Open Access Article
Open Access ArticleVisual detection of aflatoxin B1 and zearalenone via activating a new catalytic reaction of “naked” DNAzyme†
Qinrui Luab,
Yue Liub,
Qiao Liub,
Jun Liu b,
Qin Yang
b,
Qin Yang b,
Jiancai Tangc,
Zhijun Menga,
Qiang Sude,
Shengmao Li*b and
Yingping Luo
b,
Jiancai Tangc,
Zhijun Menga,
Qiang Sude,
Shengmao Li*b and
Yingping Luo *ab
*ab
aMedical Imaging Key Laboratory of Sichuan Province, North Sichuan Medical College, Nanchong 637000, P. R. China
bDepartment of Pharmacology, North Sichuan Medical College, Nanchong 637100, P. R. China. E-mail: lsm9110@163.com; lyingping@163.com
cDepartment of Basic Medical Sciences, North Sichuan Medical College, Nanchong 637100, P. R. China
dDepartment of Pharmacy, The Second Clinical Medical College of North Sichuan Medical College, Nanchong Central Hospital, Nanchong 637000, P. R. China
eNanchong Key Laboratory of Individualized Drug Therapy, Nanchong 637000, P. R. China
First published on 9th November 2022
Abstract
Here, we have found for the first time that the catalytic activity of “naked” DNAzyme, a single-stranded G-quadruplex DNAzyme (S.DNAzyme), can be modulated by aflatoxin B1 (AFB1) and zearalenone (ZEN). In fact, S.DNAzyme can mimic the activity of horseradish peroxidase to perform oxidation of 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) to colored ABTS˙+ (dark green). This catalytic activity can be inhibited upon addition of AFB1, leading to color fade of the solution. But with ZEN, the solution color will change to a yellowish-brown or yellow color. Thus, we have exploited this finding to achieve label-free, naked-eye detection of AFB1 or ZEN, and have explored the possible detection mechanisms. This approach is able to detect AFB1 and ZEN concentrations as low as 0.18 μM and 0.29 μM, respectively. Even in real samples, the limit of detection values of these two analytes are lower than 3 μM. Notably, S.DNAzyme cross-reacted with three other tested aflatoxins, including aflatoxin B2, aflatoxin G1 and aflatoxin M1, revealing the potential for a broadly applicable method to identify the family of aflatoxins. The AFB1- or ZEN-triggered new catalytic reaction between “naked” DNAzyme and ABTS may offer new opportunities for on-site visual detection of mycotoxins.
Introduction
Mycotoxins, resulting from secondary metabolites of toxic fungal species,1,2 are widespread contaminants of traditional Chinese medicines (TCMs) and foodstuffs, leading to safety concerns because of their high toxicity, carcinogenicity and biological persistence.3 Owing to their strong thermal and chemical stability,4 aflatoxins (AFs) and zearalenone (ZEN) are the two most intensively researched groups of mycotoxins. AFs are difuranocoumarin derivatives with four major members including aflatoxin B1, B2, G1 and G2 (AFB1, AFB2, AFG1 and AFG2).5 Of the known AFs, AFB1 with high toxic potency and occurrence has been well-characterized and shown to lead to the development of hepatocellular carcinoma in humans and animals.6,7 The toxicity of AFB1 is 10 times that of potassium cyanide and 68 times that of arsenic, showing considerable threats to public health safety.8 ZEN, also known as F-2 toxin, is a non-steroidal estrogenic fungus produced by several species of Fusarium spp., and it is extremely toxic to the human reproductive system and lymphoid organs.9 For these reasons, AFB1 and ZEN are classified by the International Agency for Research on Cancer (IARC) as a group I and group III carcinogens, respectively.10,11 An efficient method via detecting contaminated TCMs and foods and, then, removing them would ensure human health and reduce the economic loss caused by AFB1 or ZEN. Consequently, developing a fast and reliable approach of AFB1 and ZEN detection is a necessity.Though high-performance liquid chromatography (HPLC) and ultra-high performance liquid chromatography coupled with tandem mass spectrometry (UHPLC/MS) have high selectivity and sensitivity for AFB1 or ZEN analysis,12–15 they are time-consuming and require high instrument costs, complicated sample pre-treatment and skilled personnel. In recent decades, various antibody-based immunoassay assays have been developed such as enzyme-linked immunosorbent assays,16 immunochips,17 fluorescent immunoassays,18 lateral flow immunoassays,19 chemiluminescent immunoassays,20 and immunosensors.21 Given the limitations of antibody, most of them require strict storage conditions and are inconvenience for daily use. Therefore, it is still a challenge to construct a highly desirable platform to achieve on-site detection for AFB1 or ZEN. Aptamers are chemical antibodies that can be selected in vitro by systematic evolution of ligands by exponential enrichment, which form diverse structures to combine with target molecular under confined conditions.22,23 Compared with traditional antibody, the selected aptamers not only possess similar specificity and affinity to antibody, but also have numerous advantages including decent stability, easy synthesis and modification, and low cost.24 Hence, aptamers are widely used for developing biosensors to overcome the limits of antibodies.25–27 Among abundant aptamer-based biosensors described to date, visual sensors are especially suitable for on-site supervision of TCMs or foods and have attracted great interest, because they enable the naked eye to observe the results without the requirement of sophisticated instruments. However, there are little literatures that using aptamer instead of antibody as a sensing element to construct visual sensor for AFB1 or ZEN detection. On the basis of unique horseradish peroxidase (HRP)-like catalytic activity, easiness of synthesis, and excellent chemical stability,28 G-quadruplex-structured DNA enzymes (DNAzymes) have become one of the most widely used signal indicators in naked-eye detection. Kim group constructed a structure-switchable sensor for AFB1 detection using AFB1 aptamer and split DNAzyme,29 however, they did not explore that whether the peroxidase activity of complete DNAzyme would be modulated by AFB1. Although DNAzyme has broadly been employed as signal producer, no report of “naked” DNAzyme, a single-stranded DNAzyme (S.DNAzyme) that only is composed of four GGG repeats, being used as AFB1- or ZEN-detection sensor.
