Open Access Article
Open Access ArticleA terpene cyclase from Aspergillus ustus is involved in the biosynthesis of geosmin precursor germacradienol†
Marlies Peter,
Yiling Yang and
Shu-Ming Li *
*
Institut für Pharmazeutische Biologie und Biotechnologie, Fachbereich Pharmazie, Philipps-Universität Marburg, Robert-Koch-Straße 4, 35037 Marburg, Germany. E-mail: shuming.li@staff.uni-marburg.de
First published on 3rd October 2022
Abstract
The earthy odor of geosmin with a C12 skeleton is known from bacteria, fungi and plants. The sesquiterpenoid germacradien-11-ol (germacradienol) is a crucial intermediate in the biosynthesis of geosmin. A bifunctional terpene cyclase for germacradienol formation and its degradation to geosmin had been described in bacteria. Terpene cyclases were also suggested for geosmin formation in basidiomycetes, but not reported for ascomycetes. We identified a putative terpene cyclase in Aspergillus ustus with low sequence homology to N-termini of the bacterial germacradienol/geosmin synthases. Heterologous expression in Aspergillus nidulans and biochemical characterization led to the identification of the geosmin precursor germacradienol as the sole detected enzyme product. Germacradienol synthase (GdlS) uses strictly farnesyl diphosphate as substrate for cyclization and requires Mg2+ for its reaction. Multiple sequence alignments with known enzymes indicate the presence of the highly conserved catalytic residues including the DDXXD motif for Mg2+ binding. Phylogenetic analysis suggests different clades of bacterial germacradienol/geosmin synthases and terpene cyclases from fungi.
Introduction
Only a few natural products are as widespread as the powerful odiferous volatile geosmin, which is easily smelled after rain fall and in freshly dug earth.1 Geosmin production is strongly conserved in bacteria including actino-, cyano- and myxobacteria.2 Other organisms like mould fungi, arthropods, non-vascular plants (liverworts), vascular plants (red beet, cactus flower), basidiomycetes and amoebae were also reported to be able to produce geosmin.3–5 Geosmin is found as a common contaminant of drinking water, wine and bioaccumulates in fish leading to a musty taste. Geosmin in water and fish usually originates from microbes. Mould fungi attack grapes after cropping leading to rot and associated development of volatile compounds like geosmin, which gives the grapes an earthy and mouldy taste.6 Animals, including humans, can detect it at very low concentrations (5–7 ng L−1). Since geosmin is dispersed throughout almost all kingdoms of life with the exception of archaea,5 the question of its biological activity becomes increasingly important. Recent studies discuss its possible functions as a microbial signalling compound for the production of toxic secondary metabolites.7 It acts also as a signalling molecule between fungi and their host plants in ectomycorrhiza.4The non-canonical terpenoid geosmin with a C12 skeleton was first identified in Streptomyces griseus.1 The sesquiterpene alcohol germacradien-11-ol, shortened as germacradienol in this paper, with a C15 skeleton was isolated from Streptomyces citreus thirty years later.8 The relationship of both compounds was first described in 1996 and later confirmed in Streptomyces coelicolor.9 In the following years, the biosynthetic pathways were intensively investigated by enzyme characterization and mechanistic elucidation, particularly in actinobacteria.2
Formation of geosmin in actino-, cyano- and myxobacteria is catalysed by bifunctional sesquiterpene cyclases, the germacradienol/geosmin synthases. These enzymes share the first described αα domain architecture in bacteria.10 Both active sites are essential for the catalysis. For example, the N-terminal domain (amino acid residues 1–319) of the enzyme SCO6073 from Streptomyces coelicolor (accession number Q9X839.3) catalyses the ionisation-dependent cyclization of farnesyl diphosphate (FPP) to generate the germacradienyl cation A. Proton-initiated capture of water leads to the formation of the key intermediate germacradienol, which is subsequently released from the protein and rebound to its C-terminus, e.g. residues 374–726 aa in the case of SCO6073 from Streptomyces coelicolor (Fig. 1). A metal ion-dependent mechanism is responsible for the transannulation and fragmentation of germacradienol to geosmin.2
In contrast to bacteria, the formation of geosmin in eukaryotes was only studied in recent years. Interpretation of the results obtained from gene deletion experiments led to the hypothesis that a cytochrome P450 monooxygenase might be involved in the geosmin formation in Penicillium expansum by terpene hydroxylation.11 The first eukaryotic geosmin synthase was identified in 2021 in the basidiomycetous fungus Tricholoma vaccinum.4 SiRNA knock-down of the terpene cyclase candidate gene ges1 led to a reduced transcription and less accumulation of geosmin. However, deletion experiments and biochemical characterization were not reported, so that the mechanistic details remain unsolved. Terpene cyclase as a germacradienol synthase was identified in the social amoebae Dictyostelium purpureum.12 In this study, we identified a putative terpene cyclase in Aspergillus ustus and proved its function as a germacradienol synthase (GdlS) without a geosmin-forming activity. To the best of our knowledge, this is the first report for a germacradienol synthase in ascomycetous fungi.
Experimental
Genome sequencing and sequence analysis
The genome of Aspergillus ustus 3.3904 was sequenced by Genewiz (Suzhou, China) using Nova-seq6000/Xten (Illumina). Biosynthetic gene cluster prediction and analysis were performed with antiSMASH (http://antismash.secondarymetabolites.org/)13 and 2ndFind (https://biosyn.nih.go.jp/2ndfind/). The genomic DNA sequence of the gdlS gene reported in this study is deposited at GenBank under the accession number OP095378.To create the phylogenetic trees and alignments, amino acid sequences were collected from NCBI database, using BLASTp program (http://www.ncbi.nlm.nih.gov) for comparing protein sequences. Construction of the phylogenetic trees was carried out with MEGA version 11 software (http://www.megasoftware.net) by using the maximum-likelihood method (Fig. 2 and S1†). ClustalW and ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgibin/ESPript.cgi) were used for multiple sequence alignments (Fig. S2†).
Strains, media and growth conditions
Strains used in this study are listed in Table S1.† Aspergillus ustus 3.3904 was purchased from China General Microbiological Culture Collection Centre (Beijing, China) and cultivated in liquid or on solid PDB (potato dextrose broth, Sigma) at 30 °C. Aspergillus nidulans strains were grown at 37 °C on solid LMM (GMM) medium [1% (w/v) glucose, 50 mg L−1 salt solution, 1 mL L−1 trace element solution, and 0.5% (w/v) yeast extract with 1.5% (w/v) agar] for sporulation and transformation supplemented with uracil (1 g L−1), uridine (1.2 g L−1), riboflavin (2.5 mg L−1) and pyridoxine (0.5 mg L−1). Salt and trace element solutions are as given in the previous publication.14Escherichia coli strains were cultivated at 37 °C in liquid or on solid Luria-Bertani (LB) medium, supplemented with 50 μg mL−1 carbenicillin or 50 μg mL−1 kanamycin for selection. E. coli DH5α and BL21 (DE3) were used for DNA propagation and protein overproduction, respectively.
For homologous recombination in yeast, Saccharomyces cerevisiae HOD114-2B was cultivated at 30 °C in liquid or on solid YPD medium [1% (w/v) yeast extract, 2% (w/v) peptone, and 2% (w/v) glucose]. Synthetic complete medium without uracil (SC-Ura) [6.7 g L−1 yeast nitrogen base with ammonium sulfate, 650 mg L−1 CSM-His-Leu-Ura (MP Biomedicals), histidine and leucine] was used as selection medium.
Gene cloning and transformation
Plasmids and primers used in this study are listed in Tables S2 and S3,† respectively. Primers were synthesised by Seqlab GmbH (Göttingen, Germany). PCR amplification was performed by using Phusion®High-Fidelity DNA polymerase from New England Biolabs (NEB). PCR reaction mixtures and thermal profiles were set as recommended by the manufacturer's instruction.Heterologous expression of gdlS in A. nidulans LO8030 was performed using the vector pJN017,15 which contains flanking sequences of the A. nidulans wA gene for site-specific integration. The gdlS gene was amplified via PCR from the genomic DNA of A. ustus and cloned via homologous recombination in yeast16 into the pJN017 vector, which was linearized by SfoI. Fungal protoplastation and PEG-mediated transformation were performed according to the protocols described previously.17,18 The integrity of the transformant MP02 was verified by PCR amplification (Fig. S3†).
