Open Access Article
Open Access ArticleUltrasensitive detection of β-lactamase-associated drug-resistant bacteria using a novel mass-tagged probe-mediated cascaded signal amplification strategy†
Jianhua
Zhu‡
a,
Yunfei
Bai‡
 d,
Xiuyu
Chen
a,
Linlin
Hu
ef,
Wenjun
Zhang
a,
Chunyan
Liu
a,
Hua
Shao
*e,
Jianguo
Sun
*g and
Yun
Chen
d,
Xiuyu
Chen
a,
Linlin
Hu
ef,
Wenjun
Zhang
a,
Chunyan
Liu
a,
Hua
Shao
*e,
Jianguo
Sun
*g and
Yun
Chen
 *abc
*abc
aSchool of Pharmacy, Nanjing Medical University, 818 Tian Yuan East Road, Nanjing, 211166, China. E-mail: ychen@njmu.edu.cn; Fax: +86-25-86868467; Tel: +86-25-86868326
bState Key Laboratory of Reproductive Medicine, 210029, China
cKey Laboratory of Cardiovascular & Cerebrovascular Medicine, Nanjing, 211166, China
dState Key Laboratory of Bioelectronics, School of Biological Sciences and Medical Engineering, Southeast University, Nanjing, 210096 China
eDepartment of Pharmacy, Zhongda Hospital, School of Medicine, Southeast University, Nanjing, 210009, China. E-mail: gycsh@163.com; Fax: +86-25-83262630; Tel: +86-25-83262630
fOffice of Clinical Trial Institution, Zhongda Hospital, School of Medicine, Southeast University, Nanjing, 210009, China
gState Key Laboratory of Natural Medicines, China Pharmaceutical University, Nanjing, 210009, China. E-mail: jgsun@cpu.edu.cn; Fax: +86-25-83271176; Tel: +86-25-83271176
First published on 25th October 2022
Abstract
The emergence and spread of drug-resistant bacteria (DRB) is a global health threat. Early and accurate detection of DRB is a critical step in the treatment of DRB infection. However, traditional assays for DRB detection are time-consuming and have inferior analytical sensitivity and quantification capability. Herein, a mass-tagged probe (MP-CMSA)-mediated enzyme- and light-assisted cascaded signal amplification strategy was developed for the ultrasensitive detection of β-lactamase (BLA), an enzyme closely associated with most DRB. Each MP-CMSA probe contained multiple poly(amidoamine) (PAMAM) dendrimer molecules immobilized on a streptavidin agarose bead via a BLA-cleavable linker, and each dendrimer was modified with multiple mass tags via a photo-cleavable linker. In BLA detection, BLA could cleave the BLA-cleavable linker, leading to dendrimers shedding from the MP-CMSA probe to achieve enzyme-assisted signal amplification. Then, each dendrimer can further release mass tags under UV light to achieve light-assisted signal amplification. After this cascaded signal amplification, the released mass tags were ultimately quantified by mass spectrometry. Consequently, the sensitivity of BLA detection can be significantly enhanced by four orders of magnitude with a detection limit of 50.0 fM. Finally, this approach was applied to the blood samples from patients with DRB. This platform provides a potential strategy for the sensitive, rapid and quantitative detection of DRB infection.
1. Introduction
The increase in drug-resistant bacteria (DRB) infection has become one of the greatest dangers to public health.1 In addition, it is estimated that the mortality due to DRB infection will be significantly higher than cancer mortality in the near future.2 More importantly, DRB infection is particularly concerning during the current coronavirus pandemic because it can exacerbate coronavirus symptoms, especially in geriatric patients, and thus coronavirus-infected patients must be extremely careful to avoid secondary infection of DRB.3 On the other hand, it is always of the utmost importance to diagnose the onset of DRB as early as possible.4 There is much evidence indicating that early and appropriate combating of DRB can significantly improve the clinical outcomes.5,6 However, false diagnosis of DRB infection at the initial stage can easily miss the optimal therapeutic window and promote the transmission of DRB.7 Furthermore, the number of infectious bacteria in individuals often determines the dosing regimens.8 Suboptimal dosing regimens could also cause poor clinical outcomes and further promote drug resistance.9 Therefore, early and quantitative detection of DRB is important for efficient diagnosis and treatment of DRB infection.Currently, antibacterial susceptibility testing (AST), such as disk diffusion, broth dilution and E-test, is the gold standard for DRB detection.10 Although AST provides reliable results, these culture-based assays are time-consuming (>two days) and labor-intensive due to very few bacterial strains being available at an early stage of infection.11 In recent years, alternative techniques for early DRB detection have been developed; however, significant efforts are still required to make them quantitative.12 In the case of β-lactamase (BLA), an enzyme closely associated with most DRB, its rapid detection is critical for circumventing resistance to β-lactam antibiotics.13 Several new methods, such as the PCR and fluorescence in situ hybridization (FISH), have been attempted for its detection in recent years.14,15 However, these methods cannot cover all BLA genes and mutations given that nearly 2800 types of BLA have been identified to date.13 To achieve better sensitivity and quantitative capability, alternative methods based on smart techniques are always urged.
Recently, mass spectrometry (MS) has become an indispensable tool for target molecule detection, primarily due to its great quantitative capability.16 MS-based methods have been applied to bacterial detection in laboratory medicine.17 However, the population of DRB in patients at the early stage of infection may be much lower than the detection limit of MS (>105 colony-forming units (CFU) per mL).18 Thus, a lengthy culture-enrichment step is often required prior to MS detection.19 Fortunately, among various MS-based approaches, the one relying on mass-tagged probes and liquid chromatography-tandem mass spectrometry (LC-MS/MS) can convert and amplify the signal of the target molecule into that of mass tags such as peptides, which has significantly expanded the application of MS.20 Moreover, mass-tagged probes can convert the signal of each target molecule into that of multiple mass tag molecules using signal amplification strategies.21 Notably, multiple signal amplification strategies may be required for the detection of ultralow abundant target molecules such as DRB.
In the current work, a novel mass-tagged probe (MP-CMSA) was developed for BLA detection. The MP-CMSA probe can convert the amount of BLA to the mass tag signal via enzyme- and light-assisted cascaded signal amplification. The released mass tag was ultimately quantified by MS. After further optimization and validation, the new assay was applied for BLA detection in blood samples of patients who were diagnosed with bacterial infection.
2. Experimental section
2.1 Synthesis of biotin-poly(ethylene glycol)-7-aminocephalosporanic acid (BPEG-7ACA)
First, 200 mg of biotin-poly(ethylene glycol)-succinimidyl-carboxymethyl-ester (BPEG-NHS) and 34.5 mg of hydromethyl-7-aminocephalosporanic acid (7ACA) were dissolved in 40 mL of deionized water at a final concentration of 2.50 mM BPEG-NHS and 3.64 mM 7ACA, and then 4 μL of triethylamine (TEA) was added to adjust the pH of the mixture to 8.0. The reaction mixture was stirred at 600 rpm at 30 °C for 24 h. Thereafter, the mixture was placed into a dialysis bag with a molecular weight cutoff (MWCO) of 1.0 kDa and dialyzed against deionized water for 72 h. BPEG-7ACA was obtained through lyophilization.2.2 Synthesis of biotin-poly(ethylene glycol)-7-aminocephalosporanic acid-poly(ethylene glycol)-azido (BPEG-7ACA-N3)
For BPEG-7ACA-N3 synthesis, 110 mg of BPEG-7ACA and 150 mg of azido-poly(ethylene glycol)-succinimidyl-carboxymethyl-ester (N3-PEG-NHS) were dissolved in 40 mL of dimethyl sulfoxide (DMSO) at a final concentration of 1.25 mM BPEG-7ACA and 1.88 mM N3-PEG-NHS, and then 4 μL of TEA was added. After stirring at 600 rpm at 30 °C for 24 h, the mixture was dialyzed with a MWCO of 3.5 kDa against deionized water for 72 h. Finally, BPEG-7ACA-N3 was obtained through lyophilization.2.3 Synthesis of poly(amidoamine)-dibenzocyclooctyne (PAMAM-DBCO)
First, 100 mg of poly(amidoamine) (PAMAM) and 268 mg of dibenzocyclooctyne-N-hydroxysuccinimidyl ester (DBCO-NHS) were dissolved in 60 mL of DMSO at a final concentration of 0.0578 mM PAMAM and 11.1 mM DBCO-NHS and stirred at 600 rpm at 30 °C for 18 h. Subsequently, the mixture was placed into a dialysis bag with a MWCO of 10.0 kDa and dialyzed against DMSO for two days and then against deionized water for another two days. Finally, PAMAM-DBCO was obtained through lyophilization.2.4 Synthesis of a dual-functional molecule (DM)
BPEG-7ACA-N3 and azido-modified photo-cleavable peptide AVLGDPFR-PL-GFK-N3 (MP-PL-N3) were conjugated to PAMAM-DBCO to obtain a DM. For DM preparation, 10 mL of 10.0 mM MP-PL-N3 in PBS (equiv. to 10.0 mM mass tag) and 4 mL of 10.0 mM BPEG-7ACA-N3 in PBS (equiv. to 10.0 mM 7ACA) were mixed with 10 mL of 0.100 mM PAMAM-DBCO and stirred at 600 rpm at 30 °C for 12 h. Subsequently, the mixture was purified by dialysis with a MWCO of 10.0 kDa in deionized water. The DM product was obtained by lyophilization. Notably, the number of conjugated mass tags can be estimated from the released mass tag after the DM was exposed to UV light. The number of mass tags per DM was estimated as the number of conjugated mass tags divided by the amount of DM, which was equivalent to that of the reactant PAMAM-DBCO (please see the ESI†).2.5 Preparation of the MP-CMSA probe
Streptavidin agarose beads were immobilized with the DM to obtain the MP-CMSA probe. In brief, streptavidin agarose beads were washed with PBS three times. Then, 1 mL of streptavidin agarose beads was mixed with 1 mL of 12.0 μM DM and incubated at 37 °C for 2 h. Subsequently, the beads were washed with PBS three times and stored at 4 °C in PBS. The supernatant was exposed to UV light, and the mass tag released from the unimmobilized DM was detected by MS. The amount of the immobilized DM can be estimated from the released mass tag in the supernatant. The number of immobilized DMs per bead was calculated as the amount of immobilized DMs divided by the number of beads in the reaction (please see the ESI†).2.6 Evaluation of enzymatic-cleavage and photo-cleavage efficiencies of the MP-CMSA probe
To evaluate the photo-cleavage efficiency, 100 μL of 100 nM MP-CMSA probe was irradiated under UV light at 365 nm at various times. Then, the sample was centrifuged at 1000 rpm for 5 min, and the pellets were washed with 100 μL of PBS three times. The collected supernatants were combined and subjected to mass spectrometry.To evaluate the enzymatic-cleavage efficiency, 100 μL of 10.0 μM MP-CMSA probe was incubated with 100 μL of 100 pM BLA in PBS for various times or 100 μL of BLA at different concentrations for 2 h. Then, the sample was centrifuged at 1000 rpm for 5 min, and the pellets were washed with 100 μL of PBS three times. The collected supernatants were combined and exposed to UV light for 40 min and then subjected to LC-MS/MS analysis.