In this work, we have found for the first time that AFB1 and ZEN can induce a new reaction happening on the S.DNAzyme catalyzed 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) oxidation platform. In addition, this S.DNAzyme have been successfully employed as a colorimetric sensor to accomplish visual detection for two mycotoxins, namely, AFB1 and ZEN. Without analytes, S.DNAzyme was capable of binding hemin and performing HRP-mimicking catalytic activity to oxidize ABTS to dark green colored ABTS˙+. But in presence of AFB1, the process of ABTS˙+ production was dramatically inhibited and the action of ABTS˙+ consumption was active at the same time, making the reaction solution change from dark green to colorless. Once S.DNAzyme was incubated with ZEN, a newly produced absorbance peak at 472 nm was obtained and the solution color would change into yellowish-brown or yellow. S.DNAzyme was able to realize label-free, naked-eye detection for AFB1 and ZEN with detection limits of 0.18 μM and 0.29 μM, respectively. Furthermore, RGB-based analysis was used to increase color discrimination and we were able to observe a noticeable color change in the presence of 0.05 μM AFB1 or 10 μM ZEN. Impressively, S.DNAzyme also enables to determine other tested AFs including AFB2, AFG1 and AFM1, which showed a great potential for the analysis of multiple mycotoxins.
Materials and methods
Materials
2,2′-Azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) and hemin were purchased from Macklin Biochemical Co., Ltd (Shanghai, China). 30% hydrogen peroxide (H2O2) was purchased from Chengdu Kelong Chemical Co. Ltd (Chengdu, China). All other chemicals and DNA oligonucleotides were purchased from Sangon Biotechnology Co. Ltd (Shanghai, China). Oligonucleotides were dissolved in ultrapure water and DNA concentrations were measured with a NanoDrop 2000 (Thermo Scientific). Ultrapure water with 18.25 MΩ cm was made by a ULUPURE system (Sichuan ULUPURE Technology Co., Ltd, China).Aptamer-based hairpin constructs (ABHCs) platform for AFB1 detection
ABHCs were prepared by mixing 0.5 μM H-1 and H-3 in the 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (HEPES) buffer (pH 7.0) containing 20 mM KCl and 100 mM NaCl. AFB1 was freshly prepared in 10% DMSO. Various concentrations of AFB1 (final concentration: 0, 0.1, 10, 50, 100 μM) were added to the mixture and the solution was incubated for 30 min. Freshly prepared 1 μM hemin and 0.3% (v/v) TritonX-100 were added to the mixture, and the solution was incubated for another 30 min. Then, the reaction solution was transferred into a well of a 384-well plate (CORNING), then, 2 mM H2O2 and 1.5 mM ABTS were added to initiate the reaction. The absorption intensity at 418 nm (ABTS˙+) was recorded every minute using a Spectra Max M2 microplate reader. It's worth noting that the final concentration of DMSO in this assay was 2% (v/v). ABTS˙+ was calculated based on the extinction coefficient of ABTS˙+ at λ = 418 nm (ε = 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1).30,31 The concentration of ABTS˙+ was plotted as a function of time and the initial reaction rates (V0) were determined by calculating the slope of the linear portion of the plot.
000 M−1 cm−1).30,31 The concentration of ABTS˙+ was plotted as a function of time and the initial reaction rates (V0) were determined by calculating the slope of the linear portion of the plot.
Colorimetric assay for AFB1 and ZEN using S.DNAzyme
S.DNAzyme were prepared in 40 mM HEPES buffer (pH 7.0) containing 50 mM KCl and 10 mM NaCl. AFB1 and ZEN were freshly prepared in 10% DMSO. Different concentration of AFB1 (final concentration: 0, 0.05, 0.5, 1, 5, 10, 15, 20, 50, 80, 100 μM) or ZEN (final concentration: 0, 0.05, 0.5, 1, 5, 10, 20, 50, 80, 100, 200, 300 μM) were added to the S.DNAzyme mixture. Freshly prepared 1 μM hemin and 0.3% (v/v) TritonX-100 were added, and the solution was incubated for 30 min at room temperature. The mixture was then added to a 384-well plate, then, 2 mM H2O2 and 1.5 mM ABTS were added to initiate the reaction. The absorption intensity at 418 nm was recorded every minute. The concentration of S.DNAzyme for AFB1 and ZEN detection was 0.75 and 1 μM, respectively. The signal gain was calculated by (A0 − A)/A0 × 100%, where A and A0 are the absorbance of reaction solution with and without target, respectively.Preparation of AFB1 or ZEN in real samples
To investigate the feasibility of visual detection for AFB1 and ZEN using S.DNAzyme, some TCMs and foods easily contaminated by AFB1 (adlay, platyclade semen, tangerine peel and milk) or by ZEN (fritillariae cirrhosae bulbus, angelica sinensis, amygdalin, and rice) were employed to be used as complex matrices. Solid samples were finely grounder by the electric grinder. Then, 10 mL methanol–deionized water (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, v/v) was added to 0.5 g of TCMs or foodstuffs and shaked them for 30 min. After centrifugation for 10 min (12
30, v/v) was added to 0.5 g of TCMs or foodstuffs and shaked them for 30 min. After centrifugation for 10 min (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g min−1), the supernatant was filtered through a 0.22 μm membrane and stored at 4 °C for later analysis. For AFB1 or ZEN detection in these matrices, known concentrations of AFB1 or ZEN were added into these supernatants and performed visual detection.
000 g min−1), the supernatant was filtered through a 0.22 μm membrane and stored at 4 °C for later analysis. For AFB1 or ZEN detection in these matrices, known concentrations of AFB1 or ZEN were added into these supernatants and performed visual detection.
RGB-based analysis
All images were captured by a smartphone (iPhone xs). To further improve the intuitiveness of visual observation, we used a color detector in order to process the information of red (R), green (G), and blue (B) channels. The color is synthesized by combining R, G, and B components. Each channel is represented in the range 0 to 255, where 0 represents no contribution to the color and 255 represents the maximum contribution of that component. As is typical in this model, black is identified as the lowest contribution of the component, and white is the maximum contribution. Since S.DNAzyme can detect AFB1 and ZEN by a color change from dark green to colorless and from dark green to yellow, respectively, two algorithms were employed for this assay: G/255 for AFB1 and R/G ratio for ZEN. The black background of the paper matrix is determined as the minimum value for all components during the calibration process, and the increased color is represented as an increase in that value. Thus, a lower target concentration produced a low-intensity color, described as a low RGB value, while a higher target concentration produced a high-intensity change, denoted as a high RGB value. To achieve an accurate description, we considered the total distribution of RGB values and used the average value for calculation.32–35Results and discussion
Inspiration from aptamer-based hairpin constructs platform for AFB1 detection
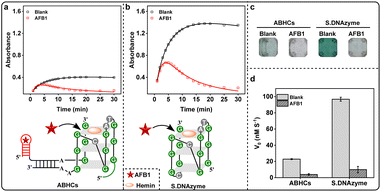
We recently developed aptamer-based hairpin constructs (ABHCs), containing H-1 and H-3, which involve the use of functional nucleic acid that upon recognizing AFB1 trigger the formation of the hemin/G-quadruplex DNAzyme to enable the colorimetric readout of sensing process (Fig. S1 and Table S1†). More specifically, H-1 includes 5′-end of AFB1 aptamer (domain I), linker DNA (domain II) and one-fourth G-quadruplex DNAzyme sequence (one GGG repeat, domain III), while the 5′-end of H-3 has a caged three-fourths of G-quadruplex DNAzyme sequence (three GGG repeats, domain III*) and a programed DNA (domain II*). The fact that H-1 and H-3 cannot self-assemble to active G-quadruplex DNAzyme without AFB1. We expected that AFB1 could trigger the configuration change of H-1 by binding to its domain I, thereby exposing domain II and domain III of H-1. The released domain II would further open H-3 by hybridizing with domain II*, and finally allow the G-quadruplex subunits (domain III and domain III*) self-assemble into the G-quadruplex structure. Then, the assembled G-quadruplex DNAzyme, in presence of hemin, was anticipated to catalyze the oxidation of ABTS by H2O2 to ABTS˙+ (λmax = 418 nm), generating a dark green color for naked eye detection. The catalytic reactivity of G-quadruplex DNAzyme assembled by ABHCs was characterized by determining the initial rate (V0) of ABTS˙+. Interestingly, the V0 value was decreased (Fig. S2a†) and the color of solution clearly faded after reacting for 15 min (Fig. S2b†) upon increasing the concentration of AFB1, which were contrary to our prediction and indicated AFB1 might directly affect the catalytic reactivity of G-quadruplex DNAzyme (Fig. S2c†).To further confirm this hypothesis, a “naked” DNAzyme (S.DNAzyme) with G-rich sequence was employed. In the presence of AFB1, the absorbance of ABTS˙+ at 418 nm was obviously decreased in both ABHCs and S.DNAzyme (Fig. 1a and b) and no recognizable green color was obtained (Fig. 1c). In the absence of AFB1, S.DNAzyme demonstrated a 4-fold increase of V0 value compared to ABHCs (Fig. 1d), because S.DNAzyme with four GGG repeats is more able to spontaneously fold into secondary structures, G-quadruplexes, via intramolecular hydrogen-bonding interactions. Excitingly, AFB1 is the first time to be found, to our knowledge, that it can reduce the activity of G-quadruplex DNAzyme. These results inspired us to construct a label-free, simple, fast, sensitive, and naked-eye detection of AFB1 using S.DNAzyme, and to explore the reaction mechanism of designed approach.
Optimization of experimental conditions
To maximize the signal gain generated by S.DNAzyme, we investigated few experimental conditions including KCl, NaCl and DNA concentration. As previously reported, the monovalent cations, K+ or Na+, have a significant effect on enzymatic activity of G-quadruplex DNAzyme, because they can stabilize the G-quadruplex structure.36 The effect of different concentration of K+ and Na+ was investigated. The signal gain reached the maximum when the K+ concentration increased to 50 mM, while it declined at higher K+ concentration (Fig. S3a†). Moreover, the signal gain increased slightly after reacting with Na+ and reached a plateau at the concentration of 10 mM (Fig. S3b†). Thus, 50 mM K+ and 10 mM Na+ were selected as the optimum ion conditions. In view that the enzymatic activity would depend on S.DNAzyme concentration, the concentration of S.DNAzyme was also studied. The signal gain was achieved the maximum while using 0.75 μM S.DNAzyme (Fig. S3c†). Hence, 0.75 μM S.DNAzyme was chosen as optimum for further study.Colorimetric detection of AFB1 using S.DNAzyme
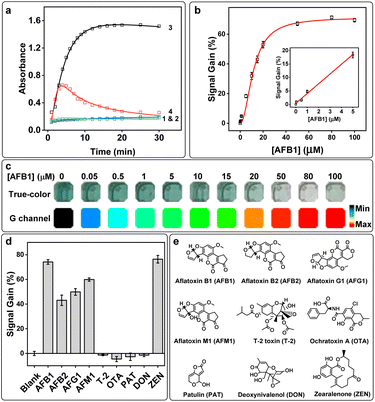
To prove the catalytic activity of S.DNAzyme was only changed in the presence of AFB1, a series of control experiments were set up: group 1, reaction buffer with AFB1; group 2, reaction buffer alone; group 3; reaction buffer with S.DNAzyme and AFB1; group 4, reaction buffer with S.DNAzyme (S.DNAzyme alone) (Fig. 2a and S4a†). No obvious absorbance increase was observed without S.DNAzyme, and further addition of AFB1 did not generate any absorbance change (Fig. 2a). When S.DNAzyme was added into AFB1-free solution (S.DNAzyme alone), a dark green color and a large change in absorbance were yielded within 15 min (Fig. S4b† and 4c). But the addition of AFB1 (group 3) produced a dramatically decrease of absorbance and a rapid development of color fade. These results confirmed the capability of S.DNAzyme for AFB1 determination.Under the optimized conditions, the S.DNAzyme was performed for the visual detection of standard solution with various AFB1 concentrations. The absorbance of ABTS˙+ at 418 nm after 15 min of reaction changed significantly over 0–100 μM AFB1, which achieved a detectable limit of detection (LOD) of 0.18 μM (S/N = 3) with a linear range from 0 to 5 μM AFB1 (Fig. 2b). The color of reaction solution was gradually fade as the AFB1 concentration increased (Fig. 2c). To enhance the observability of color change, these true-color pictures collected by smartphone were further split into red (R), green (G), and blue (B) channels and the G channel intensities were used to quantify the AFB1 concentration. It was able to achieve visual AFB1 detection as low as 0.05 μM (Fig. 2c).
For further application in TCMs or food matrices, an investigation was implemented to evaluate the selectivity of S.DNAzyme toward AFB1. Three aflatoxins with similar structures (AFB2, AFG1, and AFM1) and other toxins, including T-2 toxin (T-2), ochratoxin (OTA), patulin (PAT), deoxynivalenol (DON) and ZEN, were examined using S.DNAzyme. A high signal gain and an obvious color change were observed for AFB1, AFB2, AFG1, and AFM1 (Fig. 2d and S5†), demonstrating that S.DNAzyme possesses brilliant cross-reactivity to structurally similar aflatoxins (Fig. 2e). In addition, no measurable signal gain was detected in the presence of T-2, OTA, PAT, or DON, and none of these toxins caused noticeable color change compared to blank experiment, illustrating the specificity of S.DNAzyme toward aflatoxins over these toxins. It is worth noting that though a high signal gain was obtained in the presence of ZEN (Fig. 2d), the color of this reaction solution was yellowish-brown (Fig. S5†), which was totally different from that of the AFB1 reaction solution. All these results exhibited an excellent characteristic of S.DNAzyme that could achieve visual deterioration of various aflatoxins and ZEN by using a single assay.
