Open Access Article
Open Access ArticleEffect of the array of amines on the transfection efficiency of cationic peptidomimetic lipid molecules into neural cells†
Xiang Yu‡
,
Hai Xiao‡ ,
Muqier Muqier,
Shuqin Han* and
Huricha Baigude
,
Muqier Muqier,
Shuqin Han* and
Huricha Baigude *
*
School of Chemistry & Chemical Engineering, Inner Mongolia University, Hohhot, Inner Mongolia 010020, P.R. China. Fax: +86 471 4992511; Tel: +86 471 4992511E-mail: XiangYu@mail.imu.edu.cn; haixiao@mail.imu.edu.cn; 2770561016@qq.com; chem-hshq@imu.edu.cn; hbaigude@imu.edu.cn
First published on 3rd August 2022
Abstract
The amino groups in the head group of a cationic lipid play a determinative role regarding the nucleic acid delivery efficiency of the LNP formulated from lipids. Herein, we designed four types of lipid bearing different amine-containing branched head groups to investigate the influence of type and number of amines on the neural cell targeted nuclei acid delivery. Conjugation of an ethylamino group at selected positions of a lysine-based cationic lipid resulted in 4 distinct lipids with 3 (denoted N3 lipid), 4 (denoted N4 lipid), 5 (denoted N5 lipid) and 6 (denoted N6 lipid) amino groups, respectively. Comparative analysis by flow cytometry revealed that the N3 lipid had the highest nucleic acid (plasmid and siRNA) transfection efficiency to neural cell lines (BV2 cells and N2a cells). Furthermore, the N3 lipid mediated delivery of siRNA against Toll Like Receptor 4 (TLR4) into oxygen glucose deprivation (OGD)-treated BV2 cells resulted in remarkable silencing of TLR4, inducing alternative polarization (M2) of the cells. Collectively, our data suggest that the N3 lipid is a promising siRNA delivery agent in neural cells.
1. Introduction
RNA interference (RNAi) is an RNA-dependent gene silencing process controlled by the RNA-induced silencing complex (RISC) and activated by small interfering RNA (siRNA) molecules.1–4 In 2006, Andrew Fire and Craig Mello won the Nobel prize for their discovery of the mechanism of RNAi in C. elegans, and the field has grown rapidly since.5 siRNA has huge potential as a therapeutic agent in the treatment of diseases such as gene disorders,6 viral infections7 and cancers.8 siRNA has unique advantages over traditional small molecular drugs, and protein-based or antibody-based drugs.9 Currently, four siRNA-based drugs have been approved by the US FDA, namely Onpattro (Patisiran),10–12 Givlaari (Givosiran),13–15 Oxlumo (Lumasiran)16,17 and Leqvio (Inclisiran).18,19 However, the nuclease susceptibility20 and off-target effect21 of siRNA are still unsolved problems in siRNA delivery. Appropriate chemical modification22–24 or vector protection25–27 of siRNA can increase its circulation time and targeting ability.28 For example, a cationic polymeric delivery system can increase siRNA circulation time in vivo by 100 times.29 Moreover, recent studies have shown that lipid nanoparticles (LNP) delivered siRNA with a serum half-life of more than 90 h and have no negative effect on gene silencing efficiency.25 Nevertheless, chemical modification may cause increased toxicity and reduced gene silencing efficiency.23,30 Viral and non-viral vectors are widely used as siRNA delivery systems. Amongst these, viral vectors may induce gene mutation and fatal defects in the delivery process, with hidden dangers of triggering immunogenicity and inflammatory response.30 On the other hand, non-viral vectors are relatively non-immunogenic, non-infective, non-capacity limited, and low cost. At present, natural lipids or synthetic lipids are commonly used as materials for siRNA delivery.31,32 In particular, the FDA approval of Onpattro has led to increased focus on its lipid nanoparticle delivery systems.However, it is difficult to achieve gene silencing in organs other than liver,33,34 particularly central nervous system35–37 using the lipid delivery system. Therefore, it is necessary to develop an efficient delivery system that can maintain the integrity of siRNA and promote the cellular uptake efficiency of siRNA. And LNP delivery systems can increase their biocompatibility and transfection efficiency by adjusting the amino array and number of amino groups in the hydrophilic head.38,39 In this paper, four kinds of cationic lipid molecules with different amino arrangement and number were designed. We have explored the effects of amino group arrangement and number on biological activity by transfection on nerve cells.