2.7 BLA detection in the blood
One milliliter of blood was centrifuged at 800 rpm for 3 min to obtain the supernatant. The collected supernatant was further centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 8 min. The obtained precipitate was resuspended in 100 μL of PBS and sonicated for 20 s using an XL-2000 Series probe sonicator set at 5 W and 22 kHz. Subsequently, 100 μL of lysate was mixed with 100 μL of 10.0 μM MP-CMSA probe. The mixture was incubated in the dark at 37 °C for 2 h and then centrifuged at 1000 rpm for 5 min. The pellets were washed with 100 μL of PBS three times. The collected supernatants were combined and exposed to UV light for 40 min and then desalted using a MicroSpin C18 column (The Nest Group, Inc., MA, USA) after the addition of 10 μL of internal standard peptide solution. The obtained mass tag sample was measured by MS. This process was operator-blinded and the LC-MS/MS analysis was conducted by two trained operators.
000 rpm at 4 °C for 8 min. The obtained precipitate was resuspended in 100 μL of PBS and sonicated for 20 s using an XL-2000 Series probe sonicator set at 5 W and 22 kHz. Subsequently, 100 μL of lysate was mixed with 100 μL of 10.0 μM MP-CMSA probe. The mixture was incubated in the dark at 37 °C for 2 h and then centrifuged at 1000 rpm for 5 min. The pellets were washed with 100 μL of PBS three times. The collected supernatants were combined and exposed to UV light for 40 min and then desalted using a MicroSpin C18 column (The Nest Group, Inc., MA, USA) after the addition of 10 μL of internal standard peptide solution. The obtained mass tag sample was measured by MS. This process was operator-blinded and the LC-MS/MS analysis was conducted by two trained operators.
3. Results and discussion
3.1 Probe design and working principle
In this study, the MP-CMSA probe played both detection and amplification roles. To achieve the detection function, the probe should basically have a recognition moiety for targeting and a probe tag for detection. Correspondingly, (1) 7ACA, the parent core of β-lactam antibiotics that can be easily recognized and cleaved by BLA as a substrate,22 was employed as the recognition part of the probe. (2) A peptide with the sequence AVLGDPFR was selected as the mass tag. For the amplification feature of the probe, there are a variety of strategies for signal amplification. Among these strategies, highly branched molecules such as dendrimers with easy modification, good chemical and physical stabilities, and simple preparation have attracted widespread attention.23 There is evidence indicating that tag molecules can be conjugated on the surface of dendrimers to amplify the signal exponentially, depending on the series of generations of the dendrimer.24 Thus, a fifth-generation PAMAM dendrimer with 128 terminal amino groups