Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Two dimensional nanosheets as immunoregulator improve HIV vaccine efficacy†
Ye
Liu‡
 *b,
Yekkuni L.
Balachandran‡
a,
Zulan
Li‡
c,
Yulong
Cong
c,
Yiming
Shao
d and
Xingyu
Jiang
*b,
Yekkuni L.
Balachandran‡
a,
Zulan
Li‡
c,
Yulong
Cong
c,
Yiming
Shao
d and
Xingyu
Jiang
 *a
*a
aShenzhen Key Laboratory of Smart Healthcare Engineering, Department of Biomedical Engineering, Southern University of Science and Technology, No. 1088 Xueyuan Rd, Nanshan District, Shenzhen, Guangdong 518055, P. R. China. E-mail: jiang@sustech.edu.cn
bInstitute of Medical Biology, Chinese Academy of Medical Sciences and Peking Union Medical College, Kunming, Yunnan 650000, P. R. China. E-mail: liuye@imbcams.com.cn
cClinical Laboratory of South Building, Chinese P. L. A. General Hospital, No. 28 Fuxing Road, Beijing 100853, P. R. China
dState Key Laboratory of Infectious Disease Prevention and Control, National Center for AIDS/STD Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, P. R. China
First published on 7th December 2021
Abstract
Two-dimensional (2D) nanosheets as carriers have shown promising potential for surface-displaying or loading various drugs. Nevertheless, developing sheet-like materials themselves into an immunoregulator has never been realized so far. In this study, we take advantage of the immunoregulatory effects of rare earth elements themselves and develop water-soluble erbium–dysprosium 2D nanosheets (2D NSs). Such 2D NSs can target lymph nodes and activate macrophages to improve vaccine efficacy in mice significantly. Transcriptome analysis further reveals that six critical molecules (Msr1, Ccr2, Serpinb9, Klrk1, Klrd1, Klrc1) closely correlate with 2D NS-mediated immunoregulation in vivo. For the first time, the present work realizes a proof-of-concept for designing immunoregulatory 2D NSs and shows a promising potential of 2D NSs for improving the immunoprophylaxis/immunotherapy of vaccines.
Introduction
2D materials as carriers have shown their great potential for effectively loading and delivering various drugs in vivo due to their unique morphological diversity, structural porosity, and compositional tenability.1–4 Nevertheless, developing 2D materials into immunoregulatory drugs has never been realized so far. One of the biggest challenges is introducing applicable immunoregulatory building blocks to endow planar materials with an inherent immunoregulatory capability.5,6Of note, a few rare earth elements or rare earth element-based complexes have shown their unique effects on regulating the functions or behaviors of macrophages, one type of critical immune cell for presenting antigen, eliminating inflammation, and regulating immunity in vivo.7,8 For instance, erbium (Er) and dysprosium (Dy) can specifically regulate the activation of macrophages,9–12 instead of other immune cells such as dendritic cells,13via neutralizing the reactive oxygen species (ROS) or affecting the production of nitric oxide (NO). Lanthanum (La) can induce bone marrow-derived macrophages to release robust IL-1β to regulate the proliferation, differentiation, and apoptosis of multiple lymphoid cells (T cells, natural killer cells, dendritic cells).14,15 Neodymium (Nd) stimulates pattern recognition receptors on the surface of macrophages to regulate intracellular ROS production, which further affects the balance of the immune system.16 We, therefore, hypothesize that introducing these rare earth elements as building blocks might endow planar materials themselves with an inherent immunoregulatory capability.
Considering that multiple types of rare-earth elements (e.g. Er, Dy, La, Nd) have immunoregulatory effects on the functions or behaviors of immune effector cells, how to select rare-earth elements rationally is a core question for the design of immunoregulatory 2D NSs. In comparison to La and Nd which would cause cellular membrane permeabilization to damage the structure of cells,12 as well as over-induce inflammasome activation to disturb the balance of Ca2+ influx and cause cell death,16 Er and Dy exhibit better biosafety in vivo.12,17,18 Moreover, employed Er and Dy 2D NSs can more comprehensively use the immunoregulation functions of both Er (ROS-based mechanism)10 and Dy (NO-based mechanism),11 in comparison with either Er- or Dy-alone 2D NSs.
In the current study, we design and synthesize a new type of 2D erbium–dysprosium nanosheets with good water solubility and satisfactory biosafety in vivo. We use such 2D NSs as an immunoregulator to significantly enhance humoral and cellular responses induced by the HIV vaccine in mice. We further explore multiple critical genes, which can regulate 2D NSs-mediated immunoregulation in vivo. These results provide insights into the mechanism of such a new immunoregulator and better guide us on optimizing the design of other 2D materials with immunoregulatory effects. Overall, our study realizes a proof-of-concept for developing new immunoregulatory materials and shows promising potential for using artificially constructed 2D materials to improve vaccine-induced immunities.
Results and discussion
Characterization of 2D NSs
An alkaline aqueous solution of erbium chloride, dysprosium chloride, and 2-methylimidazole was incubated at 93 °C for 48 hours to form 2D NSs. We have tried a series of the ratio of Er ions to Dy ions (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 7.9
0, 7.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1, 7.5
0.1, 7.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5, 7
0.5, 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and obtained the size-controlled 2D NSs only at a ratio of 7
1), and obtained the size-controlled 2D NSs only at a ratio of 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 of Er
1 of Er![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
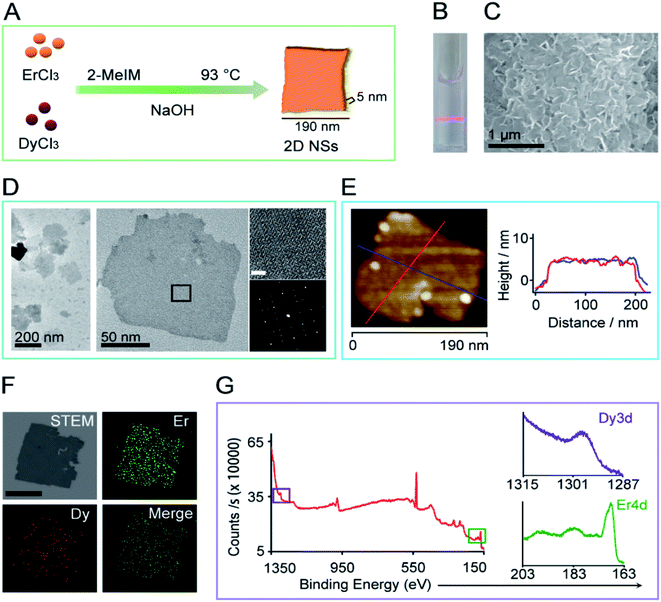
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Dy (Fig. 1A, ESI Fig. S1†). The 2D NSs appear finely suspended, as shown by the Tyndall scattering effect of the 2D NSs solution (Fig. 1B). Fig. 1C shows a typical scanning electron microscopy (SEM) image of the 2D NSs deposited on a silicon wafer. Transmission electron microscopy (TEM) shows the finely dispersed sheet-like morphology measuring edge length less than 200 nm (measured along the longest edges of NSs) (Fig. 1D). The left in Fig. 1D is the aberration-corrected high-resolution TEM (JEM-ARMF200) image and the corresponding FFT (below) validating the single crystallinity of the 2D NSs. Atomic force microscopy (AFM) image of single 2D NSs detected the uniform thickness of ∼5 nm in either direction marked with different color lines (Fig. 1E). Energy dispersive spectroscopy (EDS) mapping showed the chemical makeup of 2D NSs. The distribution of Er and Dy elements are homogeneous throughout the sheet (Fig. 1F, ESI Fig. S2†) and the ICP-MS measured 78% of Er and 22% of Dy elements (ESI Fig. S3†). Furthermore, X-ray photon spectroscopy (XPS) analysis confirmed the presence of Er and Dy elements with binding energies at 169 eV (Er4d) and 1297 eV (Dy3d) (Fig. 1G). To study the fine structure we obtained the spectra using AlKα X-ray source. The binding energy (BE) scale is corrected with reference to carbon 1s peak at BE of 284.5 eV. The Dy 3d5/2 spectra exhibited a single component at BE around 1299 eV, which correspond to metallic Dy (Dy0) chemical state (ESI Fig. S4A†). The Er 4d5/2 spectra exhibited two components at BEs around 170 and 168 eV. The former component is related to the metallic Er (Er0) chemical state and the latter to Er-oxide (Er3+), respectively (ESI Fig. S4B†).19 The presence of the oxide component is due to surface oxidation of the Er/Dy NSs. The oxidation is predominant in Er due to the larger composition of Er in the Er/Dy NSs. 2D NSs can maintain their morphology and zeta value (+27.4 mV) in an aqueous solution for more than one month at 4 °C (ESI Fig. S5†).
Dy (Fig. 1A, ESI Fig. S1†). The 2D NSs appear finely suspended, as shown by the Tyndall scattering effect of the 2D NSs solution (Fig. 1B). Fig. 1C shows a typical scanning electron microscopy (SEM) image of the 2D NSs deposited on a silicon wafer. Transmission electron microscopy (TEM) shows the finely dispersed sheet-like morphology measuring edge length less than 200 nm (measured along the longest edges of NSs) (Fig. 1D). The left in Fig. 1D is the aberration-corrected high-resolution TEM (JEM-ARMF200) image and the corresponding FFT (below) validating the single crystallinity of the 2D NSs. Atomic force microscopy (AFM) image of single 2D NSs detected the uniform thickness of ∼5 nm in either direction marked with different color lines (Fig. 1E). Energy dispersive spectroscopy (EDS) mapping showed the chemical makeup of 2D NSs. The distribution of Er and Dy elements are homogeneous throughout the sheet (Fig. 1F, ESI Fig. S2†) and the ICP-MS measured 78% of Er and 22% of Dy elements (ESI Fig. S3†). Furthermore, X-ray photon spectroscopy (XPS) analysis confirmed the presence of Er and Dy elements with binding energies at 169 eV (Er4d) and 1297 eV (Dy3d) (Fig. 1G). To study the fine structure we obtained the spectra using AlKα X-ray source. The binding energy (BE) scale is corrected with reference to carbon 1s peak at BE of 284.5 eV. The Dy 3d5/2 spectra exhibited a single component at BE around 1299 eV, which correspond to metallic Dy (Dy0) chemical state (ESI Fig. S4A†). The Er 4d5/2 spectra exhibited two components at BEs around 170 and 168 eV. The former component is related to the metallic Er (Er0) chemical state and the latter to Er-oxide (Er3+), respectively (ESI Fig. S4B†).19 The presence of the oxide component is due to surface oxidation of the Er/Dy NSs. The oxidation is predominant in Er due to the larger composition of Er in the Er/Dy NSs. 2D NSs can maintain their morphology and zeta value (+27.4 mV) in an aqueous solution for more than one month at 4 °C (ESI Fig. S5†).
