Open Access Article
Open Access ArticleSalivary SARS-CoV-2 antibody detection using S1-RBD protein-immobilized 3D melt electrowritten poly(ε-caprolactone) scaffolds†
Pingping Han‡
 ab,
Chun Liu‡ab,
Reuben Staplesab,
Corey S. Moranab,
Srinivas Sulugodu Ramachandraab,
Maria Natividad Gómez-Cerezo*ab and
Sašo Ivanovski
ab,
Chun Liu‡ab,
Reuben Staplesab,
Corey S. Moranab,
Srinivas Sulugodu Ramachandraab,
Maria Natividad Gómez-Cerezo*ab and
Sašo Ivanovski *ab
*ab
aThe University of Queensland, School of Dentistry, Brisbane, QLD 4006, Australia. E-mail: s.ivanovski@uq.edu.au
bThe University of Queensland, School of Dentistry, Center for Oral-facial Regeneration, Rehabilitation and Reconstruction (COR3), Brisbane, QLD 4006, Australia
First published on 31st August 2022
Abstract
Sensitive detection of immunoglobulin antibodies against SARS-CoV-2 during the COVID-19 pandemic is critical to monitor the adaptive immune response after BNT162b2 mRNA vaccination. Currently employed binding antibody detection tests using 2D microplate-based enzyme-linked immunosorbent assays (ELISA) are limited by the degree of sensitivity. In this study, a 3D antibody test was developed by immobilizing the receptor-binding domain on Spike subunit 1 (S1-RBD) of SARS-CoV-2 onto engineered melt electrowritten (MEW) poly(ε-caprolactone) (PCL) scaffolds (pore: 500 μm, fiber diameter: 17 μm) using carbodiimide crosslinker chemistry. Protein immobilization was confirmed using X-ray photoelectron spectroscopy (XPS) by the presence of peaks corresponding with nitrogen. Self-developed indirect ELISA was performed to assess the functionality of the 3D platform in comparison with a standard 2D tissue culture plate (TCP) system, using whole unstimulated saliva samples from 14 non-vaccinated and 20 vaccinated participants (1- and 3- weeks post-dose 1; 3 days, 1 week and 3 weeks post-dose 2) without prior SARS-CoV-2 infection. The three-dimensional S1-RBD PCL scaffolds, while demonstrating a kinetic trend comparable to 2D TCP, exhibited significantly higher sensitivity and detection levels for all three immunoglobulins assayed (IgG, IgM, and IgA). These novel findings highlight the potential of MEW PCL constructs in the development of improved low-cost, point-of-care, and self-assessing diagnostic platforms for the detection and monitoring of SARS-CoV-2 antibodies.
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic is attributed to a novel RNA single-stranded coronavirus – SARS-CoV-2.1 The spike (S) glycoprotein plays an important role in viral binding to human host cells and antibodies against S protein can neutralize the infection, thus it is a major target for vaccine development.2 The messenger RNA (mRNA) Pfizer–BioNTech BNT162b2 vaccine comprises a full-length coding sequence of the SARS-CoV-2 S protein (including the receptor-binding domain on the S1 subunit – S1-RBD), which has been proven to generate immunoglobulin (Ig) antibodies against SARS-CoV-2 virus.3–7 Detection of antibodies against S1-RBD protein following the BNT162b2 vaccine is very important as an indicator of the sustainability of the anti-RBD antibody response post-vaccine.Material science8 and 3D printing9 technologies incorporating polymers such as poly(ε-caprolactone) (PCL) have significantly contributed to the response against the COVID-19 pandemic through the manufacture of personal protective equipment, ventilators, and nasopharyngeal swab test devices.9–13 Another potential application of this technology is in the development of sensitive, and cost-effective antibody tests for the detection of antibodies against the SARS-CoV-2 virus, utilizing non-invasive biosamples, such as saliva, for the detection of IgG antibodies against S protein generated by BNT162b2 vaccination.14–18 Currently, laboratory-based surveillance of antibody response following infection or vaccination is mainly performed on a 2D polystyrene tissue culture plate (TCP) by enzyme-linked immunosorbent assay (ELISA),19–24 whose design limitations (i.e. limited surface area), insensitivity, detection performance and long assay time (∼6 hours for sandwich ELISA).25 Other platforms have been also investigated, such as gold nanoparticles26 and other 2D materials.27 Current antibody detection tests have been reviewed in ref. 25. For instance, the advantage of using laboratory 2D ELISA assays, gold nanoparticle-associated lateral flow assay (LFA) and 2D platforms showed reliable results to detect immunity against SARS-CoV-2 either following vaccination or post-viral infection. The disadvantage of traditional sandwich ELISA is time-consuming (more than 24 hours to perform in the lab). Rapid detection is feasible in LFA devices; however, extensive research on SARS-CoV-2 antigen detection, with limited effort for immunity for people after vaccination. Variability and antibody cross-reactivity between batches, sensitivity limitations, and the fact that its analysis results are qualitative or semi-quantitative remain challenges for the LFA test in terms of accurate diagnosis. Recent studies suggest that improved sensitivity to detect antibodies against the SARS-CoV-2 virus may be achieved by 3D-printed immunoassays;28,29 however, these electrochemical sensors require specific training to utilize. It is of considerable importance to develop a 3D antibody test for laboratory settings. Several studies have shown that high-resolution melt electrowritten (MEW) poly(ε-caprolactone) (PCL) scaffolds are suitable for cell growth, drug delivery and tissue regeneration.30–38 Given their advantageous properties, including precise architecture, high porosity and greater surface area, the potential of MEW PCL scaffolds as 3D immunodetection platforms for COVID-19 antibodies warrants investigation.
The present study describes the development and validation of a 3D antibody test against SARS-CoV-2 using functionalized MEW PCL scaffolds. The hypothesis was that this highly porous, high surface area platform would be more sensitive than traditional 2D platforms within 2 hours of assay time using indirect ELISA. The aim was to assess the MEW 3D PCL constructs and compare them with traditional 2D ELISA for immunoglobulin (Ig) G, IgM, and IgA detection in saliva samples from 20 vaccinated and 14 non-vaccinated (NV) participants without prior SARS-CoV-2 infection.
2. Materials and methods
2.1 Manufacture of 3D PCL MEW scaffolds
All scaffolds were manufactured by melt electrowriting (MEW) using medical-grade PCL as previously described.30,32 Briefly, polymer pellets (PC12, Corbion, Amsterdam, The Netherlands) were heated to 74 °C and 83 °C at the cartridge and needle locations within a blunt 23 G 2 mL syringe, respectively. The polymer was then extruded onto a programmable x–y stage at a pressure of 1.2 bar, a voltage of 8.2 kV, a translational speed of 850 mm min−1 and a distance of 7.4 mm between the collector plate and the spinneret. The scaffolds were subsequently printed to consist of 50 layers in a 0/90° layering pattern. After fabrication, scaffolds were sectioned into 5 mm discs to fit inside wells of a 96-well tissue culture plate (TCP).2.2 Scaffold SARS-CoV-2 S1-RBD protein immobilization and characterization
SARS-CoV-2 (2019-nCoV) Spike RBD-His recombinant protein (catalogue number: 40592-V08H-100ug, Sino Biological) was chemically conjugated to scaffolds via carbodiimide crosslinker chemistry described previously (ESI Fig. 1†).39–42 First, MEW PCL scaffolds were treated with 1.5 M sodium hydroxide (NaOH) for 1.5 hours at 37 °C to increase scaffold hydrophilicity, then washed with distilled water until the pH of the water reached 7. Scaffolds were then covered with conjugation solution in 0.1 M 2-(N-morpholino) ethanesulfonic acid (MES, pH 4.5–5.5) buffer containing 0.2 M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC, ThermoFisher Scientific) and 0.2 M N-hydroxysuccinimide (NHS, Sigma Aldrich) for 20 minutes at room temperature to activate COOH groups. NH2 covalent grafting was carried out by covering scaffolds with 2 μg mL−1 SARS-COV-2 S1-RBD protein in 0.1 M 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, pH = 7.4) buffer at 4 °C overnight. Finally, scaffolds were washed in HEPES buffer and MilliQ water twice and dried under vacuum prior to characterization. Functionalized scaffolds were labelled with S1-RBD-PCL scaffolds in the following sections.Scanning electron microscopy (SEM, Hitachi SU3500, Tokyo, Japan) operating at a voltage of 8 kV was used to evaluate the morphology of the 3D MEW scaffolds. Prior to imaging, samples were immersed in 100% ethanol and sectioned using a scalpel blade and a 5 mm biopsy punch. 15 nm platinum coating was conducted on all the samples (Q150T Plus, Quorum, East Sussex, United Kingdom). Pore size was measured from fiber centre-to-centre (n = 15) from three different samples using ImageJ (National Institutes of Health (NIH), Bethesda, MD, USA).
Surface compositional analysis (survey scans) was performed using a Kratos Axis Ultra 165 X-ray photoelectron spectroscopy (XPS) system equipped with a hemispherical analyzer. Areas of 1 mm × 0.5 mm for each scaffold were irradiated with a 100 W monochromatic Al Kα (1486.7 eV) X-ray beam at a takeoff angle of 90°. During the experiment, the chamber pressure was kept around 1 × 10−9 and 1 × 10−10 Torr. Narrow scans were conducted using 20 eV pass energy for the C 1s, O 1s and N 1s core levels. A value of 284.6 eV was used as a reference for the methylene component of the C 1s spectrum, being the calibration energy for the binding energy scale. The rest of the spectra were corrected accordingly. After the experiment, data were processed using Casa XPS software v 2.3.5. All reported atomic percentages are the average of three independent measurements, each on a minimum of five replicate fiber mat samples.
2.3 Human saliva sample collection
Staff and student volunteers (n = 20) from the University of Queensland School of Dentistry who received the BNT162b2 vaccine between May and October 2021 were recruited to the study under Metro North Hospital and Health Service (MNHHS) and The University of Queensland Human Ethics approval (approval numbers: 65509 and 2020/HE002629, respectively). This observational project was conducted in accordance with the principles embodied in the Declaration of Helsinki following local statutory requirements. Consecutive participants who were older than 18 years were included, with no exclusion criteria being applied. All the participants in this study provided written informed consent prior to enrolment in the study. Demographic data are presented in Table 1. There was an interval of 3 weeks between the two vaccine doses and a total follow-up time of 6 weeks: 1 week and 3 weeks after the first dose, and, 3 days, 1 week, and 3 weeks after the second dose. One participant withdrew from the study after the second dose, and for a second participant, there was an insufficient sample for testing at the last time point. A total of 14 unstimulated saliva samples from non-vaccinated participants without previous COVID19 infection were also included as a control. All participants reported having no prior SARS-CoV-2 infection and no underlying systemic disease.| Non-vaccinated | Post-dose 1 | Post-dose 2 | ||
|---|---|---|---|---|
| a p values were calculated against non-vaccinated participants. | ||||
| n | 14 | 20 | 19 | |
| Age in years (mean ± SD) | 31.3 ± 9.6 (23–51) | 30.6 ± 5.5 (23–43) p = 0.782 | 29.9 ± 4.8 (23–43) p = 0.934 | |
| Gender, n (%) | Female | 4 (28.57%) | 9 (45%) | 9 (47.37%) |
| Male | 10 (71.43%) | 11 (55%) | 10 (52.63%) | |
| Ethnicity, n (%) | Caucasian | 5 (35.71%) | 3 (15%) | 3 (15.79%) |
| Asian | 8 (57.14%) | 15 (75%) | 15 (78.95%) | |
| Other | 1 (7.14%) | 2 (10%) | 1 (5.3%) | |
Prior to saliva collection, all participants were asked to refrain from food and drink for at least 1 hour and unstimulated whole saliva samples using the spitting method were collected between 9.00 AM and 12.00 PM as described previously.43–46 The participants rinsed their mouths to remove any food debris using 10 mL of water before saliva collection. Participants were asked to spit their saliva into a sterile 50 mL falcon tube, which was immediately kept on ice and later aliquoted for storage at −80 °C until assayed. A volume of 10 μL whole saliva was used for the ELISA test.
2.4 Salivary antibody detection using 3D S1-RBD-PCL scaffolds and 2D TCP
A self-developed enzyme-linked immunosorbent assay (ELISA) was produced by combining PeproTech's TMB ELISA Development Kit and RayBio® COVID-19 S1 RBD Protein Human IgG ELISA Development Kit protocols. For 2D TCP ELISA, empty wells of a 96-well TCP were coated with 2 μg mL−1 SARS-CoV-2 Spike 1-RBD Protein at 4 °C overnight. After blocking with 1% BSA/PBS for 1 hour, 10 μL saliva samples were added to both the coated 2D TCP wells and the wells containing S1-RBD protein conjugated 3D MEW PCL scaffolds (separate 96 well TCP) and incubated at 37 °C for 1 hour. Following 4 washes with wash buffer, biotinylated Anti-Human IgG Antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150) was added to each well and incubated for 30 minutes. After a further 4 washes with wash buffer, diluted streptavidin-HRP conjugated HRP (1
150) was added to each well and incubated for 30 minutes. After a further 4 washes with wash buffer, diluted streptavidin-HRP conjugated HRP (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800) was added to each well and incubated at 37 °C for 30 minutes. 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate was used for colour rendering and the reaction terminated with a 1 M HCl prior to spectrophotometric measurement of a sample (absorbance) at 450 nm. The OD450 values of negative controls from the kits were used as internal controls, with cut-off values calculated as the average of 14 negative samples plus the standard deviations (SD), as described in previous reports.20,47,48 Since empty TCP well and S1-RBD PCL scaffolds (background groups) have background noise, our data are presented as OD450 − ODblank. Controls and samples were assayed in duplicates as described previously.20,49 The intraassay variability was evaluated by determining the standard deviation and percent coefficient of variation (CV%). The variability (CV%) for the majority of data was smaller than 15% CV% (91%). Standard curves were generated for both 3D MEW S1-RBD-PCL scaffolds and 2D TCP using 50 μL of positive controls at 8 serial concentrations of a positive sample which contains SARS-CoV-2 S1 RBD protein human IgG antibody provided by RayBio® COVID-19 S1 RBD Protein Human IgG ELISA. A sample was considered IgG/IgM/IgA positive if the absorbance reading value was greater than the cut-off value and internal negative control. To determine the detection limit for IgG, additional 8 different concentrations of positive controls were applied. For specificity, we coated the 2D TCP and 3D PCL scaffold with 2 μg mL−1 of bovine serum albumin (BSA) with 15 concentrations of positive controls (in Fig. 1g).
800) was added to each well and incubated at 37 °C for 30 minutes. 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate was used for colour rendering and the reaction terminated with a 1 M HCl prior to spectrophotometric measurement of a sample (absorbance) at 450 nm. The OD450 values of negative controls from the kits were used as internal controls, with cut-off values calculated as the average of 14 negative samples plus the standard deviations (SD), as described in previous reports.20,47,48 Since empty TCP well and S1-RBD PCL scaffolds (background groups) have background noise, our data are presented as OD450 − ODblank. Controls and samples were assayed in duplicates as described previously.20,49 The intraassay variability was evaluated by determining the standard deviation and percent coefficient of variation (CV%). The variability (CV%) for the majority of data was smaller than 15% CV% (91%). Standard curves were generated for both 3D MEW S1-RBD-PCL scaffolds and 2D TCP using 50 μL of positive controls at 8 serial concentrations of a positive sample which contains SARS-CoV-2 S1 RBD protein human IgG antibody provided by RayBio® COVID-19 S1 RBD Protein Human IgG ELISA. A sample was considered IgG/IgM/IgA positive if the absorbance reading value was greater than the cut-off value and internal negative control. To determine the detection limit for IgG, additional 8 different concentrations of positive controls were applied. For specificity, we coated the 2D TCP and 3D PCL scaffold with 2 μg mL−1 of bovine serum albumin (BSA) with 15 concentrations of positive controls (in Fig. 1g).
2.5 Statistical analysis
Data were analysed using GraphPad Prism (v9.0.0, San Diego, CA, USA). Scatter plots display mean ± standard deviation (SD) unless otherwise stated. Quantile–quantile plots were used to analyze whether data are normally distributed. Since there are some missing clinical samples, a mixed-effect model with Dunnett's multiple comparison test was used to analyze Ig levels at each time point against 1 week post-dose 1. The Wilcoxon matched-pairs signed-rank test (for abnormally distributed data) and paired T-test (for normally distributed data) was applied for the comparison of S1-RBD Ig concentration between 2D and 3D assays at each time point, with missing data being excluded in this analysis. A p-value < 0.05 was considered statistically significant.3. Results and discussion
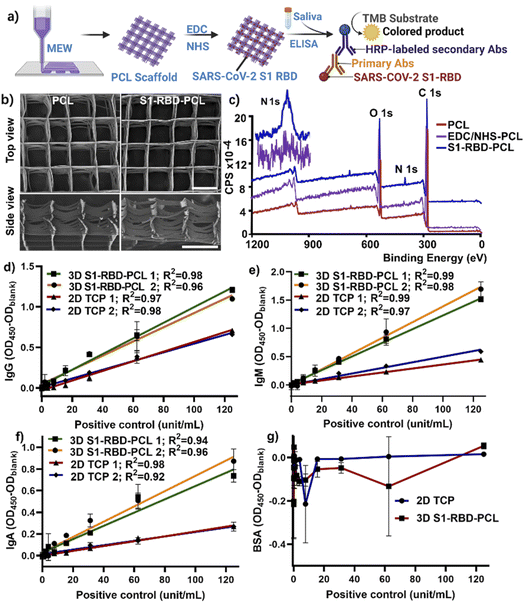
3.1 Characterization of S1-RBD-PCL scaffolds
Highly porous MEW PCL scaffolds have been shown to promote cellular behaviour in vitro and in vivo for tissue engineering applications.30–36 However, there is limited knowledge in regard to the effective utilization of these constructs in antibody detection against SARS-CoV-2, and this represents an important gap in the literature. This study developed a 3D platform to detect antibody response in participants following BNT162b2 mRNA vaccination, utilizing SARS-CoV-2 S1-RBD protein-conjugated MEW PCL scaffolds (Fig. 1a). Saliva samples from non-vaccinated and vaccinated participants were assayed by indirect enzyme-linked immunosorbent assay (ELISA) to evaluate the efficacy of the 3D S1-RBD-PCL constructs for immunoglobulin (IgG, IgM, IgA) detection in comparison with a standard 2D TCP assay.We employed carbodiimide chemistry to conjugate S1-RBD protein (ESI Fig. 1†). SEM imaging demonstrated that the MEW technique successfully produced controlled layer-by-layer fibre stacking with high precision (pore size 499 ± 17.3 μm) and fibre resolution (diameter was 17.5 ± 1.3 μm), consistent with our previous work.32 In addition, images taken pre- and post-immobilisations demonstrated that the morphology of the scaffold was not compromised by the S1-RBD protein immobilisation process (Fig. 1b). XPS was performed to further confirm protein conjugation to the PCL scaffolds, as demonstrated by the presence of nitrogen after immobilisation of S1-RBD protein and its absence in blank PCL scaffolds (Fig. 1c). It is noted EDC/NHS treated PCL scaffolds showed no nitrogen peak, further confirming the successful S1-RBD immobilization in S1-RBD-PCL scaffolds (Fig. 1c). The relative atomic% of N found with XPS for S1-RBD functionalised PCL scaffolds was 1.83 ± 0.43% (ESI Table 1†). The XPS results demonstrated the effectiveness of EDC/NHS carbodiimide chemistry as a technique to graft proteins to the PCL scaffold.39–41
To confirm the function of S1-RBD-functionalised PCL scaffolds, 2 representative standard curves were generated for 3D S1-RBD-PCL scaffolds and 2D TCP using 8 different concentrations of positive control (Fig. 1d–f). Our data demonstrated robust curves for both platforms (R2 > 0.9), however, OD450 values for the 3D constructs were ∼2-fold higher than those for 2D TCP, indicating the former to be potentially more sensitive for antibody detection. Of note, as expected, 3D S1-RBD-PCL scaffolds generated higher background noise than 2D TCP (0.078 ± 0.002 vs. 0.36 ± 0.011 at OD450) for a blank sample with 0 unit per mL of positive control. To remove this bias, our data has been displayed as OD450 − ODblank for all data. To determine the specificity, 2 μg mL−1 of BSA was coated on the 2D TCP wells and used to functionalize the 3D PCL scaffolds to detect IgG. The results show that all data were negative after removing the background (Fig. 1 g). The results shown in Fig. 1d and g indicate that S1-RBD is specific for detecting IgG against SARS-CoV-2 S1-RBD. We further examined the detection limit by including an additional 8 concentrations of positive controls (ranging from 1.9 to 0.015 unit per mL). The data showed that the detection limit was 0.061 and 0.015 unit per mL for 2D TCP and 3D S1-RBD-PCL platforms, respectively (ESI Fig. 2†), indicating our 3D S1-RBD-PCL platform showed 4 times more sensitivity compared to 2D TCP.
3.2 Performance of the 3D S1-RBD-PCL platform in antibody detection
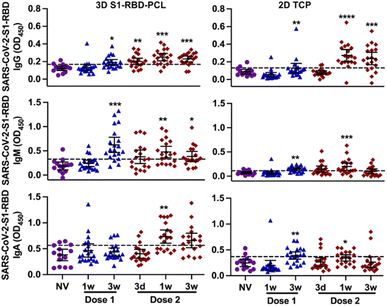
None of the participants in this study was previously infected by SARS-CoV-2 at the time of participant recruitment. There was no statistically significant difference between the average ages of the non-vaccinated and vaccinated groups (Table 1). A total of 111 unstimulated saliva samples were collected for the study (ESI Table 2†).Participant salivary antibody levels to S1-RBD were measured using the functionalised PCL scaffold assay and compared with 2D TCP. Both 3D and 2D platforms demonstrated a peak for all three Ig subtypes at 1 week post-dose 2 (Fig. 2). However, levels of IgG and IgM tended to be higher at all time points for the 3D S1-RBD-PCL assay compared to 2D TCP, with IgA antibody levels detected at higher levels from 1 week post-dose 2 (Fig. 2). In terms of antibody response trends, these tended to be similar in general for IgG subtypes across both 3D and 2D platforms (ESI Fig. 3†). IgM and IgA showed a slightly different trend between 3D and 2D platforms, with IgM peaking at 3 weeks post dose 1 and IgA peaked at 1 week post dose 2 in 3D S1-RBD-PCL. While in 2D TCP, both IgM and IgA peaked at 3 weeks post dose 1. However, we are uncertain about the factors that are contributing to this phenomenon. Increasing the sample size may be able to justify this.
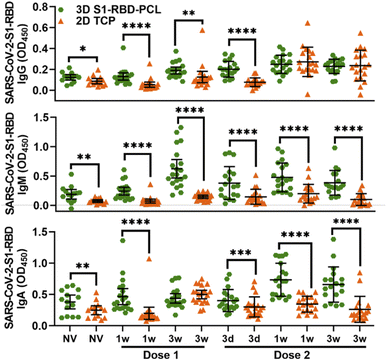
When comparing matched data between 3D and 2D platforms, OD450 readings in the 3D assay were 1.5 to 4-fold higher than the 2D assay for all three Ig subtypes (Fig. 3), with IgG at 1- and 3 weeks post-dose 2 and IgA at 3 week post-dose 1 as exceptions. The same was found when we evaluated the percentage of participants exhibiting a positive immune response (higher than cut-off values) to S1-RBD protein (ESI Table 2†). This shows that S1-RBD-functionalised PCL scaffolds have higher sensitivity for detecting Ig immune response against SARS-CoV-2 S1-RBD in subjects post BNT162b2 vaccination.
The levels of salivary IgG, IgM and IgA against SARS-CoV-2 S1-RBD protein observed in the current study are consistent with recently published data on antibodies against the spike glycoprotein being present in the saliva of BNT162b2 vaccine recipients.14,15 Of note, 3D S1-RBD-PCL scaffolds showed higher detection values than 2D TCP; this is consistent with other research that demonstrates increased bioactivity of functionalized 3D MEW constructs, such as fibronectin- and laminin-immobilized PCL nanofiber scaffolds exhibit higher neuronal tissue regeneration.39 Since there is a limited 3D PCL platform for COVID-19 antibody detection in the field, we speculate that the larger surface area available for increased antigen protein binding, together with greater sample perfusion enabled by the interconnected porous network in the 3D construct (i.e., more space for sample binding42), may account for this finding. It is worth noting that 3D S1-RBD-PCL scaffolds have higher background noise compared to 2D TCP. This is expected given they may have non-specific binding due to their increased surface area; however, this requires further studies to explore how to remove background noise.
There are several limitations to our study. It is noted that the positive sample provided by the kit is blood samples that may compromise the sensitivity of saliva samples; however, all samples showed higher OD values than internal negative controls. Additionally, we didn't use the standard curve to analyze our original OD values. The second limitation is the absence of patient-matched pre-vaccination data and serum. Future studies investigating functionalized MEW PCL scaffold in detecting COVID-19 antibodies are required to validate antigen-conjugated 3D MEW PCL platform can further provide higher sensitivity compared to a standard 2D TCP system. A limitation of MEW technology is its limited large-scale high throughput for mass commercial viability.50 Another limitation is a lack of data on the stability performance of our developed S1-RBD-PCL scaffold system. Thus, future experiments should be carried out using S1-RBD-PCL scaffolds at different temperatures (room temperature, 4 °C and 37 °C) to explore the function of the 3D platform after storage at various conditions for 1, 3, 7 days, 2 and 4 weeks. Despite limitations, the use of MEW PCL scaffolds provides a highly porous, interconnected and structurally defined 3D environment which enabled increased binding potential for the S1-RBD antigen leading to improved platform sensitivity with a 4-fold increased detection limit (ESI Fig. 2†). With this technology undergoing rapid advancement to address such limitations it could be suited for self-assessment or specific point-of-care applications, providing a low-cost alternative with improved sensitivity.
Overall, this study has revealed new insights for the enhancement of current diagnostic platforms for the detection and monitoring of SARS-CoV-2 antibodies following the mRNA vaccine, using 3D platforms with increased binding potential compared to a 2D microwell plate.
4. Conclusions
We developed a 3D binding antibody test using highly porous MEW PCL scaffolds comprising SARS-CoV-2 S1-RBD antigen immobilized to the PCL fibre surface by carbodiimide crosslinker chemistry. Our 3D platform exhibited enhanced detection of immunoglobulin antibodies compared to a standard 2D TCP platform, as evidenced by the consistently higher OD450 values determined by indirect ELISA. These findings support the utilization of 3D MEW PCL platforms for the detection of SARS-CoV-2 antibodies through a non-invasive biofluid, saliva, over more traditional 2D techniques. From the authors' perspective, although the cost of this 3D system is relatively low (∼$1 per 5 mm disc of S1-RBD scaffold), utilizing current 3D S1-RBD-PCL scaffolds as commercialized products is still in its infancy since MEW and S1-RBD functionalization can only be performed by trained researchers. It may be feasible to combine 3D S1-RBD-PCL and LFA techniques to enable rapid immunity detection; this however, requires further investigation.Data availability statement
All data that support the findings of this study are included within the article (and any ESI files†).Author contributions
P. H. M. G. C. and S. I. conceived the research; C. L., P. H., M. G. C. and performed the experiments and analyzed the data; R. S. manufactured MEW scaffolds; S. S. R., C. L. and P. H. recruited participants and sample collection; P. H. and C. L. prepared the manuscript under the supervision of M. G. C. and S. I.Conflicts of interest
The authors declare that they have no competing interestAcknowledgements
This study was supported by the Association for Dental Infection Control (ADIC), the Australasian Osseointegration Society Limited (AOS, Qld Branch, Brisbane, Australia), the Australian and New Zealand Academy of Periodontists (ANZAP, Sydney, Australia) and UQ HaBS Early Career Academic Research Accelerator Award (to P. H.). The authors acknowledge the facilities as well as the scientific and technical assistance of the Australian Microscopy & Microanalysis Research Facility at the Centre for Microscopy and Microanalysis, The University of Queensland.References
- Y. Chen, Q. Liu and D. Guo, J. Med. Virol., 2020, 92, 418–423 CrossRef CAS PubMed.
- L. Dai and G. F. Gao, Nat. Rev. Immunol., 2021, 21, 73–82 CrossRef CAS PubMed.
- G. A. Poland, I. G. Ovsyannikova and R. B. Kennedy, Lancet, 2020, 396, 1595–1606 CrossRef CAS.
- E. E. Walsh, R. W. Frenck Jr, A. R. Falsey, N. Kitchin, J. Absalon, A. Gurtman, S. Lockhart, K. Neuzil, M. J. Mulligan, R. Bailey, K. A. Swanson, P. Li, K. Koury, W. Kalina, D. Cooper, C. Fontes-Garfias, P. Y. Shi, Ö. Türeci, K. R. Tompkins, K. E. Lyke, V. Raabe, P. R. Dormitzer, K. U. Jansen, U. Şahin and W. C. Gruber, N. Engl. J. Med., 2020, 383, 2439–2450 CrossRef CAS.
- D. M. Skowronski and G. De Serres, N. Engl. J. Med., 2021, 384, 1576–1577 CrossRef CAS.
- P. Jalkanen, P. Kolehmainen, H. K. Häkkinen, M. Huttunen, P. A. Tähtinen, R. Lundberg, S. Maljanen, A. Reinholm, S. Tauriainen, S. H. Pakkanen, I. Levonen, A. Nousiainen, T. Miller, H. Välimaa, L. Ivaska, A. Pasternack, R. Naves, O. Ritvos, P. Österlund, S. Kuivanen, T. Smura, J. Hepojoki, O. Vapalahti, J. Lempainen, L. Kakkola, A. Kantele and I. Julkunen, Nat. Commun., 2021, 12, 3991 CrossRef CAS PubMed.
- P. Naaber, L. Tserel, K. Kangro, E. Sepp, V. Jürjenson, A. Adamson, L. Haljasmägi, A. P. Rumm, R. Maruste, J. Kärner, J. M. Gerhold, A. Planken, M. Ustav, K. Kisand and P. Peterson, Lancet Reg. Health Eur., 2021, 10, 100208 CrossRef PubMed.
- Z. Tang, N. Kong, X. Zhang, Y. Liu, P. Hu, S. Mou, P. Liljeström, J. Shi, W. Tan, J. S. Kim, Y. Cao, R. Langer, K. W. Leong, O. C. Farokhzad and W. Tao, Nat. Rev. Mater., 2020, 5, 847–860 CrossRef CAS PubMed.
- Y. Y. C. Choong, H. W. Tan, D. C. Patel, W. T. N. Choong, C.-H. Chen, H. Y. Low, M. J. Tan, C. D. Patel and C. K. Chua, Nat. Rev. Mater., 2020, 5, 637–639 CrossRef CAS PubMed.
- K. P. A. Kumar and M. Pumera, Adv. Funct. Mater., 2021, 31, 2100450 CrossRef CAS PubMed.
- M. Li, H. Wen, H. Li, Z. C. Yan, Y. Li, L. Wang, D. Wang and B. Z. Tang, Biomaterials, 2021, 276, 121007 CrossRef CAS PubMed.
- L. Wang, Y. Gao, J. Xiong, W. Shao, C. Cui, N. Sun, Y. Zhang, S. Chang, P. Han, F. Liu and J. He, J. Colloid Interface Sci., 2022, 606, 961–970 CrossRef CAS PubMed.
- J. Xiong, A. Li, Y. Liu, L. Wang, X. Qin and J. Yu, Small, 2021, e2105570, DOI:10.1002/smll.202105570.
- T. J. Ketas, D. Chaturbhuj, V. M. C. Portillo, E. Francomano, E. Golden, S. Chandrasekhar, G. Debnath, R. Díaz-Tapia, A. Yasmeen, K. D. Kramer, T. Munawar, W. Leconet, Z. Zhao, P. J. M. Brouwer, M. M. Cushing, R. W. Sanders, A. Cupo, P. J. Klasse, S. C. Formenti and J. P. Moore, Pathog. Immun., 2021, 6, 116–134 CrossRef PubMed.
- I. Lapić, D. Šegulja and D. Rogić, J. Med. Virol., 2021, 93, 5257–5259 CrossRef PubMed.
- M. W. Keuning, M. Grobben, A. C. de Groen, E. P. Berman-de Jong, M. W. Bijlsma, S. Cohen, M. Felderhof, F. de Groof, D. Molanus, N. Oeij, M. Rijpert, H. W. M. van Eijk, G. Koen, K. van der Straten, M. Oomen, R. Visser, F. Linty, M. Steenhuis, G. Vidarsson, T. Rispens, F. B. Plotz, M. J. van Gils and D. Pajkrt, Microbiol. Spectr., 2021, 9, e0073121 CrossRef PubMed.
- K. K. To, O. T. Tsang, W. S. Leung, A. R. Tam, T. C. Wu, D. C. Lung, C. C. Yip, J. P. Cai, J. M. Chan, T. S. Chik, D. P. Lau, C. Y. Choi, L. L. Chen, W. M. Chan, K. H. Chan, J. D. Ip, A. C. Ng, R. W. Poon, C. T. Luo, V. C. Cheng, J. F. Chan, I. F. Hung, Z. Chen, H. Chen and K. Y. Yuen, Lancet Infect. Dis., 2020, 20, 565–574 CrossRef CAS PubMed.
- P. Han, C. S. Moran, S. Sulugodu Ramachandra, L. J. Walsh and S. Ivanovski, J. Periodontol., 2022 DOI:10.1002/jper.22-0152.
- P. Fathi-Hafshejani, N. Azam, L. Wang, M. A. Kuroda, M. C. Hamilton, S. Hasim and M. Mahjouri-Samani, ACS Nano, 2021, 15, 11461–11469 CrossRef CAS PubMed.
- B. Isho, K. T. Abe, M. Zuo, A. J. Jamal, B. Rathod, J. H. Wang, Z. Li, G. Chao, O. L. Rojas, Y. M. Bang, A. Pu, N. Christie-Holmes, C. Gervais, D. Ceccarelli, P. Samavarchi-Tehrani, F. Guvenc, P. Budylowski, A. Li, A. Paterson, F. Y. Yue, L. M. Marin, L. Caldwell, J. L. Wrana, K. Colwill, F. Sicheri, S. Mubareka, S. D. Gray-Owen, S. J. Drews, W. L. Siqueira, M. Barrios-Rodiles, M. Ostrowski, J. M. Rini, Y. Durocher, A. J. McGeer, J. L. Gommerman and A. C. Gingras, Sci. Immunol., 2020, 5(52), eabe5511 CrossRef PubMed.
- S. E. Faustini, S. E. Jossi, M. Perez-Toledo, A. M. Shields, J. D. Allen, Y. Watanabe, M. L. Newby, A. Cook, C. R. Willcox, M. Salim, M. Goodall, J. L. Heaney, E. Marcial-Juarez, G. L. Morley, B. Torlinska, D. C. Wraith, T. V. Veenith, S. Harding, S. Jolles, M. J. Ponsford, T. Plant, A. Huissoon, M. K. O'Shea, B. E. Willcox, M. T. Drayson, M. Crispin, A. F. Cunningham and A. G. Richter, Immunology, 2021, 164, 135–147 CrossRef CAS PubMed.
- F. Mehdi, S. Chattopadhyay, R. Thiruvengadam, S. Yadav, M. Kumar, S. K. Sinha, S. Goswami, P. Kshetrapal, N. Wadhwa, U. Chandramouli Natchu, S. Sopory, B. Koundinya Desiraju, A. K. Pandey, A. Das, N. Verma, N. Sharma, P. Sharma, V. Bhartia, M. Gosain, R. Lodha, U. Lamminmäki, T. Shrivastava, S. Bhatnagar and G. Batra, Front. Microbiol., 2021, 11, 618097 CrossRef PubMed.
- C. Zhou, G. Bu, Y. Sun, C. Ren, M. Qu, Y. Gao, Y. Zhu, L. Wang, L. Sun and Y. Liu, J. Med. Virol., 2021, 93, 2857–2866 CrossRef CAS PubMed.
- K. Ditte, T. A. Nguyen Le, O. Ditzer, D. I. Sandoval Bojorquez, S. Chae, M. Bachmann, L. Baraban and F. Lissel, ACS Biomater. Sci. Eng., 2021 DOI:10.1021/acsbiomaterials.1c00727.
- G. Liu and J. F. Rusling, ACS Sens., 2021, 6, 593–612 CrossRef CAS PubMed.
- Z. Fu, W. Zeng, S. Cai, H. Li, J. Ding, C. Wang, Y. Chen, N. Han and R. Yang, J. Colloid Interface Sci., 2021, 604, 113–121 CrossRef CAS PubMed.
- P. Ranjan, V. Thomas and P. Kumar, J. Mater. Chem. B, 2021, 9, 4608–4619 RSC.
- M. A. Ali, C. Hu, S. Jahan, B. Yuan, M. S. Saleh, E. Ju, S. J. Gao and R. Panat, Adv. Mater., 2021, 33, e2006647 CrossRef PubMed.
- J. Muñoz and M. Pumera, Chem. Eng. J., 2021, 425, 131433 CrossRef PubMed.
- P. Han, C. Vaquette, A. Abdal-Hay and S. Ivanovski, Nanomaterials, 2021, 2943 CrossRef CAS PubMed.
- C. Vaquette, J. Mitchell, T. Fernandez-Medina, S. Kumar and S. Ivanovski, Biomaterials, 2021, 269, 120671 CrossRef CAS PubMed.
- F. Blaudez, S. Ivanovski, D. Ipe and C. Vaquette, Mater. Sci. Eng., C, 2020, 117, 111282 CrossRef CAS PubMed.
- H. P. Dang, T. Shabab, A. Shafiee, Q. C. Peiffer, K. Fox, N. Tran, T. R. Dargaville, D. W. Hutmacher and P. A. Tran, Biofabrication, 2019, 11, 035014 CrossRef CAS PubMed.
- J. He, F. Xu, R. Dong, B. Guo and D. Li, Biofabrication, 2017, 9, 015007 CrossRef PubMed.
- J. F. M. Ribeiro, S. M. Oliveira, J. L. Alves, A. J. Pedro, R. L. Reis, E. M. Fernandes and J. F. Mano, Biofabrication, 2017, 9, 025015 CrossRef PubMed.
- N. Sears, P. Dhavalikar, M. Whitely and E. Cosgriff-Hernandez, Biofabrication, 2017, 9, 025020 CrossRef PubMed.
- N. Abbasi, A. Abdal-Hay, S. Hamlet, E. Graham and S. Ivanovski, ACS Biomater. Sci. Eng., 2019, 5, 3448–3461 CrossRef CAS PubMed.
- P. Han, G. A. Gomez, G. N. Duda, S. Ivanovski and P. S. P. Poh, Acta Biomater., 2022 DOI:10.1016/j.actbio.2022.01.020.
- M. A. Elnaggar, H. A. N. El-Fawal and N. K. Allam, Mater. Sci. Eng., C, 2021, 119, 111550 CrossRef CAS PubMed.
- M. J. E. Fischer, in Surface Plasmon Resonance: Methods and Protocols, ed. N. J. Mol and M. J. E. Fischer, Humana Press, Totowa, NJ, 2010, pp. 55–73, DOI:10.1007/978-1-60761-670-2_3.
- N. E. Zander, J. A. Orlicki, A. M. Rawlett and T. P. Beebe Jr, ACS Appl. Mater. Interfaces, 2012, 4, 2074–2081 CrossRef CAS PubMed.
- H. Poli, A. L. Mutch, A. Ali, S. Ivanovski, C. Vaquette, D. G. Castner, M. N. Gomez-Cerezo and L. Grondahl, Biointerphases, 2020, 15, 061010 CrossRef CAS PubMed.
- P. Han, P. M. Bartold, C. Salomon and S. Ivanovski, Int. J. Mol. Sci., 2020, 21, 2809 CrossRef CAS PubMed.
- P. Han and S. Ivanovski, Int. J. Mol. Sci., 2019, 20, 4729 CrossRef CAS PubMed.
- P. Han, A. Lai, C. Salomon and S. Ivanovski, Int. J. Mol. Sci., 2020, 21, 5273 CrossRef CAS PubMed.
- P. Han, P. M. Bartold, C. Salomon and S. Ivanovski, Int. J. Mol. Sci., 2021, 22, 2423 CrossRef CAS PubMed.
- N. Pisanic, P. R. Randad, K. Kruczynski, Y. C. Manabe, D. L. Thomas, A. Pekosz, S. L. Klein, M. J. Betenbaugh, W. A. Clarke, O. Laeyendecker, P. P. Caturegli, H. B. Larman, B. Detrick, J. K. Fairley, A. C. Sherman, N. Rouphael, S. Edupuganti, D. A. Granger, S. W. Granger, M. H. Collins and C. D. Heaney, J. Clin. Microbiol., 2020, 59 Search PubMed.
- A. M. Shields, S. E. Faustini, C. A. Kristunas, A. M. Cook, C. Backhouse, L. Dunbar, D. Ebanks, B. Emmanuel, E. Crouch, A. Kröger, J. Hirschfeld, P. Sharma, R. Jaffery, S. Nowak, S. Gee, M. T. Drayson, A. G. Richter, T. Dietrich and I. L. C. Chapple, J. Dent. Res., 2021, 220345211020270, DOI:10.1177/00220345211020270.
- Y. T. Pinilla, C. Heinzel, L.-F. Caminada, D. Consolaro, M. Esen, P. G. Kremsner, J. Held, A. Kreidenweiss and R. Fendel, Front. Immunol., 2021, 12 Search PubMed.
- W. E. King 3rd and G. L. Bowlin, Polymers, 2021, 13 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra03979f |
| ‡ Co-first author. |
| This journal is © The Royal Society of Chemistry 2022 |