was employed in the MP-CMSA probe. As illustrated in Fig. 1, BPEG-7ACA-N3 with the BLA-cleavable linker 7ACA and MP-PL-N3 with the photo-cleavable linker PL were both conjugated to the DBCO-modified PAMAM dendrimer (PAMAM-DBCO) to obtain a DM. Multiple DMs were then anchored on a streptavidin agarose bead to acquire the MP-CMSA probe. In BLA detection, BLA molecules could cleave 7ACA, leading to DM shedding from the MP-CMSA probe. If there was sufficient time, each BLA molecule could cleave multiple 7ACA linkers sequentially, resulting in more DM shedding. Then, each dendrimer can further release mass tags under UV light to achieve signal amplification. Therefore, the cascaded signal amplification actually relied on the bearing capability of the bead and the branched structure of the dendrimer.3.2 Probe preparation and characterization
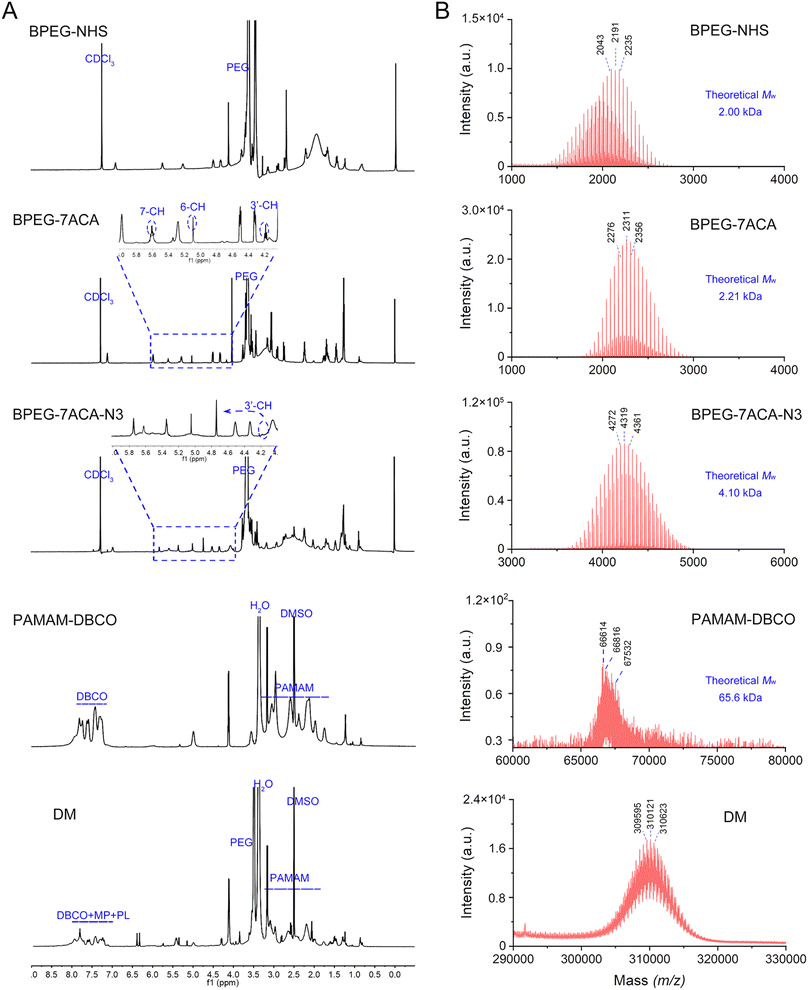
As described earlier, the MP-CMSA probe was produced by immobilizing the DM on streptavidin agarose beads. The DM preparation mainly contained four steps (Fig. S1†): (1) BPEG-NHS was conjugated to 7ACA through a condensation reaction to produce BPEG-7ACA, (2) BPEG-7ACA reacted with N3-PEG-NHS to form BPEG-7ACA-N3 through another condensation reaction, (3) DBCO-NHS was conjugated to PAMAM to form PAMAM-DBCO, and (4) both BPEG-7ACA-N3 and MP-PL-N3 reacted with PAMAM-DBCO through a copper-free click reaction to generate the DM.1H nuclear magnetic resonance spectroscopy (1H NMR) and matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) were used to verify the structure and molecular weight (Mw) of the reaction products, including BPEG-7ACA, BPEG-7ACA-N3, PAMAM-DBCO and DM. As shown in Fig. 2A, the characteristic signals at 4.17, 5.09 and 5.62 ppm in the 1H NMR spectrum of BPEG-7ACA can be attributed to the 3′-CH, 6-CH and 7-CH protons of 7ACA, confirming the formation of BPEG-7ACA. Then, the 3′-CH proton peak shifted from 4.1 ppm to 4.76 ppm in the spectrum of BPEG-7ACA-N3, demonstrating the occurrence of an esterification reaction between the hydroxyl group of 7ACA and the carboxyl group of N3-PEG-NHS. According to the spectrum of PAMAM-DBCO, the multiple peaks between 7 and 8 ppm were derived from the hydrogen protons on the benzene ring of DBCO. Finally, the majority of characteristic peaks of PAMAM, DBCO, MP-PL-N3 and BPEG-7ACA-N3 appeared in the spectrum of the DM, suggesting that the DM was successfully synthesized. The changes in the size distribution and zeta potential of PAMAM, PAMAM-DBCO and DM also confirmed this point (Fig. S2†). Moreover, the Mw values of BPEG-NHS, BPEG-7ACA-N3, BPEG-7ACA, PAMAM-DBCO and DM in the MALDI-TOF MS spectra were consistent with their theoretical values (Fig. 2B). Finally, the conjugation number of mass tags for each DM was estimated to be approximately 75 (please see the ESI†).
