Open Access Article
Open Access ArticleSupercritical CO2 assisted extraction of essential oil and naringin from Citrus grandis peel: in vitro antimicrobial activity and docking study†
Thanh-Chi Mai*ab,
Ngoc-Thinh Trana,
Dinh-Tri Maiab,
Tran Thi Ngoc Maic,
Nguyen Hong Thuc Duyen d,
Tran Nguyen Minh An*d,
Mahboob Alame,
Chi-Hien Dangab and
Thanh-Danh Nguyen
d,
Tran Nguyen Minh An*d,
Mahboob Alame,
Chi-Hien Dangab and
Thanh-Danh Nguyen *ab
*ab
aInstitute of Chemical Technology, Vietnam Academy of Science and Technology, 1A, TL29, District 12, Ho Chi Minh City, Vietnam. E-mail: maithanhchi@yahoo.com.vn; ntdanh@ict.vast.vn
bGraduate University of Science and Technology, Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet, Cau Giay, Hanoi, Vietnam
cInstitute of Applied Sciences, HUTECH University, 475A Dien Bien phu Street, Ward 25, Binh Thanh District, Ho Chi Minh City, Vietnam
dFaculty of Chemical Engineering, Industrial University of Ho Chi Minh City, Ho Chi Minh City 71420, Vietnam. E-mail: trannguyenminhan@iuh.edu.vn
eDepartment of Safety Engineering, Dongguk University, 123 Dongdae-ro, Gyeongju-si 780714, Gyeongsangbuk-do, Republic of Korea
First published on 13th September 2022
Abstract
The extraction of bioactive compounds, including essential oils and flavonoids, using organic solvents is a significant environmental concern. In this work, waste C. grandis peel was the ingredient used to extract essential oil and naringin by conducting a supercritical CO2 technique with a two stage process. In the first stage, the extraction with only supercritical CO2 solvent showed a significant enhancement of the D-limonene component, up to 95.66% compared with the hydro-distillation extraction (87.60%). The extraction of naringin using supercritical CO2 and ethanol as a co-solvent was done in the second stage of the process, followed by evaluating in vitro antimicrobial activity of both the essential oil and naringin. The essential oil indicated significant activity against M. catarrhalis (0.25 mg ml−1), S. pyogenes (1.0 mg ml−1), S. pneumoniae (1.0 mg ml−1). Whilst naringin gave good inhibition towards all tested microbial strains with MIC values in the range of 6.25–25.0 μM. In particular, naringin exhibited high antifungal activity against T. rubrum, T. mentagrophytes, and M. gypseum. The molecular docking study also confirmed that D-limonene inhibited bacterium M. catarrhalis well and that naringin possessed potential ligand interactions that proved the inhibition effective against fungi. Molecular dynamics simulations of naringin demonstrated the best docking model using Gromacs during simulation up to 100 ns to explore the stability of the complex naringin and crystal structure of enzyme 2VF5: PDB.
1. Introduction
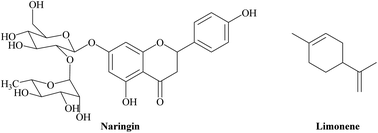
The Citrus grandis species that is distributed worldwide is the largest citrus fruit from the family Rutaceae and is consumed mostly in Southeast Asia. C. grandis fruits are not only served as food but are also considered as a traditional medicine around the world.1 Although the peel of C. grandis fruits mainly contains flavonoids and essential oils that possess beneficial bioactivities for human health, it is considered to be agricultural waste.2 Thus, the utilization of the C. grandis peel for the manufacture of natural products is an interesting topic that is related to pharmaceutical and environmental industries.Naringin is a natural flavonoid compound; naringenin-7-O-glycoside (Fig. 1) is responsible for the bitter taste of C. grandis fruits. The medicinal value of naringin has been majorly investigated for its antioxidant, anti-inflammatory, and antimicrobial functions. The pharmaceutical applications of naringin can be used to treat various diseases, such as: obesity, diabetes, and heart disease.3,4 The biological activities of C. grandis essential oil, including anti-inflammatory, antibacterial, and cytotoxicity, are considered for commercial applications such as perfume, cosmetic, and medical industries. The main composition of the essential oil is well-known as monoterpene compounds, with an abundant content of limonene (Fig. 1).5
Despite the increasing demand for C. grandis essential oils and naringin for human health applications, the lack of economical and environmental-friendly extraction methods has induced the preclusion of commercial development of these natural products. Thus, the enhancement of effective and safe extraction methods is of interest to the market for the products for human health benefits. The C. grandis peel is a good source of essential oils and naringin—up to 2%![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 and 3%,7 respectively. However, most of the extractions were investigated using a common heating extraction, organic solvents, and enzymatic methods. Regarding the biological property of natural products and environmental concerns, the alternative extraction technique using supercritical CO2 has been of particular interest.8 Our previous report pointed out that the supercritical CO2 assisting the extraction could further enhance the efficiency and property of the essential oil compared to the ordinary extraction methods.9 Quispe-Fuentes et al.10 used supercritical CO2 and ethanol for the extraction of oils that contain the major content of phenolics. Su et al. extracted 45 organic compounds from five different pomelo cultivars using the supercritical CO2 method.11 Moreover, the supercritical CO2 technique has been effectively used for the extraction of essential oils and naringin from other genus citrus.12–15 The content of naringin obtained in orange, tangerine and lemon peel extracts using the supercritical CO2 technique was determined to be 35.26, 44.05 and 19.86 mg g−1, respectively.16 However, to the best of our knowledge, the extraction of both essential oil and naringin, when using supercritical fluid techniques, is rarely reported and any further bioactivity research for them has not yet been evaluated.
6 and 3%,7 respectively. However, most of the extractions were investigated using a common heating extraction, organic solvents, and enzymatic methods. Regarding the biological property of natural products and environmental concerns, the alternative extraction technique using supercritical CO2 has been of particular interest.8 Our previous report pointed out that the supercritical CO2 assisting the extraction could further enhance the efficiency and property of the essential oil compared to the ordinary extraction methods.9 Quispe-Fuentes et al.10 used supercritical CO2 and ethanol for the extraction of oils that contain the major content of phenolics. Su et al. extracted 45 organic compounds from five different pomelo cultivars using the supercritical CO2 method.11 Moreover, the supercritical CO2 technique has been effectively used for the extraction of essential oils and naringin from other genus citrus.12–15 The content of naringin obtained in orange, tangerine and lemon peel extracts using the supercritical CO2 technique was determined to be 35.26, 44.05 and 19.86 mg g−1, respectively.16 However, to the best of our knowledge, the extraction of both essential oil and naringin, when using supercritical fluid techniques, is rarely reported and any further bioactivity research for them has not yet been evaluated.