2. Materials and methods
2.1 Chemicals
Boc-L-Glu-OH, oleylamine, H-Lys(Z)-OMe·HCl, N-(allyloxycarbo-nyloxy) succinimide, triethylamine, N,N-diisopropylethylamine (DIPEA), palladium-tetrakis(triphenylphosphine) (Pd[P(C6H3)3]4, trifluoroacetic acid (TFA), di-tert-butyl decarbonate, N,N-dimethylformamide (DMF), and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDCI) were purchased from Aladdin Chemicals (Shanghai, China). 2-(Boc-amino) ethyl bromide was purchased from J&K Chemicals (Beijing, China).2.2 Synthesis of N3 lipid
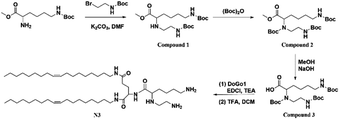
Through the modification of lysine, the amide reaction with DoGo1 (ref. 40) was carried out to synthesize N3 with three free amino groups. The specific synthesis methods and corresponding characterization are shown in ESI.†2.3 Synthesis of N4 lipid
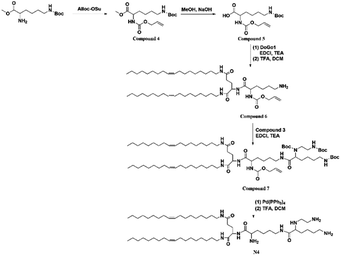
N4 containing four free amino groups is synthesized by modifying N3. The synthetic methods and corresponding characterization are shown in ESI.†2.4 Synthesis of N5 lipid
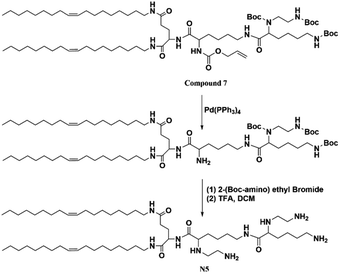
An additional amino group is added to N4 to synthesize N5 with five free amino groups. The synthesis route and characterization were shown in ESI.†2.5 Synthesis of N6 lipid
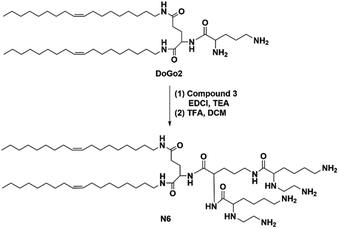
Amide reaction of modified lysine with DoGo2 (ref. 40) to synthesize N6 with six free amino groups. The synthesis schemes and characterization were shown in ESI.†2.6 Electrophoretic mobility shift assay
The complexes of siRNA and the lipids were formed at various nitrogen/phosphate (N/P) ratios (1, 2, 3, 4, 5, 6) in a gel mobility shift assay, respectively. The mixtures were incubated at r.t. for 20 min and then subjected to electrophoresis on agarose gel (2%) for 30 min at 100 V to confirm the siRNA complexation in 1 × Tris/borate/ethylenediaminetetraacetic acid (EDTA) running buffer. After the gel was stained using 0.5 μg mL−1 ethidium bromide, the banding pattern was obtained using a Gel Logic 212 PRO imaging system (Carestream, Toronto, Canada).2.7 Cell culture and cytotoxicity assay
BV2 and N2a cells (ATCC, Manassas, VA, USA) were maintained at 37 °C (with 5% CO2) in DMEM supplemented (10% fetal bovine serum) with 100 mg mL−1 streptomycin and 100 U mL−1 penicillin. For cytotoxicity assay, cells were plated onto 96-well plates at 1 × 104 cells per well in 100 μL of culture medium. Twenty-four hours after plating, lipid stock solutions (4 mg mL−1) at different final concentrations at 20, 40, 60, 100, 140 μg mL−1 were added to the cells in triplicate and incubated as described above. Lipofectamine 2000 (1 mg mL−1) and DoGo4 (4 mg mL−1) were used as control. The final concentration of lipofectamine 2000 were 5, 10, 15, 25, 35 μg mL−1, and the concentration of DoGo4 was similar with above lipids. After a 24 h of incubation, cell viability was assayed using the MTT test.2.8 Cellular uptake of LNPs by neural cells