Afterward, the DM was immobilized on the surface of streptavidin agarose beads through biotin-streptavidin interactions to produce the MP-CMSA probe. The immobilization efficiency was optimized (Fig. S3†) and estimated to be 7.33 × 103 DM per bead (please see the ESI†). The obtained MP-CMSA probe exhibited excellent stability under both storage (PBS, pH 7.4 at 4 °C) and detection conditions (PBS, pH 7.4 at 37 °C). The probe degradation percentage was only (1.52 ± 0.08)% after 7 days under the storage conditions and (1.02 ± 0.06)% after 48 h under the detection conditions (Fig. S4†).
3.3 Optimization of signal amplification conditions
The amplification conditions, including photo-assisted and enzymatic-assisted reaction conditions, were carefully optimized and characterized in this study.3.4 Ultrasensitive and specific detection of BLA-associated DRB
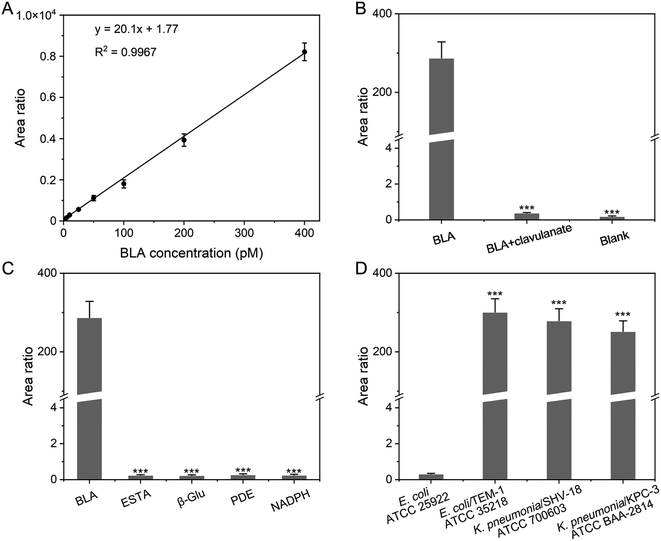
The ultrasensitive detection of BLA-associated DRB in this study was achieved by cascaded signal amplification. The mass-tagged probe was employed to convert and amplify the information of BLA to the mass tag signal. During the mass spectrometric analysis, the MRM transitions with the best signal to noise ratio (S/N) of the mass tag AVLGDPFR (i.e., m/z 437.9 → m/z 704.4, m/z 437.9 → m/z 419.3 and m/z 437.9 → m/z 171.2) were employed (Fig. S8 and S9†). The stable isotope-labeled peptide AV*LGDPFR ([D8]Val) served as an internal standard (Fig. S9†). The relative peak area ratio of MRM transitions of the mass tag and the internal standard was plotted against the BLA concentration to establish a calibration curve and used for BLA quantification (Fig. 4A). The linear relationship between the MS signal of the MP-CMSA probe and the concentration of BLA ranged from 50.0 fM to 400 pM (R2 = 0.9967). The lower limit of quantification (LLOQ) of BLA was determined to be 50.0 fM (S/N > 10, Fig. S10†), which was significantly lower than that of the standard chromogenic BLA probe (4.00 nM).34 This LLOQ was roughly equivalent to the concentration of typical BLA-associated DRB strains as low as 103 CFU mL−1.7 In addition, no significant signal was observed for the matrix blank (i.e., 5% BSA) and thus the background was low. Notably, the sample with a positive signal means that its mean signal of three measurement replicates was greater than LLOQ with a p-value less than 0.05 by the t-test.35 In addition, we also compared the MS signals of the synthetic mass tag and the released mass tag in our probe-mediated approach with BLA at an equivalent concentration. The result indicated that the sensitivity was significantly enhanced by four orders of magnitude after signal amplification. Moreover, the precision and accuracy of QCs were ≤±15% (LLOQ ≤ ±20%), (Table S1†). The precision of the approach was also confirmed by spiking the mass tag instead of the MP-CMSA probe before and after signal amplification. As a result, there is no significant difference in the MS signal variations between the sample groups.Furthermore, the specificity of the approach was also validated. First, the signal of the mass tag could not be detected in the absence of BLA (Fig. 4B). Then, the signal of the mass tag significantly decreased with pretreatment of the BLA inhibitor potassium clavulanate.36 The selectivity of the MP-CMSA probe against BLA was also confirmed using several common enzymes in bacteria including esterase (ESTA), β-glucosidase (β-Glu), phosphodiesterase (PDE), and nicotinamide adenine dinucleotide phosphate (NADPH) (Fig. 4C).37–40 Additionally, no significant signal was observed after the MP-CMSA probe was incubated with non-BLA-expressing E. coli ATCC 25922, that is the Clinical and Laboratory Standards Institute (CLSI) recommended negative quality control strain.41 Comparatively, a remarkable MS signal can be detected in three clinically prevalent BLA-expressing bacterial strains including K. pneumonia/KPC-3 ATCC BAA-2814, K. pneumonia/SHV-18 ATCC 700603 and E. coli/TEM-1 ATCC 35218 (Fig. 4D).42–44 These results confirmed the specificity of the MP-CMSA probe-mediated approach.