A molecular docking model was developed to predict the most stable binding conformation of ligand to a protein or enzyme and test for its QSAR model. It was developed based on a new scoring function that calculates the change of free Gibbs energy and the inhibition constant by using the Lamarckian model of genetics. There are three principal research methods, including the Monte Carlo simulated annealing, the traditional genetic algorithm, and the Lamarckian genetic algorithm. The Autodock and Lamarckian genetic algorithm were used to prove that the molecular docking model is valid.17 This method has successfully predicted many biological activities in silico, such as antimicrobial activity,18 SARS-CoV-2 resistance of compounds in garlic essential oil,19 anticancer,20 inhibition α-glucosidase enzyme,21 antioxidant,22 anti-inflammatory,23 tyrosinase enzyme inhibition,24 interactions between protein and nanoparticles25 and acetylcholinesterase inhibition.26 The validation of the molecular docking model was evaluated by the RMSD value,27 while the strengths of ligand interactions between ligand and target enzyme or protein were performed by the MD method.28 Some docking programs and many scoring functions were used to assess the critical docking programs and Scoring Functions.29 Some protein data bank identifications were classified via protein–ligand complexes for docking.30 Although there are many kinds of molecular docking models, very few report utilising the in silico docking model to explain the biological activities of natural products.
In this work, the supercritical CO2 technique was used to extract essential oil and naringin from Citrus grandis peels and their in vitro antimicrobial activity, as well as evaluating the in silico molecule docking model.
2. Experimental
2.1. Materials
Chemicals, including DMSO, NaOH, and anhydrous Na2SO4, were purchased from Acros Co. (Belgium) and ethanol was purchased from Chemsol Co. (Vietnam). Waste Citrus grandis peels were collected in BenTre Provine, Vietnam. The peels were cut with to a particle size of 1–2 mm using a slicer machine. Deionised water was used throughout the experiment.2.2. Extraction methods of essential oils
2.3. Extraction of naringin
Naringin was extracted using supercritical CO2 and ethanol as a co-solvent. The essential oil required 100 g of peels in 200 ml of different concentrations of ethanol solution (50, 80, 90, and 96%) and put into a stainless-steel extraction cell – 0.5 L. Extraction conditions were optimized with changes in temperature (30–70 °C), pressure (100–130 bar) and time (1–4 hours); the extract was evaporated to remove the ethanol and give the residue. 300 ml of NaOH solution (1 M) was then added to the residue to remove the polysaccharides and impurities. The solid was washed with ethanol (100 ml × 3) and oven-dried at 60 °C; the collected product was dissolved in 200 ml of H2O at 75 °C for 10 minutes. After the drying step, the hot mixture was filtered, 100 ml of ethanol solution (20%) was then added into the extraction. The solution's pH was verified to be 4, after that, it was cooled at 15 °C for 24 hours. The pure naringin was filtered and dried at 50 °C for 6 hours. The product, after this final drying process, was comprised of two epimers at position C-2 and was a white amorphous powder which could be observed in 1H and 13C NMR spectra. [α]20D = −82.4° (ethanol, 1 g ml−1). ESI-HRMS (m/z): 579.1714 [M + H]+ calcd for C27H31O14 ([M + H]+ = 579.1714).The extraction yield (%) was calculated as the weight of obtained naringin over the weight of dry peels used at the beginning.
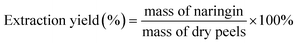 | (1) |
2.4. Measurement
Gas chromatography (GC) and Gas chromatography-mass spectrometer (GC-MS) measurements of EOs were carried out, as stated in the previous report.32 Briefly, the EOs (25 μL) were dissolved in hexane (1 ml) and analysed on GC (Agilent 6890 N) coupled with HP-5MS Capillary Column (30.0 length × 0.25 mm i.d. × 0.25 μm film thickness). The retention index was calculated based on the standard sample containing homologous series of n-alkanes (C8–C30, Sigma Co.). To identify the essential oils, their retention index was compared with the published data. For the measurement of infrared spectra, the A BRUKER EQUINOX 55 IR spectrophotometer was used during this process. 1H and 13C NMR (BRUKER AVANCE 500 NMR spectrometer) analysis was recorded with DMSO-d6 as the solvent and tetramethyl silane (TMS) played the role of the internal standard. The optical rotation was measured on the A. Kruss polarimeter P8000-T using ethanol solvent. The HRMS spectrum was measured on the Agilent 6200 series TOF.2.5. Antibacterial assay
2.6. In silico molecular docking model for anti-microbial activities
The in silico docking model for essential oil and naringin was conducted by a general procedure in Scheme 1. For essential oil, D-limonene docked to a receptor of bacterium, M. catarrhalis, code 4ZQG: PDB. The active centre of bacterium M. catarrhalis was determined at coordinates of X = −2.685, Y = −1.666, Z = −32.403. The grid parameters were installed by spacing and the numbers of points in the (X, Y, Z) axis, which were 0.5 Å and (50, 50, 50), respectively in dock.gpf file. The number of models was 200 models, while the results of the docking model were calculated by the Discovery Studio 2021 client and Molegro Molecular Viewer. Additionally, the validations of the models were calculated by the PyMOL package in values of RMSD, as reported 30. The ADMET model, or drug-likeness of the compound, were determined based on ADMETlab 2.0—an online platform solution.34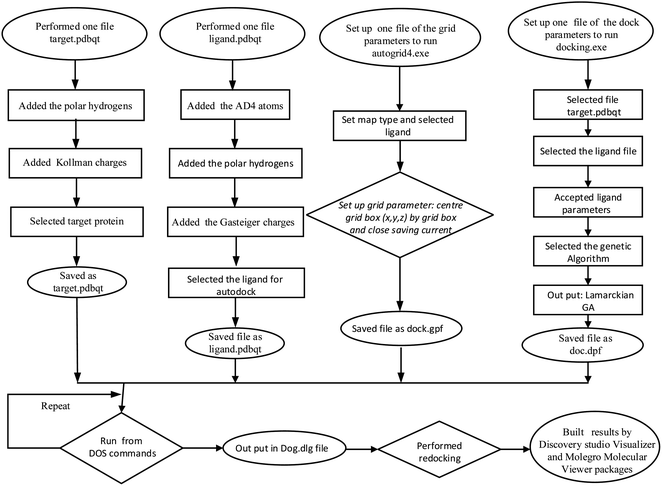 | ||
| Scheme 1 The general procedure for the docking of naringin or the main components of the essential oil to enzyme 2VF5 or bacterium: 4ZQG: PDB. | ||
The naringin ligand was docked to one receptor, an enzyme with the code 2VF5 from the protein data bank (PDB file). As the chosen enzyme, 2VF5 was determined to be the catalyst for the synthesis of glucosamine-6-phosphate (GlcN-6-P). Hence, the short-time inhibition of GlcN-6-P synthase harmed and restrained the growth of the fungal cells, or fungi.35 The active centre of 2VF5: PDB was detected at coordinates of (X = 26.579, Y = 22.731, Z = 8.113). The grid parameters were set up in dock.gpf file by the spacing of 0.5 Å, the numbers of points in (X, Y, Z) axis of (50, 50, 50), respectively, and the centre of grid box of X = 26.579, Y = 22.731, Z = 8.113. The input and output were installed by a generic algorithm with 200 models and Lamarckian in dock.dpf file, respectively.
2.7. Molecular dynamics simulation of the best docked model complex
Based on docking results, molecular dynamics (MD) simulation of naringin was performed on the selected lowest energy and the best docking complex using the GROMACS 2020 series package (gromacs.org) with the CHARMM force field (https://www.charmm.org). The SwissParam server was used to generate ligand topology files (swissparam.ch). Three-point transferable intermolecular potential (TIP3P) was chosen as a solvent model for protein–ligand complexes (triclinic water box with a size of 50 × 75 × 70 Å).36 Before the minimization process, the charge system was applied to neutralize by the addition of 0.15 M NaCl at constant temperature (300 K) and pressure (1.0 bar). Energy minimization was performed using the steepest descent approach for the protein–ligand complex. In the final step, MD simulation with 100 ns was performed for compound naringin, pose 60 and receptor, crystal structure of enzyme 2VF5: PDB in periodic boundary condition and 1000 frames were used in each simulation. Findings such as Root Mean Square Deviation and Fluctuation (RMSD/F), hydrogen bond counting, and radius of gyration (Rg) were analyzed using Xmgrace software (plasma-gate.weizmann.ac.il/Grace) and other tools such as OriginLab and VMD.3. Results and discussion
The application of the green technique is particularly important for the production of natural compounds in pharmaceutical fields. In this work, the supercritical CO2 technique was applied to extract the essential oil from waste C. grandis peel source, followed by producing naringin using the EO spent C. grandis peels. For detailed information, the study strategy is fully provided in Scheme 2. To confirm the efficiency of the extraction, the hydro-distilled essential oil was used as a reference sample. The chemical composition of essential oils was identified using GC-MS spectra, then compared with the reference. The supercritical CO2 extraction of naringin was optimized within experimental conditions; then, its purity and structure were identified using 1D and 2D NMR spectra. The pharmaceutical applications of both SCEO and naringin were evaluated via antimicrobial activity as well as a docking study.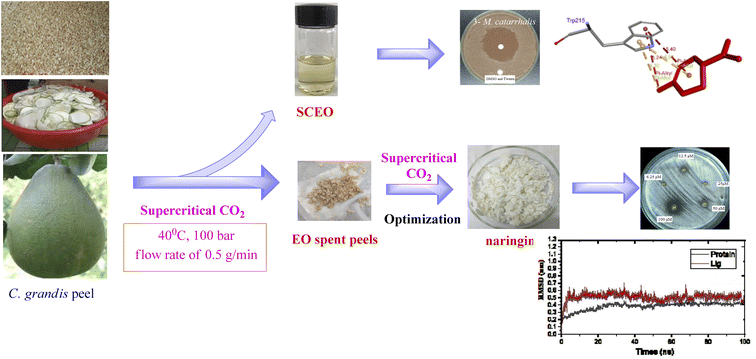 | ||
| Scheme 2 Study strategy for supercritical CO2-assisted extraction of essential oil and naringin and their applications in the pharmaceutical field. | ||
3.1. Volatile compositions, antimicrobial activity, and docking study of essential oils
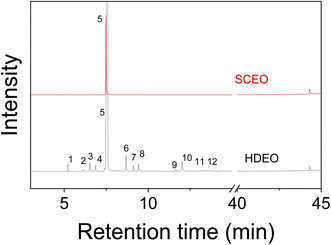 | ||
| Fig. 2 GC-MS spectra of C. grandis essential oils extracted by hydro-distillation (HDEO) and Supercritical CO2 (SCEO) methods. | ||
| No. | Compounds | RIa | RIb | % GC | Method | |
|---|---|---|---|---|---|---|
| HDEO | SCEO | |||||
| a Retention index was collected from ref. 42 and NIST database.b Retention index was calculated from the experimental data. | ||||||
| 1 | α-Pinene | 932 | 927 | 0.90 | — | RI, MS |
| 2 | β-Pinene | 974 | 980 | 1.51 | — | RI, MS |
| 3 | α-Phellandrene | 1003 | 996 | 1.13 | — | RI, MS |
| 4 | o-Cymene | 1014 | 1015 | 0.46 | — | RI, MS |
| 5 | D-Limonene | 1028 | 1022 | 87.60 | 95.66 | RI, MS |
| 6 | (Z)-Linalool oxide | 1066 | 1061 | 2.30 | — | RI, MS |
| 7 | (E)-Linalool oxide | 1076 | 1077 | 0.94 | — | RI, MS |
| 8 | Linalool | 1090 | 1089 | 1.39 | — | RI, MS |
| 9 | 4-Terpineol | 1170 | 1166 | 0.38 | — | RI, MS |
| 10 | α-Terpineol | 1187 | 1180 | 1.92 | — | RI, MS |
| 11 | β-Caryophyllene | 1413 | 1406 | 0.31 | — | RI, MS |
| 12 | α-Caryophyllene | 1438 | 1439 | 0.10 | — | RI, MS |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Monoterpenes | ||||||
| Hydrocarbons | 91.60 | 95.66 | ||||
| Alcohols | 6.93 | — | ||||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Sesquiterpenes | ||||||
| Hydrocarbons | 0.41 | — | ||||
| Not identified | 1.07 | 4.34 | ||||
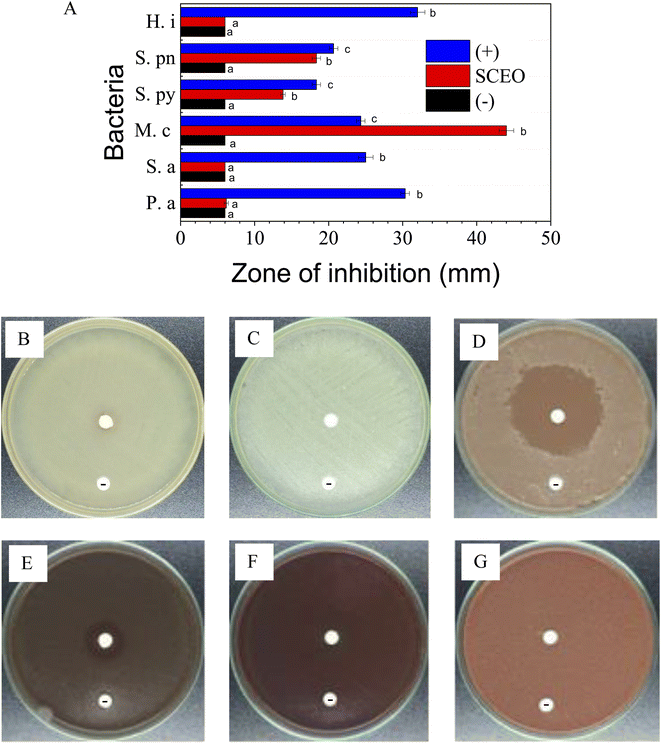
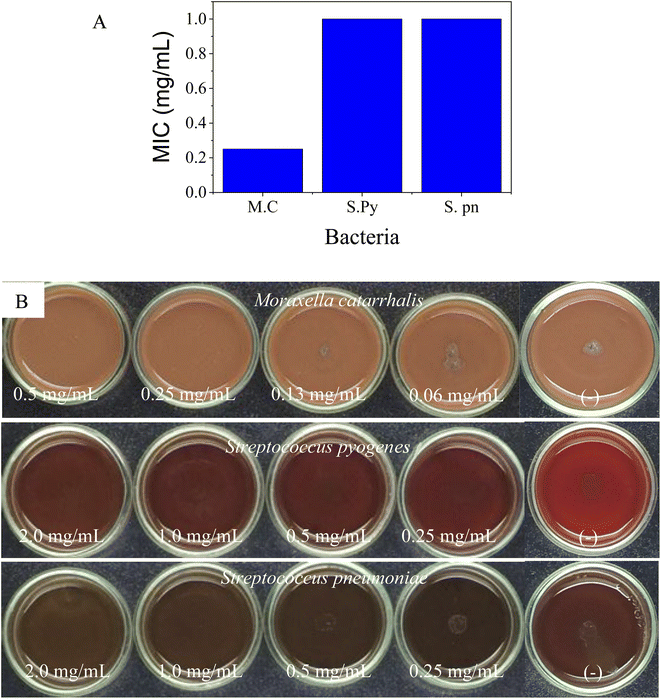
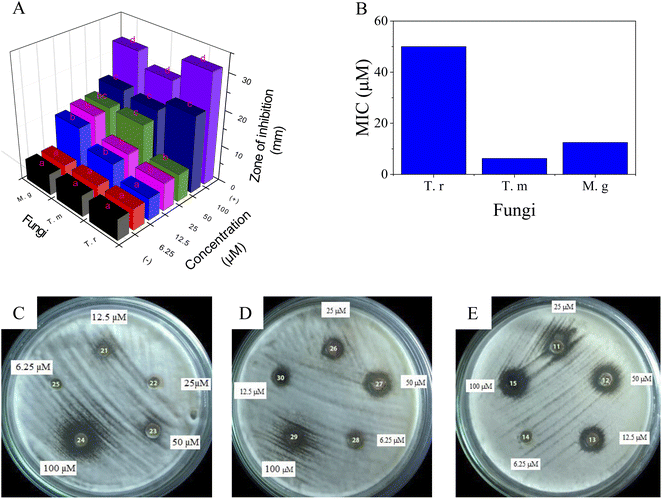
MIC values of SCEO against bacteria M. c, S. py, and S. pn were determined via the dilution method. The test concentrations were used for strain M. c in the range of 0.06–0.5 mg ml−1 while the concentration series for the inhibition of both S. py and S. pn were performed in the range of 0.25–2.0 mg ml−1, followed by plotting the antibacterial activity in Fig. 4A, while test images are shown in Fig. 4B. The result was identified and showed that the MIC values of SCEO against all bacteria strains were very low. MIC values inhibited both strains, S. py and S. pn, were determined to be 1.0 mg ml−1 while the value of M. c inhibition was low, at a value of 0.25 mg ml−1. These values showed a better activity than was found for essential oils, with an abundant content of D-limonene reported previously. Wang et al.47 found MIC of Citrus medica essential oil with D-limonene of ∼50% against strain S. pn was 24 mg ml−1. Salama46 showed that Myoporum acuminatum essential oil that contained approximately 34% of D-limonene possessed MIC value against S. pn at about 3.0 mg ml−1. Doughari and Bazza showed essential oil isolated from Citrus sinensis possessed MIC of 200 mg ml−1 against strain S. py. Meanwhile, the essential oils containing an abundant content of D-limonene were rarely used for testing against strain M. c, which may cause infections of the eye, middle ear, and central nervous system of humans.48
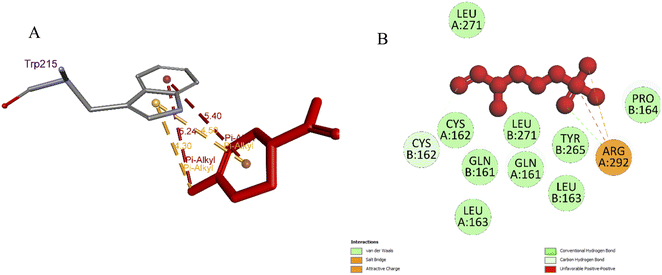 | ||
| Fig. 5 (A) Pose 66 docked to residual amino acid Trp 215 of 4ZQG: PDB; (B): small ligand, pose 18 (fosmidomycin) in crystal structure of 4ZQG: PDB was redocked to one crystal structure of protein 4ZQG: PDB. | ||
The pharmacokinetic model of D-limonene was predicted in Tables S1–S8.† As seen in Table S1,† the physicochemical properties of compound D-limonene were detected in a permission range. The significant parameters were TPSA, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S, log
S, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, and log
P, and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D. The medicinal parameters were determined as shown in Table S2,† most of them were in range as Lipinski Rule, Sascore, Fsp3, Npscore, PAINS, ALARM NMR, BMS, and Chelator Rule. The absorption properties were detected in range as HIA, Caco-2 permeability in Table S3.† As shown in Table S4,† compound D-limonene was proved to have good distribution properties, except for BBB Penetration parameter while the drug metabolism, drug excretion, drug toxicity, and toxicophoric rules were indicated in Tables S5–S8,† respectively. They were confirmed as a drug-likeness. Therefore, compound D-limonene was indicated to be the potential drug properties.
D. The medicinal parameters were determined as shown in Table S2,† most of them were in range as Lipinski Rule, Sascore, Fsp3, Npscore, PAINS, ALARM NMR, BMS, and Chelator Rule. The absorption properties were detected in range as HIA, Caco-2 permeability in Table S3.† As shown in Table S4,† compound D-limonene was proved to have good distribution properties, except for BBB Penetration parameter while the drug metabolism, drug excretion, drug toxicity, and toxicophoric rules were indicated in Tables S5–S8,† respectively. They were confirmed as a drug-likeness. Therefore, compound D-limonene was indicated to be the potential drug properties.
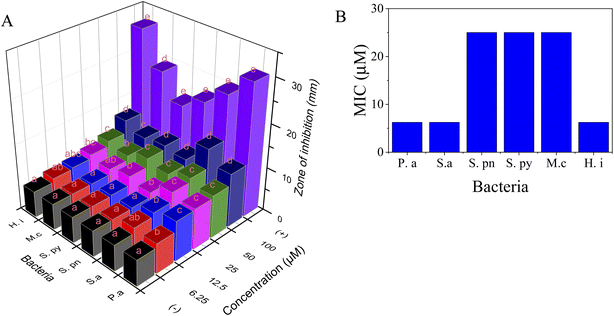
3.2. Extraction, antimicrobial activity, and docking study of naringin
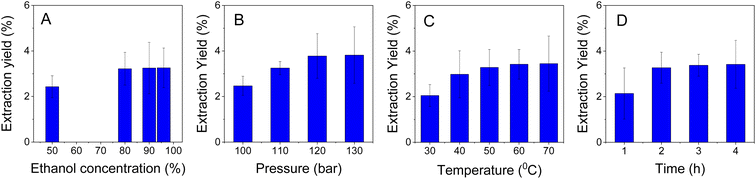 | ||
| Fig. 6 Influence of conditions on naringin extraction yields: (A) ethanol concentration; (B) pressure; (C) temperature; and (D) time. | ||
Extraction performance is strongly influenced by ethanol concentrations, as clearly observed in Fig. 6A. The extraction yield was seen to be 2.4% with an ethanol concentration of 50%. The yield was significantly improved when the ethanol concentration used was 80%. However, no significant change in the extraction yield was seen when higher concentrations of ethanol were observed. Due to the requirement of production cost efficiency, the concentration of ethanol solution (80%) was used for naringin extraction.
For the effect of pressure, the extraction was investigated in a range from 100 to 130 bar. The pressure-dependent extraction of naringin is obviously seen in Fig. 6B. The extraction yield obtained only 2.5% at the low pressure (100 bar) and increased proportionally with the pressure. The maximum yield achieved (3.8%) was at a pressure of 120 bar and did not change with a higher pressure (130 bar).
Extraction temperature was performed in a range from 30 to 70 °C (Fig. 6C). The result indicated that the extraction yield was ineffective at a low temperature (30 °C) while a high temperature (more than 50 °C) insignificantly affected the extraction of naringin. Thus, the optimum temperature was determined to be 50 °C, with a yield of 3.3%.
The time-dependent extraction of naringin was optimized at a range within 1 to 4 hours. A low extraction efficiency was observed for the initial hour and the higher yields (3.3%) were obtained after 2 hours of extraction. Thus, the result given showed that the best experimental condition for supercritical CO2-assisted extraction of naringin was carried out at a concentration of 80% ethanol solution, 120 bar pressure, a temperature of 50 °C and a time of 2 hours. The structure of the obtained compound was clearly identified as naringin by 1H and 13C NMR spectra. From the data of 2D NMR (HSQC, COSY and HMBC), the structure of the obtained naringin was confirmed to be an epimer mixture at position C-2.
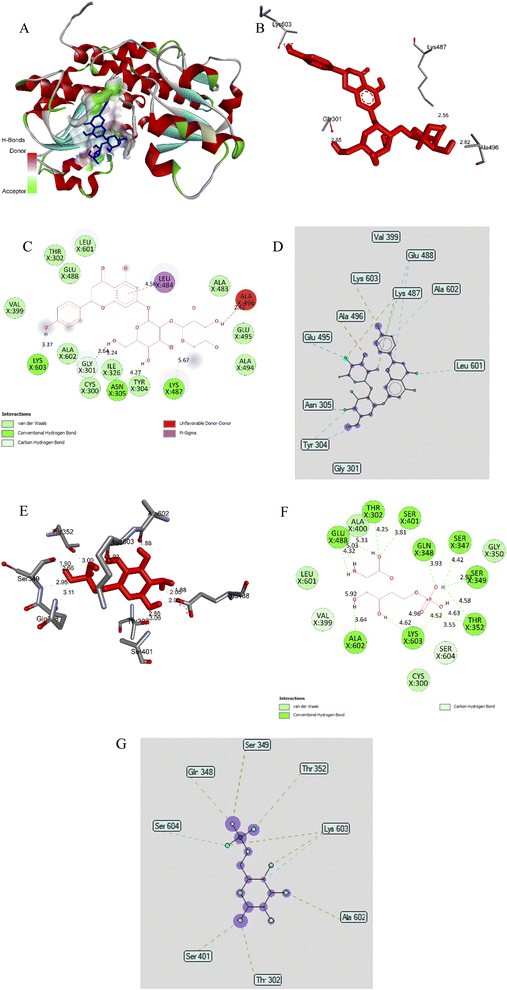 | ||
| Fig. 9 (A) The best docking pose 60 of ligand naringin docked to an enzyme pocket at an active center of enzyme 2VF5 (PDB) synthesised glucosamine-6-phosphate present in cell wall of bacteria or fungi; (B) the hydrogen bonds linked from active atom on pose 60, ligand naringin to active residual amino acids on enzyme 2VF5; (C) a 2D diagram described the significant ligand interactions between pose 60 and enzyme 2VF5; (D) the ligand map exposed the secondary interactions between the best docking pose 60 and enzyme 2VF5; (E) the hydrogen bonds formed from active residual amino acids on enzyme 2VF5 to active atom on pose 83, the small ligand available in enzyme 2VF5; (F) a 2D diagram presented the important ligand interactions between pose 83 and enzyme 2VF5; (G) the ligand map exposed the secondary interactions between pose 83 and enzyme 2VF5. | ||
The ligand interactions between pose 60 and 2VF5 could be observed on the 2D diagram (Fig. 9C). The results showed that this pose did not have good interaction with enzyme 2VF5. Ligand naringin, pose 60, interacted well with the enzyme because three parts of this pose (capping group, connecting unit, and functional group) were well detected with enzyme 2VF5.30 The capping group was identified via a π–σ interaction, which linked from Leu 484 to the system of π electrons of the aromatic ring. The connecting unit (CU) was shown to have no ligand interaction, and the functional group of pose 60 formed hydrogen bonds from Lys 603, Asn 305, Lys 487 to the hydrogen atom of phenolic hydroxyl groups of phenyl ring, the hydrogen atom of the alcohol group, and the oxygen atom of oxygen heterocyclic, respectively. As shown in Fig. 9D, the ligand map exposed the secondary interactions that formed from active residual amino acids on the enzyme to the active atom on pose 60. They included hydrogen bonds (yellow lines) from Asn 305, Ala 496, Lys 487, and Lys 603 to active atoms on pose 60 and the steric interactions from Tyr 304, Asn 305, Lys 487, Glu 488, Glu 495, Leu 601, and Ala 602. More steric forming around pose 60, the strength of interactions between this pose and the active residual amino acids, were stronger in the processing of a docking pose such as pose 60. The overlap interactions between pose 60 and 2VF5 enzyme were depicted by the violet circles on the atoms of pose 60 and the size of the violet circles were bigger and stronger in the overlapping interactions.
Small ligand in 2VF5 enzyme: ranked pose 83 docked to 2VF5 enzyme as seen in Fig. 9A–G and Table S9.† Pose 83 was the best docking pose among the 200 poses—it docked to active atoms on enzyme 2VF5, with the values of Free Gibbs energy and inhibition constant of −5.38 kcal mol−1 and 114 μM, respectively, as shown in Table S9.† Pose 83 formed 13 hydrogen bonds with active residual amino acids of enzymes such as Thr302, Glu 348, Ser 349, Thr 352, Ser 401, Glu 488, Ala 602, and Lys 603 and proved pose 83 had more hydrophilic properties (Fig. 9E). As shown in Fig. 9F, pose 83 did not interact well with the enzyme because the capping group and connecting unit were not identified via ligand interactions, they were hydrophobic interactions. As seen in Fig. 9G, a ligand map indicated secondary interactions that linked active atoms on pose 83 to active residual amino acids on the enzyme were weak interactions. Based on thermodynamic site and ligand interaction model, the interaction between ligand naringin and pose 60 was determined to be stronger than that of pose 83. Pose 60 was analysed as having a weak interaction with enzyme 2VF5 and this was also compatible with the in vitro test on the antifungal activity of naringin.
In this work, fluconazole (pose 87, the most stable conformation of the ligand fluconazole) was used as a reference drug and used in anti-fungi activity anchored to the target enzyme 2VF5, with the values of free Gibbs energy and inhibition constant of −5.55 kcal mol−1 and 86.10 μM, respectively, as shown in Fig. 10A and Table S9.† This pose formed four hydrogen bonds from residual of amino acids on enzymes such as Thr 302, Ser303, Ser 349, and Cys 300 to the active atom on pose 87. The significant ligand interactions between pose 87 and residual amino acids of enzyme indicated that Fluconazole interacted well with the enzyme because three parts of this pose, including the capping group (identification protein via aromatic or heterocyclic), connecting unit (linker, the aliphatic chains, and hydrophobic interactions) and functional groups (polar groups and hydrophilic interactions) were well identified via ligand interactions (Fig. 10B). The capping group detected by π-alkyl interactions from Ala 299 to triazole ring, π-sulphur from Cys 300 to triazole ring, π-donor hydrogen bonds from Thr 352, Ser 303 to triazole ring, another π-donor hydrogen from Gln 348, Ser 604, Lys603 to another triazole ring. The connecting unit (CU) was identified by a π-donor hydrogen bond from Ser 303 to the methylene group of the pose, a hydrophobic interaction. The functional groups of pose 87 were detected by hydrogen bonds from Cys 300 to the hydrogen atom and Thr 302 to the oxygen atom of the alcohol group and halogen interactions, fluorine interactions from Glu 488, Val 399 to fluorine atoms on aromatic rings. Fig. 10C indicated that the essential ligand interactions between pose 87 and enzyme 2VF5 (3D diagram) were determined to be similar to 2D interactions in Fig. 10B. The ligand map exposed the secondary interactions such as hydrogen bonds (brown lines), from Gln 348, Thr 302, Ser 303, Thr 352, and Cys 300 to active atoms on pose 87, steric interactions, green coloured lines from Glu 488, Val 399, Cys 300, Thr 352, Ser 303, Gln 348, Ser 349, and Lys 603 to active atom on pose 87, overlap interactions, and violet circles, indicating that the bigger violet circles were the stronger overlap interactions of the poses (Fig. 10D). In this case, many amino acids were witnessed forming around this pose and proved that the ligand interactions between pose and enzyme were very strong in conformation processing of ligand fluconazole or pose 87. Based on thermodynamic site and ligand interactions, the enzyme inhibition ability was arranged as the following ranking: pose 87 (fluconazole) > pose 60 (naringin) > pose 83 (small ligand).
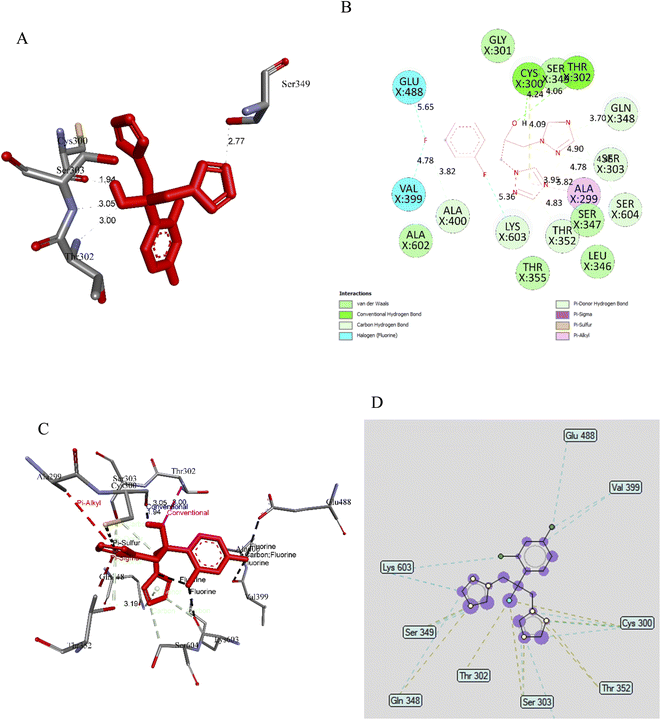 | ||
| Fig. 10 (A) Pose 87, ligand fluconazole formed hydrogen bonding with active atoms on enzyme target, 2VF5; (B) a 2D diagram showed the significant ligand interactions between pose 87 and residual amino acids on enzyme 2VF5; (C) a 3D diagram showed the significant ligand interactions between pose 97 and residual amino acids on enzyme 2VF5; (D) ligand map indicated the secondary interactions between residual active amino acids of enzyme 2VF5 and active atoms on pose 87. | ||
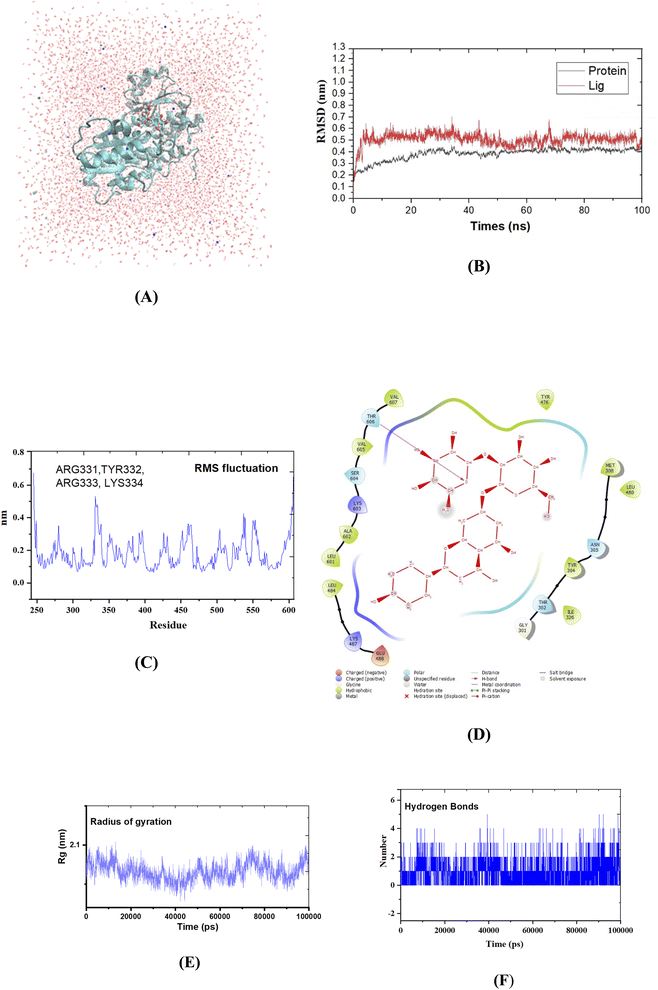 | ||
| Fig. 11 (A) Protein–naringin complex (pose 60) in three-point transferable intermolecular potential water box. (B) The graph of RMSD studies for the protein (2VF5.pdb) in the presence of pose 60; (C) the root mean square fluctuation (RMSF); (D) the significant ligand interactions between pose 60 and enzyme 2VF5; (E) the complex-pose 60 radius of gyration (Rg); (F) the number of H bonds formed between pose 60 and crystal structure of enzyme 2VF5 was determined using 100 ns simulation trajectories. | ||
4. Conclusion
The supercritical CO2 technology was effectively used for the extraction of essential oils and naringin from waste pomelo peels. The composition of D-limonene in essential oil was significantly enhanced via this technique. Both natural products were evaluated in vitro—antimicrobial activity, giving the essential oil revealed its high activity against three bacterial strains, M. catarrhalis, S. pyogenes, and S. pneumoniae. Meanwhile, naringin indicated good inhibition towards all tested microbial strains. Particularly, naringin appeared to be a high inhibition compound against fungal strains, T. rubrum, T. mentagrophytes, and M. gypseum. For the computational study, the molecular docking model and the validation of the docking model confirmed that D-limonene inhibited well against the bacterium M. catarrhalis. The molecular docking model of naringin and its MD simulation showed that this compound possessed potential ligand interactions, which then confirmed it as an effective inhibiting factor against fungi.Conflicts of interest
No potential conflict of interest was reported by the authors.Acknowledgements
This work was funded by Ho Chi Minh City Department of Science and Technology (HCMC-DOST) under the grant number 37/2020/HĐ-QPTKHCN.References
- R. J. Anmol, S. Marium, F. T. Hiew, W. C. Han, L. K. Kwan, A. K. Y. Wong, F. Khan, M. M. R. Sarker, S. Y. Chan and N. Kifli, J. Evidence-Based Integr. Med., 2021, 26, 2515690X211043741 CrossRef CAS.
- H. Hussain, N. Z. Mamadalieva, A. Hussain, U. Hassan, A. Rabnawaz, I. Ahmed and I. R. Green, Curr. Issues Mol. Biol., 2022, 44, 1960–1994 CrossRef CAS PubMed.
- Y. Zhao and S. Liu, Pharmazie, 2021, 76, 359–363 CAS.
- R. Chen, Q.-L. Qi, M.-T. Wang and Q.-Y. Li, Pharm. Biol., 2016, 54, 3203–3210 CrossRef CAS.
- S. Mahanta, B. Khanikor and R. Sarma, J. Entomol. Zool. Stud., 2017, 5, 803–809 Search PubMed.
- N.-G. Tao and Y.-J. Liu, Int. J. Food Prop., 2012, 15, 709–716 CrossRef CAS.
- K. Sudto, S. Pornpakakul and S. Wanichwecharungruang, Int. J. Food Sci. Technol., 2009, 44, 1737–1742 CrossRef CAS.
- P. A. Uwineza and A. Waśkiewicz, Molecules, 2020, 25, 3847 CrossRef CAS.
- T. T.-T. Huynh, T.-C. Mai, C.-H. Dang, T. T.-T. Vo, D.-T. Nguyen, V.-S. Dang, K. D. V. Nguyen, V.-T. Tran and T.-D. Nguyen, Arabian J. Chem., 2020, 13, 7289–7301 CrossRef.
- I. Quispe-Fuentes, E. Uribe, J. López, D. Contreras and J. Poblete, J. Food Process. Preserv., 2022, 46, e16116 CAS.
- B. Su, J. Tian, M. Liu, K. Wang, W. Yang, J. Ning, Y. Li and G. Zheng, J. Sep. Sci., 2022, 45(15), 3031–3042 CrossRef CAS PubMed.
- A. N. Giannuzzo, H. J. Boggetti, M. A. Nazareno and H. T. Mishima, Phytochem. Anal., 2003, 14, 221–223 CrossRef CAS PubMed.
- J. Yu, D. V. Dandekar, R. T. Toledo, R. K. Singh and B. S. Patil, Food Chem., 2007, 105, 1026–1031 CrossRef CAS.
- M. Hoshino, T. Suetsugu, H. Iwai, A. Takamizu, M. Tanaka, A. Quitain, M. Sasaki and M. Goto, Trans. Mater. Res. Soc. Jpn., 2014, 39, 309–311 CrossRef CAS.
- S. Rajput, S. Kaur, P. S. Panesar and A. Thakur, Biomass Convers. Biorefin., 2022, 1–10 Search PubMed.
- R. Romano, L. De Luca, A. Aiello, D. Rossi, F. Pizzolongo and P. Masi, Int. J. Food Sci. Technol., 2022, 57, 3826–3837 CrossRef CAS.
- D. W. Ingersoll, P. M. Bronstein and J. Bonventre, J. Comp. Physiol. Psychol., 1976, 90, 198 CAS.
- T. T. A. My, H. T. P. Loan, N. T. T. Hai, L. T. Hieu, T. T. Hoa, B. T. P. Thuy, D. T. Quang, N. T. Triet, T. T. V. Anh and N. T. X. Dieu, ChemistrySelect, 2020, 5, 6312–6320 CrossRef CAS PubMed.
- B. T. P. Thuy, T. T. A. My, N. T. T. Hai, L. T. Hieu, T. T. Hoa, H. Thi Phuong Loan, N. T. Triet, T. T. V. Anh, P. T. Quy and P. V. Tat, ACS Omega, 2020, 5, 8312–8320 CrossRef CAS PubMed.
- B. L. Staker, M. D. Feese, M. Cushman, Y. Pommier, D. Zembower, L. Stewart and A. B. Burgin, J. Med. Chem., 2005, 48, 2336–2345 CrossRef CAS PubMed.
- T.-B.-N. Dao, T.-M.-T. Nguyen, V.-Q. Nguyen, T.-M.-D. Tran, N.-M.-A. Tran, C. H. Nguyen, T.-H.-T. Nguyen, H.-H. Nguyen, J. Sichaem and C.-L. Tran, Molecules, 2021, 26, 2531 CrossRef CAS.
- M. Aliye, A. Dekebo, H. Tesso, T. Abdo, R. Eswaramoorthy and Y. Melaku, Sci. Rep., 2021, 11, 1–12 CrossRef PubMed.
- S. Singh, A. Datta, A. Schmidtchen, A. Bhunia and M. Malmsten, Sci. Rep., 2017, 7, 1–14 CrossRef PubMed.
- K. Hałdys, W. Goldeman, M. Jewgiński, E. Wolińska, N. Anger, J. Rossowska and R. Latajka, Bioorg. Chem., 2018, 81, 577–586 CrossRef PubMed.
- G. Brancolini, D. B. Kokh, L. Calzolai, R. C. Wade and S. Corni, ACS Nano, 2012, 6, 9863–9878 CrossRef CAS PubMed.
- M. N. Sibanyoni, S. K. Chaudhary, W. Chen, H.-R. Adhami, S. Combrinck, V. Maharaj, D. Schuster and A. Viljoen, Fitoterapia, 2020, 146, 104650 CrossRef CAS PubMed.
- E. W. Bell and Y. Zhang, J. Cheminf., 2019, 11, 1–9 CAS.
- T.-H. Duong, A. P. Devi, N.-V. Huynh, J. Sichaem, H.-D. Tran, M. Alam, T.-P. Nguyen, H.-H. Nguyen, W. Chavasiri and T.-C. Nguyen, Bioorg. Med. Chem. Lett., 2020, 30, 127359 CrossRef CAS.
- G. L. Warren, C. W. Andrews, A.-M. Capelli, B. Clarke, J. LaLonde, M. H. Lambert, M. Lindvall, N. Nevins, S. F. Semus and S. Senger, J. Med. Chem., 2006, 49, 5912–5931 CrossRef CAS PubMed.
- P. Agrawal, H. Singh, H. K. Srivastava, S. Singh, G. Kishore and G. P. Raghava, BMC Bioinf., 2019, 19, 105–124 CrossRef PubMed.
- F. Maltese, C. Erkelens, F. van der Kooy, Y. H. Choi and R. Verpoorte, Food Chem., 2009, 116, 575–579 CrossRef CAS.
- T. N.-T. Nguyen, T. N.-N. Huynh, V.-T. Tran, C.-H. Dang, T. K.-D. Hoang and T.-D. Nguyen, J. Essent. Oil Res., 2018, 30, 285–292 CrossRef CAS.
- M. Tuncbilek, T. Kiper and N. Altanlar, Eur. J. Med. Chem., 2009, 44, 1024–1033 CrossRef CAS PubMed.
- G. Giannini, W. Cabri, C. Fattorusso and M. Rodriquez, Future Med. Chem., 2012, 4, 1439–1460 CrossRef CAS PubMed.
- F. U. Eze, U. C. Okoro, D. I. Ugwu and S. N. Okafor, Front. Chem., 2019, 634 CrossRef CAS PubMed.
- H. Berendsen, J. Grigera and T. Straatsma, J. Phys. Chem., 1987, 91, 6269–6271 CrossRef CAS.
- A. B. Hsouna, M. Trigui, R. B. Mansour, R. M. Jarraya, M. Damak and S. Jaoua, Int. J. Food Microbiol., 2011, 148, 66–72 CrossRef.
- Y. Han, Z. Sun and W. Chen, Molecules, 2019, 25, 33 CrossRef.
- L. Yu, J. Yan and Z. Sun, Mol. Med. Rep., 2017, 15, 2339–2346 CrossRef CAS PubMed.
- J. A. Miller, P. A. Thompson, I. A. Hakim, H.-H. S. Chow and C. A. Thomson, Oncol. Rev., 2011, 5, 31–42 CrossRef.
- V. M. Grinevicius, K. S. Andrade, F. Ourique, G. A. Micke, S. R. Ferreira and R. C. Pedrosa, J. Supercrit. Fluids, 2017, 128, 94–101 CrossRef CAS.
- R. P. Adams, Identification of essential oil components by gas chromatography/mass spectrometry, Allured publishing corporation, Carol Stream, 2007 Search PubMed.
- V.-D. Doan, V.-T. Le, T.-L. Phan, T. L.-H. Nguyen and T.-D. Nguyen, J. Cluster Sci., 2021, 32, 1673–1682 CrossRef CAS.
- M.-T. Tran, L.-P. Nguyen, D.-T. Nguyen, L. Cam-Huong, C.-H. Dang, T. T. K. Chi and T.-D. Nguyen, Res. Chem. Intermed., 2021, 47, 4613–4633 CrossRef CAS.
- G. A. Subramenium, K. Vijayakumar and S. K. Pandian, J. Med. Microbiol., 2015, 64, 879–890 CrossRef CAS PubMed.
- M. T. I. Salama, J. Essent. Oil-Bear. Plants, 2017, 20, 233–239 CrossRef CAS.
- F. Wang, H. You, Y. Guo, Y. Wei, P. Xia, Z. Yang, M. Ren, H. Guo, R. Han and D. Yang, Ind. Crops Prod., 2020, 147, 112172 CrossRef CAS.
- A. Tan, L. V. Blakeway, Y. Yang, Y. Zhou, J. M. Atack, I. R. Peak and K. L. Seib, PLoS One, 2020, 15, e0234306 CrossRef CAS PubMed.
- S. Andrade-Ochoa, K. F. Chacón-Vargas, L. E. Sánchez-Torres, B. E. Rivera-Chavira, B. Nogueda-Torres and G. V. Nevárez-Moorillón, Membranes, 2021, 11, 405 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra04068a |
| This journal is © The Royal Society of Chemistry 2022 |