimated by calculating the log
O⋯H–N bonding interactions. Two such one-dimensional sheets would bundle up via intermolecular hydrogen bonding interaction through hydroxyl groups (from the diol subunit) and carbonyl groups (from the peptide subunit) with the water molecules to form a water channel (Fig. 1B). The chirality of the phenylalanine α-carbon was varied for generating two different series. The octyl and alkyl chains were introduced onto the monomers to tune the membrane permeability,43 which was estimated by calculating the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values of each compound using the Marvin Sketch program (Fig. 1A).48 Moreover, we expected that the dimeric rosette assembly would offer adequate channel pore and strong noncovalent interactions for water recognition by excluding the salts and hence increasing the water–salt permselectivity.
P values of each compound using the Marvin Sketch program (Fig. 1A).48 Moreover, we expected that the dimeric rosette assembly would offer adequate channel pore and strong noncovalent interactions for water recognition by excluding the salts and hence increasing the water–salt permselectivity.
Results and discussion
Synthesis of peptide 1,2-diols
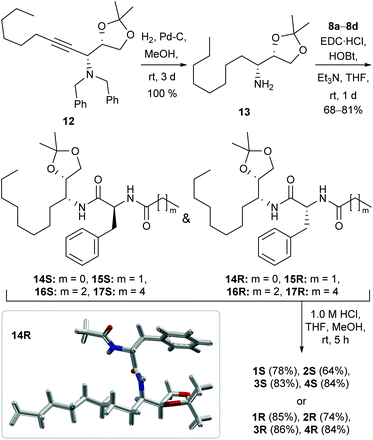
The synthesis of the channel forming molecules was started from L-phenylalanine and D-mannitol. At first, the L-phenylalanine was converted to its amide analogues 8a–8d by reacting with different aliphatic carboxylic acids following a reported literature protocol (Scheme S1†).49 On the other hand, D-mannitol was converted to acetonide protected dibenzyl-amine derivative 12 in three steps following our previously reported protocol (Scheme S2†).50,51 The hydrogenation of dibenzyl derivative 12 in the presence of Pd–C led to the debenzylation and reduction of the triple bond for the formation of acetonide-protected amine 13 in quantitative yield. Then, amine 13 reacted with different L-phenylalanine derivatives 8a–8dvia an EDC·HCl assisted coupling reaction to form amide derivatives 14S–17S. Surprisingly, the diastereomeric compounds 14R–17R were also formed almost in equal amounts due to the inversion of configuration at phenylalanine α-carbon. S- and R-configuration products were isolated separately by column chromatography. The formation of the R-configuration product was confirmed from the crystal structure of 14R (Scheme 1). Subsequently, all the acetonide-protected amides were separately treated with dilute HCl for the deprotection of the acetonide group to furnish the desired diol compounds 1S–4S and 1R–4R in 64–86% yield (Scheme 1).Solid-state morphology analysis
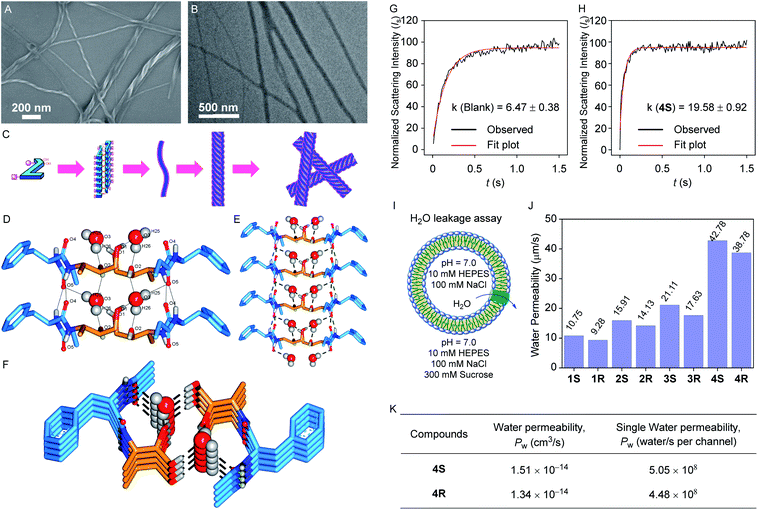
After synthesizing all the desired derivatives, we performed the experiments for morphological studies. At first, field emission scanning electron microscopy (FESEM) images for 4S and 4R were recorded (while preparing the samples in chloroform, Fig. S1A and S2A†), where no specific self-aggregated pattern was observed. However, when we performed the same experiments for these compounds by preparing the sample in moist methanol (2% water in methanol solvent), self-aggregated left-handed helical fibril morphology was observed for each compound (Fig. 2A and S1B† for 4S and Fig. S2B† for 4R). These observations indicate that the designed systems can form a self-aggregated assembly, where a polar protic solvent assists the assembled states. Such a fibril formation can be rationalized by considering the intermolecular hydrogen bonding interactions between the peptide backbones to form sheet structures followed by the self-aggregation via water molecules bridging to create a continuous chain, where multiple chains of this type bundled up for the formation of a left-handed helical fibril (Fig. 2A and C).52,53 The fibril formation was further confirmed by transmission electron microscopy (TEM) studies for 4S and 4R, where the samples were prepared in moist methanol (2% water in methanol solvent, Fig. 2B and S3†).Solid-state crystal structure analysis
A good quality crystal of 2R, suitable for solid-state single crystal X-ray analysis, was obtained by dissolving the compound in moist methanol (2% water in methanol solvent) followed by slow evaporation. In the solid-state, multiple monomer units participate in the parallel self-aggregation process via intermolecular amide C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–N hydrogen bonding interactions to form one-dimensional stacked sheets. Further, two such one-dimensional sheets bundle up antiparallelly through water-bridged hydrogen bonds. In this assembly, each water molecule forms one C
O⋯H–N hydrogen bonding interactions to form one-dimensional stacked sheets. Further, two such one-dimensional sheets bundle up antiparallelly through water-bridged hydrogen bonds. In this assembly, each water molecule forms one C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–OH hydrogen bond with the phenylalanine carbonyl of one monomer and a second C
O⋯H–OH hydrogen bond with the phenylalanine carbonyl of one monomer and a second C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯H–OH hydrogen bond with the terminal ethylamide carbonyl of the next monomer of the same sheet (Fig. 2D). Moreover, it also forms two C–O⋯H–OH hydrogen bonds, one with the secondary hydroxyl carbon of the first monomer and the other with a monomer from the second sheet C–O⋯H–OH hydrogen bond with the primary hydroxyl carbon of a monomer from the second sheet. Interestingly, the presence of two distinct water channels was evident within the cavity created by the two sheets. Moreover, the water molecules of each channel (i.e., in the same water array) are well separated (4.93 Å) to restrict the proton transport as per our proposed design. Finally, two antiparallel aggregated sheets are ensembled in a way that allows all the polar groups to either face inwardly or interact with each other to favour the assembly, whereas the hydrophobic phenyl and alkyl units are outwardly directed, which can help it to be inserted inside the lipid bilayer easily.
O⋯H–OH hydrogen bond with the terminal ethylamide carbonyl of the next monomer of the same sheet (Fig. 2D). Moreover, it also forms two C–O⋯H–OH hydrogen bonds, one with the secondary hydroxyl carbon of the first monomer and the other with a monomer from the second sheet C–O⋯H–OH hydrogen bond with the primary hydroxyl carbon of a monomer from the second sheet. Interestingly, the presence of two distinct water channels was evident within the cavity created by the two sheets. Moreover, the water molecules of each channel (i.e., in the same water array) are well separated (4.93 Å) to restrict the proton transport as per our proposed design. Finally, two antiparallel aggregated sheets are ensembled in a way that allows all the polar groups to either face inwardly or interact with each other to favour the assembly, whereas the hydrophobic phenyl and alkyl units are outwardly directed, which can help it to be inserted inside the lipid bilayer easily.
IR studies of channel-forming molecules in the lipid membrane
To check the channel formation behaviour of the channel-forming molecules, IR spectra were recorded after incorporating the compound in the lipid membrane.54,55 The change in the IR spectrum of the vesicles in the presence of the channel-forming molecule (5 mol% with respect to the lipid) compared to the free vesicles indicated the insertion of the channel-forming compound inside the bilayer membrane (Fig. S6†). Additionally, the broadening of the diol O–H peak (3268 cm−1) and shifting of the amide C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peak (1628 cm−1 to 1640 cm−1) of the compound in the presence of the lipid indicated the formation of a channel inside the bilayer membrane.
O peak (1628 cm−1 to 1640 cm−1) of the compound in the presence of the lipid indicated the formation of a channel inside the bilayer membrane.
Water permeability studies across the lipid bilayer membrane
The surprising results from the solvent dependent morphology studies and water channel formation by 2R in the solid-state encouraged us to assess the water transport permeability and selectivity by the transporter molecules across the phospholipid bilayer membrane. For this, a stopped-flow light scattering instrument was used for measuring the light scattering intensity of the vesicle suspension at a millisecond scale. At first, large unilamellar vesicles (LUVs) with a diameter of ∼150 nm were prepared using the DOPC lipid by incorporating each channel forming compound (molar channel-to-lipid ratio, mCLRs = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50) in 100 mM NaCl solution buffered to 7.0 using HEPES (Fig. 2I).40,41 When such a vesicle suspension is exposed suddenly to an equal volume of 300 mM sucrose solution, it leads to the shrinkage of the vesicles by outwardly directed water leakage through the water channels. This event subsequently leads to the change in light scattering intensity of the vesicle suspension. For each compound, a significant change in light scattering intensity compared to the blank sample was observed, and the rate constant value was also calculated (Fig. 2G, H and S7A–S7K†). Further, the rate constant values were used to calculate the water permeability values.39,56
50) in 100 mM NaCl solution buffered to 7.0 using HEPES (Fig. 2I).40,41 When such a vesicle suspension is exposed suddenly to an equal volume of 300 mM sucrose solution, it leads to the shrinkage of the vesicles by outwardly directed water leakage through the water channels. This event subsequently leads to the change in light scattering intensity of the vesicle suspension. For each compound, a significant change in light scattering intensity compared to the blank sample was observed, and the rate constant value was also calculated (Fig. 2G, H and S7A–S7K†). Further, the rate constant values were used to calculate the water permeability values.39,56
All our molecules 1S–4S and 1R–4R were found to transport the water across the bilayer membrane (Fig. 2H, J, and K), and the permeability values calculated using the conventional model are 85.7 (±2.8%), 100.3 (±3.5%), 115.1 (±7.3%), 176.5 (±4.7%), 81.5 (±0.6%), 95.2 (±5.8%), 105.2 (±5.9%), and 164.8 (±6.9%) μm s−1, respectively. We have also recalculated the permeability values for 1S–4S and 1R–4R using a recently updated model,39,56,57 considering the osmolyte concentration correction factor, and the values are 30.2 (±2.8%), 35.4 (±3.5%), 40.6 (±7.3%), 62.3 (±4.7%), 28.8 (±0.6%), 33.6 (±5.8%), 37.1 (±5.9%), and 58.17 (±6.9%) μm s−1, respectively. We assume that the 8 layers of the self-assembled system (2 × 8 layers) form a single channel structure and further considering the channel insertion efficiency and the corrected mCLRs values (Fig. S8 and S9†), the water permeability (unit = cm3 s−1) and single channel water permeability (unit = water molecules per s per channel) values were calculated for 4S and 4R channels. The 4S channel exhibited the highest water transport rate with a permeability value of 1.51 × 10−14 cm3 s−1 compared to the 4R channel, for which the estimated permeability value is 1.34 × 10−14 cm3 s−1. Further, these water permeability values were used to calculate the single channel water permeability, and the values are 5.05 × 108 and 4.48 × 108 water molecules per s per channel for 4S and 4R, respectively, indicating 4S as the most efficient water channel forming molecule (Fig. 2J, S7 and Table S2†). The formation of two distinct water channels within a single aggregate could be a prominent reason for the excellent single channel water permeability of 4S and 4R. Moreover, the data indicate that the efficiency of the water channels increases when a longer hydrophobic alkyl tail is present, indicating that the higher log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value contributes to better membrane permeability of the channel. It is also apparent that the activity of an S-configuration product is higher than that of the corresponding R-configuration product. This may be attributed to the better self-assembly of the S-configuration molecules.
P value contributes to better membrane permeability of the channel. It is also apparent that the activity of an S-configuration product is higher than that of the corresponding R-configuration product. This may be attributed to the better self-assembly of the S-configuration molecules.
Water permselectivity studies across the lipid bilayer membrane
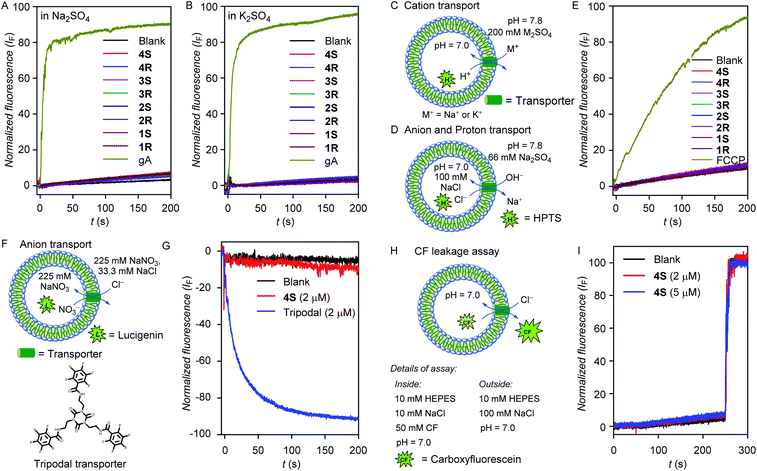
As discussed earlier, the salt exclusion is the essential criteria for water purification application of the water channel system. The salt exclusion behavior of the channel molecules while transporting the water was confirmed by performing the water leakage assay in different osmolytes other than sucrose.40,58 Here, we have calculated the reflection coefficient value for our system, where the reflection coefficient is the water permeability ratio of the channel in NaCl and sucrose solutions. We observed the reflection coefficient value 0.98 (Fig. S10†), which is close to 1, indicating that the water permeability value remains unaltered while shifting from sucrose solution to NaCl solution. This observation also confirmed the salt exclusion capability of our water channel system.Ion transport studies across the lipid bilayer membrane
The cation transport activity of the water channel-forming molecules across the bilayer membrane was evaluated following the standard ion transport experiment protocol.40,59–61 For this, we have prepared large unilamellar vesicles using the EYPC lipid by entrapping pH-sensitive 8-hydroxypyrene-1,3,6-trisulfonic acid trisodium salt (HPTS, 1.0 mM), a fluorescent dye, in 10 mM HEPES buffer (pH = 7.0). The vesicles were then suspended in 200 mM Na2SO4 solution, and a pH gradient (ΔpH = 0.8) was applied between the intra- and the extra-vesicular medium (Fig. 3C). Then the fluorescence intensity change of the HPTS dye was monitored with time after the addition of transporter molecules. None of our diol molecules showed any increment in the fluorescence intensity at 2.0 μM of the transporter concentration (Fig. 3A), whereas gramicidin A (gA, a natural cationophore) furnished a significant transport activity (∼90%) even at 20 times diluted concentration (0.1 μM). These results indicate that our water channel molecules do not transport Na+ across the bilayer membrane. When a similar experiment was performed for each compound by suspending the vesicles in K2SO4 solution, no significant transport activity was observed (Fig. 3B and C), which indicates that the 1,2-diol systems do not transport the K+ ions either.Next, anion and proton transport activities of the 1,2-diol compounds across the bilayer membrane were evaluated using HPTS assay (Fig. 3D). Here, no significant transport activity by these compounds were observed, whereas, carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP, a proton carrier) showed a significant transport activity (∼92%) even at the one-tenth concentration (Fig. 3E). These observations indicate that the 1,2-diol compounds are incapable of transporting anions and protons. Additionally, the anion influx by 4S was also evaluated across large unilamellar EYPC vesicles by entrapping lucigenin dye.47,62 Under intravesicular NaNO3 (225 mM) and extravesicular NaNO3 (225 mM) and NaCl (33.3 mM) conditions, no destruction of the chloride gradient in the presence of 4S was observed (Fig. 3F and G). In contrast, a reported tripodal chloride transporter60 showed excellent chloride influx activity (∼88%) under comparable conditions. Therefore, the inability of 4S for transporting anions was further established. In the above-mentioned anion, cation, and proton transport experiments, the intra- and extra-vesicular osmolyte concentration balance has not been maintained. To check for any ion transport in the iso-osmolar salt concentration, the ion transport activity was evaluated using standard HPTS assay, where 100 mM NaCl solution was used as intra- and extra-vesicular salt. In this assay, no transport activity was observed for the 4S channel (Fig. S14†), which ruled out the possibility of ion transport by the channel-forming molecule.
The carboxyfluorescein (CF) leakage assay,63 using 2 and 5 μM concentrations of 4S resulted in no release of CF dye (Fig. 3H and I), which indicates that the membrane integrity is preserved in the presence of the compounds and also rules out any possibility of pore formation.
Molecular dynamics simulation studies of the water channel in the lipid bilayer model
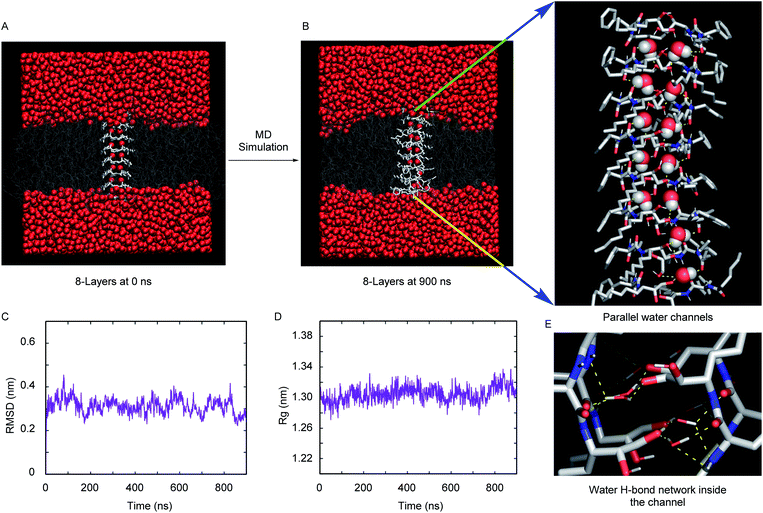
The excellent water channel properties of the 1,2-diol system encouraged us to carry out computational studies to get atomistic insight into the channel architecture and water transport mechanism. The comparable water transport properties between 4S and 4R and, most importantly, the availability of crystallographic coordinates of the R-configuration product with water, prompted us to focus on simulating the 4R superstructure and water permeation through it. The self-assembly pattern obtained from the crystal structure of 2R with water was used to generate the 2 × 8 layered (i.e., 8 layers of 2 molecules) assemblies of 4R. The assembly of 4R was generated by elongating each ethyl chain with a pentyl one.Fig. 4A and B, respectively, demonstrate the representative snapshot of the membrane-embedded 2 × 8 layered assembly of 4R at the initial point and at completion of the computer simulation after about microsecond (900 ns) long time-evolution. The visual inspection of the snapshot indicates that the simulations maintain a stable superstructure during the time period at ambient pressure (1 bar) and temperature (298 K). Unlike previous investigations, which required introduction of an order of magnitude larger lateral pressure or restraining forces for maintaining a stable synthetic channel,36,37 the current simulation was found to produce a structurally integrated channel inside the phospholipid bilayer under the ambient conditions of 1 bar pressure. This is further demonstrated by a steady time profile of root-mean-squared deviation (RMSD) of the 2 × 8 layered assembly relative to the crystal structure (Fig. 4C). A low RMSD of 0.3 nm from the crystal structure and a steady trend radius of gyration (Rg) profile (Fig. 4D) with an average Rg fluctuating around 1.3 nm further provide credence to the stability of the 2 × 8 layered superstructure.
We had also explored the effect of the number of layers on the overall integrity of the superstructure. Fig. S15A and S15B† compare the representative snapshot of superstructures across 2 × 7 layers and 2 × 9 layers, respectively. As depicted from the steady RMSD and Rg profiles (Fig. S15C and S15D†), the overall assembly remains stable in these two cases as well. However, when compared across all three cases (2 × 7 layers, 2 × 8 layers and 2 × 9 layers), the registry and overall orientation of the 2 × 8 layered assembly inside the membrane are found to be relatively optimal. Hence, we proceeded with simulation trajectories of 2 × 8 layers for subsequent analysis of water permeation dynamics inside the channel.
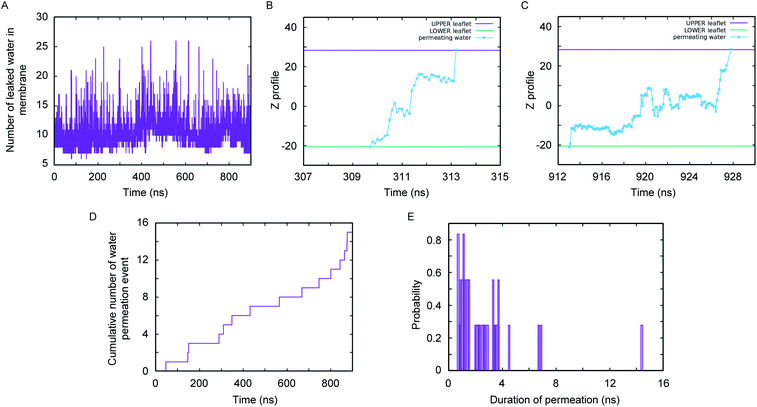
A close inspection of the synthetic channel (see the enlarged snapshot of Fig. 4B) highlighted the distinct presence of two parallel, non-intersecting water wires inside the synthetic channel. A molecular-level characterization of the inner architecture of water inside the simulated channel indicated that the water molecule forms an intricate hydrogen bond network (Fig. 4E) with the phenylalanine carbonyl oxygen, the diol (or hydroxyl) moiety and also with the amide nitrogen of the channel-forming molecules. The organization of inner water molecules is mainly guided by the integrity of the superstructure. However, there is no hydrogen bond between the water molecules present inside the channel. The visual inspection of the MD simulation trajectory indicated significant water leakage inside the phospholipid bilayer, as induced by the synthetic channel. A quantitative analysis of water leakage events (see method) provided further insights into the dynamical behavior of water molecules in a lipid bilayer environment. Fig. 5A shows the time profile of the number of water molecules leaked inside the membrane, suggesting significant water transport via the channel at ambient pressure and temperature. Very interestingly, tracking the movement of leaked water molecules revealed multiple water permeation events during the period of simulation. Fig. 5B and C depict two representative water-permeation events in which the water molecules make a full pass across the membrane from one of the leaflets to the other leaflet. The characterization of the simulation trajectory indicated a regular occurrence of water permeation events during the simulation period (Fig. 5D). However, the durations of such water permeation events are not uniform. In fact, as shown in Fig. 5E, the duration of water permeation is quite heterogeneous, ranging from as low as 2 ns to as large as 14 ns. Overall, the extent of water leakage and water permeation via maintaining two distinct files of water wires inside a single superstructure dictates a highly efficient synthetic water channel. The observation of two coexisting parallel water wires inside the channel formed by 4R is considerably distinct, when compared with previous investigations, which have reported either a single water wire or more disordered water structure inside the channel.
The quantitative value of water/salt selectivity was estimated by MD simulation studies40 in the presence of NaCl salt, where 7247 water molecules in total were present on both sides of the lipid bilayer, and the salt concentration was maintained at 0.10 M by adding 12 atoms of Na+ and 12 atoms of Cl− ions. In the transport simulation, a total of 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 223
223![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 223 water entry events up to the 1/8th (the first layer; till 4.79 Å) of the water channel was recorded on a time scale of 900 ns. Out of this, 16 molecules of water were found to be successfully transported across the bilayer membrane within this time scale. Additionally, we have not observed any transport of either of the Na+ and Cl− ions across the bilayer membrane. The transport selectivity (water/Na+ or water/Cl−) was calculated from the data for Na+ and Cl− ions, and is 5
223 water entry events up to the 1/8th (the first layer; till 4.79 Å) of the water channel was recorded on a time scale of 900 ns. Out of this, 16 molecules of water were found to be successfully transported across the bilayer membrane within this time scale. Additionally, we have not observed any transport of either of the Na+ and Cl− ions across the bilayer membrane. The transport selectivity (water/Na+ or water/Cl−) was calculated from the data for Na+ and Cl− ions, and is 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 223
223![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 223/(7247/12), i.e. 8649.
223/(7247/12), i.e. 8649.
The 4R channel has shown efficient experimental water permeability. Therefore, we have chosen this derivative for MD simulation studies as mentioned above, where the initial coordinates were generated from the solid-state crystal structure of the 2R channel by elongating the alkyl chain length. However, similar MD simulation studies (Fig. S16A and S16B†) were also performed for the 2R channel as well, where the initial channel structure was generated directly from the crystal structure. Here, we observed the maintenance of a stable channel structure similar to the 4R channel, with a slight deviation of RMSD (Fig. S16C†) and Rg values (Fig. S16D†) without applying any lateral pressures and restrain. Moreover, the salt-exclusion water permeation events were observed in these experiments (Fig. S16E and S16F†), and it was seen that for a particular configuration product, the elongation of alkyl chain length (facing outside) does not make any difference in the channel-forming mode.
In the quest for the channel structure of 4S, the 2 × 8 layered 4S channel assembly was generated by inverting the stereochemistry of the chiral phenylalanine centre and elongating the peptide attached methyl group to pentyl on each molecule of the 2 × 8 layered 2R channel, and subsequently, MD simulation studies were performed. However, stable channel formation was not observed in the case of 4S after applying similar simulation conditions (Fig. S17†). Perhaps some other type of channel assembly is formed in the case of the 4S channel for the transport of water across the bilayer membrane. However, as the single-channel permeabilities of 4R (4.48 × 108 water molecules per s) and 4S (5.05 × 108 water molecules per s) do not differ significantly, the channel structures of 2R and 4R are sufficient to represent the peptide 1,2-diol-based water channel systems.
Conclusions
In conclusion, we have designed and synthesized peptide diol-based water channel systems, which form two distinct water channels in a single self-assembled structure by employing the intermolecular hydrogen bonding interactions of water molecules with the hydroxyl groups and amide side chains. The experimental analysis revealed that the molecules are able to transport water across the lipid bilayer membrane with the highest permeability rate of 5.05 × 108 water molecules per s per channel, observed for the 4S derivative. Most importantly, a precise single-channel diameter of 2.5 Å was expected to allow only water molecules to pass selectively through the pore by excluding Na+, Cl−, and H+ ions, and this was evidenced from experimental and computational studies. The MD simulation studies for 4R and 2R superstructures confirmed the formation of a stable water channel assembly inside the bilayer membrane at ambient temperature and pressure, and more importantly, they show the transport of water molecules through its channel pore by forming two distinct water arrays. The superfast water transport rate and salt exclusion behaviour of the self-assembled channels can be applied in various applications, including the development of biomimetic RO membranes.Data availability
The detailed experimental procedures and characterization data are provided in the ESI.† The solid state single crystal X-ray analysis data have been deposited to Cambridge Crystallographic Data Center (CCDC) with the deposition codes: 2122390 (for 2R) and 2122389 (for 14R).Author contributions
P. T. conceived the project and directed the experimental studies. J. M. directed the computational studies. D. M. synthesized and characterized the compounds. B. R. D. performed the computational studies. D. M., M. A., and A. M. performed the transport studies. D. M., B. R. D., J. M., and P. T. wrote the paper. All authors approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by SERB, Govt. of India (DIA/2018/000017 for PT and CRG/2019/001219 for JM), and DST, Govt. of India (under the Indo-Russian Collaborative Project Scheme, DST/INT/RUS/RSF/P-24 for PT). BRD and JM acknowledge the support of the Department of Atomic Energy, Government of India, under Project Identification No. RTI 4007. DM and MA thank UGC, Govt. of India, for research fellowships. AM is thankful for the Prime Minister's Research Fellowship.Notes and references
- H. Genda, Geochem. J., 2016, 50, 27–42 CrossRef CAS.
- A. Czajkowski, L. Remiorz, S. Pawlak, E. Remiorz, J. Szyguła, D. Marek, M. Paszkuta, G. Drabik, G. Baron, J. Paduch and O. Antemijczuk, Appl. Sci., 2021, 11, 4081 CrossRef CAS.
- J. Eliasson, Nature, 2015, 517, 6 CrossRef CAS PubMed.
- R. F. Service, Science, 2006, 313, 1088–1090 CrossRef CAS PubMed.
- E. J. Byker and V. Ezelle-Thomas, J. Res. Child. Educ., 2021, 35, 268–280 CrossRef.
- A. Prüss-Ustün, J. Wolf, J. Bartram, T. Clasen, O. Cumming, M. C. Freeman, B. Gordon, P. R. Hunter, K. Medlicott and R. Johnston, Int. J. Hyg. Environ. Health, 2019, 222, 765–777 CrossRef PubMed.
- D. Mara, J. Lane, B. Scott and D. Trouba, PLoS Med., 2010, 7, e1000363 CrossRef PubMed.
- M. A. Shannon, P. W. Bohn, M. Elimelech, J. G. Georgiadis, B. J. Mariñas and A. M. Mayes, Nature, 2008, 452, 301–310 CrossRef CAS PubMed.
- K. Park, J. Kim, D. R. Yang and S. Hong, J. Membr. Sci., 2020, 595, 117607 CrossRef CAS.
- R. J. Petersen, J. Membr. Sci., 1993, 83, 81–150 CrossRef CAS.
- L. F. Greenlee, D. F. Lawler, B. D. Freeman, B. Marrot and P. Moulin, Water Res., 2009, 43, 2317–2348 CrossRef CAS PubMed.
- M. Barboiu and A. Gilles, Acc. Chem. Res., 2013, 46, 2814–2823 CrossRef CAS PubMed.
- R. Semiat, Environ. Sci. Technol., 2008, 42, 8193–8201 CrossRef CAS PubMed.
- G. Pérez-Zuñiga, R. Rivas-Perez, J. Sotomayor-Moriano and V. Sánchez-Zurita, Processes, 2020, 8, 1100 CrossRef.
- Y. M. El-Sayed, Desalination, 2001, 134, 129–158 CrossRef CAS.
- B. Peñate and L. García-Rodríguez, Desalination, 2012, 284, 1–8 CrossRef.
- E. Tajkhorshid, P. Nollert, M. Jensen, L. J. Miercke, J. O'Connell, R. M. Stroud and K. Schulten, Science, 2002, 296, 525–530 CrossRef CAS PubMed.
- K. Murata, K. Mitsuoka, T. Hirai, T. Walz, P. Agre, J. B. Heymann, A. Engel and Y. Fujiyoshi, Nature, 2000, 407, 599–605 CrossRef CAS PubMed.
- P. Agre, Angew. Chem., Int. Ed., 2004, 43, 4278–4290 CrossRef CAS PubMed.
- M. J. Borgnia, D. Kozono, G. Calamita, P. C. Maloney and P. Agre, J. Mol. Biol., 1999, 291, 1169–1179 CrossRef CAS PubMed.
- C. Hélix-Nielsen, Membranes, 2018, 8, 44–59 CrossRef PubMed.
- A. S. Verkman, Curr. Biol., 2013, 23, R52–R55 CrossRef CAS PubMed.
- Y. Le Duc, M. Michau, A. Gilles, V. Gence, Y.-M. Legrand, A. van der Lee, S. Tingry and M. Barboiu, Angew. Chem., Int. Ed., 2011, 50, 11366–11372 CrossRef CAS PubMed.
- M. Barboiu, Angew. Chem., Int. Ed., 2012, 51, 11674–11676 CrossRef CAS PubMed.
- X.-B. Hu, Z. Chen, G. Tang, J.-L. Hou and Z.-T. Li, J. Am. Chem. Soc., 2012, 134, 8384–8387 CrossRef CAS PubMed.
- M. S. Kaucher, M. Peterca, A. E. Dulcey, A. J. Kim, S. A. Vinogradov, D. A. Hammer, P. A. Heiney and V. Percec, J. Am. Chem. Soc., 2007, 129, 11698–11699 CrossRef CAS PubMed.
- Y.-x. Shen, W. Si, M. Erbakan, K. Decker, R. De Zorzi, P. O. Saboe, Y. J. Kang, S. Majd, P. J. Butler, T. Walz, A. Aksimentiev, J.-l. Hou and M. Kumar, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 9810–9815 CrossRef CAS PubMed.
- H. Zhao, S. Sheng, Y. Hong and H. Zeng, J. Am. Chem. Soc., 2014, 136, 14270–14276 CrossRef CAS PubMed.
- I. Kocsis, M. Sorci, H. Vanselous, S. Murail, S. E. Sanders, E. Licsandru, Y. M. Legrand, A. van der Lee, M. Baaden, P. B. Petersen, G. Belfort and M. Barboiu, Sci. Adv., 2018, 4, eaao5603 CrossRef PubMed.
- W. Song, Y.-x. Shen, C. Lang, P. Saha, I. V. Zenyuk, R. J. Hickey and M. Kumar, Faraday Discuss., 2018, 209, 193–204 RSC.
- H. Tunuguntla Ramya, Y. Henley Robert, Y.-C. Yao, A. Pham Tuan, M. Wanunu and A. Noy, Science, 2017, 357, 792–796 CrossRef CAS PubMed.
- R. H. Tunuguntla, R. Y. Henley, Y. C. Yao, T. A. Pham, M. Wanunu and A. Noy, Science, 2017, 357, 792–796 CrossRef CAS PubMed.
- Y. Li, Z. Li, F. Aydin, J. Quan, X. Chen, Y. C. Yao, C. Zhan, Y. Chen, T. A. Pham and A. Noy, Sci. Adv., 2020, 6, eaba9966 CrossRef CAS PubMed.
- R. H. Tunuguntla, F. I. Allen, K. Kim, A. Belliveau and A. Noy, Nat. Nanotechnol., 2016, 11, 639–644 CrossRef CAS PubMed.
- W. Song and M. Kumar, Curr. Opin. Chem. Eng., 2019, 25, 9–17 CrossRef.
- E. Licsandru, I. Kocsis, Y.-x. Shen, S. Murail, Y.-M. Legrand, A. van der Lee, D. Tsai, M. Baaden, M. Kumar and M. Barboiu, J. Am. Chem. Soc., 2016, 138, 5403–5409 CrossRef CAS PubMed.
- L.-B. Huang, A. Hardiagon, I. Kocsis, C.-A. Jegu, M. Deleanu, A. Gilles, A. van der Lee, F. Sterpone, M. Baaden and M. Barboiu, J. Am. Chem. Soc., 2021, 143, 4224–4233 CrossRef CAS PubMed.
- S. Murail, T. Vasiliu, A. Neamtu, M. Barboiu, F. Sterpone and M. Baaden, Faraday Discuss., 2018, 209, 125–148 RSC.
- W. Song, H. Joshi, R. Chowdhury, J. S. Najem, Y.-x. Shen, C. Lang, C. B. Henderson, Y.-M. Tu, M. Farell, M. E. Pitz, C. D. Maranas, P. S. Cremer, R. J. Hickey, S. A. Sarles, J.-l. Hou, A. Aksimentiev and M. Kumar, Nat. Nanotechnol., 2020, 15, 73–79 CrossRef CAS PubMed.
- J. Shen, R. Ye, A. Romanies, A. Roy, F. Chen, C. Ren, Z. Liu and H. Zeng, J. Am. Chem. Soc., 2020, 142, 10050–10058 CrossRef CAS PubMed.
- J. Shen, J. Fan, R. Ye, N. Li, Y. Mu and H. Zeng, Angew. Chem., Int. Ed., 2020, 59, 13328–13334 CrossRef CAS PubMed.
- A. Roy, J. Shen, H. Joshi, W. Song, Y.-M. Tu, R. Chowdhury, R. Ye, N. Li, C. Ren, M. Kumar, A. Aksimentiev and H. Zeng, Nat. Nanotechnol., 2021, 16, 911–917 CrossRef CAS PubMed.
- L.-B. Huang, M. Di Vincenzo, M. G. Ahunbay, A. van der Lee, D. Cot, S. Cerneaux, G. Maurin and M. Barboiu, J. Am. Chem. Soc., 2021, 143, 14386–14393 CrossRef CAS PubMed.
- B. L. de Groot, T. Frigato, V. Helms and H. Grubmüller, J. Mol. Biol., 2003, 333, 279–293 CrossRef CAS PubMed.
- U. Kosinska Eriksson, G. Fischer, R. Friemann, G. Enkavi, E. Tajkhorshid and R. Neutze, Science, 2013, 340, 1346–1349 CrossRef CAS PubMed.
- D. Marx, ChemPhysChem, 2006, 7, 1848–1870 CrossRef CAS PubMed.
- T. Saha, S. Dasari, D. Tewari, A. Prathap, K. M. Sureshan, A. K. Bera, A. Mukherjee and P. Talukdar, J. Am. Chem. Soc., 2014, 136, 14128–14135 CrossRef CAS PubMed.
- Marvin, ver. 5.8.0, ChemAxon, 2012, https://www.chemaxon.com Search PubMed.
- B. Sadhasivam, M. F. Rigana, B. Rukmanikrishnan and S. Muthusamy, Polym. Bull., 2018, 75, 829–849 CrossRef CAS.
- S. C. Deshmukh and P. Talukdar, J. Org. Chem., 2014, 79, 11215–11225 CrossRef CAS PubMed.
- S. C. Deshmukh, A. Roy and P. Talukdar, Org. Biomol. Chem., 2012, 10, 7536–7544 RSC.
- D. Yang, P. Duan, L. Zhang and M. Liu, Nat. Commun., 2017, 8, 15727 CrossRef CAS PubMed.
- Z. Shen, Y. Jiang, T. Wang and M. Liu, J. Am. Chem. Soc., 2015, 137, 16109–16115 CrossRef CAS PubMed.
- L. Chen, W. Si, L. Zhang, G. Tang, Z.-T. Li and J.-L. Hou, J. Am. Chem. Soc., 2013, 135, 2152–2155 CrossRef CAS PubMed.
- Y. Li, J. Dong, W. Gong, X. Tang, Y. Liu, Y. Cui and Y. Liu, J. Am. Chem. Soc., 2021, 143, 20939–20951 CrossRef CAS PubMed.
- Y. D. Yuan, J. Dong, J. Liu, D. Zhao, H. Wu, W. Zhou, H. X. Gan, Y. W. Tong, J. Jiang and D. Zhao, Nat. Commun., 2020, 11, 4927 CrossRef CAS PubMed.
- C. Hannesschläger, T. Barta, C. Siligan and A. Horner, Sci. Rep., 2018, 8, 8516 CrossRef PubMed.
- A.-K. Meinild, D. A. Klaerke and T. Zeuthen, J. Biol. Chem., 1998, 273, 32446–32451 CrossRef CAS PubMed.
- S. J. Moore, C. J. E. Haynes, J. Gonzalez, J. L. Sutton, S. J. Brooks, M. E. Light, J. Herniman, G. J. Langley, V. Soto-Cerrato, R. Perez-Tomas, I. Marques, P. J. Costa, V. Felix and P. A. Gale, Chem. Sci., 2013, 4, 103–117 RSC.
- D. Mondal, A. Sathyan, S. V. Shinde, K. K. Mishra and P. Talukdar, Org. Biomol. Chem., 2018, 16, 8690–8694 RSC.
- R. Ye, C. Ren, J. Shen, N. Li, F. Chen, A. Roy and H. Zeng, J. Am. Chem. Soc., 2019, 141, 9788–9792 CrossRef CAS PubMed.
- N. Busschaert, M. Wenzel, M. E. Light, P. Iglesias-Hernández, R. Pérez-Tomás and P. A. Gale, J. Am. Chem. Soc., 2011, 133, 14136–14148 CrossRef CAS PubMed.
- A. Roy, A. Gautam, J. A. Malla, S. Sarkar, A. Mukherjee and P. Talukdar, Chem. Commun., 2018, 54, 2024–2027 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2122389 and 2122390. For ESI and crystallographic data in CIF or other electronic format see https://doi.org/10.1039/d2sc01737g |
| This journal is © The Royal Society of Chemistry 2022 |