3.5 Detection of BLA-associated DRB in the blood
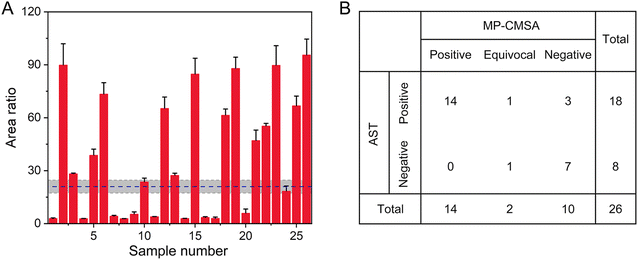
We further investigated the potential utility of this system to reveal BLA-associated DRB directly from blood samples (n = 35) of patients (age: ≥65 years (n = 13), <65 years (n = 13)) who were diagnosed with bacterial infections. The samples were originally defined by hospital pathologists using gold-standard AST (disc diffusion test, K–B method) following the CLSI.41 On the other hand, BLA detection using the mass-tagged probe was performed in the laboratory. This process was operator-blinded. Moreover, the blood samples were collected and analyzed by two trained operators. The analysis time of each sample was around 3 h. As shown in Fig. 5A, positive MS signals were observed in 26 samples using the MP-CMSA probe-mediated approach. The other 9 samples without positive MS signals were also assessed as BLA-negative using AST. Passing–Bablok regression analysis showed a good agreement between the results from the two operators (y = 0.992x − 0.034, p = 0.53) (Fig. S12†).To further evaluate the approach performance, a classification strategy using a three-zone partition was employed for patient grouping in 26 samples.45 Specifically, an optimal assay threshold was defined as 21.0 relying on Youden's index.46 Then, an intermediate grey zone with an area ratio range of 16.8–25.2 was established using the 20% tolerance range of the cutoff value and defined as “equivocal”. The zone with an area ratio of > 25.2 was defined as “positive” and that with an area ratio of < 16.8 was defined as “negative”. The BLA-positive patients should be resistant to the β-lactam antibiotic treatment. Using this three-zone classification, the sensitivity and specificity of the MP-CMSA assay for the positive and negative samples were 83.3% and 100%, compared to the AST results (Fig. 5B). Notably, there were two “equivocal” samples revealed by the MP-CMSA assay, among which one was positive and the other was negative in AST. This inconsistency deserves further analysis in the future. Moreover, due to the limitation of the sample size, the cutoff value and the three-zone classification determined here needed further investigation with a large sample size. Moreover, more BLA-associated DRB infections were observed in patients aged over 65 (61.1%, data not shown), confirming that geriatric patients are more easily colonized or infected with BLA-associated DRB. This clinical phenomenon may be associated with nutritional deprivation or immunological impairment in those patients.47
4. Conclusions
In summary, we have developed a mass-tagged probe, MP-CMSA, with enzyme- and light-assisted cascaded signal amplification to convert and amplify the level of BLA into the signal of the mass tag. In this probe, the BLA substrate was used as the recognition part, and the peptide was used as the mass tag. The dendrimer structure and photo- and enzyme-cleavable characteristics of the MP-CMSA probe were employed for signal amplification. After cascaded amplification, the detection sensitivity of BLA can be effectively enhanced by four orders of magnitude, and as low as 50.0 fM BLA in blood samples can be detected. The results also demonstrated that geriatric patients were easily colonized or infected with BLA-associated DRB. Furthermore, this assay enabled the measurement of BLA directly from patient blood samples within a few hours without the need for bacteria culturing and thus could advance the diagnostic report by two to three days. Following the same strategy, other DRB or other types of pathogens could also be detected only by replacing the BLA recognition moiety with carbapenemase, extended-spectrum BLA-specific substrates, or other enzyme-specific substrates. Overall, the mass-tagged probe-mediated cascaded signal amplification strategy provides a potential approach for facilitating the sensitive and rapid diagnosis of DRB and timely selection of suitable therapeutic regimens.Ethical approval
This study was approved by the Institutional Review Board of Nanjing Medical University, Nanjing, China.Data availability
All the data supporting this article have been included in the main text and the ESI material.†Author contributions
Jianhua Zhu: conceptualization, methodology, investigation, writing-original draft. Yunfei Bai: methodology, investigation, writing-original draft. Xiuyu Chen: methodology, validation, formal analysis. Linlin Hu: resources. Wenjun Zhang: methodology, validation, formal analysis. Chunyan Liu: methodology, validation, formal analysis. Hua Shao: resources, visualization, writing-review & editing. Jianguo Sun: data curation, writing-review & editing. Yun Chen: conceptualization, project administration, funding acquisition, writing-review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this article.Acknowledgements
The National Natural Science Foundation of China (22174068 and 21722504), the Natural Science Foundation of Jiangsu Province (BK20221303), the Jiangsu Provincial Key Research and Development Program (BE2022796), the SEU-NJMU-CPU Cooperation Project (2242019K3DNZ2), the Open Foundation of State Key Laboratory of Reproductive Medicine (SKLRM-2022BP1), and the Science and Technology Development Fundation of NJMU (NMUB2019014, NJMUQY2022003) awarded to Dr Chen, the Leading Technology Foundation Research Project of Jiangsu Province (BK20192005) awarded to Dr Sun, and the Changzhou Siyao Hospital Pharmaceutical Foundation of Nanjing Pharmaceutical Association (2019YX012) awarded to Dr Shao are gratefully acknowledged.Notes and references
- S. B. Levy and B. Marshall, Nat. Med., 2004, 10, S122–S129 CrossRef CAS.
