Open Access Article
Open Access ArticleAntibacterial activities against Staphylococcus aureus and Escherichia coli of extracted Piper betle leaf materials by disc diffusion assay and batch experiments
Pimploy Ngamsurachab and
Pornsawai Praipipat *ab
*ab
aDepartment of Environmental Science, Khon Kaen University, Khon Kaen, 40002, Thailand. E-mail: pornprai@kku.ac.th; pimploy@kkumail.com; Tel: +66 818774991
bEnvironmental Applications of Recycled and Natural Materials (EARN) Laboratory, Khon Kaen University, Khon Kaen, 40002, Thailand
First published on 16th September 2022
Abstract
The use of contaminated water by bacteria may cause many diseases, and thus clean water is needed. Chlorine is normally used for the disinfection of wastewater treatment; however, it produces unwanted odors. Using extracted Piper betle (P. betle) is an interesting choice because it is a good chemical compound for bacterial inhibitions. This study attempted to extract P. betle leaf and synthesize P. betle beads (PBB) to characterize materials and investigate antibacterial efficiencies by disc diffusion assay, batch tests, adsorption isotherms, kinetics, and material reusability. The results demonstrated the successful extraction and synthesis of the materials of P. betle. P. betle powder (PBP) had porous and rough surfaces, whereas PBB had a spherical shape with a coarse surface. The four main chemical elements and functional groups of PBP and PBB were carbon, oxygen, calcium, chlorine, and O–H, C–H, N–H, C–O, respectively. The extraction yield and total phenolic, flavonoid, and tannin contents of P. betle were 11.30%, 201.55 ± 0.31 mg GAE per g, 56.86 ± 0.14 mg RE per g, and 41.76 ± 1.32 mg CE per g, respectively. The six main compounds of eugenol, quercetin, apigenin, kaempferol, ascorbic acid, and hydroxychavicol were detected by HPLC analysis. The results of the disc diffusion assay confirmed antibacterial efficiencies of PBB, and the batch tests examined high antibacterial efficiencies of PBB for 100% on Staphylococcus aureus and Escherichia coli. The adsorption isotherms and kinetics of PBB corresponded to Freundlich model and pseudo-second order kinetic model, and the desorption experiments confirmed the reusability of PBB. Therefore, PBB can be possibly applied for an antibacterial purpose in wastewater treatment systems.
Introduction
Water is one of main factors for living organisms; thus, access to clean water is necessary for a safe life. Because of increase in water consumption through many human activities such as household, industry, agriculture, and transportation, water pollution including bacterial contamination in water can occur.1,2 Generally, bacteria are classified in two types, namely, Gram-positive and Gram-negative bacteria. Staphylococcus aureus (S. aureus) is normally represented as a Gram-positive bacteria, and it is also widely used for antibacterial experiments because of its high possibility of causing human diseases.3 Escherichia coli (E. coli) is a common Gram-negative bacteria found in contaminated water as a pathogenic microorganism that causes human diseases, and it is also used as an indicator of water quality.4 As a result, using water contaminated by bacteria may cause many diseases such as diarrhea, dysentery, typhoid fever, and septicemia. Therefore, water treatment is required before its safe use.Ultraviolet light (UV), ozone, and chlorine are generally used for antibacterial treatment in a wastewater system with effective methods against many types of bacteria; however, these methods have the disadvantages of high operating costs, complicated maintenances, and unwanted odor after treatments.5 Therefore, alternative methods have been studied to solve these problems. Many previous studies have investigated various alternatively antibacterial materials against S. aureus and E. coli in the form of nanomaterials with metal oxides of TiO2, ZnO, CuO, MgO, iron oxide, silver nanoparticles (AgNPs), and green materials of various plant extractions with or without metal oxides as shown in Table 1. In Table 1, the antibacterial results of nanomaterials and green materials with metal oxides were closely related values. For green material comparison, their antibacterial activities of green materials with or without metal oxides had close values except that some studies of green materials with Ag or ZnO had higher values than others. However, the antibacterial materials of nanomaterials with metal oxides and green materials with metal oxides have the disadvantage of metal oxides harming the environment after treatment. As a result, an alternative of using green plant extractions without metal oxides may be better than nanomaterials and green materials with metal oxides for the antibacterial activities of Gram-positive and Gram-negative bacteria without a dangerous concern to the ecosystem because they are currently applied in many applications of food preservation, skincare products, hair, dental, cosmetics, and biopesticides.6–11
| Methods/materials | S. aureus/E. coli | Applications | Ref. | |||
|---|---|---|---|---|---|---|
| Agar well diffusion (mm) | Disc diffusion (mm) | MIC (mg mL−1) | MBC (mg mL−1) | |||
| Nanomaterials with metal oxides | ||||||
| Chemical | ||||||
| Ag NPs | — | 12.1/13.5 | 0.075/0.05 | — | Antibacterial activity | 18 |
| ZnO NPs | 12.4 ± 0.1–17.0 ± 0.5 (S. aureus) | 13.1 ± 0.7 to 17.7 ± 0.1 | — | — | Antibacterial and anticancer activity | 19 |
| Mechanochemical | ||||||
| CuO NPs | 11.3–12.4/10.6–11.2 | — | 2.5/3.75 | 5/7.5 | Antibacterial activity | 20 |
| Wet chemical reduction | ||||||
| CuO NPs | — | — | 0.25/1 | — | Antibacterial activity | 21 |
| ZnO NPs | — | — | 0.1 (S. aureus) | — | ||
| Iron-oxide NPs | 13–29/10–26 | — | — | — | Antibacterial activity | 22 |
| Sol–gel | ||||||
| Boron-doped TiO2NPs | 4.5–8.5 (E. coli) | — | — | — | Antibacterial activity | 23 |
| TiO2–ZnO–MgO | 12.00 ± 0.94 to 15.16 ± 0.30/14.00 ± 0.89 to 14.91 ± 0.66 | — | — | — | Antibacterial activity | 24 |
| TiO2NPs | 0.8–2.55 (S. aureus) | — | — | — | Antibacterial activity and dye degradation (methylene blue) | 25 |
| CuO-doped TiO2NPs | 3.25–4.85 (S. aureus) | — | — | — | ||
| Co-precipitation | ||||||
| CuO–NiO–ZnO NPs | 7.2/9 | — | — | — | Antibacterial activity | 26 |
| Refluxing precursor | ||||||
| ZnO NPs | 1–14, 1–7 (S. aureus) | — | — | — | Antibacterial activity | 27 |
| Hydrothermal | ||||||
| MgO NPs | 7–11/8–12 | — | 0.0125–0.05/0.0125–0.06 | — | Antibacterial activity and dye degradation (methyl orange) | 28 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Green materials | ||||||
| Biosynthesis | ||||||
| CuO NPs using actinomycetes | 11.6 ± 0.57 to 19.6 ± 0.57 (S. aureus) | — | — | — | Antibacterial activity | 29 |
| Ag2ONPs using Lippia citriodora | 19 ± 0.02 (S. aureus) | — | — | — | Antibacterial activity and dye degradation (acid orange 8) | 30 |
| Green synthesis | ||||||
| Ag NPs using Carica papaya | — | 14 ± 0.31/15 ± 0.12 | — | — | Antibacterial activity and dye degradation (blue CP and yellow 3RS) | 31 |
| Carica papaya extract | — | 2 ± 0.42/5 ± 0.22 | — | — | ||
| CuO NPs using Ruellia tuberosa | — | 15.5/11 | — | — | Antibacterial activity and dye degradation (crystal violet) | 32 |
| TiO2NPs using Artocarpus heterophyllus | — | 17/23 | — | — | Antibacterial activity | 33 |
| ZnO NPs using Ulva lactuca | 24.0 ± 1.0 (E. coli) | — | — | — | Antibacterial activity, photocatalytic dye (methylene blue) and mosquito larvicides | 34 |
| MgO NPs using Phyllanthus emblica | 6.0 ± 0.29 to 14.0 ± 0.19/8.0 ± 0.24 to 15.0 ± 0.21 | — | — | — | Antibacterial activity and degradation dye (evans blue) | 35 |
| Syzygium polyanthum L. | — | 9.33 ± 0.52/7.00 ± 0.28 | 0.63/1.25 | 1.25/2.5 | Antibacterial activity and food sanitizer | 36 |
| Syzygium antisepticum | — | — | 0.125 (S. aureus) | 0.5 (S. aureus) | Antibacterial activity (application in cooked chicken) | 37 |
| Lawsonia inermis | 15.5 ± 0.5 to 21.3 ± 1.5/8.1 ± 0.4 to 12.1 ± 0.6 | — | 6.25 ± 0.0/1.5 ± 1.4 | 25.0 ± 0.0/12.00 ± 0.0 | Antibacterial activity (application in wound infections) | 38 |
| Azadirachta indica | 6.2 ± 0.3 to 8.2 ± 0.7/6.3 ± 0.3 to 7.4 ± 0.4 | — | 25.0 ± 0.0/83.3 ± 28.9 | 50.0 ± 0.0/100.00 ± 0.0 | ||
| Achyranthes aspera | 6.3 ± 0.6 to 7.0 ± 1.0/6.8 ± 0.8 to 7.6 ± 0.5 | — | 50.0 ± 0.0/100.0 ± 0.0 | 100.0 ± 0.0/200.0 ± 0.0 | ||
Many plants such as Sechium edule, Piper sarmentosum, Mentha cordifolia, Limnophila aromatica, Polygonum odoratum, Garcinia cowa, and Piper betle have chemical compounds such as rutin, catechin, and hydroxylchavicol; thus, they can possibly inhibit both Gram-positive bacteria (Staphylococcus aureus, Staphylococcus epidermidis, Bacillus cereus) and Gram-negative bacteria (Escherichia coli, Klebsiella pneumonia, Pseudomonas aeruginosa).12,13 In particular, Piper betle (P. betle) is an interesting choice for an antibacterial agent for both bacteria types because its leaf has active chemical compounds such as phenols, alkaloids, carbohydrates, flavonoids, chavibetol, and eugenol.14–17 Therefore, this study attempted to extract P. betle from powdered P. betle leaf, and then modify the extracted P. betle to a bead form as a novel antibacterial material from the applied engineering aspect. Moreover, their antibacterial efficiencies on S. aureus and E. coli were investigated through a series of batch experiments for possible disinfectant applications in wastewater treatment.
This study aimed to extract P. betle leaf for antibacterial activities against S. aureus and E. coli to synthesize and characterize P. betle powder (PBP) and beads (PBB), to identify the extraction yield and total phenolic, flavonoid, and tannin contents of extracted P. betle (EPB), to determine compounds of EPB, to examine disc diffusion assay of extracted EPB and PBB, and to investigate the antibacterial efficiencies of PBB by a series of batch experiments, adsorption isotherms, and kinetics. Finally, the material reusability was explored by desorption experiments.
Materials and methods
Raw material
Piper betle (P. betle) leaves in Piperaceae were used as raw materials that were collected from a plantation from Sakon Nakhon province, Thailand between March and April 2019, and then they were kept at room temperature.Chemicals
All chemical reagents were of analytical grade without the purification, which were nutrient agar and nutrient broth (HiMedia, India), 99.9% ethanol (C2H5OH) (RCI Labscan, Thailand), 65% nitric acid (HNO3) (Merck, Germany), sodium hydroxide (NaOH) (RCI Labscan, Thailand), 99.5% dimethyl sulfoxide (C2H6OS or DMSO) (SDFCL, India), sodium alginate (Merck, Germany), calcium chloride (CaCl2) (KEMAUS, New Zealand), Folin-Ciocalteu reagent (Sigma-Aldrich, Germany), gallic acid (Sigma-Aldrich, Germany), rutin (Sigma-Aldrich, Germany), vanillin (Sigma-Aldrich, Germany), catechin (Sigma-Aldrich, Germany), sodium carbonate (Na2CO3) (Sigma-Aldrich, Germany), aluminum chloride hexahydrate (AlCl3) (Sigma-Aldrich, Germany), and hydrochloric acid (HCl) (Sigma-Aldrich, Germany). For HPLC analysis, the methanol (CH3OH), ethanol, and standard chemicals of eugenol, quercetin, apigenin, kaempferol, ascorbic acid, hydroxychavicol, rutin, syringic acid, catechin, sinapic acid, p-coumaric acid, caffeic acid, ferrulic acid, myrecetin, and gallic acid were HPLC grade (Sigma-Aldrich, Germany). 1% sodium hydroxide (NaOH) (RCI Labscan, Thailand) and 1% nitric acid (HNO3) (Merck, Germany) were used for pH adjustments.Microorganisms
Two bacterial strains, namely, Staphylococcus aureus (DMST 562) and Escherichia coli (DMST 4212), which were procured from Department of Medical Sciences, Ministry of Public Health, Bangkok, Thailand (DMST), were used in this study.Preparation of bacterial water sample
The water samples were prepared by diluting S. aureus or E. coli of 108 CFU mL−1 in sterile deionized water to obtain initial S. aureus or E. coli concentrations in the range from 104 to 107 CFU mL−1.Synthesis of materials
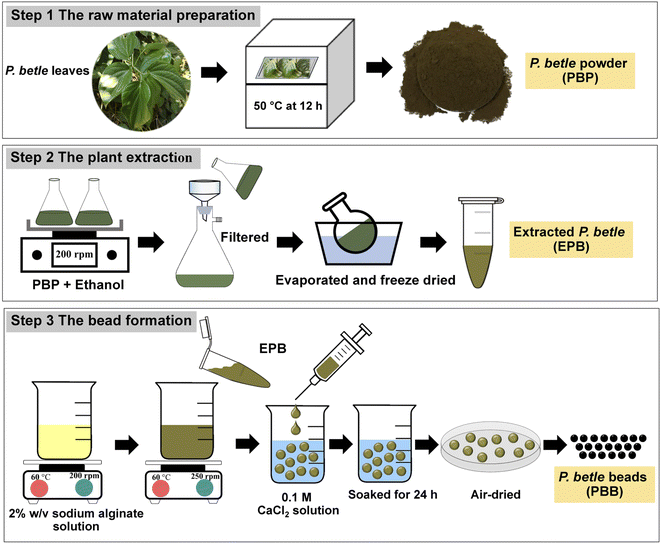
Fig. 1 demonstrated the synthesized steps of three materials, which were P. betle powder (PBP), the extracted P. betle (EPB), and P. betle beads (PBB). The three steps included raw material preparation, plant extraction, and bead formation, and their details are clearly explained below.Note: The amount of EPB that was added into 2% w/v sodium alginate depended upon the concentration of the plant extract to the volume of the solvent solution prepared. For example, 0.1 g EPB was added into 1 mL 2% w/v sodium alginate for a concentration of 100 mg mL−1.
Characterization of materials
Field Emission Scanning Electron Microscopy and Focus Ion Beam (FESEM-FIB) with Energy Dispersive X-Ray Spectrometer (EDX) (FEI, Helios NanoLab G3 CX, USA) were used to investigate the surface morphologies and chemical elements of materials, on which the samples were mounted on aluminum stubs by double-side carbon tapes and coated using a gold-coater for 4 min using a 108 auto Sputter Coater with thickness controller MTM-20 model (Cressington, Ted Pella Inc, USA) by analyzing at 10 kV accelerating voltage. The chemical functional groups of the materials were identified by Fourier Transform Infrared Spectroscopy (FTIR) (Bruker, TENSOR27, Hong Kong) in the range of 4000–600 cm−1 with a resolution of 4 cm−1 and 16 scans over the entire covered range.39Determination of the extraction yield and total phenolic, flavonoid, and tannin contents of P. betle
The extraction yield in percentage was calculated using eqn (1).| Yield (%) = Wc/Wr × 100 | (1) |
For the total phenolic content, a Folin-Ciocalteu assay was used in this study by modifying from the study of Singleton, V. L. et al.40 0.5 mg of the extracted P. betle, 2.5 mL of 10% Folin-Cioaclteu, and 2 mL of 7.5% Na2CO3 were added to a test tube. Then, it was vortexed and left at room temperature to react for 30 min. After that, the total phenolic content was measured by a UV-Vis spectrophotometer (Hitachi, UH5300, Japan) with a wavelength of 760 nm. Triplicate experiments were conducted for confirming the result. The average value was reported in mg gallic acid equivalent per gram of extracted P. betle (mg GAE per g).
For the total flavonoid content, a colorimetric assay was used in this study by modifying from Zhishen, J. et al.41 0.5 mg of the extracted P. betle and 2 mL of 2% AlCl3 were added to a test tube. Then, it was vortexed and left at room temperature to react for 30 min. After that, the total flavonoid content was measured by a UV-Vis spectrophotometer with a wavelength of 420 nm. Triplicate experiments were conducted for confirming the result. The average value was reported in mg rutin equivalent per gram of extracted P. betle (mg RE per g).
For the total tannin content, a vanillin assay was used in this study by modifying from the study of Sun, B. et al.42 0.5 mg of the extracted P. betle and 5 mL of the mixed solution containing 4% AlCl3 and 8% HCI were added to a test tube. Then, it was vortexed and left at room temperature to react for 30 min by keeping in the dark. After that, the total tannin content was measured by a UV-Vis spectrophotometer with a wavelength of 500 nm. Triplicate experiments were conducted for confirming the result. The average value was reported in mg catechin equivalent per gram of extracted P. betle (mg CE per g).
Determination compounds in extracted P. betle by HPLC analysis
For high-performance liquid chromatography (HPLC) (Agilent Technology, series 1220, USA), the HPLC column used was Discovery C18, 250 × 4.6 mm, with 5 μm of particle size (Sigma-Aldrich, Germany). 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 methanol–water solution was applied for setting the mobile phase with a flow rate of 1 mL min−1 for 20 min, and the UV detector was set at 280 nm. The stock standards of each compound were prepared from a ratio of 1 mg of standard
30 methanol–water solution was applied for setting the mobile phase with a flow rate of 1 mL min−1 for 20 min, and the UV detector was set at 280 nm. The stock standards of each compound were prepared from a ratio of 1 mg of standard![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mL of methanol. For sample preparation, 1 mg of extracted P. betle was dissolved in 1 mL ethanol, and then the sample was filtered by a 0.22 μm filter for injecting into the HPLC system. For HPLC analysis, 20 μL of the sample was injected into the HPLC system, triplicate experiments were applied, and the average was reported. Each standard was used under the same conditions with sample analysis for each compound.
1 mL of methanol. For sample preparation, 1 mg of extracted P. betle was dissolved in 1 mL ethanol, and then the sample was filtered by a 0.22 μm filter for injecting into the HPLC system. For HPLC analysis, 20 μL of the sample was injected into the HPLC system, triplicate experiments were applied, and the average was reported. Each standard was used under the same conditions with sample analysis for each compound.
Disc diffusion assay
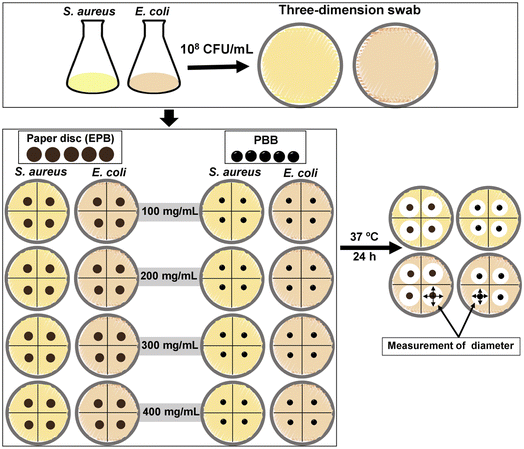
EPB and PBB were used for disc diffusion assay, and the test details are shown in the diagram in Fig. 2. Three steps were applied, whose details are explained below; however, PBB skipped step 2.Step 3.1: EPB. For each plate test, the bacteria were applied to nutrient agar using the three-dimensional swab technique. Then, four pieces of paper disc, which were soaked into the test solution in step 2, were put in a plate test.