For protein overproduction in E. coli BL21, the coding region of gdlS of 1236 bp was amplified by PCR from cDNA of the gdlS overexpression strain A. nidulans MP02 (wA-PKS::gpdA-gdlS-Afribo) and cloned into pGEM-T-Easy vector (Promega). The sequence integrity and the correct splicing were confirmed by sequencing. Subsequently, the gene was released with the restriction endonucleases BamHI and NdeI from pGEM-T-Easy vector and ligated into the expression vector pET-28a(+), which was linearized by the same restriction endonucleases. The resulted plasmid pMP014 (Fig. S4†) was used for protein overproduction.
Extraction and isolation of germacradienol
Aspergillus nidulans strains generated in this study were cultivated on 20 g of jasmine rice (Royal Tiger) with 30 mL of distilled H2O supplemented with uracil (1 g L−1), uridine (1.2 g L−1), and pyridoxine (0.5 mg L−1) at 25 °C for 7 days for monitoring the secondary metabolite profiles. Aspergillus ustus 3.3904 was cultivated on 20 g of jasmine rice (Royal Tiger) with 30 mL of distilled H2O without supplements for secondary metabolite production (Fig. S5†).To isolate germacradienol, A. nidulans strain MP02 was cultivated in 1 kg of jasmine rice (Royal Tiger) in ten 2L flasks, supplemented with 0.5 g L−1 uracil, 0.5 g L−1 uridine, and 0.5 mg L−1 pyridoxine for 7 days at 25 °C. The cultures were extracted three times with petroleum ether, which was evaporated at 25 °C under slightly reduced pressure to obtain a dry crude extract. The crude extract was subjected to silica gel column chromatography and eluted with petroleum ether:ethyl acetate (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to yield 22 fractions. Fractions 10–17 contained 124 mg of germacradienol in high purity, as confirmed by LC-MS.
1) to yield 22 fractions. Fractions 10–17 contained 124 mg of germacradienol in high purity, as confirmed by LC-MS.
Germacradienol: white powder; 1H NMR (400 MHz, CDCl3, see ESI† for numbering) δ 4.97 (1H, ddd, J = 16.0, 9.8, 1.9 Hz, H-1), 5.66 (1H, dd, J = 16.0, 3.5 Hz, H-2), 2.41 (overlap, H-3), 1.71 (1H, t, J = 3.3 Hz, H-4), 1.51 (1H, dd, J = 8.1, 4.8 Hz, H-4), 2.41 (overlap, H-5), 1.90 (1H, d, J = 14.1 Hz, H-5), 5.03 (1H, d, J = 11.6 Hz, H-6), 2.25 (overlap, H-8), 1.16 (1H, m, H-9), 1.25 (1H, m, H-9), 2.25 (overlap, H-10), 1.07 (3H, s, H-12), 1.16 (3H, s, H-13), 1.10 (3H, d, J = 6.9 Hz, H-14), 1.54 (3H, s, H-15); 13C NMR (100 MHz, CDCl3) δ 123.9 (C-1), 143.3 (C-2), 34.0 (C-3), 32.9 (C-4), 23.9 (C-5), 130.7 (C-6), 131.3 (C-7), 41.5 (C-8), 22.2 (C-9), 59.1 (C-10), 71.9 (C-11), 26.4 (C-12), 27.0 (C-13), 14.9 (C-14), 16.9 (C-15); HRESIMS m/z 223.2054 [M + H]+ (calculated for C15H27O, 223.2056). These data correspond very well to those of germacradien-11-ol described in the literature.19
Feeding experiments
A. ustus culture was grown for 2 days in 10 mL liquid PDB medium in a 50 mL flask at 30 °C and 230 rpm. Germacradienol in DMSO (2 × 25 μmol) was added to the culture at day 3 and 4 after inoculation. A. ustus culture without feeding and PDB medium fed with the same amount of germacradienol served as controls. Samples were taken on day 1, 3 and 5 after feeding and extracted three times with ethyl acetate and resuspended in 50 μL MeOH for LC-MS analysis (Fig. S6†).Overproduction and purification of GdlS
The gdlS expression plasmid pMP014 was transformed into E. coli BL21 (DE3). Protein overproduction and purification were carried out as described in a previous study.20 The recombinant protein was then analysed on 12% (w/v) SDS-PAGE (Fig. 4A) and stored at −80 °C.In vitro assays and determination of the kinetic parameters
To test the enzyme activity of GdlS in vitro, reaction mixtures (50 μL) containing 50 mM Tris–HCl (pH 7.5), 1 mM FPP, 5 mM MgCl2 and 2.5 μg of the purified recombinant protein were incubated at 37 °C for 30 min. After addition of 50 μL H2O, the reaction mixtures were extracted three times with 100 μL of ethyl acetate each. After evaporation under slightly reduced pressure, the samples were dissolved in 25 μL methanol and 5 μL were used for LC-MS analysis (Fig. 5). Assay with heat-inactivated GdlS was used as negative control. Assays with other substrates and additives were performed in the same manner (Fig. 5, Table 1). Enzyme assays for determination of the GdlS kinetics (50 μL) contained 50 mM Tris–HCl pH 7.5, 5 mM MgCl2, 0.5 μg of the purified GdlS and FPP at final concentrations from 0.01 to 2 mM. The reaction mixtures were incubated at 37 °C for 15 min. After addition of 50 μL H2O, the reaction mixtures were extracted with 100 μL of ethyl acetate for three times. The extracts were evaporated, dissolved in 25 μL MeOH and analysed on LC-MS. The isolated germacradienol served as an authentic standard for quantification.LC-MS analysis
LC-MS analysis were performed as described previously.18 To monitor the secondary metabolite profile, separation was performed at a flow rate of 0.25 mL min−1 with a 40 min linear gradient from 5 to 100% ACN in H2O (long elution profile). For evaluation of enzyme assays, a linear gradient from 70 to 100% ACN in H2O in 10 min and a flow rate of 0.3 mL min−1 was used (short elution profile).NMR-analysis
The isolated germacradien-11-ol was dissolved in CDCl3 for NMR analysis. The NMR spectra including 1H NMR, 13C NMR, 1H–1H–COSY, HSQC and HMBC (Fig. S7–S11†) were recorded on a JOEL ECZ-400S spectrometer (JOEL, Tokyo, Japan) at room temperature. All spectra were processed with MestReNov 6.1.0 (Mestrelab Research, Santiago de Compostella, Spain).Results and discussion
Identification and sequence analysis of the putative terpene cyclase gene gdlS in the genome of Aspergillus ustus
The genome of Aspergillus ustus 3.3904 was sequenced in 2015.21 To get a sequence with better quality, we resequenced it20 and identified indeed additional genes. These include a putative terpene cyclase gene, termed gdlS in this study, which was not detected in the sequence published by Pi et al.21 The coding sequence of gdlS of 1236 bp comprises 9 exons with lengths of 161 bp, 124 bp, 19 bp, 195 bp, 157 bp, 90 bp, 34 bp, 224 bp and 232 bp, which are interrupted by 8 introns of 69 bp, 53 bp, 57 bp, 60 bp, 52 bp, 59 bp, 58 bp and 59 bp, respectively. PCR amplification of cDNA and sequencing of the PCR product confirmed this exon-intron structure. The deduced protein comprises 411 amino acid residues with a calculated molecular weight of 46.7 kDa. BLASTp search revealed that GdlS shares only very low sequence homologies with known proteins, e.g. 21% on the amino acid level with the N-terminal sequence (AA 1–319) of the germacradienol/geosmin synthase SCO6073 from Streptomyces coelicolor.22 GdlS homologues with unknown function are found in Aspergillus calidoustus (CEL10922.1), Aspergillus bertholletiae (KAE8381259.1) and Aspergillus bombycis (XP_022394871.1) with sequence identities of 72, 56 and 53%, respectively.Since GdlS shares low sequence similarity with the N-terminal sequence of the germacradienol/geosmin synthase SCO6073 from S. coelicolor, we used both N-terminal (AA 1–319) and C-terminal (AA 374–726) sequences of this enzyme as queries to get homologues from databases. The N-terminal domain sequences of bacterial proteins and homologues from eukaryotes have lengths between 320–510 amino acid residues and those of C-terminus between 320–500 residues. Bacterial proteins share 65–99% sequence identity with each other, whereas eukaryotic candidates show only 20–25% sequence identities with SCO6073, among them 21% between GdlS and SCO6073.
Phylogenetic analysis (Fig. 2 and S1†) revealed clearly the presence of distinct clades for bacteria, ascomycetes and basidiomycetes, for both N- and C-terminal regions of the bacterial germacradienol/geosmin synthases. More homologues of N- than C-terminal sequences of germacradienol/geosmin synthases were found in fungi. Based on sequence alignments and the published data,2 eukaryotic organisms expose no common bifunctional germacradienol/geosmin synthases. Basidiomycetes and a few ascomycetes contain both N-terminal and C-terminal sequences as two distinct proteins. It can be speculated that these proteins catalyse the conversion of FPP to geosmin via germacradienol. Some ascomycetous fungi only have homologues of the N-terminal region, among them the germacradienol synthase GdlS identified in this study. A homologue of the C-terminal sequences of bacterial germacradienol/geosmin synthases could not be found in the genome of A. ustus. We speculate therefore that fungi produce germacradienol more frequently than geosmin. However, it could not be excluded that conversion of germacradienol to geosmin is catalysed in some fungi by other enzymes, as hypothesised for terpene hydroxylation in Penicillium expansum by a cytochrome P450 monooxygenase.11
No germacradienol accumulation in Aspergillus ustus and no conversion of the externly fed germacradienol to geosmin
After identification of the putative germacradienol synthase GdlS, we tried to detect germacradienol and/or geosmin in A. ustus 3.3904 after cultivation under different conditions. No characteristic geosmin smell was realized during the cultivation process. No peaks for germacradienol and geosmin were detected on LC-MS, as examplified for the ethyl acetate extract of a 7 days-old rice culture (Fig. S5†). This indicates that gdlS may be silent in the native strain Aspergillus ustus 3.3904 under the tested laboratory conditions. However, it cannot be excluded that other processes like translation are responsible for the absence of the product.As the next, we proved the possible conversion of germacradienol to geosmin or other metabolites by feeding experiments in A. ustus. The isolated germacradienol was fed to the fungal culture (experimental details see above). However, neither geosmin nor other metabolites could be detected with LC-MS (Fig. S6†). Therefore, we assumed that A. ustus is not able to consume germacradienol.
Heterologous expression of gdlS in Aspergillus nidulans led to the accumulation of germacradienol
To explore the function of the putative terpene cyclase, we initially carried out gene deletion experiments in A. ustus. Unfortunately, genetic approach could not be successfully performed in the producer and we used therefore heterologous expression in A. nidulans LO8030![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 23 for functional proof of the target gene.
23 for functional proof of the target gene.
For this purpose, the gene of interest without upstream and downstream sequences was amplified from genomic DNA of A. ustus and cloned into the expression vector pJN017![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 behind the strong gpdA promoter via homologous recombination in Saccharomyces cerevisiae to create the expression construct pMP008 (Table S2†). After PEG-mediated transformation into A. nidulans protoplasts,17 potential transformants were selected by riboflavine autotrophy and verified by PCR amplification (Table S3 and Fig. S3†). The obtained transformant MP02 was cultivated, extracted and analysed by LC-MS. In comparison to the control strain BK06 with the empty vector pJN017 in A. nidulans LO8030,18 an additional peak at 26.4 min with an absorption maximum at 220 nm was detected (Fig. 3). Detailed inspection revealed that this peak had ions at m/z = 223.2056 (6% intensity) and 205.1951 (100% intensity). For structural elucidation, the accumulated product was isolated from a large-scale fermentation and subjected to NMR analysis. Interpretation of its 1H and 13C NMR spectra including 1H–1H COSY, HSQC and HMBC (Fig. S7–S11†) and comparison with the data in the literature19 confirmed the structure to be germacradien-11-ol (Fig. 1).
15 behind the strong gpdA promoter via homologous recombination in Saccharomyces cerevisiae to create the expression construct pMP008 (Table S2†). After PEG-mediated transformation into A. nidulans protoplasts,17 potential transformants were selected by riboflavine autotrophy and verified by PCR amplification (Table S3 and Fig. S3†). The obtained transformant MP02 was cultivated, extracted and analysed by LC-MS. In comparison to the control strain BK06 with the empty vector pJN017 in A. nidulans LO8030,18 an additional peak at 26.4 min with an absorption maximum at 220 nm was detected (Fig. 3). Detailed inspection revealed that this peak had ions at m/z = 223.2056 (6% intensity) and 205.1951 (100% intensity). For structural elucidation, the accumulated product was isolated from a large-scale fermentation and subjected to NMR analysis. Interpretation of its 1H and 13C NMR spectra including 1H–1H COSY, HSQC and HMBC (Fig. S7–S11†) and comparison with the data in the literature19 confirmed the structure to be germacradien-11-ol (Fig. 1).
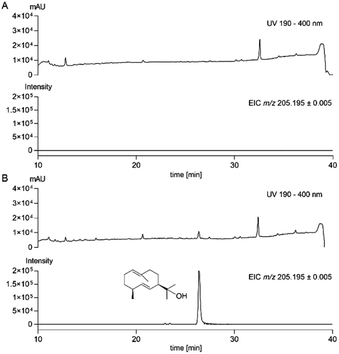 | ||
| Fig. 3 LC-MS monitoring on metabolite production in the control strain A. nidulans BK06 (A) and the transformant MP02 (B). Germacradienol was detected at 26.4 min with the long elution profile. | ||
Biochemical investigations proved germacradienol to be the GdlS product
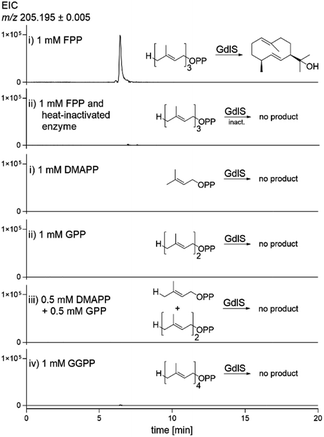
To provide evidence that germacradienol is indeed the product of the terpene cyclase from A. ustus, biochemical investigations were carried out with the recombinant and purified GdlS. The coding sequence of gdlS was amplified from cDNA and inserted into the expression vector pET-28a(+) (Fig. S4†) and transformed into Escherichia coli BL21 (DE3) for protein overproduction. The recombinant protein was purified on Ni-NTA agarose resin to near homogeneity and gave a yield of 2.9 mg per litre culture (Fig. 4A). The purified protein was then incubated with farnesyl diphosphate (FPP) in the presence of Mg2+ at 37 °C for 30 min. Analysis of the reaction mixture after termination with MeOH showed a minor product peak (data not shown). To improve the analytic method, we extracted the reaction mixture with ethyl acetate. After evaporation, the ethyl acetate extract was subsequently analysed by LC-MS. In this way, the sensibility of the detection was significantly increased. As shown in Fig. 5, a product peak was clearly detected, which was absent in the assay with heat-inactivated protein. Comparison of UV, MS and MS2 fragmentation pattern confirmed germacradienol as the enzyme product of GdlS in vitro. No geosmin in the incubation mixture was smelled.To get more insights into substrate specificity of GdlS and the importance of metal ions for its reaction, enzyme assays were performed with different isoprenoid precursors and various monovalent and divalent ions under the same conditions as mentioned above. GdlS showed a strict substrate specificity toward FPP and did not accept any other prenyl diphosphates alone or in combination such as dimethylallyl diphosphate, geranyl diphosphate, dimethylallyl diphosphate/geranyl diphosphate and geranylgeranyl diphosphate (Fig. 5). This feature is comparable with that of SCO6073 from Streptomyces coelicolor.9
Terpene cyclases require Mg2+ ions for their reactions and also use Mn2+ at low concentrations.24 This was confirmed for GdlS in this study (Table 1). Almost no conversion of FPP to germacradienol was detected in the absence of Mg2+ ions and after addition of EDTA. 6.2 and 1.7% of relative activities to that of Mg2+ were calculated for Mn2+ at final concentrations of 0.1 and 5 mM, respectively. About 10% of relative activity was detected in the assay with Co2+. Assays with other ions showed less than 5% of relative activities of that with Mg2+ ions. Substrate inhibition of FPP conversion to germacradienol by GdlS was observed at 0.2 mM or higher concentrations (Fig. 4B). Half maximal reaction velocity was calculated at 0.08 mM FPP.
| Additive | Relative activity [%] |
|---|---|
| a Three independent experiments were carried out and standard deviations are given as ± values. The product yield of 60% with Mg2+ was set to 100% of relative activity. With the exception for Mn2+, other metal ions and EDTA were added to final concentrations of 5 mM. | |
| Mg2+ | 100 |
| No additive | 0.92 ± 0.01 |
| EDTA | 0.12 ± 0.01 |
| Na+ | 1.12 ± 0.06 |
| K+ | 1.14 ± 0.04 |
| Ca2+ | 1.40 ± 0.02 |
| Mn2+ (0.1 mM) | 6.21 ± 0.20 |
| Mn2+ (5 mM) | 1.68 ± 0.03 |
| Fe2+ | 4.53 ± 0.08 |
| Co2+ | 10.50 ± 0.62 |
| Ni2+ | 1.01 ± 0.02 |
| Cu2+ | 0.06 ± 0.01 |
| Zn2+ | 0.41 ± 0.01 |
Conclusions
In summary, a terpene cyclase GdlS, being responsible for the formation of the geosmin precursor germacradienol, was identified in an ascomycete and proven experimentally. Phylogenetic analysis of homologues of GdlS and N-terminal as well as C-terminal regions of the bacterial germacradienol/geosmin synthases revealed distinct clades for bacteria, ascomycetes and basidiomycetes. The characterized GdlS shares only a sequence identity of 21% with the N-terminus of the germacradienol/geosmin synthase from Streptomyces coelicolor, which catalyses the formation of germacradienol. Heterologous expression of gdlS featured germacradienol to be the sole product. Neither germacradienol nor geosmin, even after feeding of germacradienol, could be detected in the native strain. Biochemical investigations confirmed germacradienol as the enzyme product and exposed substrate specificity in the presence of Mg2+ ions. This study provides evidence for different strategies of germacradienol/geosmin formation in bacteria and ascomycetous fungi.Author contributions
M. P. conducted genetic manipulation, biochemical studies, and compound isolation. Y. Y. elucidated the structure. M. P. and S.-M. L. designed the experiments, analyzed the data and wrote the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We thank S. Newel and L. Ludwig-Radtke from the Philipps-Universität Marburg for taking NMR and MS, respectively. This project was financially funded in part by the Deutsche Forschungsgemeinschaft (INST 160/620-1). Y. Y. (201808530447) is a scholarship recipient from the China Scholarship Council.Notes and references
- N. N. Gerber and H. A. Lechevalier, Appl. Microbiol., 1965, 13, 935–938 CrossRef CAS.
- J. S. Dickschat, Nat. Prod. Rep., 2016, 33, 87–110 RSC.
- S. J. Hanson, J. C. Dawson and I. L. Goldman, G3: Genes, Genomes, Genet., 2021, 11, jkab344 CrossRef CAS PubMed.
- O. Abdulsalam, K. Wagner, S. Wirth, M. Kunert, A. David, M. Kallenbach, W. Boland, E. Kothe and K. Krause, Mycorrhiza, 2021, 31, 173–188 CrossRef CAS PubMed.
- X. Chen, T. G. Köllner, Q. Jia, A. Norris, B. Santhanam, P. Rabe, J. S. Dickschat, G. Shaulsky, J. Gershenzon and F. Chen, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 12132–12137 CrossRef CAS PubMed.
- S. La Guerche, S. Chamont, D. Blancard, D. Dubourdieu and P. Darriet, Antonie van Leeuwenhoek, 2005, 88, 131–139 CrossRef.
- L. Zaroubi, I. Ozugergin, K. Mastronardi, A. Imfeld, C. Law, Y. Gelinas, A. Piekny and B. L. Findlay, Appl. Environ. Microbiol., 2022, 88, e0009322 CrossRef.
- D. Ganßer, F. C. Pollak and R. G. Berger, J. Nat. Prod., 1995, 58, 1790–1793 CrossRef.
- D. E. Cane and R. M. Watt, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 1547–1551 CrossRef CAS PubMed.
- G. G. Harris, P. M. Lombardi, T. A. Pemberton, T. Matsui, T. M. Weiss, K. E. Cole, M. Koksal, F. V. Murphy, L. S. Vedula, W. K. Chou, D. E. Cane and D. W. Christianson, Biochemistry, 2015, 54, 7142–7155 CrossRef CAS PubMed.
- M. H. Siddique, T. Liboz, N. Bacha, O. Puel, F. Mathieu and A. Lebrihi, Afr. J. Microbiol. Res., 2012, 6, 4122–4127 CAS.
- H. Xu, J. Rinkel, X. Chen, T. G. Köllner, F. Chen and J. S. Dickschat, Org. Biomol. Chem., 2021, 19, 370–374 RSC.
- K. Blin, S. Shaw, A. M. Kloosterman, Z. Charlop-Powers, G. P. Van Wezel, M. H. Medema and T. Weber, Nucleic Acids Res., 2021, 49, W29–W35 CrossRef CAS PubMed.
- S. A. Stierle and S.-M. Li, J. Fung, 2022, 8, 493 CrossRef CAS.
- F. Kindinger, J. Nies, A. Becker, T. Zhu and S.-M. Li, ACS Chem. Biol., 2019, 14, 1227–1234 CrossRef CAS PubMed.
- L. Mojardín, M. Vega, F. Moreno, H. P. Schmitz, J. J. Heinisch and R. Rodicio, Fungal Genet. Biol., 2018, 111, 16–29 CrossRef PubMed.
- W. B. Yin, Y. H. Chooi, A. R. Smith, R. A. Cacho, Y. Hu, T. C. White and Y. Tang, ACS Synth. Biol., 2013, 2, 629–634 CrossRef CAS PubMed.
- J. Zhou and S.-M. Li, Appl. Microbiol. Biotechnol., 2021, 105, 9181–9189 CrossRef CAS.
- X. He and D. E. Cane, J. Am. Chem. Soc., 2004, 126, 2678–2679 CrossRef CAS.
- L. Zheng, H. Wang, A. Fan and S.-M. Li, Nat. Commun., 2020, 11, 4914 CrossRef CAS PubMed.
- B. Pi, D. Yu, F. Dai, X. Song, C. Zhu, H. Li and Y. Yu, PLoS One, 2015, 10, e0116089 CrossRef PubMed.
- J. Jiang, X. He and D. E. Cane, Nat. Chem. Biol., 2007, 3, 711–715 CrossRef CAS PubMed.
- Y. M. Chiang, M. Ahuja, C. E. Oakley, R. Entwistle, A. Asokan, C. Zutz, C. C. Wang and B. R. Oakley, Angew. Chem., Int. Ed. Engl., 2016, 55, 1662–1665 CrossRef CAS PubMed.
- A. Vattekkatte, S. Garms, W. Brandt and W. Boland, Org. Biomol. Chem., 2018, 16, 348–362 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Strains and verification, primers, plasmids, phylogenetic analysis, NMR data and spectra. See https://doi.org/10.1039/d2ra05033a |
| This journal is © The Royal Society of Chemistry 2022 |