- World Health Organization, Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2021, available at: https://creativecommons.org/licenses/by-nc-sa/3.0/igo Search PubMed.
- T. M. Rawson, D. Ming, R. Ahmad, L. S. P. Moore and A. H. Holmes, Nat. Rev. Microbiol., 2020, 18, 409–410 CrossRef CAS.
- G. Quero, S. Zuppolini, M. Consales, L. Diodato, P. Vaiano, A. Venturelli, M. Santucci, F. Spyrakis, M. P. Costi, M. Giordano, A. Borriello, A. Cutolo and A. Cusano, Sens. Actuators, B, 2016, 230, 510–520 CrossRef CAS.
- N. G. Bonine, A. Berger, A. Altincatal, R. Wang, T. Bhagnani, P. Gillard and T. Lodise, Am. J. Med. Sci., 2019, 357, 103–110 CrossRef.
- A. P. Macgowan and BSAC Working Parties on Resistance Surveillance, J. Antimicrob. Chemother., 2008, 62(2), 105–114 Search PubMed.
- J. Xie, R. Mu, M. Fang, Y. Cheng, F. Senchyna, A. Moreno, N. Banaei and J. Rao, Chem. Sci., 2021, 12, 9153–9161 RSC.
- I. C. Gyssens, Int. J. Antimicrob. Agents, 2011, 38, 11–20 CrossRef CAS PubMed.
- R. Sugden, R. Kelly and S. Davies, Nat. Microbiol., 2016, 1, 16187 CrossRef CAS.
- A. van Belkum, C.-A. D. Burnham, J. W. A. Rossen, F. Mallard, O. Rochas and W. M. Dunne, Nat. Rev. Microbiol., 2020, 18, 299–311 CrossRef CAS.
- C. Hu, S. Kalsi, I. Zeimpekis, K. Sun, P. Ashburn, C. Turner, J. M. Sutton and H. Morgan, Biosens. Bioelectron., 2017, 96, 281–287 CrossRef CAS.
- H. B. Thai, J. K. Yu, B. S. Park, Y. J. Park, S. J. Min and D. R. Ahn, Biosens. Bioelectron., 2016, 77, 1026–1031 CrossRef CAS.
- Y. Ding, Z. Li, C. Xu, W. Qin, Q. Wu, X. Wang, X. Cheng, L. Li and W. Huang, Angew. Chem., Int. Ed. Engl., 2021, 60, 24–40 CrossRef CAS.
- L. Cerqueira, R. M. Fernandes, R. M. Ferreira, M. Oleastro, F. Carneiro, C. Brandao, P. Pimentel-Nunes, M. Dinis-Ribeiro, C. Figueiredo, C. W. Keevil, M. J. Vieira and N. F. Azevedo, J. Clin. Microbiol., 2013, 51, 1887–1893 CrossRef PubMed.
- M. F. Anjum, E. Zankari and H. Hasman, Microbiol. Spectrum, 2017, 5, 33–50 Search PubMed.
- F. Xu, W. Zhou, J. Cao, Q. Xu, D. Jiang and Y. Chen, Theranostics, 2017, 7, 2849–2862 CrossRef CAS PubMed.
- M. Oviaño and G. Bou, Clin. Microbiol. Rev., 2018, 32, e00037–e00018 CrossRef PubMed.
- M. Oviano, C. L. Ramirez, L. P. Barbeyto and G. Bou, J. Antimicrob. Chemother., 2017, 72, 1350–1354 CAS.
- CLSI M58, methods for the identification of cultured microorganisms using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry, Clinical and Laboratory Standards Institute, 1st edn, 2017 Search PubMed.
- Y. Hu, Z. Wang, L. Liu, J. Zhu, D. Zhang, M. Xu, Y. Zhang, F. Xu and Y. Chen, Chem. Sci., 2021, 12, 7993–8009 RSC.
- Y. Kuang, J. Cao, F. Xu and Y. Chen, Anal. Chem., 2019, 91, 8820–8826 CrossRef CAS PubMed.
- H. Gröger, M. Pieper, B. König, T. Bayer and H. Schleich, Sustainable Chem. Pharm., 2017, 5, 72–79 CrossRef.
- S. Zhou, L. Yuan, X. Hua, L. Xu and S. Liu, Anal. Chim. Acta, 2015, 877, 19–32 CrossRef CAS.
- M. A. Swiderska and J.-L. Reymond, Nat. Chem., 2009, 1, 527–528 CrossRef CAS PubMed.
- W. Zhou, F. Xu, D. Li and Y. Chen, Clin. Chem., 2018, 64, 526–535 CrossRef CAS.
- Y. Kuang, L. Liu, Z. Wang and Y. Chen, Talanta, 2020, 211, 120726 CrossRef CAS.
- S. Xu, W. Ma, Y. Bai and H. Liu, J. Am. Chem. Soc., 2019, 141, 72–75 CrossRef CAS.
- P. Klan, T. Solomek, C. G. Bochet, A. Blanc, R. Givens, M. Rubina, V. Popik, A. Kostikov and J. Wirz, Chem. Rev., 2013, 113, 119–191 CrossRef CAS PubMed.
- T. Kakiyama, K. Usui, K.-y. Tomizaki, M. Mie, E. Kobatake and H. Mihara, Polym. J., 2013, 45, 535–539 CrossRef CAS.
- L. Liu, Y. Kuang, Z. Wang and Y. Chen, Chem. Sci., 2020, 11, 11298–11306 RSC.
- M. Rinnová, M. Nováková, V. Kašička and J. Jiráček, J. Pept. Sci., 2000, 6, 355–365 CrossRef.
- R. Satchi-Fainaro, H. Hailu, J. W. Davies, C. Summerford and R. Duncan, Bioconjugate Chem., 2003, 14, 797–804 CrossRef CAS.
- A. M. Egorov, M. M. Ulyashova and M. Y. Rubtsova, Biochemistry, 2020, 85, 1292–1309 CAS.
- T. R. deBoer, N. J. Tarlton, R. Yamaji, S. Adams-Sapper, T. Z. Wu, S. Maity, G. K. Vesgesna, C. M. Sadlowski, P. t. DePaola, L. W. Riley and N. Murthy, Chembiochem, 2018, 19, 2173–2177 CrossRef CAS.
- W. Zhou, L. Hu, L. Ying, Z. Zhao, P. K. Chu and X. F. Yu, Nat. Commun., 2018, 9, 5012 CrossRef PubMed.
- P. S. Saudagar, S. A. Survase and R. S. Singhal, Biotechnol. Adv., 2008, 26, 335–351 CrossRef CAS PubMed.
- B. D. Bennett, E. H. Kimball, M. Gao, R. Osterhout, S. J. Van Dien and J. D. Rabinowitz, Nat. Chem. Biol., 2009, 5, 593–599 CrossRef CAS PubMed.
- U. T. Bornscheuer, FEMS Microbiol. Rev., 2002, 26, 73–81 CrossRef CAS.
- N. Singh and D. Bhattacharyya, Microbiol. Immunol., 2015, 59, 311–321 CrossRef CAS PubMed.
- M. Dabek, S. I. McCrae, V. J. Stevens, S. H. Duncan and P. Louis, FEMS Microbiol. Ecol., 2008, 66, 487–495 CrossRef CAS.
- M. Frieri, K. Kumar and A. Boutin, Journal of Infection and Public Health, 2017, 10, 369–378 CrossRef.
- P. T. Cherian, M. N. Cheramie, R. K. R. Marreddy, D. M. Fernando, J. G. Hurdle and R. E. Lee, Bioorg. Med. Chem. Lett., 2018, 28, 3105–3112 CrossRef CAS.
- T. Tian, J. Yi, Y. Liu, B. Li, Y. Liu, L. Qiao, K. Zhang and B. Liu, Biosens. Bioelectron., 2022, 197, 113778 CrossRef CAS PubMed.
- H. S. Sader, R. E. Mendes, S. J. R. Arends, C. G. Carvalhaes and M. Castanheira, Eur. J. Clin. Microbiol. Infect. Dis., 2022, 41, 477–487 CrossRef CAS.
- T. Mazzu-Nascimento, G. G. Morbioli, L. A. Milan, F. C. Donofrio, C. A. Mestriner and E. Carrilho, Anal. Chim. Acta, 2017, 950, 156–161 CrossRef CAS PubMed.
- R. Fluss, D. Faraggi and B. Reiser, Biom. J., 2005, 47, 458–472 CrossRef.
- N. S. Ku, Y. C. Kim, M. H. Kim, J. E. Song, D. H. Oh, J. Y. Ahn, S. B. Kim, H.-w. Kim, S. J. Jeong, S. H. Han, C. O. Kim, Y. G. Song, J. M. Kim and J. Y. Choi, Arch. Gerontol. Geriatr., 2014, 58, 105–109 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: ESI table (S1), ESI figures (S1–S13) and ESI experimental information (S1–S14) including materials and reagents, instruments, MS analysis, synthesis routes for chemicals, characterization of products, optimization of experimental conditions, bacterial strains and analysis, preparation of stock solutions, calibration standards and quality controls (QCs), BLA inhibition assay, validation results of the assay, representative AST results, and LC-MS/MS chromatograms of calibration standards and the patient samples. See DOI: https://doi.org/10.1039/d2sc01530g |
| ‡ J. Z. and Y. B. contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |