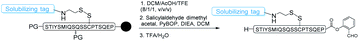Open Access Article
Open Access ArticleEnabling chemical protein (semi)synthesis via reducible solubilizing tags (RSTs)†
Jiamei
Liu
,
Tongyao
Wei
,
Yi
Tan
,
Heng
Liu
and
Xuechen
Li
 *
*
Department of Chemistry, State Key Lab of Synthetic Chemistry, The University of Hong Kong, Hong Kong. E-mail: xuechenl@hku.hk
First published on 28th December 2021
Abstract
Chemical synthesis of proteins with poor solubility presents a challenging task. The existing solubilizing tag strategies are not suitable for the expressed protein segment. To address this issue, we report herein that solubilizing tags could be introduced at the side chain of the peptide and C-terminal peptide salicylaldehyde esters via a disulfide linker. Such reducible solubilizing tags (RSTs) are compatible with peptide salicylaldehyde ester-mediated Ser/Thr ligation and Cys/Pen ligation for purifying and ligating peptides with poor solubility. This strategy features operational simplicity and readily accessible materials. Both the protein 2B4 cytoplasmic tail and FCER1G protein have been successfully synthesized via this strategy. Of particular note, the RST strategy could be used for solubilizing the expressed protein segment for protein semi-synthesis of the HMGB1 protein.
Introduction
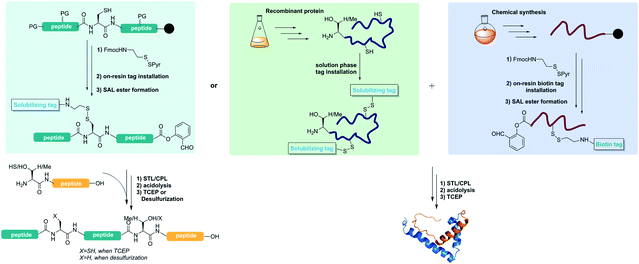
Protein chemical synthesis can be used to generate proteins which are difficult to generate by biologically controlled methods, such as proteins with site-specific modifications or unnatural amino acids, mirror image proteins and so on. Contemporary chemical protein synthesis involves mainly two steps: (1) preparation of short peptide segments by solid phase peptide synthesis (SPPS). SPPS normally works well for the synthesis of peptides with less than 50 amino acids but becomes difficult for longer peptides due to its linear synthesis feature; and (2) chemoselective peptide ligation joining the short peptide segments together to form longer peptides of protein domain sizes.1,2 The peptide ligation allows for peptide assembly with side chain unprotected peptides specifically at the C-/N-terminus with high chemo-, regio- and stereo-selectivity.3,4 With the advancement in the ligation toolbox over the past few decades, including native chemical ligation (NCL) and its variant,5 KAHA ligation,6 Ser/Thr ligation (STL)7–17 and Cys/Pen ligation (CPL),18 there are many options for designing protein retrosynthetic strategies. One of the often-met problems is the poor solubility of the peptide segments for ligation.19–21 Generally, the ligation can proceed smoothly when the peptides are well dissolved in the ligation solution. However, some peptides with hydrophobic residues or a tendency for self-assembly are very difficult for reversed phase HPLC purification.22–24 Furthermore, they have poor solubility in ligating buffers, making the ligation difficult, if not impossible. Of course, membrane proteins are extremely difficult for total synthesis and handling.25To overcome the problems of insoluble peptides during chemical protein synthesis, different methods have been developed over the past few decades to introduce solubilizing tags into the peptides,26–31 including C-terminal thioesters with solubilizing tags,32–34 removable backbone modifications,35 Lys-side chain helping hands,36 and so on. After the peptide synthesis and ligation assembly, these solubilizing tags can be removed to provide native primary proteins. In addition, the disulfide bond linkage with polyethylene glycol or other solubilizing tags was used for solid phase peptide synthesis with the transmembrane segment.37–41 For instance, Deber's group developed a PEG-α-Cys reagent and successfully introduced it into the ALC peptide via the disulfide bond.37 All these reported methods for incorporating solubilizing tags have their own scope and limitations for chemical protein synthesis. They were only used for transiently adding solubilizing tags. However, the stability of these linkers during the chemical ligation step remains unclear and the application of this method in the protein synthesis needs more exploration. Of particular note, the incorporation of these solubilizing tags via the abovementioned C-terminal thioesters, peptide backbone or Lys side chain relies on the de novo peptide synthesis, thus it is not compatible with expressed protein segments. Herein, we report a reducible solubilizing tag (RST) strategy which is compatible with Ser/Thr ligation and Cys/Pen ligation for chemical protein synthesis and semisynthesis.
Results and discussion
General design of the reducible solubility tag strategy
The RST strategy allows for the late-stage installing of solubilizing tags, making it possible for solubilizing expressed protein segments. Ser/Thr ligation and Cys/Pen ligation conditions do not involve reducing agents,17,18 therefore we envisaged that the solubilizing tags could be incorporated into the peptide via a disulfide linker which would be stable during the ligation process. After the ligation step, the solubilizing tags could be readily removed upon TCEP treatment or desulfurization,42 regenerating the free cysteine or alanine (Fig. 1). Towards this goal, FmocHN-Cys(StBu)-COOH or FmocHN-Cys(SMe)-COOH was used as the building block for Fmoc-Solid Phase Peptide Synthesis (SPPS). After peptide assembly, the StBu or SMe protecting group was selectively removed on the resin with EDT/DIEA/DMF (2/1/7, v/v/v) for 4–12 h. Next, the resin was treated with a solution of (9H-fluoren-9-yl)methyl 2-(pyridin-2-yldisulfanyl)ethylcarbamate,43 which reacted with the thiol group of Cys on resin-bound peptides in DCM for 4–12 h to install the disulfide linker. After removing the Fmoc group on the linker, different solubilizing tags, PEG tag,37,44 polyLys tag33,38,39,45,46 and polyArg tag,29,34,47–49 could be introduced into peptides successfully by Fmoc SPPS. After the ligation, the reducible solubilizing tags were detached with 0.2 M TCEP in pH ∼7 PB buffer (0.2 M PB solution, containing 6 M Gn·HCl) or ACN/H2O for 30 min to 1 h. It should be noted that the solubilizing tags could also be removed during the free radical based desulfurization step, highlighting the simplicity of this method by combining with CPL-desulfurization (Fig. 1).Installation of various solubilizing tags into difficult peptides by the RST strategy
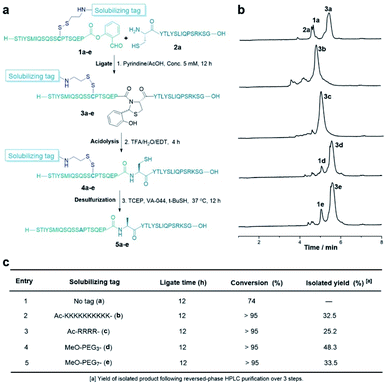
First, we used a model hydrophobic peptide STIYSMIQSQSSAPTSQEP taken from the protein 2B4 sequence to explore this strategic design. This sequence does not contain a cysteine residue, so the alanine residue was mutated into cysteine which is used to introduce the solubilizing tag. As shown in Table 1, the synthesis of the peptide salicylaldehyde ester 1a without a solubilizing tag was very difficult, mainly due to the limited solubility during the purification step, and only 6% yield was obtained after purification. As a contrast, incorporating the tags such as Lys10, Arg4, PEG3 or PEG7 into the peptides could dramatically improve the recovery yield of the peptides 1b–e (15–24%).Furthermore, all these tags improved the solubility of peptide SAL esters (1b–e) in the ligation buffer, which ligated with a peptide with N-terminal Cys to form the desired peptide under the Cys/Pen ligation conditions (Fig. 2a). Surprisingly, the disulfide linkage was stable under the CPL conditions, not leading to the disulfide exchange with the N-terminal Cys residue. As analyzed by liquid chromatography mass spectrometry (LCMS), the CPL of the tagged peptides gave better conversions than that using untagged peptide (Fig. 2b). After ligation and desulfurization, the desired peptides were obtained in 25–48% after HPLC purification over 3 steps (Fig. 2c).
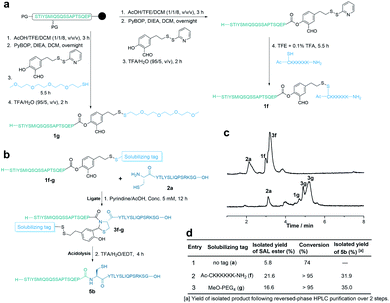
To overcome the limitation of this strategy requiring Cys or Ala residue side chains for incorporating RSTs, we designed the second generation, in which the solubilizing tag was incorporated into the peptide via the salicylaldehyde ester (Fig. 3a). First, 2-hydroxy-5-(2-(pyridin-2-yldisulfaneyl)ethyl)benzaldehyde was synthesized (see details in the ESI†) and used for the preparation of peptide salicylaldehyde esters. As shown in Fig. 3a, the fully protected crude peptide was dissolved in dry DCM and coupled with 2-hydroxy-5-(2-(pyridin-2-yldisulfaneyl)ethyl)benzaldehyde in the presence of PyBOP and DIEA. Then, the PEG4 tag (tetra-ethylene glycol monomethyl ether thiol) was added into the reaction mixture and reacted for 5.5 h. Finally, after global deprotection and HPLC purification, 1g was obtained in a 17% isolated yield. In a similar way, peptide salicylaldehyde ester 1f with the polyLys-tag (Ac-CKKKKKK-NH2) could also be prepared. However, due to the poor solubility of the polyLys tag in DCM, it was attached to the peptide salicylaldehyde ester in TFE (containing 0.1% TFA) after global deprotection with TFA. Overall, solubilizing tags with the thiol group can be successfully installed into the peptide salicylaldehyde ester before the HPLC purification step. The peptide salicylaldehyde ester with the PEG4 tag and Lys6 tag showed a remarkably improved isolated yield (Fig. 3d). Similar to the side chain tagging strategy, the solubilizing tag at the salicylaldehyde ester also improved the ligation process and purification (Fig. 3). After acidolysis, the salicylaldehyde with the solubilizing tags was removed to afford the native peptide sequence (Fig. 3b and c). Although the solubilizing tags could be introduced into the salicylaldehyde ester via other linkages (e.g., esters and amides), the usage of the disulfide linkage here allows for late-stage incorporation of different tags.
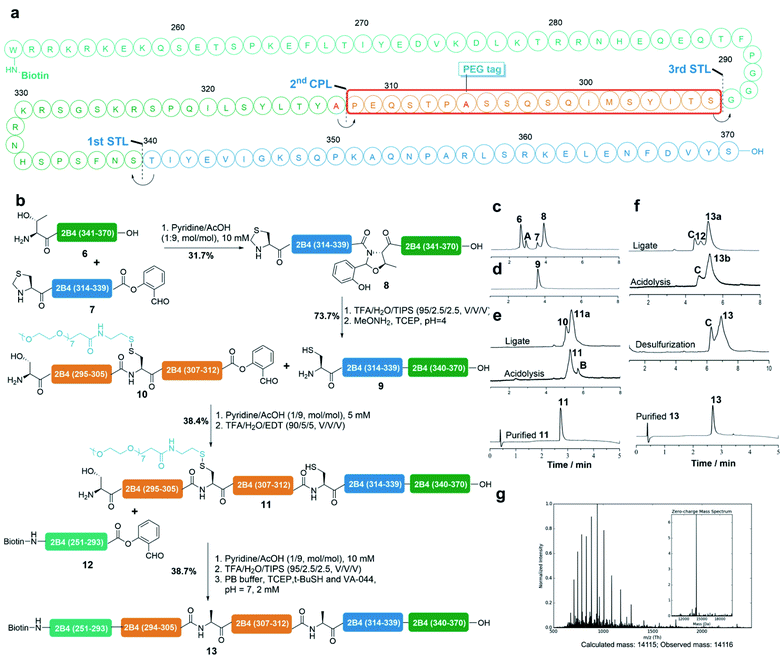
Synthesis of protein 2B4
We demonstrated the application of this strategy for the total synthesis of the protein 2B4 cytoplasmic tail. 2B4 (CD244) is a human protein encoded by the CD244 gene and it is also known as natural killer cell receptor 2B4.50–52 As displayed in Fig. 4a, our synthetic plan involves three sequential ligations from the C-terminus to the N-terminus: STL at the S339-T340 site, CPL at the Pro312-Ala313(mutated to Cys) site and STL at the G293-S294 site. Four peptide fragments (6, 7, 10 and 12) were synthesized by Fmoc-SPPS (see the ESI† for details). Notably, we installed the reducible PEG7 tag at the Ala296 (mutated to Cys) site via our reducible solubilizing tag strategy and successfully obtained peptide 10 in a 21% isolated yield by HPLC purification. The 1st STL between peptide 6 and 7 at the S339-T340 site occurred smoothly in pyridine/acetic acid (1/9, mol mol−1) at a concentration of 5 mM at room temperature and was completed in 12 h (Fig. 4c). However, the unreacted 7 was coeluted with peptide 9 during the subsequent Thz (thia-zolidine) opening step, thus peptide 8 was purified before acidolysis and then subjected to the Thz-opening buffer (Fig. 4d). Product 9 was purified by HPLC in a 74% isolated yield and identified by ESI-MS (Fig. S27†). The subsequent 2nd CPL of 9 (1 equiv., 5 mM) and 10 (1 equiv., 5 mM) was also performed in pyridine/acetic acid (1/9, mol mol−1) for 12 h at room temperature. It was noted that peptide 10 has good solubility in the ligation buffer (pyridine/acetic acid) and as a result, the CPL occurred very well and generated the ligated intermediate 11a with high conversion. The crude ligate intermediate was then treated with TFA/EDT/H2O for 4 h to generate peptide 11 with the Pro-Cys linkage, which after HPLC purification was obtained in a 38% isolated yield (Fig. 4e). The last STL between peptide 11 (1 equiv., 5 mM) and 12 (2 equiv., 10 mM) was conducted in pyridine/acetic acid (1/9, mol mol−1) at room temperature for 12 h, which generated the ligated intermediate 13a with very high conversion. Without purification peptide 13a was subjected to acidolysis with TFA/H2O/TIPS for 30 min, regenerating the peptide 13b native Gly-Ser linkage. Finally, the PEG7 tag was removed in situ under the desulfurization conditions (TCEP, VA-044 and tBuSH), affording the native 2B4 protein tail (13) in a 39% isolated yield over one pot 3 steps (Fig. 4f). Peptide 13 was characterized by ESI-MS with its high purity (Fig. 4g).Synthesis of protein FCER1G
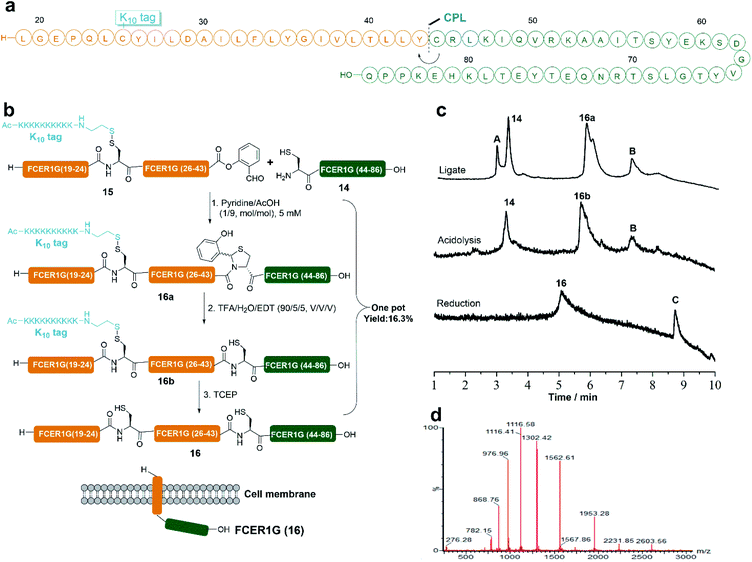
To showcase the power of our RST strategy, we also applied it in the synthesis of membrane protein FCER1G. FCER1G (human) serves as an adaptor protein and can signal allergic inflammation.53 To synthesize the protein FCER1G, the main difficulty is the hydrophobic nature of its membrane peptide fragment (Leu19 to Tyr43), which brought huge challenges to the preparation of this fragment by HPLC purification. Indeed, it was found that transmembrane peptide fragment could not even be detected by LC-MS without the tag modification due to its hydrophobic nature (Fig. S35†). We then envisioned to disconnect FCER1G at the Tyr43-Cys44 site and synthesized the protein by CPL between peptide 14 and 15 (Fig. 5a). As expected, attachment of the Lys10 tag at the Cys25 site of the transmembrane peptide fragment made the purification of the peptide fragment by HPLC become feasible. Peptide 15 was obtained in a 7.3% HPLC isolated yield. The CPL of 14 (1 equiv., 5 mM) and 15 (1 equiv., 5 mM) was performed in pyridine/acetic acid (1/9, mol mol−1) for 12 h at room temperature and 16a was produced successfully. The crude ligate intermediate 16a was treated with TFA/EDT/H2O for 1 h to generate peptide 16b with the Tyr-Cys linkage, which without HPLC purification was subjected to TCEP reduction to remove the PolyLys tag. FCER1G (16) was obtained in a 16.3% HPLC isolated yield in one pot over 3 steps (Fig. 5).Semi-synthesis of protein HMGB1
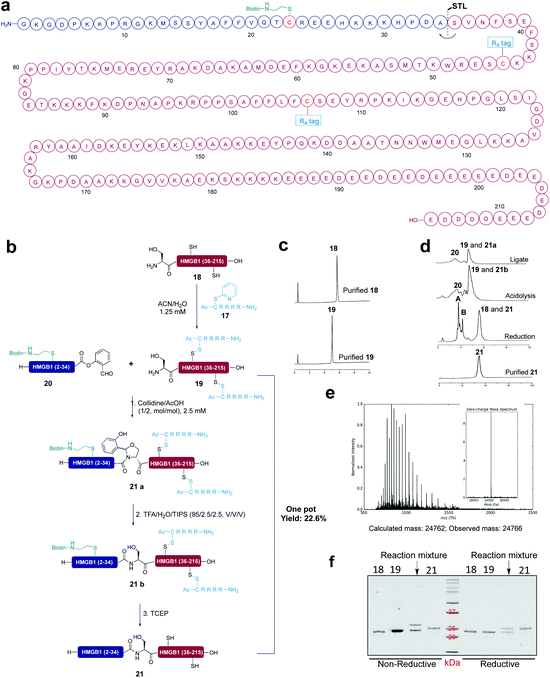
All the existing peptide solubilizing strategies based on the backbone and side chain modification cannot be used for the peptide segments obtained from expression systems. It will be advantageous to incorporate the solubilizing tag into the expressed protein segment for the following ligation. To this end, we chose high mobility group protein B1 (HMGB1) (human) as the semi-synthetic target. HMGB1 is a non-histone protein related to chromatin, which plays important roles as DNA chaperone and extracellular inflammatory cytokine.54 To realize the semi-synthesis of protein HMGB1, we divided HMGB1 at Ala34-Ser35 into two fragments and ligated them by STL (Fig. 6a). First, we conducted the control experiment. Expressed protein segment 18 (1 equiv., 5.0 mM) and segment 20 (5 equiv., 25 mM) were dissolved in the pyridine/acetic acid or collidine/acetic acid (1/2, mol mol−1) and reacted for 6 h, followed by treatment with TFA/TIPS/H2O for 30 min and 20 mM TCEP for 1 h. However, we found that expressed protein segment 18 has poor solubility in both solvent systems and only a trace of the product was detected by LC-MS (Fig. S46†). In this context, we incorporated two reducible Arg4 tags into expressed protein segment 18 at the Cys45 site and Cys106 site in the solution phase to improve the solubility of expressed protein segment 18 both in ACN/H2O and the ligation solvent, in order to improve the ligation efficiency.The expressed protein segment 18 and solubilizing tag 17 were dissolved in ACN/H2O (3/1, v/v) at room temperature and reacted for 12 h. After that, the expressed protein fragment with Arg4 solubilizing tag 19 was obtained in quantitative yield, which was successfully detected by LC-MS (Fig. S42†). The SDS-page analysis showed the band of 19, which has a higher molecular weight than expressed protein segment 18. In addition, based on our recent studies on protein-semisynthesis with Ser/Thr ligation,55 we installed the reducible biotin tag into peptide 20 at the Cys23 site to achieve the purification of HMGB1 (21) by using the streptavidin resin. The STL of 19 (1 equiv., 5.0 mM) and 20 (5 equiv., 25 mM) was performed in collidine/acetic acid (1/2, mol mol−1) for 6 h at room temperature and 21a was obtained successfully in a 43% conversion (less peptide salicylaldehyde ester hydrolysis was observed in the collidine/acetic acid mixture than in the pyridine/acetic acid mixture), followed by treatment with TFA/TIPS/H2O for 30 min to generate peptide 21b with the Ala-Ser linkage. The resulting residue was refolded by dissolving in 6 M guanidine, followed by 10 times dilution in the refolding buffer (50 mM Tris–HCl, pH 7.5, 500 mM NaCl). After purification by using the size exclusion column and then the streptavidin resin, HMGB1 (21) was successfully obtained in a 22.7% yield by directly treating the resin with 20 mM TCEP in the refolding buffer for 1 h at room temperature. The final product protein HMGB1 was detected by LC-MS (Fig. S45†) and SDS-page showed the band of 21 (Fig. 6).
Conclusions
In summary, chemical synthesis of proteins via peptide ligation approaches requires accessing suitable peptide fragments, which is often plagued with peptides having poor solubility for purification and ligation. Incorporation of solubilizing tags can effectively address this problem, which, however, demands easy-to-use tagging and de-tagging strategies. In this regard, we introduced the reducible solubilizing tag (RST) strategy for synthesis of proteins with poor solubility, in which the solubilizing tags (i.e., PEG tag, polyLys tag and polyArg tag) can be readily incorporated into the peptide via disulfide linkers of the internal cysteine or alanine (via desulfurization), and the salicylaldehyde ester. The process of incorporating RSTs into peptides is operationally simple and this strategy fits well with Ser/Thr ligation and Cys/Pen ligation, solving the problem of peptides with limited solubility for purification and ligation. More importantly, the RST strategy can be successfully used for expressed peptides containing internal cysteine residues to enable chemical protein synthesis and semi-synthesis.Data availability
All data needed to be evaluated the conclusions in the paper are available in the paper or the ESI.†Author contributions
XL conceived and supervised the project. JL, YT and HL designed the methodology and experiments. JL conducted the peptide synthesis, condition screening, solubilizing tag attachment and detachment, peptide ligation and protein synthesis. TW expressed and purified proteins. XL and JL wrote the original draft. All the authors reviewed the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Research Grants Council of Hong Kong (17302621, C7017-18G, C7147-20G, and AoE/P-705/16).References
- S. S. Kulkarni, J. Sayers, B. Premdjee and R. J. Payne, Nat. Rev. Chem., 2018, 2, 0122 CrossRef CAS.
- Y. Tan, H. Wu, T. Wei and X. Li, J. Am. Chem. Soc., 2020, 142, 20288–20298 CrossRef CAS PubMed.
- A. C. Conibear, E. E. Watson, R. J. Payne and C. F. W. Becker, Chem. Soc. Rev., 2018, 47, 9046–9068 RSC.
- P. Dawson, T. Muir, I. Clark-Lewis and S. Kent, Science, 1994, 266, 776–779 CrossRef CAS PubMed.
- V. Agouridas, O. El Mahdi, V. Diemer, M. Cargoët, J.-C. M. Monbaliu and O. Melnyk, Chem. Rev., 2019, 119, 7328–7443 CrossRef CAS PubMed.
- J. W. Bode, Acc. Chem. Res., 2017, 50, 2104–2115 CrossRef CAS PubMed.
- H. Liu and X. Li, Acc. Chem. Res., 2018, 51, 1643–1655 CrossRef CAS PubMed.
- Y. Zhang, C. Xu, H. Y. Lam, C. L. Lee and X. Li, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 6657–6662 CrossRef CAS PubMed.
- X. Li, H. Y. Lam, Y. Zhang and C. K. Chan, Org. Lett., 2010, 12, 1724–1727 CrossRef CAS PubMed.
- D. Chen, K. H. L. Po, P. Blasco, S. Chen and X. Li, Org. Lett., 2020, 22, 4749–4753 CrossRef CAS PubMed.
- C. T. T. Wong, T. Li, H. Y. Lam, Y. Zhang and X. Li, Front. Chem., 2014, 2, 28 Search PubMed.
- C. L. Lee and X. Li, Curr. Opin. Chem. Biol., 2014, 22, 108–114 CrossRef CAS PubMed.
- P. M. Levine, T. W. Craven, R. Bonneau and K. Kirshenbaum, Org. Lett., 2014, 16, 512–515 CrossRef CAS PubMed.
- H. Y. Lam, Y. Zhang, H. Liu, J. Xu, C. T. T. Wong, C. Xu and X. Li, J. Am. Chem. Soc., 2013, 135, 6272–6279 CrossRef CAS PubMed.
- C. Xu, J. Xu, H. Liu and X. Li, Chin. Chem. Lett., 2018, 29, 1119–1122 CrossRef CAS.
- Z. Sun, Z. Shang, N. Forelli, K. H. L. Po, S. Chen, S. F. Brady and X. Li, Angew. Chem., Int. Ed., 2020, 59, 19868–19872 CrossRef CAS PubMed.
- C. L. Lee, H. Liu, C. T. T. Wong, H. Y. Chow and X. Li, J. Am. Chem. Soc., 2016, 138, 10477–10484 CrossRef CAS PubMed.
- Y. Tan, J. Li, K. Jin, J. Liu, Z. Chen, J. Yang and X. Li, Angew. Chem., Int. Ed., 2020, 59, 12741–12745 CrossRef CAS PubMed.
- T. Li, H. Liu and X. Li, Org. Lett., 2016, 18, 5944–5947 CrossRef CAS PubMed.
- D.-L. Huang, C. Montigny, Y. Zheng, V. Beswick, Y. Li, X.-X. Cao, T. Barbot, C. Jaxel, J. Liang, M. Xue, C.-L. Tian, N. Jamin and J.-S. Zheng, Angew. Chem., Int. Ed., 2020, 59, 5178–5184 CrossRef CAS PubMed.
- B. Zhang, Q. Deng, C. Zuo, B. Yan, C. Zuo, X.-X. Cao, T. F. Zhu, J.-S. Zheng and L. Liu, Angew. Chem., Int. Ed., 2019, 58, 12231–12237 CrossRef CAS PubMed.
- M. Paradís-Bas, J. Tulla-Puche and F. Albericio, Chem. Soc. Rev., 2016, 45, 631–654 RSC.
- Y. Asahina, S. Komiya, A. Ohagi, R. Fujimoto, H. Tamagaki, K. Nakagawa, T. Sato, S. Akira, T. Takao, A. Ishii, Y. Nakahara and H. Hojo, Angew. Chem., Int. Ed., 2015, 54, 8226–8230 CrossRef CAS PubMed.
- C. E. Murar, M. Ninomiya, S. Shimura, U. Karakus, O. Boyman and J. W. Bode, Angew. Chem., Int. Ed., 2020, 59, 8425–8429 CrossRef CAS PubMed.
- C. Zuo, S. Tang and J.-S. Zheng, J. Pept. Sci., 2015, 21, 540–549 CrossRef CAS PubMed.
- Y.-C. Huang, Y.-M. Li, Y. Chen, M. Pan, Y.-T. Li, L. Yu, Q.-X. Guo and L. Liu, Angew. Chem., Int. Ed., 2013, 52, 4858–4862 CrossRef CAS PubMed.
- Z. Tan, S. Shang and S. J. Danishefsky, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4297–4302 CrossRef CAS PubMed.
- C. Bello, S. Wang, L. Meng, K. W. Moremen and C. F. W. Becker, Angew. Chem., Int. Ed., 2015, 54, 7711–7715 CrossRef CAS PubMed.
- S. K. Maity, G. Mann, M. Jbara, S. Laps, G. Kamnesky and A. Brik, Org. Lett., 2016, 18, 3026–3029 CrossRef CAS PubMed.
- S. Bondalapati, E. Eid, S. M. Mali, C. Wolberger and A. Brik, Chem. Sci., 2017, 8, 4027–4034 RSC.
- S. Tsuda, M. Mochizuki, H. Ishiba, K. Yoshizawa-Kumagaye, H. Nishio, S. Oishi and T. Yoshiya, Angew. Chem., Int. Ed., 2018, 57, 2105–2109 CrossRef CAS PubMed.
- J.-X. Wang, G.-M. Fang, Y. He, D.-L. Qu, M. Yu, Z.-Y. Hong and L. Liu, Angew. Chem., Int. Ed., 2015, 54, 2194–2198 CrossRef CAS PubMed.
- E. C. B. Johnson, E. Malito, Y. Shen, D. Rich, W.-J. Tang and S. B. H. Kent, J. Am. Chem. Soc., 2007, 129, 11480–11490 CrossRef CAS PubMed.
- E. C. B. Johnson and S. B. H. Kent, Tetrahedron Lett., 2007, 48, 1795–1799 CrossRef CAS PubMed.
- J.-B. Li, S. Tang, J.-S. Zheng, C.-L. Tian and L. Liu, Acc. Chem. Res., 2017, 50, 1143–1153 CrossRef CAS PubMed.
- M. T. Jacobsen, M. E. Petersen, X. Ye, M. Galibert, G. H. Lorimer, V. Aucagne and M. S. Kay, J. Am. Chem. Soc., 2016, 138, 11775–11782 CrossRef CAS PubMed.
- N. C. Pomroy and C. M. Deber, Anal. Biochem., 1999, 275, 224–230 CrossRef CAS PubMed.
- M. A. Hossain, A. Belgi, F. Lin, S. D. Zhang, F. Shabanpoor, L. D. Chan, C. Belyea, H.-T. Truong, A. R. Blair, S. Andrikopoulos, G. W. Tregear and J. D. Wade, Bioconjugate Chem., 2009, 20, 1390–1396 CrossRef CAS PubMed.
- S. A. Abboud, E. h. Cisse, M. Doudeau, H. Bénédetti and V. Aucagne, Chem. Sci., 2021, 12, 3194–3201 RSC.
- K. M. Linn, M. G. Derebe, Y. Jiang and F. I. Valiyaveetil, Biochemistry, 2010, 21, 4450–4456 CrossRef PubMed.
- Y. Marsac, J. Cramer, D. Olschewski, K. Alexandrov and C. F. W. Becker, Bioconjugate Chem., 2006, 17, 1492–1498 CrossRef CAS PubMed.
- Q. Wan and S. J. Danishefsky, Angew. Chem., Int. Ed., 2007, 46, 9248–9252 CrossRef CAS PubMed.
- A. A. Volmer and E. M. Carreira, ChemBioChem, 2010, 11, 778–781 CrossRef CAS PubMed.
- Y. Ito and S. Manabe, Chem.-Eur. J., 2002, 8, 3076–3084 CrossRef CAS.
- A. C. Baumruck, D. Tietze, L. K. Steinacker and A. A. Tietze, Chem. Sci., 2018, 9, 2365–2375 RSC.
- S. Tsuda, H. Nishio and T. Yoshiya, Chem. Commun., 2018, 54, 8861–8864 RSC.
- P. W. Harris and M. Brimble, Synthesis, 2009, 20, 3460–3466 CrossRef.
- J.-S. Zheng, M. Yu, Y.-K. Qi, S. Tang, F. Shen, Z.-P. Wang, L. Xiao, L. Zhang, C.-L. Tian and L. Liu, J. Am. Chem. Soc., 2014, 136, 3695–3704 CrossRef CAS PubMed.
- J.-S. Zheng, Y. He, C. Zuo, X.-Y. Cai, S. Tang, Z. A. Wang, L.-H. Zhang, C.-L. Tian and L. Liu, J. Am. Chem. Soc., 2016, 138, 3553–3561 CrossRef CAS PubMed.
- P. Eissmann, L. Beauchamp, J. Wooters, J. C. Tilton, E. O. Long and C. Watzl, Blood, 2005, 105, 4722–4729 CrossRef CAS PubMed.
- J. L. Cannons, S. G. Tangye and P. L. Schwartzberg, Annu. Rev. Immunol., 2011, 29, 665–705 CrossRef CAS PubMed.
- C. S. Ma, K. E. Nichols and S. G. Tangye, Annu. Rev. Immunol., 2007, 25, 337–379 CrossRef CAS PubMed.
- W. Cao, D. B. Rosen, T. Ito, L. Bover, M. Bao, G. Watanabe, Z. Yao, L. Zhang, L. L. Lanier and Y. J. Liu, J. Exp. Med., 2006, 203, 1399–1405 CrossRef CAS PubMed.
- S. L. Yuan, Z. P. Liu, Z. R. Xu, J. Liu and J. Zhang, J. Hematol. Oncol., 2020, 13, 91–109 CrossRef PubMed.
- T. Wei, J. Liu, Y. Tan, R. Wei, J. Wang, H. Wu, Y. Tang and X. Li, bioRxiv, 2021 DOI:10.1101/biorxiv.2021.10.05.463167.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc06387a |
| This journal is © The Royal Society of Chemistry 2022 |