2D NSs on regulating the function of macrophages
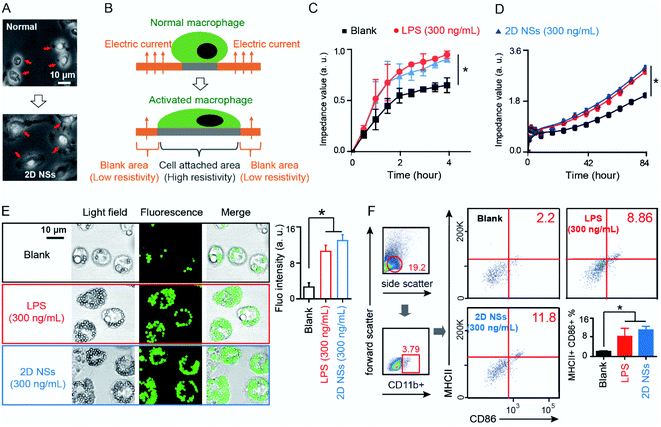
We investigated the effect of 2D NSs on regulating the function or behavior of macrophages. We isolated primary macrophages from mouse enterocoelia after three days of a 2 mL paroline injection20 and co-cultured 1 × 106 primary macrophages with 2D NSs with different concentration (30 μg mL−1, 3 μg mL, 300 ng mL−1 and 30 ng mL−1). 300 ng mL−1 is an optimized concentration for the activation of macrophages (ESI Fig. S6†). Activated macrophages will attach closer to their growth basement and have an obvious extension of cellular morphology (bigger size, pseudopods).21 In a visual field, we use an optical microscope to observe that macrophages stimulated with 300 ng mL−1 2D NSs extended their size and grow many pseudopods, which are two critical characteristics to evaluate the activation of primary macrophages according to previous studies.22 In contrast, naked macrophages have smaller sizes and exhibit the spherical shape that is a typical half-attachment growth situation of cells (Fig. 2A). We evaluated the activation of macrophages via real-time monitoring of the electrical resistivity of the macrophage-cultured plate. As shown in Fig. 2B, in comparison to normal macrophages, activated macrophages will extend their size and cover the broader area of the plate, causing increased electrical resistivity. During 4 hours, macrophages stimulated with 300 ng mL−1 2D NSs significantly increased the electrical resistivity of the plate compared to naked macrophages. A similar increase of electrical resistivity appears in the plate cultured with macrophages stimulated with 300 ng mL−1 lipopolysaccharide (LPS), a positive stimulator for macrophage activation (Fig. 2C).23 We observed a continuous increase of electrical resistivity even up to 84 hours (Fig. 2D). All these results solidly supported that 2D NSs can induce the activation of macrophages.The characteristics of our 2D NSs on activating macrophages might suggest its potential as a general immunoregulator for the enhancement of vaccine efficacy. Among various strategies for enhancing vaccine-triggered immune responses, activating macrophages to improve the uptake and presentation of antigens is a very effective means for improving vaccine efficacy.24 Macrophages participate in almost all checkpoint events during the whole process of vaccine-induced immune responses, including the uptake of vaccines, the process of antigens, the presentation of epitopes, and the regulation of the functions of immune effector cells (e.g. T cells, B cells).7
We evaluated the phagocytosis of 2D NSs-activated macrophages against green fluorescent microspheres. In this test, we selected mouse-derived RAW 264.7 macrophages as a standard cell line, which has been domesticated and widely used to evaluate the functions of macrophages.25 At Day 2, 4, 6, and 8, macrophages stimulated with either 300 ng mL−1 2D NSs or 300 ng mL−1 LPS produced significantly stronger fluorescence intensity (around 3–4 fold) than naked macrophages (Fig. 2E, ESI Table S1†). These results indicated that 2D NSs-activated macrophages enhanced their phagocytosis capability. Furthermore, we evaluated the activation of in vivo macrophages via quantifying the surface marker (CD86) on macrophages in mice.23 As shown in left part in Fig. 2F, we first gated the living splenocytes (19.2%) in all splenocytes. We further gated CD11b-positive cells (3.79%) as macrophages in all living splenocytes for analyzing the activation of macrophages. Flow cytometry analysis revealed that 300 ng mL−1 2D NSs significantly increase the expression of CD86 on the surface of macrophages in vivo. It is consistent with the result from macrophages stimulated with 300 ng mL−1 LPS (Fig. 2F). Importantly, we observed a significantly increased expression of MHC II marker for antigen presentation26 on the surface of macrophage after stimulating with 300 ng mL−1 2D NSs in vivo (Fig. 2F). Furthermore, we also evaluate macrophage-derived production of reactive oxygen species (ROS) and nitric oxide (NO) induced by 2D NSs. 300 ng mL−1 2D NSs can facilitate macrophages to significantly enhance the expression of both ROS (ESI Fig. S7†) and NO (ESI Fig. S8†). Together, these results indicated that 2D NSs activated macrophages to enhance their phagocytosis capability and bioactivity for the increased presentation of antigens.
2D NSs targeting lymph node
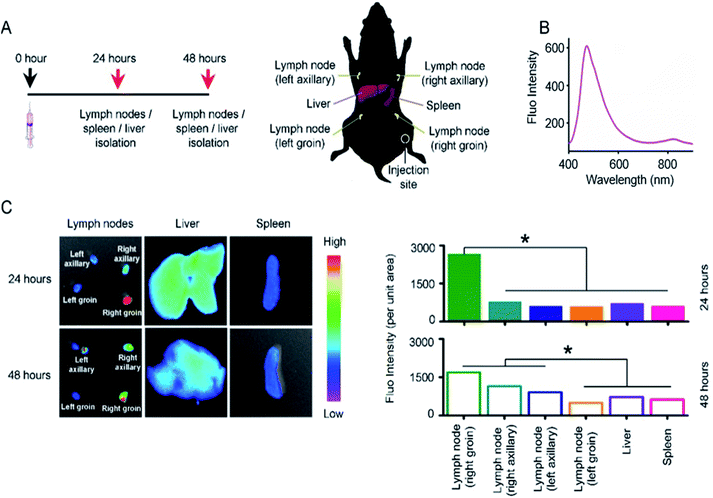
We investigated the targeting behavior of 2D NSs in vivo. We can track the targeting behavior of 2D NSs via detecting Er/Dy-derived fluorescence signals.27 Moreover, the lymph node and spleen are the most important organs/tissues for mediating immunity in vivo.28,29 The liver is the critical organ, which is in charge of the metabolism of nanomaterials in vivo.30 We, therefore, observed the targeting of 2D NSs to these organs/tissues. We injected 30 ng 2D NSs into each mouse. At the 24 and 48 hours, we harvested lymph nodes at four different positions (left axillary, right axillary, left groin, right groin), spleen, and liver from 2D NSs-injected mice (Fig. 3A). Significantly higher fluorescence intensity (2D NSs-derived fluorescence, Fig. 3B) appeared in lymph nodes at the 24 and 48 hours, in comparison with other organs (p < 0.05, Fig. 3C). These results proved that 2D NSs targets lymph nodes in vivo. Considering lymph node is a critical type of immunological tissue for mediating vaccine-induced immune responses,28 such lymph node targeting suggested that 2D NSs as an immunoregulator might improve the efficacy of other vaccines effectively.Of note, 2D NSs carry positive zeta potentials (+27.4 mV, ESI Fig. S4†) in an aqueous solution, and the surface charge of DNA vaccines is negative. For the co-administration of 2D NSs and DNA vaccines in vivo, 2D NSs can adsorb DNA vaccines by electrostatic attraction (ESI Fig. S9†) and deliver these DNA vaccines to lymph nodes. A similar phenomenon also is commonly observed between DNA molecules and other 2D nanosheets, such as graphene.31 Therefore, 2D NSs in the current study may play a dual-functional role in both activating immune cells and target delivering DNA vaccines to lymph nodes.
2D NSs facilitated immune responses HIV DNA vaccine
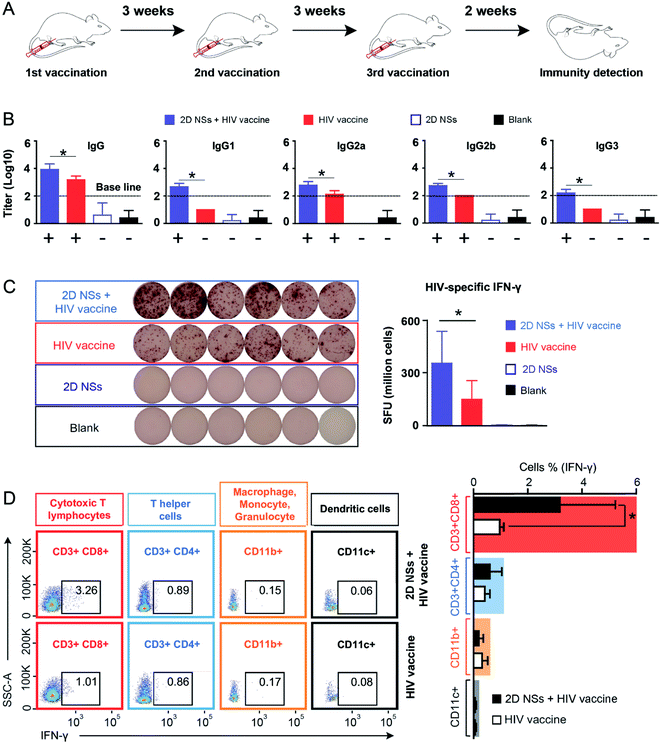
Based on our finding that 2D NSs can activate macrophages to enhance the uptake and presentation of antigens, we further investigated whether 2D NSs can facilitate the HIV DNA vaccine to trigger stronger immune responses in the mouse model (Fig. 4A). We use DNA vaccine candidates to evaluate the immunoregulation of our 2D NSs. DNA vaccines highlight the advantages of precise construction, easy production, convenient transport, and satisfactory biosafety.32 However, it suffers modest immunogenicity in vivo, which greatly limits its practical application.32,33 Enhancing DNA vaccine-triggered immune responses, therefore, becomes a long-sought aim of many immunologists. The use of our 2D NSs simultaneously enhances both humoral and cellular immune responses induced by the HIV DNA vaccine in vivo. Such a balanced enhancement of HIV-specific humoral and cellular immune responses regulated by 2D NSs might be an unparalleled advantage for realizing the neutralization (mediated by humoral response) and cytotoxicity (cellular response) against HIV, in comparison with other most of the reported immunoregulators, which only can enhance either humoral or cellular responses induced by HIV vaccines.34–41For the antibody response, we quantified the titer of HIV-specific IgG and its four subclasses (IgG1, IgG2a, IgG2b and IgG3) regulated by 2D NSs in vivo. HIV-specific IgG titer increased around 4 fold in mice vaccinated with 20 μg HIV DNA vaccine with 30 ng 2D NSs, compared to that vaccinated with 20 μg HIV DNA vaccine alone (Fig. 4B). We further analyzed which IgG subclasses majorly contribute to 2D NSs-caused enhancement of IgG response. Naked HIV DNA vaccine-induced two HIV-specific IgG subclass responses (positive IgG2a and IgG2b responses) in mice. By contrast, the introduction of 2D NSs allowed the HIV DNA vaccine not only to enhance the titer of IgG subclasses (p < 0.05) but also to broaden IgG subclass response (positive IgG1, IgG2a, IgG2b and IgG3 responses) (Fig. 4B). Notably, a broad IgG subclass response appears in HIV long-term non-progressors infected by HIV but does not or very slowly develop into AIDS.42–45 It suggests that 2D NSs might help HIV vaccine to inhibit the progress of HIV better. Of course, more solid data is necessary to confirm such a hypothesis. For T cell response, we evaluated the production of HIV-specific IFN-γ from immune effector cells in mice. The expression of HIV-specific IFN-γ significantly enhanced (2.3 fold) in mice vaccinated with 20 μg HIV DNA vaccine with 30 ng 2D NSs, compared to that vaccinated with 20 μg HIV DNA vaccine alone (Fig. 4C). This result demonstrated that 2D NSs facilitated the HIV DNA vaccine to induce a stronger T cell response in mice. Moreover, we analyzed the source of HIV-specific IFN-γ to understand the immunoregulation effect of 2D NSs on the HIV DNA vaccine. We selected multiple subgroups of immune cells, including macrophages/monocytes/granulocytes, cytotoxic T lymphocytes, T helper cells, and dendritic cells46 because these immune cells majorly produce IFN-γ in vivo.47,48 For the naked HIV DNA vaccine, both cytotoxic T lymphocytes (1.01%) and T helper cells (0.86%) produced most of HIV-specific IFN-γ (Fig. 4D). A much smaller part of HIV-specific IFN-γ derived from macrophages/monocytes/granulocytes (0.17%) and dendritic cells (0.08%). Although the introduction of 2D NSs failed to induce more T helper cells (0.89%), macrophages/monocytes/granulocytes (0.15%), and dendritic cells (0.06%) to produce HIV-specific IFN-γ, it significantly enhanced cytotoxic T lymphocytes (from 1.01% to 3.26%, p < 0.05) to produce HIV-specific IFN-γ (Fig. 4D). These results indicated that 2D NSs majorly regulated cytotoxic T lymphocytes to enhance the expression of HIV-specific IFN-γ in mice. Overall, 2D NSs as immunomodulators can significantly improve both antibody and T cell response induced by HIV DNA vaccine in mice.
Molecular mechanism of 2D NSs improving HIV DNA vaccine
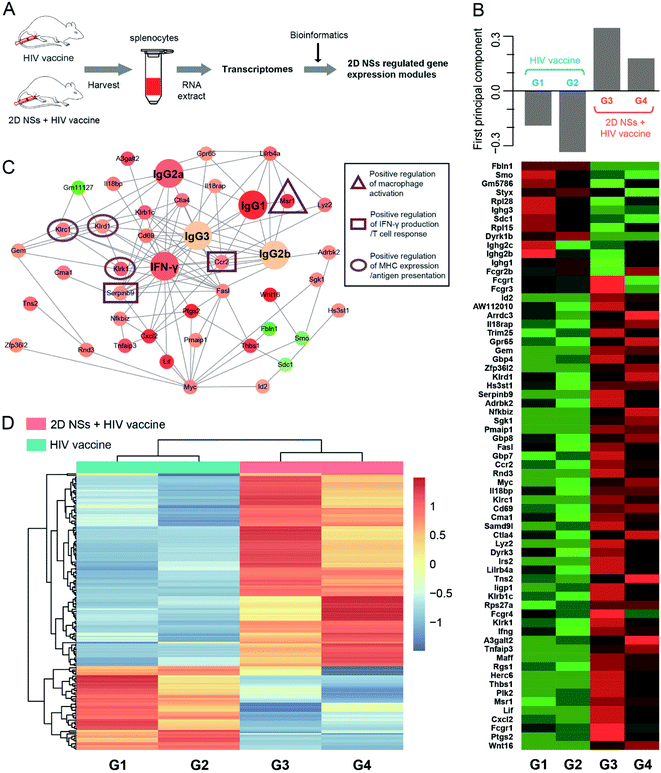
To understand the molecular mechanism of 2D NSs improving HIV DNA vaccine-triggered immune responses in vivo, we analyzed the transcriptomes49 of the splenocytes isolated from the mice treated with 30 ng 2D NSs (Fig. 5A). We employed protein–protein interaction (PPI) analysis based on the STRING database to identify the highly correlated gene modules, which have significantly up-regulated or down-regulated expression (Fig. 5B). Principal component analyses of the top 67 most variable transcripts revealed a significant up-regulation of immunomodulatory gene module in 2D NSs treated mice (Fig. 5B). Cluster Profiler gene ontology enrichment analyses of gene module revealed that six genes are at the center of this interconnected network (Msr1, Ccr2, Serpinb9, Klrk1, Klrd1, and Klrc1) showed the most related with 2D NSs-regulated HIV vaccine-triggered immune responses in vivo (Fig. 5C). According to previous reports, three natural killer cell lectin-like receptor subfamily genes (Klrk1, Klrd1, and Klrc1)50 will effectively enhance the presentation of antigens. Such enhancement is possibly caused by activating critical antigen-presenting cells (APCs), such as dendritic cells and macrophages. Further investigation also found a significantly up-regulated expression of Msr1, a critical gene to activate macrophages.51 Moreover, two other genes (Ccr2 and Serpinb9) that play critical roles in up-regulating cytokine production52,53 showed increased expression, which is consistent with the activation of macrophages. The results mentioned above showed an immunoregulation-related network involving the activation of immune cells, antigen presentation, and the production of immune effectors. Such an immunoregulatory network might explore why 2D NSs can facilitate the HIV DNA vaccine to trigger stronger immune responses. Moreover, hierarchical clustering analyses of differentially expressed genes (DEGs) (p-value < 0.05) confirmed that the significantly different transcriptional profiles between 2D NSs and HIV DNA vaccine-treated mice and HIV DNA vaccine-treated mice (Fig. 5D). Together, transcriptomes data revealed that 2D NSs could regulate Msr1, Ccr2, Serpinb9, Klrk1, Klrd1 and Klrc1 gene to enhance HIV DNA vaccine-triggered immune responses in vivo.In vitro and in vivo biosafety of 2D NSs
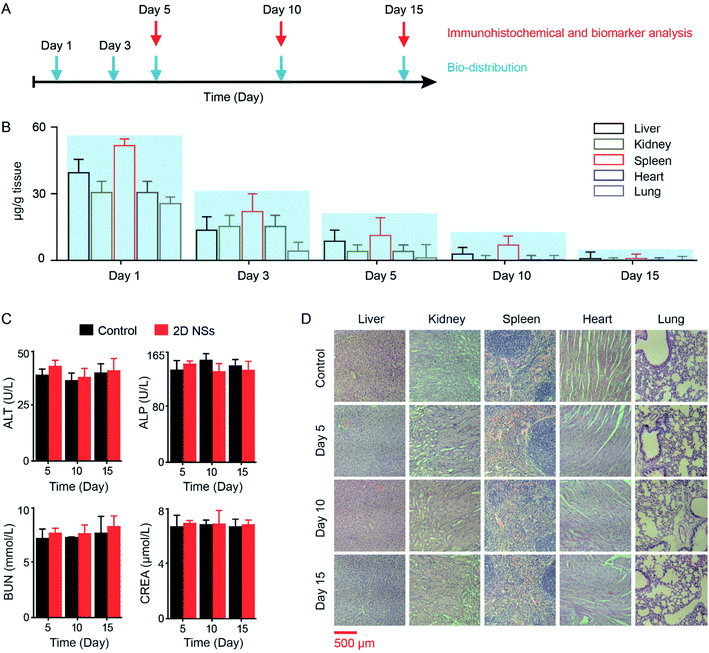
We studied the biodistribution, cytotoxicity, and in vivo toxicity of 2D NSs. We assessed the biodistribution of 2D NSs in five organs (liver, kidney, spleen, heart and lung) at different time points (Day 1, Day 3, Day 5, Day 10, and Day 15) using an inductively coupled plasma-mass spectrometry (ICP-MS) system (Fig. 6A). The residual amount of 2D NSs significantly reduced in mice from Day 1 to Day 15. On Day 15, 2D NSs in the liver, kidney, spleen, heart, and lung have been almost removed (Fig. 6B). We evaluated the biosafety of 2D NSs. 2D NSs exhibited satisfactory cytotoxicity in vitro. The viability of HeLa and HUVEC cells has not been significantly reduced after incubation of 2D NSs with a broad range of concentrations (0–300 μg mL−1) for 24 hours (ESI Fig. S10†). For the evaluation of in vivo biosafety of 2D NSs, we monitored the expression of multiple important biochemical indicators, including alanine aminotransferase (ALT), alkaline phosphatase (ALP), creatinine (CREA), and urea nitrogen (BUN) in serum from 2D NSs-injected mice for 15 days. All four indicators showed no significant changes in 2D NSs-treated mice at Day 5, Day 10, and Day 15, compared to normal mice (Fig. 6C). Furthermore, we performed the histopathologic analysis of multiple organs (heart, liver, spleen, lung, and kidney). There was no evidence to show the infiltration and necrosis of inflammatory cells in these organs from 2D NSs-injected mice. There was no visible difference between 2D NSs-treated mice and normal mice (Fig. 6D). These results prove satisfactory biosafety of 2D NSs in vitro and in vivo.Conclusions
In conclusion, we developed water-soluble 2D NSs into a new immunoregulator. Such 2D NSs can target the lymph node and effectively activate macrophages to improve HIV DNA vaccine-induced immune responses in vivo significantly. Molecular mechanism investigation revealed that six genes (Msr1, Ccr2, Serpinb9, Klrk1, Klrd1, and Klrc1) play critical roles in mediating 2D NSs-mediated immunoregulation. The realization of the concept of 2D NSs immunoregulator dramatically broadens the scope for choices to optimize the vaccination of infectious diseases, tumor immunotherapy, and other immune-based prophylaxis and therapy.Ethical statement
Animal studies were approved by the Animal Ethics Committee of the Institute of Medical Biology, Chinese Academy of Medical Sciences and Peking Union Medical College and executed according to guidelines from the Committee of Welfare and Ethics of Laboratory Animals in Kunming.Data availability
We provide all data in ESI.† All readers can contact the corresponding authors for further discussions.Author contributions
Y. L., Y. L. B., and X. J. conceived the project and designed the experiments. Y. L., Y. L. B. and Z. L. performed the experiments and analyzed the data. X. J., Y. C., and Y. S. supervised the study. Y. L., Y. L. B., Z. L., Y. C., Y. S., and X. J. discussed the data and wrote the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank financial supports from the National Natural Science Foundation of China (21874033, 21535001, 81730051, 21761142006, 81501790), the National Key R&D Program of China (2018YFA0902600, 2017YFA0205901), the Chinese Academy of Sciences (QYZDJ-SSW-SLH039, 121D11KYSB20170026, XDA16020902), Tencent Foundation through the XPLORER PRIZE, the Leading Medical Talents Program of Health Commission of Yunnan Province (L-2018013), the Young and Middle-Aged Academic and Technical Leaders Program of Yunnan Province (202005AC160010), the Basic Research Program of Yunnan Province (202101AS070051), Shenzhen Key Laboratory of Smart Healthcare Engineering (ZDSYS20200811144003009).References
- Z. Wang, Y. Fu, Z. Kang, X. Liu, N. Chen, Q. Wang, Y. Tu, L. Wang, S. Song, D. Ling, H. Song, X. Kong and C. Fan, J. Am. Chem. Soc., 2017, 139, 15784–15791 CrossRef CAS PubMed.
- K. Lu, C. He, N. Guo, C. Chan, K. Ni, R. R. Weichselbaum and W. Lin, J. Am. Chem. Soc., 2016, 138, 12502–12510 CrossRef CAS PubMed.
- Y. Zhang, F. Wang, E. Ju, Z. Liu, Z. Chen, J. Ren and X. Qu, Adv. Funct. Mater., 2016, 26, 6454–6461 CrossRef CAS.
- N. Stock and S. Biswas, Chem. Rev., 2012, 112, 933–969 CrossRef CAS PubMed.
- G. P. Boyd, Y. Lee and B. Smith, Nat. Rev. Mater., 2017, 2, 17037 CrossRef.
- P. Horcajada, T. Chalati, C. Serre, B. Gillet, C. Sebrie, T. Baati, J. F. Eubank, D. Heurtaux, P. Clayette, C. Kreuz, J.-S. Chang, Y. K. Hwang, V. Marsaud, P.-N. Bories, L. Cynober, S. Gil, G. Férey, P. Couvreur and R. Gref, Nat. Mater., 2010, 9, 172–178 CrossRef CAS PubMed.
- S. W. Kashem, M. Haniffa and D. H. Kaplan, Annu. Rev. Immunol., 2017, 26, 469–499 CrossRef PubMed.
- F. O. Martinez, L. Helming and S. Gordon, Annu. Rev. Immunol., 2009, 27, 451–483 CrossRef CAS PubMed.
- A. P. West, I. E. Brodsky, C. Rahner, D. K. Woo, H. E. Bromage, P. Tempst, M. C. Walsh, Y. Choi, G. S. Shadel and S. Ghosh, Nature, 2011, 472, 476–480 CrossRef CAS PubMed.
- Y. C. Liu and Z. Y. Yang, J. Biochem., 2010, 147, 381–391 CrossRef CAS PubMed.
- I. Kostova and T. Stefanova, J. Trace Elem. Med. Biol., 2010, 24, 7–13 CrossRef CAS PubMed.
- J. Gao, R. Li, F. Wang, X. Liu, J. Zhang, L. Hu, J. Shi, B. He, Q. Zhou, M. Song, B. Zhang, G. Qu, S. Liu and G. Jiang, Environ. Sci. Technol., 2017, 51, 13938–13948 CrossRef CAS PubMed.
- C. Portioli, M. Pedroni, D. Benati, M. Donini, R. Bonafede, R. Mariotti, L. Perbellini, M. Cerpelloni, S. Dusi, A. Speghini and M. Bentivoglio, Nanomedicine, 2016, 11, 3039–3051 CrossRef CAS PubMed.
- H. M. Russo, J. Rathkey, A. B. Tressler, M. A. Katsnelson, D. W. Abbott and G. R. Dubyak, J. Immunol., 2016, 197, 1353–1367 CrossRef CAS PubMed.
- C. Garlanda, C. A. Dinarello and A. Mantovani, Immunity, 2013, 39, 1003–1018 CrossRef CAS PubMed.
- H. Yao, Y. Zhang, L. Liu, Y. Xu and L. P. Wen, Biomaterials, 2016, 108, 143–156 CrossRef CAS PubMed.
- A. Mantion, P. Graf, I. Florea, A. Haase, A. F. Thünemann, A. Mašić, O. Ersen, P. Rabu, W. Meier, A. Luch and A. Taubert, Nanoscale, 2011, 3, 5168–5179 RSC.
- P. E. Petrochenko, Q. Zhang, H. Wang, T. Sun, B. Wildt, M. W. Betz, P. L. Goering and R. J. Narayan, Int. J. Appl. Ceram. Technol., 2012, 9, 881–892 CrossRef CAS.
- J. F. Moulder, W. F. Stickle, P. E. Sobol, and K. D. Bomben, Handbook of X-ray Photoelectron Spectroscopy, Perkin Elmer Corporation, Eden Prairie, MN, 1992 Search PubMed.
- L. Xu, Y. Liu, Z. Chen, W. Li, Y. Liu, L. Wang, L. Ma, Y. Shao, Y. Zhao and C. Chen, Adv. Mater., 2013, 25, 5928–5936 CrossRef CAS PubMed.
- M. H. Baumann, K. Heinrich, S. A. Sahn, C. Green, R. Harley and C. Strange, Exp. Lung Res., 1993, 19, 731–742 CrossRef CAS PubMed.
- A. K. Hoffmann, X. Naj and S. Linder, FASEB J., 2014, 28, 3075–3089 CrossRef CAS PubMed.
- D. M. Mosser and J. P. Edwards, Nat. Rev. Immunol., 2008, 8, 958–969 CrossRef CAS PubMed.
- M. A. Chattergoon, T. M. Robinson, J. D. Boyer and D. B. Weiner, J. Immunol., 1998, 160, 5707–5718 CAS.
- D. Bi, R. Zhou, N. Cai, Q. Lai, Q. Han, Y. Peng, Z. Jiang, Z. Tang, J. Lu, W. Bao, H. Xu and X. Xu, Int. J. Biol. Macromol., 2017, 105, 1446–1454 CrossRef CAS PubMed.
- E. R. Unanue, V. Turk and J. Neefjes, Annu. Rev. Immunol., 2016, 34, 265–297 CrossRef CAS PubMed.
- J. C. Bunzil and C. Piguet, Chem. Soc. Rev., 2005, 34, 1048–1077 RSC.
- H. Liu, K. D. Moynihan, Y. Zheng, G. L. Szeto, A. V. Li, B. Huang, D. S. Van Egeren, C. Park and D. J. Irvine, Nature, 2014, 507, 519–522 CrossRef CAS PubMed.
- V. Bronte and M. J. Pittet, Immunity, 2013, 39, 806–818 CrossRef CAS PubMed.
- Y. N. Zhang, W. Poon, A. J. Tavares, I. D. McGilvray and W. C. W. Chan, J. Controlled Release, 2016, 240, 332–348 CrossRef CAS PubMed.
- C. Xu, H. Hong, Y. Lee, K. Park, M. Sun, T. Wang, M. E. Aikins, Y. Xu and J. J. Moon, ACS Nano, 2020, 14, 13268–13278 CrossRef CAS PubMed.
- Y. Liu, F. Li, Z. Qi, Y. Hao, K. Hong, Y. Liu, Y. Cong and Y. Shao, J. Virol., 2013, 10, 297 CrossRef PubMed.
- Y. Liu, Y. L. Balachandran, D. Li, Y. Shao and X. Jiang, ACS Nano, 2016, 10, 3589–3596 CrossRef CAS PubMed.
- T. Tokatlian, D. W. Kulp, A. A. Mutafyan, C. A. Jones, S. Menis, E. Georgeson, M. Kubitz, M. H. Zhang, M. B. Melo, M. Silva, D. S. Yun, W. R. Schief and D. J. Irvine, Sci. Rep., 2018, 8, 16527 CrossRef PubMed.
- T. Tokatlian, B. J. Read, C. A. Jones, D. W. Kulp, S. Menis, J. Y. H. Chang, J. M. Steichen, S. Kumari, J. D. Allen, E. L. Dane, A. Liguori, M. Sangesland, D. Lingwood, M. Crispin, W. R. Schief and D. J. Irvine, Science, 2019, 363, 649–654 CrossRef CAS PubMed.
- M. Melo, E. Porter, Y. Zhang, M. Silva, N. Li, B. Dobosh, A. Liguori, P. Skog, E. Landais, S. Menis, D. Sok, D. Nemazee, W. R. Schief, R. Weiss and D. J. Irvine, Mol. Ther., 2019, 27, 2080–2090 CrossRef CAS PubMed.
- L. He, S. Kumar, J. D. Allen, D. Huang, X. Lin, C. J. Mann, K. L. Saye-Francisco, J. Copps, A. Sarkar, G. S. Blizard, G. Ozorowski, D. Sok, M. Crispin, A. B. Ward, D. Nemazee, D. R. Burton, I. A. Wilson and J. Zhu, Sci. Adv., 2018, 4, eaau6769 CrossRef CAS PubMed.
- L. He, N. de Val, C. D. Morris, N. Vora, T. C. Thinnes, L. Kong, P. Azadnia, D. Sok, B. Zhou, D. R. Burton, I. A. Wilson, D. Nemazee, A. B. Ward and J. Zhu, Nat. Commun., 2016, 7, 12041 CrossRef CAS PubMed.
- P. J. M. Brouwer, A. Antanasijevic, Z. Berndsen, A. Yasmeen, B. Fiala, T. P. L. Bijl, I. Bontjer, J. B. Bale, W. Sheffler, J. D. Allen, A. Schorcht, J. A. Burger, M. Camacho, D. Ellis, C. A. Cottrell, A. J. Behrens, M. Catalano, I. D. Moral-Sánchez, T. J. Ketas, C. LaBranche, M. J. van Gils, K. Sliepen, L. J. Stewart, M. Crispin, D. C. Montefiori, D. Baker, J. P. Moore, P. Johan Klasse, A. B. Ward, N. P. King and R. W. Sanders, Nat. Commun., 2019, 10, 4272 CrossRef PubMed.
- C. D. Morris, P. Azadnia, N. de Val, N. Vora, A. Honda, E. Giang, K. Saye-Francisco, Y. Cheng, X. Lin, C. J. Mann, J. Tang, D. Sok, D. R. Burton, M. Law, A. B. Ward, L. He and J. Zhu, mBio, 2017, 8, e00036 CrossRef CAS PubMed.
- R. P. Ringe, V. M. Cruz Portillo, P. Dosenovic, T. J. Ketas, G. Ozorowski, B. Nogal, L. Perez, C. C. LaBranche, J. Lim, E. Francomano, I. A. Wilson, R. W. Sanders, A. B. Ward, D. C. Montefiori, M. C. Nussenzweig, P. J. Klasse, A. Cupo and J. P. Moore, J. Virol., 2020, 94, e01883 CrossRef CAS PubMed.
- K. Banerjee, P. J. Klasse, R. W. Sanders, F. Pereyra, E. Michael, M. Lu, B. D. Walker and J. P. Moore, AIDS Res. Hum. Retroviruses, 2010, 26, 445–458 CrossRef CAS PubMed.
- G. D. Tomaras and B. F. Haynes, Curr. Opin. HIV AIDS, 2009, 4, 373–379 CrossRef PubMed.
- A. W. Chung, M. Ghebremichael, H. Robinson, E. Brown, I. Choi, S. Lane, A. Dugast, M. K. Schoen, M. Rolland, T. J. Suscovich, A. E. Mahan, L. Liao, H. Streeck, C. Andrews, S. Rerks-Ngarm, S. Nitayaphan, M. S. de Souza, J. Kaewkungwal, P. Pitisuttithum, D. Francis, N. L. Michael, J. H. Kim, C. Bailey-Kellogg, M. E. Ackerman and G. Alter, Sci. Transl. Med., 2014, 6, 228–238 Search PubMed.
- V. A. Sundqvist, A. Linde, R. Kurth, A. Werner, E. B. Helm, M. Popovic, R. C. Gallo and B. Wahren, J. Infect. Dis., 1986, 153, 970–973 CrossRef CAS PubMed.
- M. M. Kaneda, K. S. Messer, N. Ralainirina, H. Li, C. J. Leem, S. Gorjestani, G. Woo, A. V. Nguyen, C. C. Figueiredo, P. Foubert, M. C. Schmid, M. Pink, D. G. Winkler, M. Rausch, V. J. Palombella, J. Kutok, K. McGovern, K. A. Frazer, X. Wu, M. Karin, R. Sasik, E. E. Cohen and J. A. Varner, Nature, 2016, 539, 437–442 CrossRef CAS PubMed.
- Y. Liu, H. Wang, D. Li, Y. Tian, W. Liu, L. Zhang, W. Zheng, Y. Y. Hao, J. Liu, Z. Yang, Y. Shao and X. Jiang, Nanoscale Horiz., 2016, 1, 135–143 RSC.
- D. M. Frucht, T. Fukao, C. Bogdan, H. Schindler, J. J. O'Shea and S. Koyasu, Trends Immunol., 2001, 22, 556–560 CrossRef CAS PubMed.
- H. Lui, J. Zhang, S. R. Makinson, M. K. Cahill, K. W. Kelley, H. Huang, Y. Shang, M. C. Oldham, L. H. Martens, F. Gao, G. Coppola, S. A. Sloan, C. L. Hsieh, C. C. Kim, E. H. Bigio, S. Weintraub, M. Mesulam, R. Rademakers, I. R. Mackenzie, W. W. Seeley, A. Karydas, B. L. Miller, B. Borroni, R. Ghidoni, R. V. FareseJr, J. T. Paz, B. A. Barres and E. J. Huang, Cell, 2016, 165, 921–935 CrossRef CAS PubMed.
- M. M. Rahim and A. P. Makrigiannis, Immunol. Rev., 2015, 267, 137–147 CrossRef CAS PubMed.
- Y. Dai, W. Su, Z. Ding, X. Wang, F. Mercanti, M. Chen, S. Raina and J. L. Mehta, Biochem. Biophys. Res. Commun., 2013, 431, 496–500 CrossRef CAS PubMed.
- W. Sato, A. Tomita, D. Ichikawa, Y. Lin, H. Kishida, S. Miyake, M. Ogawa, T. Okamoto, M. Murata, Y. Kuroiwa, T. Aranami and T. Yamamura, J. Immunol., 2012, 189, 5057–5065 CrossRef CAS PubMed.
- A. W. Ansari, J. N. Temblay, S. H. Alyahya and P. G. Ashton-Rickardt, J. Immunol., 2010, 185, 877–883 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details including materials, synthesis and methods. ESI Fig. S1–S10 and ESI Table S1. See DOI: 10.1039/d1sc04044h |
| ‡ The authors equally contribute this work. |
| This journal is © The Royal Society of Chemistry 2022 |