Step 3.2: PBB. For each plate test, bacteria were applied to nutrient agar using the three-dimensional swab technique. Then, four pieces of PBB were put in a plate test.
Then, the plates from steps 3.1 and 3.2 were taken in an incubation oven of 37 °C for 24 h. The results were analyzed by the diameter measurement of the inhibition zones. Triplicate experiments of each bacteria were used to confirm the results, and the average of the diameter of the inhibition zones was reported.
Antibacterial batch experiments
Antibacterial batch experiments were designed to investigate the influencing factors of dose, contact time, pH, and initial bacteria concentration on the ability of PBB for inhibiting S. aureus and E. coli by a series of batch experiments. The influencing factors of material dosage of 0.1–0.4 g, contact time of 1–8 h, pH of 5, 7, and 9, and initial bacterial concentration of 104–107 CFU mL−1 were applied with the control condition of a bacterial concentration of 106 CFU mL−1, a sample volume of 100 mL, a shaking speed of 200 rpm, and a temperature of 25 °C. The optimum condition of each influencing factor was used for a next sequencing batch experiment, in which the optimum condition was chosen from the lowest value of each influencing factor, which had the highest antibacterial efficiency of PBB. The results of the antibacterial batch experiments were confirmed by triplicate experiments of each influencing factor and their average values were reported. The plate count technique was used to analyze all the samples, and the percentage of antibacterial efficiency was calculated using eqn (2).| Antibacterial efficiency (%) = ((C0 − Ce)/C0) × 100 | (2) |
Adsorption isotherms
Adsorption isotherms can be used to describe the interaction of the bacteria in the solution with PBB, which was analyzed using linear and non-linear Langmuir isotherm equations in (3) and (4) and Freundlich isotherm equations in (5) and (6), respectively.43,44Langmuir isotherm:
Linear:
| Ce/qe = 1/qmKL + Ce/qm | (3) |
Non-linear:
| qe = qmKLCe/1 + KLCe | (4) |
Freundlich isotherm:
Linear:
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) qe = log qe = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KF + 1/n KF + 1/n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ce Ce
| (5) |
Non-linear:
| qe = KFCe1/n | (6) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) qe versus log
qe versus log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ce, respectively. For non-linear plot features of both the isotherm models, they were plotted by the capacity of bacterial adsorption on PBB at equilibrium (qe) versus the equilibrium of bacterial concentration (Ce).
Ce, respectively. For non-linear plot features of both the isotherm models, they were plotted by the capacity of bacterial adsorption on PBB at equilibrium (qe) versus the equilibrium of bacterial concentration (Ce).
For the adsorption isotherm experiment, 0.3 g PBB was added to 250 mL Erlenmeyer flasks with variable bacterial concentrations in the range from 104 to 107 CFU mL−1. The control condition was a sample volume of 100 mL, a contact time of 6 h, pH 7, a temperature of 25 °C, and a shaking speed of 200 rpm.
Adsorption kinetics
Adsorption kinetics can be used to explain the mechanism of antibacterial efficiency of PBB. The characteristic constants of sorption were investigated using linear and non-linear models of a pseudo-first order kinetic model following eqn (7) and (8) and a pseudo-second order kinetic model following eqn (9) and (10), respectively.45,46Pseudo-first order kinetic model:
Linear:
ln(qe − qt) = ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) qe − k1t qe − k1t
| (7) |
Non-linear:
| qt = qe(1 − e−k1t) | (8) |
Pseudo-second order kinetic model:
Linear:
| t/qt = 1/k2qe2 + (1/qe)t | (9) |
Non-linear:
| qt = k2qe2t/(1 + qek2t) | (10) |
For adsorption kinetics experiment, 3 g PBB were added to a 1000 mL beaker with a bacterial concentration of 106 CFU mL−1. The control condition was a sample volume of 1000 mL, a contact time of 8 h, pH 7, a temperature of 25 °C, and a shaking speed of 200 rpm.
Desorption experiments
Desorption experiments were used to examine the possible reuse of PBB. After an adsorption process, PBB were desorbed using 100 mL 0.01 M HNO3 solution with a shaking speed of 200 rpm for 2 h. After that, PBB was washed with deionized water and dried at room temperature. Then, PBB was ready for the next adsorption cycle. The desorption efficiency in percentage was calculated following eqn (11).| Desorption (%) = (qd/qa) × 100 | (11) |
Results and discussion
The physical characteristics of P. betle
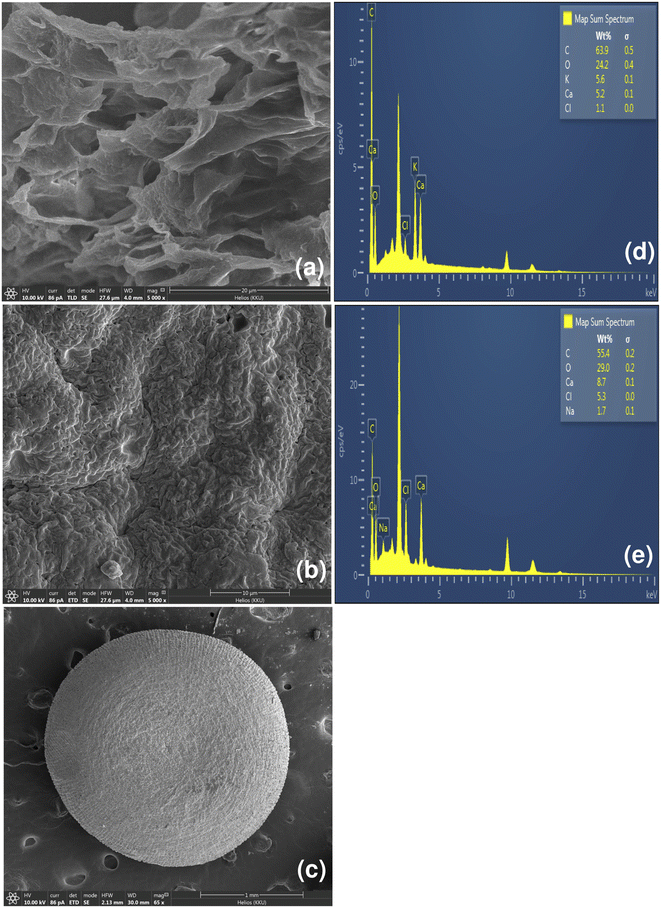
P. betle is a member of the Piperaceae family, which is commonly grown in Southeast Asia such as Indian, Malaysia, Vietnam, Myanmar, Indonesia, and Thailand. It is a perennial with the height of 20 m and a diameter of 15–20 cm, whose fruit is fully concrescent and velutinous or woolly with 3–5 cm length. P. betle leaf is simple and alternate, and the leaf color is bright or dark green.47 The physical characteristics of P. betle in leaf, powder, and bead forms used in this study are presented in Fig. 3a–c. The leaf is an ovate-shaped simple leaf and cordate leaf base with alternate leaf arrangement, as illustrated in Fig. 3a. Both sides of the leaf have smooth surfaces with a dark green color, but the upper surface has a glossier color than the lower surface. P. betle powder (PBP) has a dark green–brown color, whereas P. betle beads (PBB) has black colored beads, which are displayed in Fig. 3b and c, respectively.Material characterization
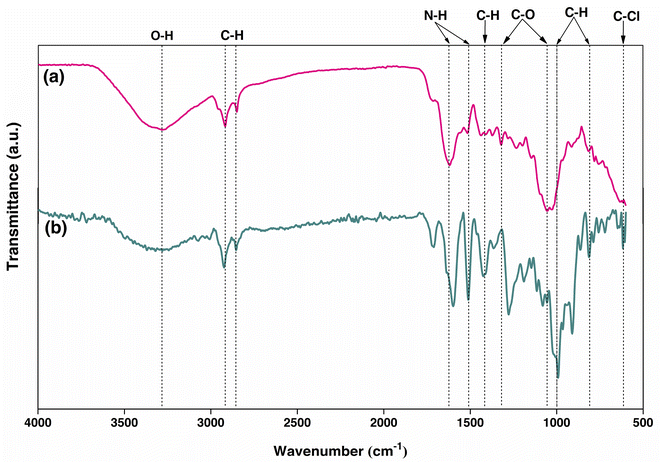
The chemical functional groups of PBP were observed for O–H at 3284.12 cm−1, C–H at 2918.29 and 2850.69 cm−1, N–H at 1618.94 cm−1, and C–O at 1319.26 and 1054.06 cm−1, which were similarly found in other studies with a little shift of the wavelengths.50,52 For PBB, the chemical functional groups were presented for O–H at 3320.44 cm−1, C–H at 2923.82, 1424.46, 991.75, 910.76, and 813.05 cm−1, N–H at 1598.79 and 1511.29 cm−1, C–O at 1277.87 and 1189.70 cm−1, and C–Cl at 617.30 cm−1. As a result, the changing form from powder to beads is affected by changing the chemical functional groups of materials by only PBB found C–Cl (chloride compound) from the main chemical functional groups.
Determination of the extraction yield and total phenolic, flavonoid, and tannin contents of P. betle
The extraction yield and total phenolic, flavonoid, and tannin contents of P. betle demonstrated in Table 2. The extraction yield of P. betle was 11.30%. The total phenolic, flavonoid, and tannin contents of P. betle were 201.55 ± 0.31 mg GAE per g, 56.86 ± 0.14 mg RE per g, and 41.76 ± 1.32 mg CE per g, respectively, and these results were closely related to other previous studies for extracted P. betle by ethanol.55–58| Extraction yield (%) | Mean ± SD | ||
|---|---|---|---|
| Total phenolic content (mg GAE per g) | Total flavonoid content (mg RE per g) | Total tannin content (mg CE per g) | |
| 11.30% | 201.55 ± 0.31 | 56.86 ± 0.14 | 41.76 ± 1.32 |
Determination compounds in extracted P. betle by HPLC analysis
Several compounds were detected in extracted P. betle by HPLC analysis, as reported in Table 3. The six main compounds of eugenol, quercetin, apigenin, kaempferol, ascorbic acid, and hydroxychavicol were observed to be similarly found in previous studies,56 whereas the nine compounds, namely, rutin, syringic acid, catechin, sinapic acid, p-coumaric acid, caffeic acid, ferrulic acid, myrecetin, and gallic acid, were commonly found in small amounts correlated with the reports of other studies.59| Compound names | Content of compounds (mg g−1) (mean ± SD) |
|---|---|
| Eugenol | 18.06 ± 0.12 |
| Quercetin | 15.88 ± 0.15 |
| Apigenin | 5.15 ± 0.22 |
| Kaempferol | 2.77 ± 0.19 |
| Ascorbic acid | 1.86 ± 0.24 |
| Hydroxychavicol | 1.51 ± 0.17 |
| Rutin | 0.81 ± 0.08 |
| Syringic acid | 0.30 ± 0.04 |
| Catechin | 0.29 ± 0.05 |
| Sinapic acid | 0.26 ± 0.03 |
| p-Coumaric acid | 0.22 ± 0.07 |
| Caffeic acid | 0.14 ± 0.02 |
| Ferrulic acid | 0.06 ± 0.04 |
| Myrecetin | 0.04 ± 0.02 |
| Gallic acid | 0.03 ± 0.01 |
Results of the disc diffusion assay
Disc diffusion assay is a method to examine the efficiency of extracted P. betle on specific organisms (S. aureus and E. coli). In this study, a paper disc was used as the preliminary test to determine the antibacterial efficiency on both bacterial types with varying concentrations of extracted P. betle in the range of 100–400 mg mL−1 before PBB was used to confirm the modified bead materials of extracted P. betle leaf. The results of disc diffusion assay are reported in Table 4 and Fig. 6.| Materials | The average diameter of inhibition zone (mm) | ||||
|---|---|---|---|---|---|
| 100 mg mL−1 | 200 mg mL−1 | 300 mg mL−1 | 400 mg mL−1 | ||
| EPB | S. aureus | 15.3 ± 1.0 | 21.2 ± 0.3 | 21.3 ± 1.3 | 21.4 ± 1.2 |
| E. coli | 22.5 ± 1.0 | 24.5 ± 1.3 | 24.6 ± 0.6 | 24.7 ± 1.0 | |
| PBB | S. aureus | 11.4 ± 0.5 | 14.5 ± 0.7 | 15.7 ± 1.2 | 15.9 ± 0.4 |
| E. coli | 12.6 ± 0.6 | 15.4 ± 0.1 | 15.8 ± 0.9 | 16.0 ± 0.7 | |
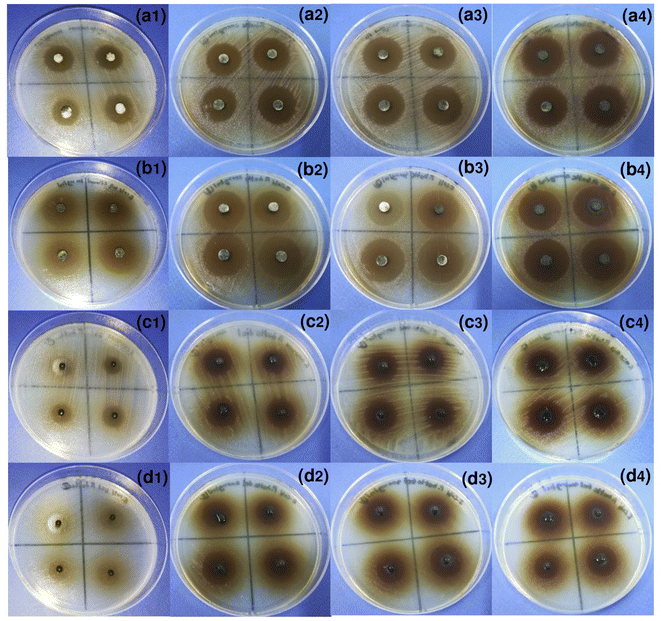 | ||
| Fig. 6 The images of disc diffusion assay on S. aureus and E. coli by (a, b) EPB and (c, d) PBB in varying concentrations of (1) 100 mg mL−1, (2) 200 mg mL−1, (3) 300 mg mL−1, and (4) 400 mg mL−1. | ||
For EPB, the average diameters of inhibition zones with varying EPB concentrations from 100 to 400 mg mL−1 on S. aureus and E. coli were 15.3 ± 1.0 and 22.5 ± 1.0, 21.2 ± 0.3 and 24.5 ± 1.3, 21.3 ± 1.3 and 24.6 ± 0.6, and 21.4 ± 1.2 and 24.7 ± 1.0 mm, respectively, and the average of four concentrations were 19.8 ± 3.0 and 24.1 ± 1.1 mm, respectively. As a result, their antibacterial efficiencies increased with increasing EPB concentrations, and they were constant at concentrations of 200–400 mg mL−1 with average values of 21.3 ± 0.1 and 24.6 ± 0.1 mm for S. aureus and E. coli, respectively. Moreover, EPB demonstrated higher antibacterial efficiency on E. coli than S. aureus. Since their average diameters of inhibition zones were the same value at 300 mg mL−1 EPB concentration, thus, 300 mg mL−1 was the optimum concentration for inhibition on both bacterial types.
For PBB, the average diameters of the inhibition zones at varying EPB concentrations from 100 to 400 mg mL−1 on S. aureus and E. coli were 11.4 ± 0.5 and 12.6 ± 0.6, 14.5 ± 0.7 and 15.4 ± 0.1, 15.7 ± 1.2 and 15.8 ± 0.9, and 15.9 ± 0.4 and 16.0 ± 0.7 mm, respectively, and the average of four concentrations were 14.4 ± 2.1 and 15.0 ± 1.6 mm, respectively. As a result, their antibacterial efficiencies were increased with the increasing EPB concentrations, and they were constant at concentrations in the range of 200–400 mg mL−1 with average values of 15.4 ± 0.8 and 15.7 ± 0.5 mm for S. aureus and E. coli, respectively. In addition, PBB demonstrated higher antibacterial efficiency on E. coli than S. aureus, which agreed with the results from paper discs. Since their average diameters of the inhibition zones were nearly the same value at 300 mg mL−1 EPB concentration, thus 300 mg mL−1 was used as the optimum concentration for inhibitions on both bacterial types by PBB.
For comparing the results of the disc diffusion assay by EPB and PBB, the results of both materials demonstrated the same tendency of increasing antibacterial efficiency with increasing EPB concentration, and the EPB concentration of 300 mg mL−1 was the appropriate concentration for inhibition on S. aureus and E. coli. In deep consideration of the test material effect, two factors of diameter of the test materials and the interference of sodium alginate in bead materials might be a reason why the antibacterial activities of paper discs (EPB) were higher than PBB. For the diameter of the test materials, the diameter of paper disc (6 mm) was larger than the diameter of the glass syringe (3 mm) in bead formation; thus, it was possible that the paper disc might get a higher volume of extracted P. betle solution than bead materials, although the EPB concentrations were the same. For the interference of sodium alginate, the bead formation of extracted P. betle by sodium alginate might be an obstacle in the release of extracted P. betle for the antibacterial activity. However, the difference in the average diameters of inhibition zones with EPB concentrations of 100–400 mg mL−1 by paper disc (EPB) and PBB were not focused on because paper disc tests were only used as a preliminary confirmation of the ability of EPB against S. aureus and E. coli, whereas this study would focus on PBB for applying it in batch experiments.
Moreover, the comparison of the average diameter of the inhibition zone of the extracted P. betle leaf against S. aureus and E. coli to other previous studies with various extraction solutions is reported in Table 5. The six extracted solvents of methanol, ethyl acetate, petroleum ether, chloroform, water, and ethanol were used for P. betle extraction to inhibit S. aureus and E. coli. For S. aureus, the ethanol solvent demonstrated a higher antibacterial activity than the other solvents. EPB presented a higher zone of inhibition on S. aureus than other studies except the studies of Valle et al. and Khan J. A. and Kumar N.16,60. On the other hand, PBB illustrated a higher or closer zone of inhibition on S. aureus than other studies except the studies of Valle et al. and Khan J. A. and Kumar N.16,60 similar to EPB. As a result, ethanol was recommended as an extraction solvent to inhibit S. aureus. For E. coli, the methanol solvent demonstrated a higher antibacterial activity than other solvents; however, it presented a little higher zone of inhibition on E. coli than the ethanol solvent. EPB presented a higher zone of inhibition on E. coli than other studies except the study of Jayalakshmi B. et al.,61 whereas PBB demonstrated a higher or closer zone of inhibition on E. coli than other studies except the studies of Jayalakshmi B. et al., Valle D. L. et al., and Kulnanan P. et al.16,61,62. Finally, since methanol is known as a more toxic solvent than ethanol, ethanol might be suitable for use as the extraction solvent of P. betle for the antibacterial activities on both S. aureus and E. coli more than methanol.
| Solvents/methods | The average diameter of inhibition zone (mm) | Reference | |
|---|---|---|---|
| S. aureus | E. coli | ||
| Methanol | |||
| Agar well diffusion | 25 | 15 | 60 |
| — | 25.5 ± 0.28 | 61 | |
| Disc diffusion | — | 23.5 ± 0.28 | |
| 6.77 ± 0.25 | 8.53 ± 0.25 | 63 | |
| — | 19 | 64 | |
| 15.02 ± 0.27 to 21.03 ± 0.79 | 16.28 ± 0.19 to 16.40 ± 0.23 | 49 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Ethyl acetate | |||
| Agar well diffusion | — | 20.25 ± 0.25 | 61 |
| Disc diffusion | — | 18.75 ± 0.47 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Petroleum ether | |||
| Agar well diffusion | — | 12.25 ± 1.03 | |
| Disc diffusion | — | 13.25 ± 0.28 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Chloroform | |||
| Agar well diffusion | — | 22.75 ± 0.47 | |
| Disc diffusion | — | 18.25 ± 0.25 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Water | |||
| Agar well diffusion | — | 18 | 65 |
| 5.4 ± 0.01–12.3 | 8.5 ± 0.10 | 66 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| Ethanol | |||
| Agar well diffusion | 9.7 ± 0.02 to 18.0 ± 0.18 | 8.9 ± 0.21 to 11.0 ± 0.12 | 66 |
| 16 | 17 | 60 | |
| 2.50 ± 5.00 to 20.38 ± 6.33 | — | 67 | |
| 9.5–15 | 11–17 | 57 | |
| 13.92–18.40 | 14.15–16.40 | 68 | |
| Disc diffusion | 30 | 16 | 16 |
| 10.5 ± 0.8 | 69 | ||
| 15.06 ± 0.57 to 19.02 ± 0.74 | 14.93 ± 0.51 to 15.01 ± 0.57 | 49 | |
| 8 ± 0.25–13 ± 0.43 | 70 | ||
| — | 20 | 64 | |
| — | 20.00–0.00 | 62 | |
| EPB | 15.3 ± 1.0–21.4 ± 1.2 | 22.5 ± 1.0–24.7 ± 1.0 | This study |
| PBB | 11.4 ± 0.5–15.9 ± 0.4 | 12.6 ± 0.6–16.0 ± 0.7 | This study |
Therefore, the results of disc diffusion assay confirmed the abilities for inhibiting both bacterial types by EPB and PBB, and then, PBB was used to investigate the antibacterial efficiency by the batch tests with several affecting factors to confirm its efficiency to remove S. aureus and E. coli in water to study its possibly application for a disinfection process in the wastewater treatment system in the future.
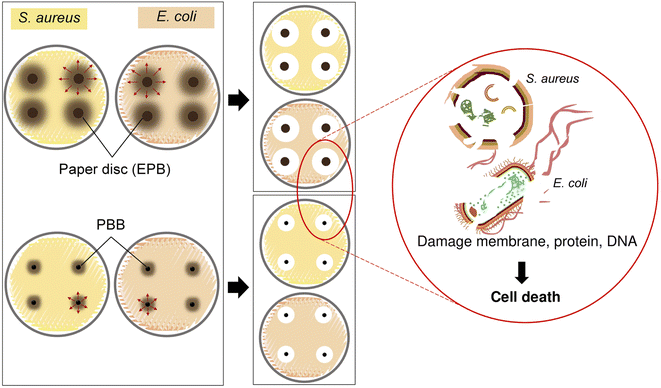
The possible mechanisms of S. aureus and E. coli inhibition by extraction or beads materials (EPB and PBB)
Fig. 7 demonstrated the possible mechanisms of S. aureus and E. coli inhibition by EPB and PBB modified from a previous study.39 For EPB, a paper disc soaked with extracted P. betle solution contacted the cell bacteria in a plate test, and then the phenolic compounds, alkaloids, and flavonoids in the extracted P. betle that penetrated into the cell resulted in cell death by damaging the membrane, protein, DNA, and the main functions of bacteria.71,72 For PBB, the same method was applied for the inhibition mechanism as that of EPB, but it only changed the material test from the paper disc to the extracted P. betle in the bead materials.Therefore, using extracted P. betle by changing it a stable bead form may help to convenient apply it for the future wastewater treatment with easy separation after treatment instead of directly using extracted P. betle, which may be separated with difficulty from the treated wastewater, while the antibacterial batch experiments were designed to preliminarily prove this idea.
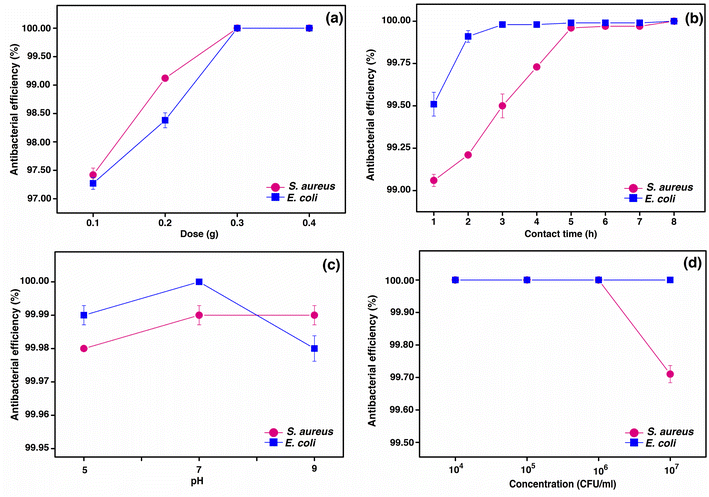
Antibacterial batch experiments
In conclusion, the material dosage had a higher influence on the bacterial removal efficiencies of PBB on both bacterial types than the contact time, pH, and concentration, and the optimum condition of PBB to inhibit S. aureus and E. coli was 0.3 g, 6 h, pH 7, and 106 CFU mL−1 for 100% bacterial removal. On comparing with a previous study,39 the optimum condition of this study had a similar material dose of 0.3 g and pH of 7 with the previous study for 100% inhibition of S. aureus and E. coli, whereas this study spent a contact time of 6 h more than the previous study, which was 3 h and 2 h for S. aureus and E. coli, respectively, possibly resulting from the use of different plants.
Adsorption isotherms
The adsorption isotherm studies of PBB on S. aureus and E. coli with the fitting of linear and non-linear Langmuir and Freundlich models are demonstrated in Fig. 9. For the linear model, the Langmuir model was plotted for Ce/qe versus Ce, whereas the Freundlich model was plotted for log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) qe versus log
qe versus log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ce. For the non-linear model, both the models were plotted as qe versus Ce. In addition, the equilibrium isotherm parameters are reported in Table 6.
Ce. For the non-linear model, both the models were plotted as qe versus Ce. In addition, the equilibrium isotherm parameters are reported in Table 6.
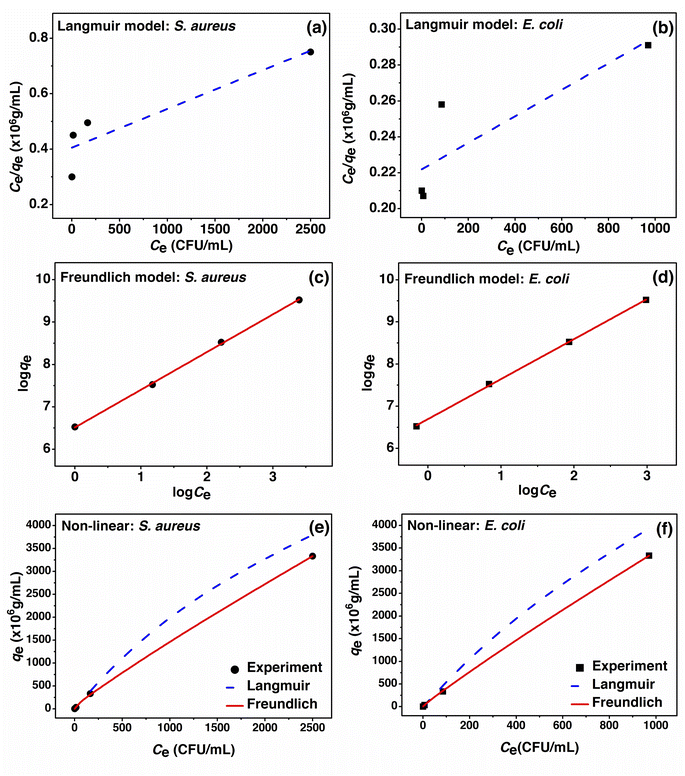 | ||
| Fig. 9 (a–d) Linear and (e, f) non-linear adsorption isotherm models of PBB on S. aureus and E. coli. | ||
| Isotherm model | Linear | Non-linear | ||
|---|---|---|---|---|
| S. aureus | E. coli | S. aureus | E. coli | |
| Langmuir model | ||||
| qm (CFU g−1) | 1 × 1010 | 1.4286 × 1010 | 1.0083 × 1010 | 1.4404 × 1010 |
| KL (L CFU−1) | 0.0003 | 0.0004 | 0.0002 | 0.0003 |
| R2 | 0.8344 | 0.7407 | 0.9714 | 0.9516 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Freundlich model | ||||
| KF (CFU g−1) (L CFU−1)1/n | 3.2546 × 106 | 4.9136 × 106 | 3.6357 × 106 | 5.4890 × 106 |
| 1/n | 0.8895 | 0.9502 | 0.8718 | 0.9318 |
| R2 | 0.9994 | 0.9997 | 1 | 1 |
For the Langmuir linear model, the maximum adsorption capacity (qm) of PBB on S. aureus and E. coli was 1 × 1010 and 1.4286 × 1010 CFU g−1, respectively, and the Langmuir linear adsorption constants (KL) of PBB were 0.0003 and 0.0004 L CFU−1 for S. aureus and E. coli, respectively. For the Langmuir non-linear model, the maximum adsorption capacity (qm) of PBB on S. aureus and E. coli was 1.0083 × 1010 and 1.4404 × 1010 CFU g−1, respectively, and the Langmuir non-linear adsorption constants (KL) of PBB were 0.0002 and 0.0003 L CFU−1 for S. aureus and E. coli, respectively. The R2 of Langmuir linear and non-linear models were 0.8344 and 0.9714 for S. aureus and 0.7407 and 0.9516 for E. coli, respectively. In the Freundlich model, 1/n is the constant depicting the adsorption intensity, and 1/n < 1 means favorable adsorption with different concentrations, where the adsorption capacity will decrease with increasing concentration.73 For the Freundlich linear model, the 1/n values of PBB on S. aureus and E. coli were 0.8895 and 0.9502, respectively, and the 1/n value of E. coli was higher than that of S. aureus, which meant that PBB had an inhibition capacity on E. coli better than that on S. aureus. Moreover, the Freundlich linear adsorption constants (KF) of PBB on S. aureus and E. coli were 3.2546 × 106 and 4.9136 × 106 (CFU g−1) (L CFU−1)1/n, respectively. For the Freundlich non-linear model, the 1/n values of PBB on S. aureus and E. coli were 0.8718 and 0.9318, respectively, and the 1/n value of E. coli was also higher than S. aureus, which meant that PBB had a better inhibition capacity on E. coli than S. aureus. Moreover, the Freundlich non-linear adsorption constants (KF) of PBB on S. aureus and E. coli were 3.6357 × 106 and 5.4890 × 106 (CFU g−1) (L CFU−1)1/n, respectively. The R2 of Freundlich linear and non-linear models were 0.9994 and 1 for S. aureus and 0.9997 and 1 for E. coli, respectively. Generally, the R2 value was used to decide which adsorption isotherm model well explains the adsorption pattern of the adsorbent; thus, a higher R2 close to 1 was suitably chosen. As a result, the adsorption isotherms of PBB on S. aureus and E. coli corresponded to Freundlich model in both the linear model (R2 of 0.9994 and 0.9997) and non-linear models at R2 equal to 1, which is related to the physiochemical adsorption process similarly found in a previous study.39 On comparing the linear and non-linear adsorption isotherm models, their results were found to have close adsorption parameters, which represented the agreement results of linear and non-linear models similarly to other studies.74–77 As a result, the plotting of the linear and non-linear adsorption isotherm models is necessary to avoid data mistranslation.78,79
Adsorption kinetics
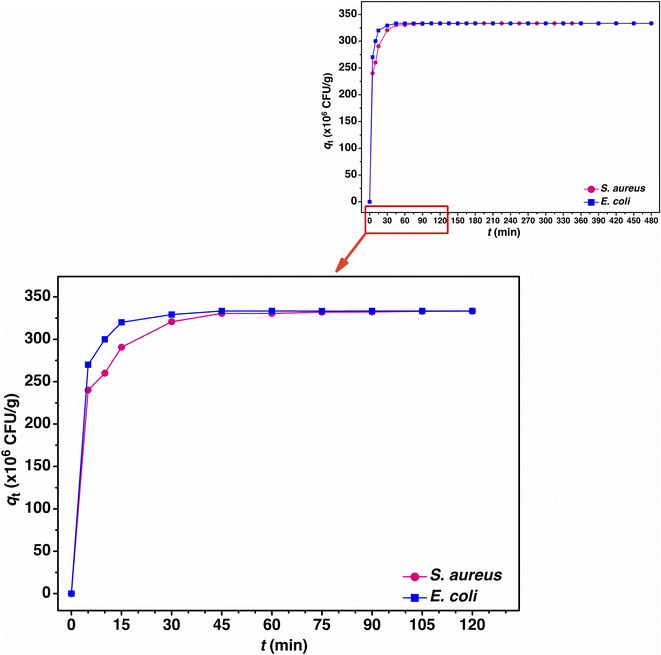
The adsorption mechanism including the rate of adsorption to time by the adsorbent can be explained by adsorption kinetics.80 For the linear model, a pseudo-first order and a pseudo-second order kinetic models were used for the adsorption kinetic studies of PBB by the plotting of ln(qe − qt) versus time (t) for the pseudo-first order linear kinetic model and t/qt versus time (t) for the pseudo-second order linear kinetic model, respectively. For the non-linear model, both kinetic models for the plotting of qt versus time (t) are shown in Fig. 10. The adsorption kinetic equilibrium of PBB on S. aureus and E. coli is represented in Fig. 11, and the adsorption kinetic parameters are reported in Table 7.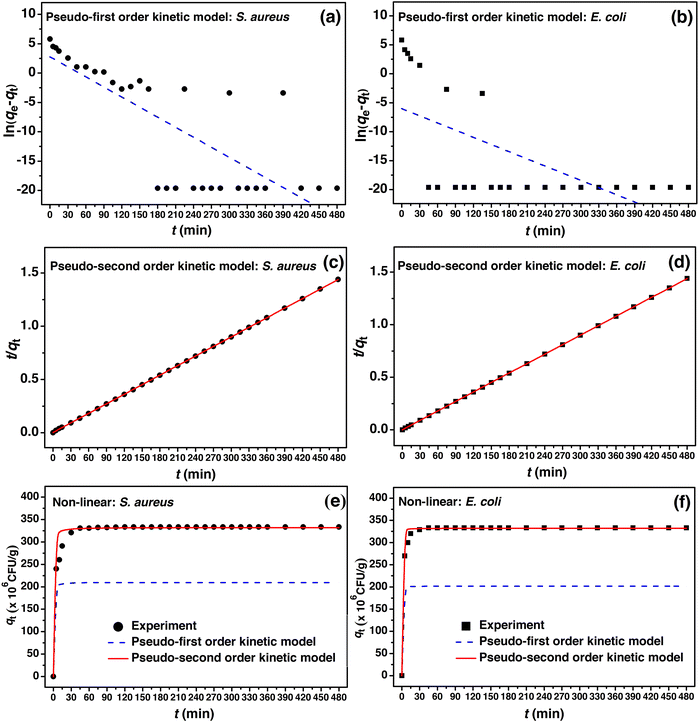 | ||
| Fig. 10 (a–d) Linear and (e, f) non-linear adsorption kinetic models of PBB on S. aureus and E. coli. | ||
| Kinetic model | Linear | Non-linear | ||
|---|---|---|---|---|
| S. aureus | E. coli | S. aureus | E. coli | |
| Pseudo-first order kinetic model | ||||
| k1 (min−1) | 0.055 | 0.033 | 0.078 | 0.046 |
| qe (×106 CFU g−1) | 8.586 | 168.662 | 9.264 | 181.989 |
| R2 | 0.571 | 0.316 | 0.588 | 0.330 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Pseudo-second order kinetic model | ||||
| k2 (×106 g CFU−1 min−1) | 0.009 | 0.045 | 0.010 | 0.050 |
| qe (×106 CFU g−1) | 333.333 | 333.333 | 332.138 | 332.138 |
| R2 | 1 | 1 | 1 | 1 |
For the linear pseudo-first order kinetic model, the adsorption capacities (qe) of PBB on S. aureus and E. coli were 8.586 × 106 and 168.662 × 106 CFU g−1, and the k1 values were 0.055 and 0.033 min−1, respectively. For the linear pseudo-second order kinetic model, the adsorption capacities (qe) of PBB on S. aureus and E. coli had the same value of 333.333 × 106 CFU g−1, and the k2 values were 0.009 × 106 and 0.045 × 106 g CFU−1 min−1, respectively. For the non-linear pseudo-first order kinetic model, the adsorption capacities (qe) of PBB on S. aureus and E. coli were 9.264 × 106 and 181.989 × 106 CFU g−1, and the k1 values were 0.078 and 0.046 min−1, respectively. For the non-linear pseudo-second order kinetic model, the adsorption capacities (qe) of PBB on S. aureus and E. coli had the same value of 332.138 × 106 CFU g−1, and the k2 values were 0.010 × 106 and 0.050 × 106 g CFU−1 min−1, respectively. Considering the R2 values of both linear and non-linear kinetic models, the results for both bacterial types corresponded to the pseudo-second order kinetic model with R2 equal to 1, which meant that the adsorption kinetics of PBB involved chemisorption and was related to the physiochemical interaction of PBB on bacteria similarly reported in a previous study.39 When a pseudo-second order kinetic model was chosen as the best fit model to the actual experiment data, the rate constant (k2) should be mentioned. In Table 6, the rate constant (k2) of PBB on S. aureus was less than E. coli in both linear and non-linear kinetics models, which meant that the period of adsorption time on S. aureus by PBB was shorter time than that on E. coli. The adsorption capacities of bacterial uptake of PBB on S. aureus and E. coli were gradually increased from 0 to 45 min, and then stable adsorption equilibrium was obtained, as shown in Fig. 11.
Since the plotting of non-linear kinetic models helped to confirm the result of linear plotting to protect the processing data,81 many previous studies have reported both linear and non-linear kinetic models to confirm their results.76,77,82,83 Similarly, this study reported and compared the results of linear and non-linear of both the kinetic models, and their results agreed closely with the kinetic parameter values.
Desorption experiments
For industrial applications, the main reason for selecting a new wastewater treatment technology is a reasonable cost with high efficiency; thus, a reusable material of adsorption technique for wastewater treatment is an essential point for long-time reuse. Therefore, the desorption experiments were investigated through several adsorption–desorption cycles to confirm material reusability.0.01 M HNO3 solution was used for bacterial desorption on PBB, and three cycles of adsorption–desorption were carried out to confirm material reusability. The results confirmed that PBB had high bacterial adsorption and desorption on both bacterial types in all the cycles, as shown in Table 8. For the adsorption process, the adsorption cycles presented high antibacterial efficiencies by PBB in the range of 90 ± 0.3 to 100 ± 0.0% on S. aureus and 93 ± 0.4 to 100 ± 0.0% on E. coli, respectively. For the desorption process, both the bacterial types in adsorbed PBB were almost desorbed by 0.01 M HNO3 solution in the range of 88 ± 0.2 to 100 ± 0.0% on S. aureus and 90 ± 0.4 to 100 ± 0.0% on E. coli, respectively. As a result, the antibacterial efficiencies on S. aureus and E. coli decreased by 10% and 7%, respectively, whereas the bacteria desorption efficiencies on S. aureus and E. coli decreased by 12% and 10%, respectively. Therefore, PBB is a potential material with reusability after three continuous adsorption–desorption cycles, and PBB is a suitable material to possibly apply for the disinfection process in wastewater treatment systems in the future.
| Bacterial | Cycle | Adsorption (%) | Desorption (%) |
|---|---|---|---|
| S. aureus | 1 | 100 ± 0.0 | 100 ± 0.0 |
| 2 | 96 ± 0.4 | 93 ± 0.4 | |
| 3 | 90 ± 0.3 | 88 ± 0.2 | |
| E. coli | 1 | 100 ± 0.0 | 100 ± 0.0 |
| 2 | 98 ± 0.3 | 95 ± 0.3 | |
| 3 | 93 ± 0.4 | 90 ± 0.4 |
Possibility of application in industrial wastewater treatment systems
PBB might be used as an antibacterial material for inhibiting S. aureus and E. coli for applying it as an adsorbent material in the filter tank for contaminated water passed through it for treatment. PBB is not in the treated water for releasing to receiving water, so they might not affect the ecosystem. In particular, the extracted P. betle was studied and applied in many applications of food preservations, skincare products, hair, dental, cosmetics, and biopesticides; thus, its use is not dangerous to the ecosystem and environment.In this study, approximately 3 g P. betle leaves were used for synthesizing PBB at a concentration of 300 mg mL−1 for treating 100 mL bacterial concentration of S. aureus or E. coli at 106 CFU mL−1 at 100% removal in batch experiments. Thus, 30 g P. betle could treat 1000 mL bacterial concentration of S. aureus or E. coli at 106 CFU mL−1. As a result, the bacterial concentration and wastewater volume might be used for a possible consideration of wastewater treatment applications and the amount of P. betle that should be used.
Moreover, the desorption results confirmed the reusability of PBB for inhibiting both bacterial types. For S. aureus, since the antibacterial efficiency of PBB on S. aureus was decreased by 10% in 3 cycles or 300 mL in volume, thus, PBB could be reused for more than 30 cycles or 3000 mL in volume for a bacterial concentration of 106 CFU mL−1. For E. coli, since the antibacterial efficiency of PBB on E. coli was decreased by 7% in 3 cycles or 300 mL in volume; thus, PBB could be reused for more than 42 cycles or 4200 mL in volume for a bacterial concentration of 106 CFU mL−1. If the industry needs to treat wastewater of 1000 L in volume, it requires 1 kg and 700 g of P. betle leaves for PBB synthesis for inhibiting S. aureus and E. coli, respectively. Therefore, not only could PBB be used against S. aureus and E. coli in wastewater but also it did not leave a big load of waste in the environment.
In addition, although P. betle is mostly grown in many countries in Southeast Asia such as Thailand, Malaysia, Vietnam, India, and Sri Lanka, it can also grow everywhere in the world with appropriate conditions of tropical wet climate and slightly acidic, sandy-loamy, or lightly damp soils. Thus, P. betle leaves have enough raw materials for PBB synthesis for use as antibacterial adsorbents in world. Furthermore, the synthesis method of PBB is not complicated and can be applied for industrial manufacturing as an alternative product of antibacterial materials for killing contaminated bacteria, especially S. aureus and E. coli in wastewater.
Conclusion
This study extracted P. betle powder (PBP) and synthesized P. betle beads (PBB) for antibacterial activity against S. aureus and E. coli. The FESEM-FIB images show that PBP had a porous and rough surface, whereas PBB had a spherical shape with coarse surfaces. EDX analysis illustrated that the four main chemical compositions of PBP and PBB were carbon (C), oxygen (O), calcium (Ca), and chlorine (Cl), and O–H, C–H, N–H, and C–O were the four main functional groups of PBP and PBB were identified by FTIR. The extraction yield of P. betle was 11.30%, and total phenolic, flavonoid, and tannin contents of P. betle were 201.55 ± 0.31 mg GAE per g, 56.86 ± 0.14 mg RE per g, and 41.76 ± 1.32 mg CE per g, respectively. The six main compounds of eugenol, quercetin, apigenin, kaempferol, ascorbic acid, and hydroxychavicol and nine trace compounds of rutin, syringic acid, catechin, sinapic acid, p-coumaric acid, caffeic acid, ferrulic acid, myrecetin, and gallic acid were detected by HPLC analysis. The results of disc diffusion assay by EPB and PBB confirmed that the extracted P. betle potentially inhibited S. aureus and E. coli. For antibacterial batch experiments, the optimum condition of PBB for 100% bacterial removal on both bacterial types was 0.3 g, 6 h, pH 7, and 106 CFU mL−1. The adsorption isotherms and kinetics of PBB on S. aureus and E. coli corresponded to the Freundlich model, and the pseudo-second order kinetic model was related to chemisorption and physiochemical interaction of PBB on both bacterial types. Moreover, the desorption experiments confirmed the material reusability. Finally, PBB was a potential material for inhibiting S. aureus and E. coli, and could possibly be applied in the disinfection process of wastewater treatment systems in the future.For future works, other bacterial types of Gram-positive and Gram-negative bacteria should be investigated by PBB as competing bacterial contaminations in real wastewater, and other contaminated compounds of organic matters or nutrients cannot be ignored. Moreover, continuous flow study is recommended to explore whether PBB can be possibly applied for the disinfection process of a real wastewater system.
Data availability
The raw/processed data required to reproduce these findings cannot be shared at this time due to legal or ethical reasons.The raw/processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.
Author contributions
Pimploy Ngamsurach: investigation, visualization, writing – original draft Pornsawai Praipipat: supervision, conceptualization, funding acquisition, validation, investigation, methodology, visualization, writing – original draft, writing – review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
The authors are grateful for the financial supported by The Office of the Higher Education Commission and The Thailand Research Fund grant (MRG6080114), Coordinating Center for Thai Government Science and Technology Scholarship Students (CSTS) and National Science and Technology Development Agency (NSTDA) Fund grant (SCHNR2016-122), and Research and Technology Transfer Affairs of Khon Kaen University.References
- L. Schweitzer and J. Noblet, Green Chem., 2018, 261–290 Search PubMed.
- A. E. Evans, J. Mateo-Sagasta, M. Qadir, E. Boelee and A. Ippolito, Curr. Opin. Environ. Sustain., 2019, 36, 20–27 CrossRef.
- A. Kozajda, K. Jezak and A. Kapsa, Environ. Sci. Pollut. Res., 2019, 26, 34741–34753 CrossRef CAS PubMed.
- M. Schaechter, S. Diego and S. Diego, Escherichia coli, Encyclopedia of Microbiology, 3rd edn, 2009, pp. 125–132 Search PubMed.
- J. Wang, J. Shen, D. Ye, X. Yan, Y. Zhang, W. Yang, X. Li, J. Wang, L. Zhang and L. Pan, Environ. Pollut., 2020, 262, 114665 CrossRef CAS PubMed.
- S. Basak and P. Guha, LWT--Food Sci. Technol., 2017, 80, 510–516 CrossRef CAS.
- A. Roy and P. Guha, J. Food Process. Preserv., 2018, 42, 1–7 Search PubMed.
- F. Rodrigues, A. Palmeira-de-Oliveira, J. das Neves, B. Sarmento, M. H. Amaral and M. B. Oliveira, Ind. Crops Prod., 2013, 49, 634–644 CrossRef CAS.
- N. Joshi, K. Patidar, R. Solanki and V. Mahawar, Int. J. Green Pharm., 2018, 12, S835–S839 CAS.
- M. A. Lugo-Flores, K. P. Quintero-Cabello, P. Palafox-Rivera, B. A. Silva-Espinoza, M. R. Cruz-Valenzuela, L. A. Ortega-Ramirez, G. A. Gonzalez-Aguilar and J. F. Ayala-Zavala, Biomedicines, 2021, 9(11), 1669 CrossRef CAS PubMed.
- P. Yushananta and M. Ahyanti, Open Access Maced. J. Med. Sci., 2021, 9, 895–900 CrossRef.
- P. Niyomkam, S. Kaewbumrung, S. Kaewnpparat and P. Panichayupakaranant, Pharm. Biol., 2010, 48, 375–380 CrossRef CAS PubMed.
- S. Nanasombat and N. Teckchuen, J. Med. Plants Res., 2009, 3, 443–449 Search PubMed.
- K. Subramani, B. K. Shanmugam, S. Rangaraj, V. Murugan, S. Srinivasan, O. K. Awitor, C. Massard and R. Venkatachalam, J. Coat. Technol. Res., 2020, 17, 1363–1375 CrossRef CAS.
- A. Ali, X. Y. Lim and P. F. Wahida, J. Herb. Med., 2018, 14, 29–34 CrossRef.
- D. L. Valle, J. I. Andrade, J. J. M. Puzon, E. C. Cabrera and W. L. Rivera, Asian Pac. J. Trop. Biomed., 2015, 5, 532–540 CrossRef.
- V. Dwivedi and S. Tripathi, J. Pharmacogn. Phytochem., 2014, 3, 93–98 Search PubMed.
- Y. Lin, K. Xiong, Z. Lu, S. Liu, Z. Zhang, Y. Lu, R. Fu and D. Wu, Chem. Commun., 2017, 53, 9777–9780 RSC.
- W. P. T. D. Perera, R. K. Dissanayake, U. I. Ranatunga, N. M. Hettiarachchi, K. D. C. Perera, J. M. Unagolla, R. T. De Silva and L. R. Pahalagedara, RSC Adv., 2020, 10, 30785–30795 RSC.
- S. Moniri Javadhesari, S. Alipour, S. Mohammadnejad and M. R. Akbarpour, Mater. Sci. Eng. C, 2019, 105, 110011 CrossRef CAS PubMed.
- R. B. Asamoah, A. Yaya, B. Mensah, P. Nbalayim, V. Apalangya, Y. D. Bensah, L. N. W. Damoah, B. Agyei-Tuffour, D. Dodoo-Arhin and E. Annan, Results Mater., 2020, 7, 100099 CrossRef.
- M. Thukkaram, S. Sitaram, S. K. Kannaiyan and G. Subbiahdoss, Int. J. Biomater., 2014, 716080 Search PubMed.
- X. Xue, Y. Wang and H. Yang, Appl. Surf. Sci., 2013, 264, 94–99 CrossRef CAS.
- L. M. Anaya-Esparza, E. Montalvo-González, N. González-Silva, M. D. Méndez-Robles, R. Romero-Toledo, E. M. Yahia and A. Pérez-Larios, Materials, 2019, 12(5), 698 CrossRef CAS.
- M. Ikram, E. Umar, A. Raza, A. Haider, S. Naz, A. Ul-Hamid, J. Haider, I. Shahzadi, J. Hassan and S. Ali, RSC Adv., 2020, 10, 24215–24233 RSC.
- D. Paul, S. Mangla and S. Neogi, Mater. Lett., 2020, 271, 127740 CrossRef CAS.
- P. P. Mahamuni, P. M. Patil, M. J. Dhanavade, M. V. Badiger, P. G. Shadija, A. C. Lokhande and R. A. Bohara, Biochem. Biophys. Rep., 2019, 17, 71–80 Search PubMed.
- M. I. Jahanger, N. A. Shad, M. M. Sajid, K. Akhtar, Y. Javed, A. Ullah, M. A. Hassan, M. H. Sarwar, M. Sarwar and M. Sillanpää, React. Kinet., Mech. Catal., 2022, 135, 499–510 CrossRef CAS.
- M. I. Nabila and K. Kannabiran, Biocatal. Agric. Biotechnol., 2018, 15, 56–62 CrossRef.
- R. Li, Z. Chen, N. Ren, Y. Wang, Y. Wang and F. Yu, J. Photochem. Photobiol., B, 2019, 199, 111593 CrossRef CAS.
- A. Jain, F. Ahmad, D. Gola, A. Malik, N. Chauhan, P. Dey and P. K. Tyagi, Environ. Nanotechnol., Monit. Manage., 2020, 14, 100337 Search PubMed.
- S. Vasantharaj, S. Sathiyavimal, M. Saravanan, P. Senthilkumar, K. Gnanasekaran, M. Shanmugavel, E. Manikandan and A. Pugazhendhi, J. Photochem. Photobiol., B, 2019, 191, 143–149 CrossRef CAS PubMed.
- A. K. M. Atique Ullah, A. N. Tamanna, A. Hossain, M. Akter, M. F. Kabir, A. R. M. Tareq, A. K. M. Fazle Kibria, M. Kurasaki, M. M. Rahman and M. N. I. Khan, RSC Adv., 2019, 9, 13254–13262 RSC.
- R. Ishwarya, B. Vaseeharan, S. Kalyani, B. Banumathi, M. Govindarajan, N. S. Alharbi, S. Kadaikunnan, M. N. Al-anbr, J. M. Khaled and G. Benelli, J. Photochem. Photobiol., B, 2018, 178, 249–258 CrossRef CAS.
- A. Ananda, T. Ramakrishnappa, S. Archana, L. S. Reddy Yadav, B. M. Shilpa, G. Nagaraju and B. K. Jayanna, Mater. Today: Proc., 2021, 49, 801–810 Search PubMed.
- S. Ramli, S. Radu, K. Shaari and Y. Rukayadi, BioMed Res. Int., 2017, 9024246 Search PubMed.
- W. Yuan and H. G. Yuk, Food Microbiol, 2018, 72, 176–184 CrossRef PubMed.
- D. Nigussie, G. Davey, B. A. Legesse, A. Fekadu and E. Makonnen, BMC Complementary Med. Ther., 2021, 21, 1–10 CrossRef PubMed.
- P. Ngamsurach and P. Praipipat, ACS Omega, 2021, 6, 32215–32230 CrossRef CAS.
- V. L. Singleton, R. Orthofer and R. M. Lamuela-Raventós, Methods Enzymol., 1999, 299, 152–178 CAS.
- J. Zhishen, T. Mengcheng and W. Jianming, Food Chem., 1999, 64, 555–559 CrossRef CAS.
- B. Sun, J. M. Ricardo-da-Silva and I. Spranger, J. Agric. Food Chem., 1998, 46, 4267–4274 CrossRef CAS.
- I. Langmuir, J. Am. Chem. Soc., 1918, 40, 1361–1403 CrossRef CAS.
- H. Freundlich, J. Phys. Chem., 1906, 57, 385–470 CAS.
- S. Lagergren, K. Sven. Vetenskapsakad. Handl., 1898, 24, 1–39 Search PubMed.
- Y. S. Ho and G. Mckay, Process Biochem., 1999, 34, 451–465 CrossRef CAS.
- V. P. B. Rekha, M. Kollipara, S. Gupta, Y. Bharath and K. K. Pulicherla, Am. J. Ethnomed., 2015, 1, 276–289 Search PubMed.
- T. Varadavenkatesan, R. Selvaraj and R. Vinayagam, J. Mol. Liq., 2016, 221, 1063–1070 CrossRef CAS.
- T. P. Singh, G. Chauhan, R. K. Agrawal and S. K. Mendiratta, J. Food Meas. Charact., 2019, 13, 466–475 CrossRef.
- L. Muruganandam, A. Krishna, J. Reddy and G. S. Nirmala, Resour. Technol., 2017, 3, 385–393 Search PubMed.
- N. H. Nguyen, T. T. Y. Nhi, N. T. Van Nhi, T. T. T. Cuc, P. M. Tuan and D. H. Nguyen, J. Nanomater., 2021, 2021, 5518389 Search PubMed.
- U. Gaware, V. Kamble and B. Ankamwar, Int. J. Electrochem., 2012, 2012, 1–6 CrossRef.
- M. M. O. Rashid, M. S. Islam, M. A. Haque, M. A. Rahman, M. T. Hossain and M. A. Hamid, Iran. J. Pharm. Res., 2016, 15, 591–597 CAS.
- N. T. Thuong, H. T. Ngoc Bich, C. N. H. Thuc, B. T. P. Quynh and L. Van Minh, ChemistrySelect, 2019, 4, 8150–8157 CrossRef CAS.
- L. Nouri, A. Mohammadi Nafchi and A. A. Karim, Ind. Crops Prod., 2014, 62, 47–52 CrossRef CAS.
- L. T. T. Nguyen, T. T. Nguyen, H. N. Nguyen and Q. T. P. Bui, Eng. Reports, 2020, 2, 2–9 Search PubMed.
- H. V. Annegowda, P. Y. Tan, M. N. Mordi, S. Ramanathan, M. R. Hamdan, M. H. Sulaiman and S. M. Mansor, Food Anal. Methods, 2013, 6, 715–726 CrossRef.
- U. Taukoorah, N. Lall and F. Mahomoodally, S. Afr. J. Bot., 2016, 105, 133–140 CrossRef.
- A. Noor, M. Al Murad, A. Jaya Chitra, S. N. Babu and S. Govindarajan, Food Biosci., 2022, 47, 101715 CrossRef CAS.
- J. A. Khan and N. Kumar, J. Pharm. Biomed. Sci., 2011, 11, 1–3 Search PubMed.
- B. Jayalakshmi, K. A. Raveesha, M. Murali and K. N. Amruthesh, Int. J. Pharm. Pharm. Sci., 2015, 7(10), 23–29 CAS.
- P. Kulnanan, J. Chuprom, T. Thomrongsuwannakij, C. Romyasamit, S. Sangkanu, N. Manin, V. Nissapatorn, M. de Lourdes Pereira, P. Wilairatana, W. Kitpipit and W. Mitsuwan, Arch. Microbiol., 2022, 204, 1–14 CrossRef.
- K. N. Akter, P. Karmakar, A. Das, S. N. Anonna, S. A. Shoma and M. M. Sattar, Avicenna J. Phytomed., 2014, 4, 320–329 CAS.
- D. L. Valle, E. C. Cabrera, J. J. M. Puzon and W. L. Rivera, PLoS One, 2016, 11, 1–14 CrossRef PubMed.
- A. A. Ali Almahdi and Y. Kumar, Int. J. Curr. Microbiol. Appl. Sci., 2019, 8, 2009–2019 CrossRef.
- B. Kaveti, L. Tan, T. S. Kuan and M. Baig, J. Pharm. Teach., 2011, 2, 129–132 Search PubMed.
- R. R. Lubis, M. Hawley and D. D. Wahyuni, Am. J. Clin. Exp. Immunol., 2020, 9, 1–5 Search PubMed.
- F. U. Ermawati, R. Sari, N. P. Putri, L. Rohmawati, D. H. Kusumawati, Munasir and Z. A. I. Supardi, J. Phys.: Conf. Ser., 2021, 1951, 012004 CrossRef CAS.
- A. Budiman and D. L. Aulifa, Pharmacogn. J., 2020, 12, 473–477 CrossRef CAS.
- A. Datta, S. Ghoshdastidar and M. Singh, Int. J. Pharma Sci. Res., 2011, 2, 104–109 Search PubMed.
- W. K. Jung, H. C. Koo, K. W. Kim, S. Shin, S. H. Kim and Y. H. Park, Appl. Environ. Microbiol., 2008, 74, 2171–2178 CrossRef CAS.
- Y. L. Ying, S. Y. Pung, M. T. Ong and Y. F. Pung, J. Ind. Eng. Chem., 2018, 67, 437–447 CrossRef CAS.
- M. Rezakazemi and Z. Zhang, Desulfurization Materials, 2018, vol. 2 Search PubMed.
- P. Ganguly, R. Sarkhel and P. Das, Surf. Interfaces, 2020, 20, 100616 CrossRef CAS.
- Q. Sun and B. Chen, Ind. Eng. Chem. Res., 2020, 59, 16838–16850 CrossRef CAS.
- A. Threepanich and P. Praipipat, J. Environ. Chem. Eng., 2021, 9, 106007 CrossRef CAS.
- A. Threepanich and P. Praipipat, Environ. Sci. Pollut. Res., 2022, 29, 46077–46090 CrossRef CAS PubMed.
- A. D. Zand and M. R. Abyaneh, Sustain. Environ. Res., 2020, 30, 1–16 CrossRef.
- R. Yang, J. Aoqi, S. He, Q. Hu, M. Sun, T. Dong, Y. Hou and S. Zhou, Chem. Eng. J., 2020, 406, 125982 CrossRef.
- A. M. Aljeboree, A. N. Alshirifi and A. F. Alkaim, Arab. J. Chem., 2017, 10, S3381–S3393 CrossRef CAS.
- P. Paluri, K. A. Ahmad and K. S. Durbha, Biomass Convers. Biorefin., 2022, 12, 4031–4048 CrossRef CAS.
- F. Cao and J. Shen, J. Chem. Eng. Data, 2019, 64, 5622–5629 CrossRef CAS.
- D. M. Salh, B. K. Aziz and S. Kaufhold, Silicon, 2020, 12, 87–99 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2022 |