Applicability of S.DNAzyme for AFB1 determination in complex analytical matrices
The TCMs and foods samples, such as adlay, platyclade semen, tangerine peel and milk, were used as substrates to evaluate the practicality of S.DNAzyme. Because these are the TCMs and foods most commonly polluted by AFB1.37,38 AFB1-positive real samples were prepared by spiking AFB1 into collected matrices and the absorbance at 418 nm was recorded after 15 min of reaction. Compared with AFB1-free samples, the color and signal gain of the AFB1-containing samples progressively changed upon the increase of AFB1 concentration (Fig. S6†). Based on the calibration curve, the LOD of AFB1 in adlay, platycladi semen, tangerine peel and milk was 1.4 μM, 0.78 μM, 1.17 μM and 2.3 μM, respectively (Table S2†). The obtained results showed the successful application of the S.DNAzyme for AFB1 detection in real samples.Mechanism investigation of AFB1 detection using S.DNAzyme
The production of ABTS˙+ was dramatically inhibited in the presence of AFB1, thus we hypothesized that there might be two potential mechanisms: (1) AFB1 might cause a conversion of ABTS˙+ back into ABTS. (2) AFB1 might induce the hemin/G-quadruplex structure of S.DNAzyme to disassemble, leading to the disappearance of catalytic reactivity of S.DNAzyme. To verify these hypotheses, several control experiments were employed. Specifically, AFB1 was added to the solution after reaction for 0 min and 15 min, respectively. At the same time, the blank experiment was added with DMSO, because AFB1 was dissolved in it. The data showed that the absorption of ABTS˙+ at 418 nm only slightly changed after reaction for 100 min when added AFB1 at the beginning of the reaction (Fig. S7†). Without AFB1, the absorbance of ABTS˙+ progressively increased, and more than 80 μM ABTS˙+ were generated in the solution after reacting for 15 min. At this time, the addition of AFB1 made a sharply decrease of absorbance within 3 min, demonstrating that reduction of ABTS˙+ has occurred and that AFB1 may possess antioxidant effect. Thus, trolox, a widely used standard antioxidant with the ability to scavenge ABTS˙+ within 5 min,39 was selected as the model antioxidant to prove our hypotheses. AFB1 exhibited the same ABTS˙+ scavenging as trolox (Fig. S8†), indicating that AFB1 has an antioxidant capacity to covert ABTS˙+ back into ABTS.Since S.DNAzyme and horseradish peroxidase (HRP) have the same behavior of catalytic activity, it was examined whether AFB1 has the same inhibitory effect on HRP. Interestingly, after incubated with AFB1, only a minimal V0 decrease in HRP-induced reaction was observed (Fig. S9†). Actually, the V0 value of S.DNAzyme was almost 2-fold higher than HRP in the absence of AFB1. But when S.DNAzyme was incubated with AFB1, the V0 was reduced 86%. The big difference between S.DNAzyme and HRP showed that AFB1 might affect the stability of the G-quadruplex structure of S.DNAzyme, which would dramatically decrease the catalytic activity of S.DNAzyme and further reduce the production of ABTS˙+. To further confirm this hypothesis, a circular dichroism (CD) experiment was employed. The CD spectrum of S.DNAzyme in the presence of hemin (S.DNAzyme + hemin) had a positive peak at 240 nm and a negative peak at approximately 270 nm (Fig. S10a†), indicating that S.DNAzyme formed a left-handed G-quadruplex (Fig. S10b†).40,41 When AFB1 was added (S.DNAzyme + hemin + AFB1), two positive peaks (∼210 nm and 275 nm) and a negative peak (235 nm) were observed, suggesting that AFB1 induced the conformation transformation of S.DNAzyme into G-triplex (Fig. S10c†).42 On the basis of the obtained results, it is believed that the AFB1 could inhibit the catalytic reactivity of S.DNAzyme by converting the ABTS˙+ back into ABTS or by reassembling S.DNAzyme into G-triplex (Fig. 3).
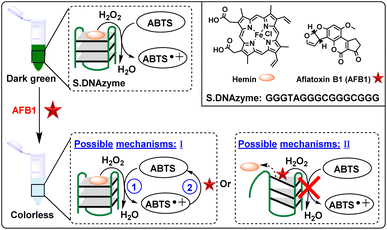 | ||
| Fig. 3 Schematic illustration of the possible working principles of the S.DNAzyme for visual detection of AFB1. | ||
In addition, another G-rich DNA, named as 1XAV,43 with the ability to form a G-quadruplex topology was used to detect AFB1 (Table S1†). Unfortunately, no measurable signal gain was observed in the presence of AFB1 (Fig. S11a†). We then removed the flanking regions of G-quadruplex structure of 1XAV, thereby generating 1XAV-2 (Table S1 and Fig. S11b†). The signal gain immediately rose to the same level as S.DNAzyme (Fig. S11a†), most likely due to the steric hindrance of flanking regions in 1XAV that significantly affected its binding to AFB1.
The capability of S.DNAzyme in ZEN detection by naked-eye
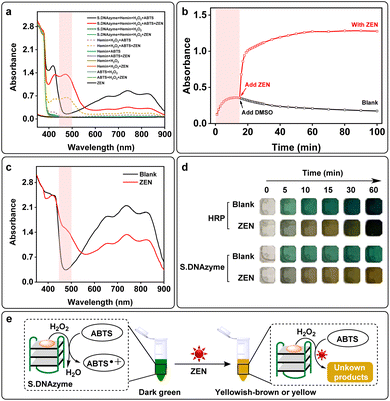
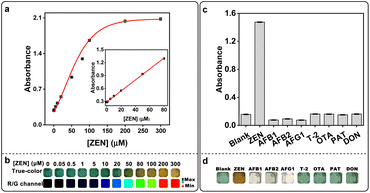
In the previous experiments, we have found that in the presence of ZEN, the solution color would turn into yellowish-brown (Fig. S5†), and its absorption at 472 nm significantly increased while the absorptions at 418 nm and 734 nm were decreased (Fig. S12a† and 4a). Upon addition of reaction time, the absorbance value at 472 nm increased gradually and reached a plateau at 30 min (Fig. S12b†). Without S.DNAzyme, ABTS was slightly oxidized by H2O2 and hemin (hemin + H2O2 + ABTS) which generated a minor increase in the absorbance at 418 nm (Fig. 4a). When ZEN was added (hemin + H2O2 + ABTS + ZEN), a new absorption peak at 472 nm was obtained. Further addition of the S.DNAzyme produced a higher absorbance at 472 nm. But without hemin, H2O2, ABTS or ZEN, this phenomenon did not observe (Fig. 4a and S13†). In addition, ABTS˙+ was produced by S.DNAzyme after reaction for 15 min, but its characteristic peaks at 418 nm and 734 nm immediately reduced after adding ZEN (Fig. S14†). Meanwhile, the absorbance at 472 nm instantly enhanced after adding ZEN with the increase of reaction time (Fig. 4b). These results suggested that the generated ABTS˙+ might be a substrate for ZEN-induced reaction that could produce a new product with a characteristic absorbance peak at 472 nm. The same phenomenon was observed in the HRP-induced oxidation of ABTS platform. Compared with the control experiment (blank), a new peak at 472 nm and a decrease of absorption at 734 nm was detected when added ZEN (Fig. 4c). Though HRP had the similar property to S.DNAzyme, the ratio was much lower (Fig. S15†). Both HRP and S.DNAzyme could identify ZEN by naked eye after reaction for 10 min or 15 min, the color of these ZEN-containing solution was yellowish-brown while it remained dark green for ZEN-free solution (Fig. 4d). Compared with HRP, S.DNAzyme was more suitable for ZEN detection by naked eye. Because there was a significant color difference between ZEN-free and ZEN-containing groups even after 1 h. By contrast, the color difference decreased with the increase of reaction time when employed HRP as a sensor for ZEN assay. This is probably due to the faster reaction velocity of HRP, so that the velocity of ABTS˙+ consumption by ZEN was lower than that of ABTS˙+ production by HRP. This drawback can be avoided by reducing the concentration of HRP. All these results demonstrated that ABTS oxidation platform using S.DNAzyme or HRP was capable of detecting ZEN by naked eye. But in this paper, we focused on introducing S.DNAzyme as sensing element for ZEN detection. Without ZEN, S.DNAzyme would perform H2O2-mediated oxidation of colorless ABTS into dark green ABTS˙+ (Fig. 4e, left). However, the obtained ABTS˙+ would be transformed into a new unknown chemical in the presence of ZEN, generating a yellowish-brown to yellow color visible to the naked eye within 15 minutes (Fig. 4e, right).When ZEN was added to the S.DNAzyme catalytic reaction, the absorbance at 472 nm increased and the solution rapidly developed a yellowish-brown color (Fig. 4d and S12a†). Thus, we generated a calibration curve using the absorbance at 472 nm after 30 min of the reaction using a microplate reader and obtained a measurable detection limit of 0.29 μM with a linear range of 0–80 μM (Fig. 5a). Real-time photos were taken by smartphone, and it enables us to detect ZEN concentrations by the naked eye within 15 min (Fig. 5b and S16†). In addition, true-color images were split into three channels (R, G and B channels) and the ratio of R- and G-channel intensities (R/G ratio) could be used to quantify ZEN concentration. Thus, we further obtained a lower visual detection limit of 10 μM using R/G ratio intensities. Importantly, this assay also displayed high specificity against the other mycotoxins including AFB1, AFB2, AFG1, T-2, OTA, PAT, and DON (Fig. 5c). Only ZEN could be detected via dark green-to-yellowish brown color change visible to the naked eye (Fig. 5d). Moreover, the color change from dark green to colorless could be used for AFs visual detection. To determine the sensitivity of our assay in real samples, ZEN spiked matrices including fritillaria cirrhosis bulbous, angelica sinensis, amygdalin and rice were used to generate calibration curves. Both the absorbance increase (472 nm) and the color change of these real samples were concentration-dependent (Fig. S17†). Furthermore, the obtained limits of detection for ZEN in all real samples were lower than 2 μM (Table S3†). Given the high sensitivity and specificity as well as its ease of use, the S.DNAzyme-based assay would be highly amenable for on-site ZEN detection.
Conclusions
In this work, we have shown for the first time that the ABTS oxidation catalyzed by “naked” DNAzyme (S.DNAzyme) will be modulated by AFB1 or ZEN in different mechanisms. AFB1 can significantly inhibit the catalytic activity of S.DNAzyme through two main ways: first, it may promote the consumption of colored products by converting ABTS˙+ back into ABTS. Second, it may reduce the generation of ABTS˙+ by reassembling S.DNAzyme into G-triplex. For ZEN, we have found it can react with ABTS˙+ to generate yellowish-brown or yellow products. Based on these observations, we directly used the S.DNAzyme as a sensor for AFB1 and ZEN visual detection. Without AFB1 or ZEN, the S.DNAzyme could catalyze the oxidation of ABTS to form the colored ABTS˙+ (dark green). However, the color of solution would immediately change into colorless in the presence of AFB1, while a yellowish-brown or yellow color would be obtained by adding ZEN. Moreover, RGB-based analysis can be used to improve the observability of the collected images. Thus, S.DNAzyme can be considered as an ideal sensing probe for AFB1 and ZEN detection, which enables fast, label-free, enzyme-free and naked-eye detection.Under the optimized conditions, the S.DNAzyme exhibited excellent dynamic linear detection of AFB1 from 0 to 5 μM and ZEN from 0 to 80 μM. This proposed assay demonstrated high detection sensitivity for both AFB1 and ZEN, gaining LOD values as low as 0.18 μM and 0.29 μM, respectively. Using RGB-based analysis, we were able to observe a visible color change after 15 min of reaction in the presence of 0.05 μM AFB1 and 10 μM ZEN, respectively. Moreover, S.DNAzyme was able to determine AFB1 and ZEN in the complex matrices, such as TCMs and foodstuffs. Notably, the S.DNAzyme cross-reacted to other three tested AFs, including AFB2, AFG1 and AFM1, revealing the potential for a broadly applicable method to identify the family of AFs in the real samples. Compared with other colorimetric methods for AFB1 or ZEN detection (Table S4†), our “naked” DNAzyme-based assay is timesaving, low cost, label-free, enzyme-free, antibody-free and naked-eye detection. More than half of reported works only focused one target anlysis (AFB1 or ZEN). However, our method could be used for detection of four tested aflatoxins (AFB1, AFB2, AFG1 and AFM1) and ZEN, demonstrating the potential for a broadly applicable method to identify the family of aflatoxins or ZEN. These advantages make it an excellent sensor for the Food and Drug Administration (FDA) or other quality control laboratories to rapid screen AFs and ZEN in real samples. Thus, once “naked” DNAzyme-based assay is used in clinical practice, it will ensure the safety of foods and drugs to a large extent. The proposed method in this work still needs to do a lot of work to improve its sensitivity to the level of pM or fM.
Although we have achieved a visual detection for AFB1 and ZEN in real samples using S.DNAzyme, and have done various experiments to investigate the reaction mechanisms of the proposed sensing platform, the new product of reaction between ZEN and ABTS˙+ is unknown. Thus, in the future, we will focus on identification of its chemical structure by other methods, such as HPLC-MS trace analysis and nuclear magnetic resonance (NMR).
Author contributions
Conceptualization, Y. L. (Yingping Luo); data curation, Q. L. (Qinrui Lu), Q. L. (Qiao Liu), S. L., and Y. L. (Yingping Luo); formal analysis, Y. L. (Yue Liu); funding acquisition, J. L. and Y. L. (Yingping Luo); investigation, Q. L. (Qinrui Lu); methodology, Q. L. (Qiao Liu) and J. L.; project administration, Y. L. (Yingping Luo); software, Y. L. (Yue Liu), Q. Y., J. T. and Q. S.; supervision, Y. L. (Yingping Luo); visualization, Q. L. (Qinrui Lu); writing – original draft, Q. L. (Qinrui Lu) and Y. L. (Yingping Luo); writing – review & editing, Z. M. and Y. L. (Yingping Luo). All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This research was supported by the Special Projects of Central Government Guiding Development of Local Science and Technology (No. 2021ZYD0051, 2021ZYD0065), National Natural Science Foundation of China (No. 82104116), Opening Project of Medical Imaging Key Laboratory of Sichuan Province (No. MIKLSP202102), and Doctor Research Foundation of North Sichuan Medical College (No. CBY19-QD03). The authors acknowledge the Nanchong Key Laboratory of Metabolic Drugs and Biological Products for providing the platform for research.References
- M. E. Flores-Flores, E. Lizarraga, A. L. de Cerain and E. González-Peñas, Food Control, 2015, 53, 163–176 CrossRef CAS.
- A. G. Marroquin-Cardona, N. M. Johnson, T. D. Phillips and A. W. Hayes, Food Chem. Toxicol., 2014, 69, 220–230 CrossRef CAS PubMed.
- Q. Liu, L. Jiang, L. Xiao and K. wei, AMB Express, 2021, 11(1), 1–10 CrossRef PubMed.
- Z. Ren, J. Luo and Y. Wan, ACS Appl. Mater. Interfaces, 2019, 11(34), 30542–30550 CrossRef CAS PubMed.
- V. Kumar, A. Bahuguna, S. Ramalingam, J. S. Lee, S. S. Han, H. S. Chun and M. Kim, J. Fungi, 2022, 8(2), 190 CrossRef CAS PubMed.
- D. Pickova, V. Ostry, J. Toman and F. Malir, Toxins, 2021, 13(6), 399 CrossRef CAS PubMed.
- A. M. Torres, G. G. Barros, S. A. Palacios, S. N. Chulze and P. Battilani, Food Res. Int., 2014, 62, 11–19 CrossRef CAS.
- P. Li, Q. Zhang and W. Zhang, TrAC, Trends Anal. Chem., 2009, 28(9), 1115–1126 CrossRef CAS.
- A. Rai, M. Das and A. Tripathi, Crit. Rev. Food Sci. Nutr., 2020, 60(16), 2710–2729 CrossRef CAS PubMed.
- V. Ostry, F. Malir, J. Toman and Y. Grosse, Mycotoxin Res., 2017, 33(1), 65–73 CrossRef CAS PubMed.
- A. Vidal, S. Marín, A. J. Ramos, G. Cano-Sancho and V. sanchis, Food Chem. Toxicol., 2013, 53, 133–138 CrossRef CAS PubMed.
- S. M. Herzallah, Food Chem., 2009, 114(3), 1141–1146 CrossRef CAS.
- H. E. Ok, S. W. Choi, M. Kim and H. S. Chun, Food Chem., 2014, 163, 252–257 CrossRef CAS PubMed.
- C. McCullum, P. Tchounwou, L. S. Ding, X. Liao and Y. M. Liu, J. Agric. Food Chem., 2014, 62(19), 4261–4267 CrossRef CAS PubMed.
- R. Wei, F. Qiu, W. Kong, J. Wei, M. Yang, Z. Luo, J. Qin and X. Ma, Food Control, 2013, 32(1), 216–221 CrossRef CAS.
- Z. Xu, L. L. Long, Y. Q. Chen, M. L. Chen and Y. H. Cheng, Food Chem., 2021, 338, 128039 CrossRef CAS PubMed.
- S. Liu, X. Liu, Q. Pan, Z. Dai, M. Pan and S. Wang, Biosensors, 2021, 11(2), 53 CrossRef CAS PubMed.
- R. Li, Y. Wen, L. Yang, A. Liu, F. Wang and P. He, Food Chem., 2022, 366, 130527 CrossRef CAS PubMed.
- Y. Wu, B. Yu, P. Cui, T. Yu, G. Shi and Z. Shen, Anal. Bioanal. Chem., 2021, 413(6), 1629–1637 CrossRef CAS PubMed.
- C. Zong, F. Jiang, X. Wang, P. Li, L. Xu and H. Yang, Biosens. Bioelectron., 2021, 177, 112998 CrossRef CAS PubMed.
- S. C. Pei, W. J. Lee, G. P. Zhang, X. F. Hu, S. A. Eremin and L. J. Zhang, Food Control, 2013, 31(1), 65–70 CrossRef CAS.
- L. Li, J. Wan, X. Wen, Q. Guo, H. Jiang, J. Wang, Y. Ren and K. Wang, Anal. Chem., 2021, 93(19), 7369–7377 CrossRef CAS PubMed.
- C. Zhu, L. Li, G. Yang and F. Qu, Anal. Chem., 2021, 93, 17030–17035 CrossRef CAS PubMed.
- L. Wu, Y. Wang, X. Xu, Y. Liu, B. Lin, M. Zhang, J. Zhang, S. Wan, C. Yang and W. Tan, Chem. Rev., 2021, 121(19), 12035–12105 CrossRef CAS PubMed.
- Y. Jia, G. Zhou, X. Wang, Y. Zhang, Z. Li, P. Liu, B. Yu and J. Zhang, Talanta, 2020, 219, 121342 CrossRef CAS PubMed.
- S. Sun, R. Zhao, S. Feng and Y. Xie, Microchim. Acta, 2018, 185(12), 1–7 CAS.
- S. Wu, L. Liu, N. Duan, Q. Li, Y. Zhou and Z. Wang, J. Agric. Food Chem., 2018, 66(8), 1949–1954 CrossRef CAS PubMed.
- E. Golub, H. B. Albada, W. C. Liao, Y. Biniuri and I. Willner, J. Am. Chem. Soc., 2016, 138(1), 164–172 CrossRef CAS PubMed.
- Y. Seok, J. Y. Byun, W. B. Shim and M. G. Kim, Anal. Chim. Acta, 2015, 886, 182–187 CrossRef CAS PubMed.
- S. Chakraborty, S. M. Shet, M. M. Pereira, S. K. Nataraj and D. Mondal, Int. J. Biol. Macromol., 2021, 183, 1784–1793 CrossRef CAS PubMed.
- P. Pullmann and M. J. Weissenborn, ACS Synth. Biol., 2021, 10(6), 1360–1372 CrossRef PubMed.
- C. Celik, G. Can Sezgin, U. G. Kocabas, S. Gursoy, N. Ildiz, W. Tan and I. Ocsoy, Anal. Chem., 2021, 93(15), 6246–6253 CrossRef CAS PubMed.
- D. C. Christodouleas, A. Nemiroski, A. A. Kumar and G. M. Whitesides, Anal. Chem., 2015, 87(18), 9170–9178 CrossRef CAS PubMed.
- N. R. Ha, I. P. Jung, S. H. Kim, A. R. Kim and M. Y. Yoon, Process Biochem., 2017, 62, 161–168 CrossRef CAS.
- M. He, Z. Li, Y. Ge and Z. Liu, Anal. Chem., 2016, 88(3), 1530–1534 CrossRef CAS PubMed.
- M. Nishio, K. Tsukakoshi and K. Ikebukuro, Biosens. Bioelectron., 2021, 178, 113030 CrossRef CAS PubMed.
- C. Sun, X. Liao, B. Jia, L. Shi, D. Zhang, R. Wang, L. Zhou and W. Kong, Microchim. Acta, 2020, 187(4), 1–9 CrossRef PubMed.
- S. P. Zhao, D. Zhang, L. H. Tan, B. Yu and W. G. Cao, Sci. Rep., 2016, 6(1), 1–9 CrossRef PubMed.
- S. M. Jia, X. F. Liu, D. M. Kong and H. X. Shen, Biosens. Bioelectron., 2012, 35(1), 407–412 CrossRef CAS PubMed.
- W. J. Chung, B. Heddi, E. Schmitt, K. W. Lim, Y. Mechulam and A. T. Phan, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(9), 2729–2733 CrossRef CAS PubMed.
- B. Bakalar, B. Heddi, E. Schmitt, Y. Mechulam and A. T. Phan, Angew. Chem., Int. Ed. Engl., 2019, 58(8), 2331–2335 CrossRef CAS PubMed.
- H. Zhou, Z. F. Wu, Q. J. Han, H. M. Zhong, J. B. Peng, X. Li and X. L. Fan, Anal. Chem., 2018, 90(5), 3220–3226 CrossRef CAS PubMed.
- A. Marchand, F. Rosu, R. Zenobi and V. Gabelica, J. Am. Chem. Soc., 2018, 140(39), 12553–12565 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra05683f |
| This journal is © The Royal Society of Chemistry 2022 |