For transfection of the plasmid, 2 μg pcDNA3-eGFP was diluted in 50 μL Opti-MEM. This solution was mixed with 50 μL Opti-MEM containing lipids (N3, N4, N5, N6, DoGo4, and lipofectamine 2000, respectively), and incubated at room temperature for 20 min before adding into cell culture. The final concentrations were as follows: N3, N4, N5, N6, DoGo4 (32 μg mL−1) and lipofectamine 2000 (4 μg mL−1). After 48 h, the GFP expression was evaluated by observation under a fluorescence microscope. For flow cytometric analysis (FCM), cells were trypsinized and resuspended in PBS and assessed on a NovoCyte Benchtop Flow Cytometer (ACEA Biosciences, Inc., San Diego, CA, USA).For siRNA transfection, FITC-siRNA transfection experiments were conducted on BV2 and N2a cells. BV2 or N2a were seeded at 70% cell confluency in a 24-well plate (Costar, Corning Inc.) and were cultured overnight. Then, the FITC-siRNA/N3 complexes were added to the cell cultures and incubated in a cell-culturing incubator. FITC-siRNA were synthesized by Takara Biotechnology Co., Ltd (Dalian, Liaoning, China). Before adding the complexes into the cell culture, the mixtures were incubated at r.t. for 20 min. The concentrations were as follows: FITC-siRNA (50 nM)/N3 (32 μg mL−1), DoGo4 (16 μg mL−1), and lipofectamine 2000 (4 μg mL−1). After incubation for 6 h, the cells were washed with 1 × PBS (PH = 7.4) and quantified the fluorescence intensity by flow cytometry.
BV2 cells were seeded in a 24-well plate at 1 × 106 cell density and cultured for 24 h. After blocking the cells with different final concentrations (10, 20, 40, 80, 100, 150, and 200 μg mL−1) of blocking solutions (mannan, zymosan, and dextran) for 2 h, FITC-siRNA/N3 complexes were added to the cell cultures and incubated for 6 h in a cell-culturing incubator. FITC-siRNA/N3 complexes were incubated at r.t. for 20 min before transfection. The results were assessed by flow cytometry. For temperature-dependent cellular uptake, the cells were transfected with FITC-siRNA/N3 complexes and incubated at 4 or 37 °C. The cells were re-suspended in 110 μL of PBS and assessed by flow cytometry.
2.9 siRNA transfection efficiency
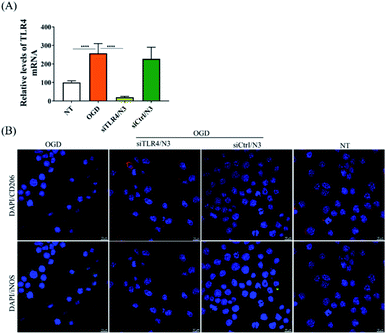
In order to verify the transfection efficiency of siRNA, siGENOME nontargeting siRNA was used as the control (Dharmacon, Lafayette, CO, USA). TLR4 siRNA was custom-synthesized by Takara Biotechnology Co., Ltd (Dalian, Liaoning, China). The sequences of the TLR4 siRNA are as follows: antisense strand: 5′-UUAUAGUCAAAUAUGGGCCtt-3′; sense strand: 5′-GGCCCAUAUUUGACUAUAAtt-3′. The BV2 cells were seeded in 24-well plate for 24 h before siRNA transfection. siTLR4 or siRNA control (100 nM in 50 μL) and N3 (32 μg mL−1 in 50 μL) were mixed in Opti-Medium, respectively, and incubated at r.t. for 20 min. The cell culture medium was replaced by fresh complete medium before adding the transfection solution into the cells. The cells were treated with OGD insult41 after the incubation of 4 h. For OGD insult, cells were washed three times in DMEM without glucose and FBS, and then, the medium was replaced to low glucose DMEM. Cells were cultured in an airtight incubation chamber flushing through with 5% CO2 and 95% N2 for 2 h, then sealing the chamber for the duration of the experiment. OGD was alleviated by removing the culture from the incubator and maintaining the cells in high glucose medium to reoxygenate for 24 h under normal conditions (37 °C, 5% CO2, and 95% air at high humidity). Total RNA was extracted with TRIzol (Invitrogen, Carlsbad, CA, USA) and the expression of endogenous TLR4 mRNA was measured using iScript reverse transcription supermix for RT-qPCR and iTaq Universal SYBR Green supermix (Bio-Rad, Hercules, CA, USA) for qPCR.2.10 In vitro microglia polarization
Immunocytochemistry (ICC) was used to detect the expression of M1 and M2 markers in BV2 microglia.42,43 BV2 cells were grown on 24-well plate with a glass slide chamber for overnight and were treated with either siTLR4/N3 or siCtrl/N3 complexes for 4 h, followed by OGD treatment. Cells were cultured for 24 h under normal cell culture conditions and fixed with 4% PFA in 0.1 M PBS. The fixed cells were rinsed using PBS, permeabilized with 0.1% Triton-X 100 for 10 min, and then incubated with 2% BSA in PBS for 1 h to block nonspecific binding. Then the cells were probed with primary antibodies for the M1 phenotype (iNOS, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400; Cell Signaling Technology, MA, USA) or the M2 phenotype (CD206, 1
400; Cell Signaling Technology, MA, USA) or the M2 phenotype (CD206, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400; Cell Signaling Technology, MA, USA). For visualization, the secondary antibodies, anti-rabbit Alexa-594 (1
400; Cell Signaling Technology, MA, USA). For visualization, the secondary antibodies, anti-rabbit Alexa-594 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Cell Signaling Technology, MA, USA), or goat anti-rabbit IgG-FITC (1
1000; Cell Signaling Technology, MA, USA), or goat anti-rabbit IgG-FITC (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Abcam, Cambridge, MA) were used along with the nuclear marker DAPI (1
1000; Abcam, Cambridge, MA) were used along with the nuclear marker DAPI (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Thermo Scientific, MA, USA). Images were obtained by a Nikon fluorescence confocal microscope (Nikon, C2+).
1000; Thermo Scientific, MA, USA). Images were obtained by a Nikon fluorescence confocal microscope (Nikon, C2+).
3. Results and discussion
We designed novel peptidomimetic based cationic lipids and investigated their efficiency in intracellular nucleic acid delivery on neural cells. Our previous work confirmed that the lipid conjugated peptidomimetics based on natural amino acids (DoGo lipids) can deliver siRNA efficiently in vivo and in vitro.40 We also proved that head group configuration remarkably decreased the cytotoxicity of peptidomimetic based cationic lipids.44 Therefore, in order to maximize the transfection efficiency and minimize the cytotoxicity, four cationic lipids were designed in this study: 2-(6-amino-2-((2-aminoethyl)amino)hexanamido)-N1,N5-di((Z)-octadec-9-en-1-yl)pentanediamide (denoted N3), 2-(2-amino-6-(6-amino-2-((2-aminoethyl)amino)hexanamido)hexanamido)-N1,N5-di((Z)-octadec-9-en-1-yl)pentanediamide (denoted N4), 2-(6-(6-amino-2-((2-aminoethyl)amino)hexanamido)-2-((2 aminoethyl) amino) hexanamido)-N1,N5-di((Z)-octadec-9-en-1-yl)pentanediamide (denoted N5), or 2-(2,5-bis(6-amino-2-((2-aminoethyl)amino)hexanamido)pentanamido)-N1,N5-di((Z)-octadec-9-en-1-yl)pentanediamide (denoted N6).3.1 Characterization of cationic lipids and siRNA binding assay
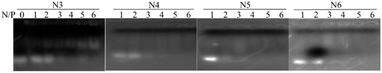
To evaluate the nuclei acids binding affinity of the novel lipids, we performed gel shift assay on all lipids at various cationic lipid (amine, N) to siRNA (phosphate, P) ratio. The positive charge of cationic lipids and the negative charge on siRNA enforce electrostatic interaction between them to form the nanoparticles. The influence of charge ratios on siRNA condensation by lipids can be estimated by the gel retardation assay. The results showed that the migration of siRNA in gel was retarded with the increasing N/P ratio in all lipids, indicating that lipids and siRNA can form a complete complex through electrostatic interactions at this condition (Fig. 1). All lipids completely bound to siRNA at N/P ratio 1, 2, 3, 4, 5, and 6, respectively. Moreover, measurement of particle size the siRNA/lipid complexes at N/P = 5 by dynamic light scattering revealed that the complexes have monodisperse peaks and high dispersion, which lay in the range of optimal size of complexes for nucleic acid delivery to various cell lines and tissues. The results indicated that siRNA/lipid complexes display an average size of 135.4 nm, 200.5 nm, 239.8 nm, 250.7 nm (PDI: 0.401, 0.403, 0.357, and 0.754), respectively (Table 1). The particle size of the siRNA/lipid complexes were in the optimal range of gene delivery. In addition, electrokinetic potential is an important parameter representing the stability of a compound. The smaller the electrokinetic potential, the more likely it is to induce polymerization. On the contrary, the larger the electrokinetic potential, the stronger the stability in solution. The zeta potential of siRNA/lipid complexes were +52.1, +39.2, +35.8 and +41.2 mV, respectively (Table 1). These results indicated that all lipids formed a stable complex with siRNA, which was not easy to generate polymerization and precipitation, and could exist stably in solution.| Sample | Particle size (d, nm) | Zeta potential (mV) | PDI |
|---|---|---|---|
| siRNA/N3 | 135.4 ± 0.16 | +52.1 ± 1.15 | 0.401 |
| siRNA/N4 | 200.5 ± 1.21 | +39.2 ± 2.19 | 0.403 |
| siRNA/N5 | 239.8 ± 1.58 | +35.8 ± 1.01 | 0.357 |
| siRNA/N6 | 250.7 ± 0.79 | +41.2 ± 1.97 | 0.754 |
3.2 Cytotoxicity measurement
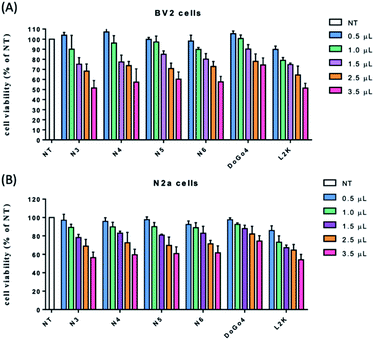
Safety is one of the primary considerations in gene delivery. To examine the cytotoxicity of the novel lipids, BV2 and N2a cells were treated with various concentration of the lipids, respectively, and the cell viability was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. DoGo4 and lipofectamine 2000 were used as control. Cells treated with all lipids and DoGo4 had higher survival rates at all the concentration ranges that were tested (Fig. 2). Impressively, the cell viabilities after all lipids and DoGo4 treatment remained around 50–90% at the highest concentration tested (140 μg mL−1) while the concentration of L2K reached 35 μg mL−1, the cell viability was comparable to that of other lipids, demonstrating that all lipids and DoGo4 have low cytotoxicity and may not induce various adverse effects in the cells after the treatment.3.3 Transfection efficiency
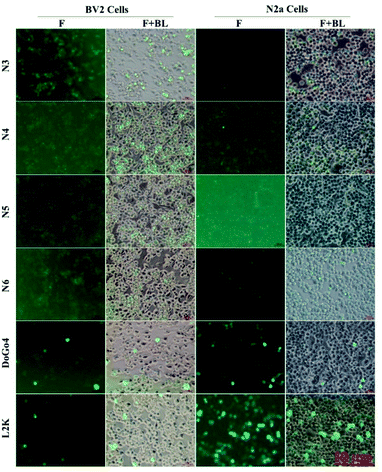
Next, we evaluated the nucleic acid transfection efficiency of all lipids. First, we conducted a test of the plasmid DNA transfection efficiency of all lipids using DoGo4 and lipofectamine 2000 as control. To do this, we complexed a plasmid encoding green fluorescence protein (GFP) (pcDNA3-eGFP) with the lipids, DoGo4 and lipofectamine 2000, respectively, and treated with BV2 cells and N2a cells with the complexes for 24 h. Subsequent observation of the cells treated with plasmid/lipid complex under a fluorescence microscope revealed that the N3 lipid has the highest efficiency followed by lipofectamine 2000 and the DoGo4 (Fig. 3). Further quantification by flow cytometry revealed that the highest GFP expressing cell population were 79% on BV2 cells (Fig. 4A) and 80% on N2a cells (Fig. 4B) after a single transfection, demonstrating that N3 lipids had remarkable transfection efficiency. Therefore, we chose N3 lipids as a nucleic acid delivery agent for the subsequent experiment. Then, we performed FITC-siRNA transfection experiment on BV2 cells and N2a cells by using N3 lipid. Flow cytometry analysis showed that the FITC-positive cell for N3 lipid was higher than L2K and DoGo4 in N2a cells (Fig. 5B), while slightly weaker than L2K and DoGo4 in BV2 cells (Fig. 5A). These data indicated that N3 lipid has high transfection efficiency of plasmid DNA or siRNA in various cell type.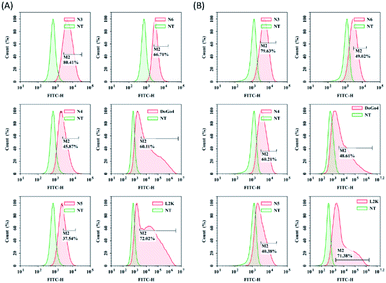 | ||
| Fig. 4 Quantification of GFP expression of BV2 and N2a cells treated with pcDNA3-eGFP/lipid complexes by flow cytometry, lipids: N3, N4, N5, N6, DoGo4 and lipofectamine 2000. | ||
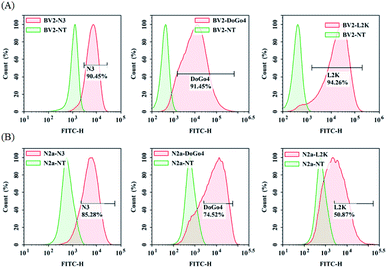 | ||
| Fig. 5 Quantification of BV2 and N2a cellular uptake of FITC-siRNA/lipid complexes by flow cytometry. | ||
4. Conclusions
Through particle size distribution, zeta potential measurement, cytotoxicity analysis and plasmid DNA or siRNA transfection in neural cells, we conclude that, when the hydrophilic head of the cationic lipid molecule has a branched structure, the zeta potential and nucleic acid transfection efficiencies will decrease as the branched chain increases; when the branched chain is modified but the number of amino groups is the same, the zeta potential and transfection efficiency (GFP, FITC-siRNA) will increase as the branch structure increases. Collectively, we successfully synthesized a novel cationic lipid (denoted N3) for plasmid DNA or siRNA delivery. N3 showed low cytotoxicity, high transfection efficiency and can regulate microglia polarization by downregulate siTLR4.Author contributions
X. Y. and H. X.: methodology, investigation, writing-original draft preparation. M. M.: software and validation. S. H. and H. B.: conceptualization, supervision, writing-original draft preparation, reviewing and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was kindly supported by the National Natural Science Foundation of China (grant number: 32160231, 21875124) and Natural Science Foundation of Inner Mongolia Autonomous Region (grant number: 2021MS02009, 2019MS02009).Notes and references
- A. Fire, S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver and C. C. Mello, Nature, 1998, 391, 806–811 CrossRef CAS PubMed.
- J. A. Moss, Radiologic Technol., 2014, 86, 155–180 Search PubMed.
- P. D. Zamore, T. Tuschl, P. A. Sharp and D. P. Bartel, Cell, 2000, 101, 25–33 CrossRef CAS.
- N. Manjunath and D. M. Dykxhoorn, Discovery Medicine, 2010, 9, 418–430 CAS.
- J. Whelan, Drug Discovery Today, 2005, 10, 1014–1015 CrossRef PubMed.
- S. D. Olson, K. Pollock, A. Kambal, W. Cary, G. M. Mitchell, J. Tempkin, H. Stewart, J. McGee, G. Bauer, H. S. Kim, T. Tempkin, V. Wheelock, G. Annett, G. Dunbar and J. A. Nolta, Mol. Neurobiol., 2012, 45, 87–98 CrossRef CAS PubMed.
- J. DeVincenzo, R. Lambkin-Williams, T. Wilkinson, J. Cehelsky, S. Nochur, E. Walsh, R. Meyers, J. Gollob and A. Vaishnaw, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 8800–8805 CrossRef CAS PubMed.
- S. I. Pai, Y. Y. Lin, B. Macaes, A. Meneshian, C. F. Hung and T. C. Wu, Gene Ther., 2006, 13, 464–477 CrossRef CAS PubMed.
- F. T. Vicentini, L. N. Borgheti-Cardoso, L. V. Depieri, D. de Macedo Mano, T. F. Abelha, R. Petrilli and M. V. Bentley, Pharm. Res., 2013, 30, 915–931 CrossRef CAS.
- S. Chakradhar, Nat. Med., 2018, 24, 1785–1787 CrossRef CAS PubMed.
- I. Urits, D. Swanson, M. C. Swett, A. Patel, K. Berardino, A. Amgalan, A. A. Berger, H. Kassem, A. D. Kaye and O. Viswanath, Neurol. Ther., 2020, 9, 301–315 CrossRef PubMed.
- S. M. Hoy, Drugs, 2018, 78, 1605–1613 CrossRef CAS PubMed.
- L. J. Scott, Drugs, 2020, 80, 1–5 CrossRef PubMed.
- G. Gonzalez-Aseguinolaza, N. Engl. J. Med., 2020, 382, 2366–2367 CrossRef CAS PubMed.
- T. Steinberg, M. Kilic, K. Fuchs, K. Hanyk, R. A. Linker, F. Schlachetzki and B. Neumann, J. Neurol. Sci., 2021, 422, 117334 CrossRef PubMed.
- L. J. Scott and S. J. Keam, Drugs, 2021, 81, 277–282 CrossRef CAS PubMed.
- S. F. Garrelfs, Y. Frishberg, S. A. Hulton, M. J. Koren and J. C. Lieske, N. Engl. J. Med., 2021, 384, 1216–1226 CrossRef CAS PubMed.
- Y. N. Lamb, Drugs, 2021, 81, 389–395 CrossRef CAS PubMed.
- K. K. Ray, R. S. Wright, D. Kallend, W. Koenig, L. A. Leiter, F. J. Raal, J. A. Bisch, T. Richardson, M. Jaros, P. L. J. Wijngaard, J. J. P. Kastelein, Orion-10 and Orion-11 Investigators, N. Engl. J. Med., 2020, 382, 1507–1519 CrossRef CAS PubMed.
- K. A. Whitehead, R. Langer and D. G. Anderson, Nat. Rev. Drug Discovery, 2009, 8, 129–138 CrossRef CAS PubMed.
- J. E. Zuckerman and M. E. Davis, Nat. Rev. Drug Discovery, 2015, 14, 843–856 CrossRef CAS PubMed.
- Y. Gao, X. Liu and X. Li, Int. J. Nanomed., 2011, 6, 1017–1025 CrossRef CAS PubMed.
- A. H. Hall, J. Wan, E. E. Shaughnessy, B. Ramsay Shaw and K. A. Alexander, Nucleic Acids Res., 2004, 32, 5991–6000 CrossRef CAS PubMed.
- L. Li and Y. Shen, Expert Opin. Biol. Ther., 2009, 9, 609–619 CrossRef CAS PubMed.
- J. Soutschek, A. Akinc, B. Bramlage, K. Charisse, R. Constien, M. Donoghue, S. Elbashir, A. Geick, P. Hadwiger, J. Harborth, M. John, V. Kesavan, G. Lavine, R. K. Pandey, T. Racie, K. G. Rajeev, I. Rohl, I. Toudjarska, G. Wang, S. Wuschko, D. Bumcrot, V. Koteliansky, S. Limmer, M. Manoharan and H. P. Vornlocher, Nature, 2004, 432, 173–178 CrossRef CAS PubMed.
- A. Rolland, Adv. Drug Delivery Rev., 2005, 57, 669–673 CrossRef CAS PubMed.
- A. David, S. Dwight, E. Andrew and Pinal C., J. Am. Chem. Soc., 2009, 131, 2072–2073 CrossRef PubMed.
- K. Bruno, Adv. Drug Deliv. Rev., 2011, 63, 1210–1226 CrossRef CAS PubMed.
- A. Kano, T. Yamano, S. W. Choi and A. Maruyama, Adv. Mater. Res., 2008, 47–50, 762–764 CAS.
- H. Liao and J. H. Wang, Oligonucleotides, 2005, 15, 196–205 CrossRef CAS.
- R. Kanasty, J. R. Dorkin, A. Vegas and D. Anderson, Nat. Mater., 2013, 12, 967–977 CrossRef CAS.
- P. Midoux and C. Pichon, Expert Rev. Vaccines, 2015, 14, 221–234 CrossRef CAS PubMed.
- V. Bitko, A. Musiyenko, O. Shulyayeva and S. Barik, Nat. Med., 2005, 11, 50–55 CrossRef CAS PubMed.
- B.-j. Li, Q. Tang, D. Cheng, C. Qin, F. Y. Xie, Q. Wei, J. Xu, Y. Liu, B.-j. Zheng, M. C. Woodle, N. Zhong and P. Y. Lu, Nat. Med., 2005, 11, 944–951 CrossRef CAS PubMed.
- S. M. Smith, S. A. Friedle and J. J. Watters, PLoS One, 2013, 8, e81584 CrossRef PubMed.
- P. M. Grace, K. M. Ramos, K. M. Rodgers, X. Wang, M. R. Hutchinson, M. T. Lewis, K. N. Morgan, J. L. Kroll, F. R. Taylor, K. A. Strand, Y. Zhang, D. Berkelhammer, M. G. Huey, L. I. Greene, T. A. Cochran, H. Yin, D. S. Barth, K. W. Johnson, K. C. Rice, S. F. Maier and L. R. Watkins, Neuroscience, 2014, 280, 299–317 CrossRef CAS PubMed.
- S. Lehnardt, C. Lachance, S. Patrizi, S. Lefebvre, P. L. Follett, F. E. Jensen, P. A. Rosenberg, J. J. Volpe and T. Vartanian, J. Neurosci., 2002, 22, 2478–2486 CrossRef CAS PubMed.
- K. Buyens, B. Lucas, K. Raemdonck, K. Braeckmans, J. Vercammen, J. Hendrix, Y. Engelborghs, S. Smedt and N. N. Sanders, J. Controlled Release, 2008, 126, 67–76 CrossRef CAS PubMed.
- J. A. Kulkarni, P. R. Cullis and R. van der Meel, Nucleic Acid Ther., 2018, 28, 146–157 CrossRef CAS PubMed.
- H. Xiao, A. Altangerel, G. Gerile, Y. Wu and H. Baigude, ACS Appl. Mater. Interfaces, 2016, 8, 7638–7645 CrossRef CAS PubMed.
- R. Milner, S. Hung, X. Wang, G. I. Berg, M. Spatz and G. J. del Zoppo, Stroke, 2008, 39, 191–197 CrossRef PubMed.
- M. Ghosh, Y. Xu and D. D. Pearse, J. Neuroinflammation, 2016, 13, 9 CrossRef PubMed.
- M. Ghosh, D. Garcia-Castillo, V. Aguirre, R. Golshani, C. M. Atkins, H. M. Bramlett, W. D. Dietrich and D. D. Pearse, Glia, 2012, 60, 1839–1859 CrossRef PubMed.
- G. Gerile, T. Ganbold, Y. Li and H. Baigude, J. Mater. Chem. B, 2017, 5, 5597–5607 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra03347j |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |