Open Access Article
Open Access ArticleEngineering colloidally stable, highly fluorescent and nontoxic Cu nanoclusters via reaction parameter optimization†
Kumar Babu Busia,
Jyothi Kothab,
Shamili Bandarua,
Jyothi Priyanka Ghantasalab,
Sheik Haseena a,
Keerti Bhamidipatic,
Nagaprasad Puvvadacd,
Mahesh Kumar Ravva
a,
Keerti Bhamidipatic,
Nagaprasad Puvvadacd,
Mahesh Kumar Ravva a,
Manjunatha Thondamalb and
Sabyasachi Chakrabortty
a,
Manjunatha Thondamalb and
Sabyasachi Chakrabortty *a
*a
aDepartment of Chemistry, SRM University AP Andhra Pradesh, Andhra Pradesh 522240, India. E-mail: sabyasachi.c@srmap.edu.in
bDepartment of Biological Sciences, SRM University AP Andhra Pradesh, Andhra Pradesh 522240, India
cApplied Biology Division CSIR-Indian Institute of Chemical Technology, Hyderabad 500007, Telangana, India
dDepartment of Chemistry, Indrashil University, Rajpur, Mehsana-382740, Gujarat, India
First published on 14th June 2022
Abstract
Metal nanoclusters (NCs) composed of the least number of atoms (a few to tens) have become very attractive for their emerging properties owing to their ultrasmall size. Preparing copper nanoclusters (Cu NCs) in an aqueous medium with high emission properties, strong colloidal stability, and low toxicity has been a long-standing challenge. Although Cu NCs are earth-abundant and inexpensive, they have been comparatively less explored due to their various limitations, such as ease of surface oxidation, poor colloidal stability, and high toxicity. To overcome these constraints, we established a facile synthetic route by optimizing the reaction parameters, especially altering the effective concentration of the reducing agent, to influence their optical characteristics. The improvement of the photoluminescence intensity and superior colloidal stability was modeled from a theoretical standpoint. Moreover, the as-synthesized Cu NCs showed a significant reduction of toxicity in both in vitro and in vivo models. The possibility of using such Cu NCs as a diagnostic probe toward C. elegans was explored. Also, the extension of our approach toward improving the photoluminescence intensity of the Cu NCs on other ligand systems was demonstrated.
1. Introduction
Noble metal nanoclusters (NCs) have attracted great attention compared to their conventional counterparts due to their ability to alter and tune properties. In particular, photoluminescence (PL) has emerged as one of the important characteristics of metal NCs.1–3 Several potential applications are envisioned in the fields of superior biological imaging agents, sensors, photocatalytic centers, light-emitting devices, photosensitizers, etc.4–6 Being extremely small with several tens of metal atoms, these clusters show properties similar to molecules without exhibiting the characteristic surface plasmon properties of relatively large metal nanoparticles (NPs).7 It is believed that the ultrasmall core size imparts quantum effects on NCs and their subsequent optical phenomenon. They can be designed to possess good photostability, a larger Stokes shift, better photocatalytic properties, superior biocompatibility, improved colloidal stability, etc., which enable their use as better diagnostic agents.8 Compared to traditional other PL materials, such as quantum dots or organic dyes, the main advantage of NCs is related to toxicity and PL stability issues.9,10 Moreover, the satisfactory renal clearance of metal NCs make them a better candidate for bioapplications.11 However, they suffer from a poor fluorescent quantum yield (QY).12 A limited number of optimized strategies are available to control their optical properties. A number of approaches such as alloy formation, aggregation-induced emission, confined fluorescence enhancement, and rigidifying the ligand shell have been employed to improve the QYs of metal NCs.13 Several factors, such as the metal composition, surface modification, hydrophilic nature, surface charge, intrinsic structure, and the local environment, may influence their PL behavior. However, a generalized concept for QY improvement that could be applied to any composition of metal NCs has not yet been established.Among all the studied metal NCs, gold NCs and silver NCs14,15 have been extensively researched in terms of their size-controlled synthesis, intermetallic reaction mechanism, structural characterization, ligand-induced optical behavior, property, biological applications, catalytic performance, etc.16,17 Comparing them with other metal NCs, their better conductivity, much lower cost, and earth abundancy have made Cu NCs a popular/alternative choice in application in many industries.18 Still, attention toward the development of Cu NCs is relatively rare owing to the difficulty in preparing small clusters and due to their susceptibility to be easily oxidized. In the literature, a series of scaffolds or capping agents, such as polymers, oligonucleotides, small molecules, proteins, and peptides, have been employed to synthesize Cu NCs successfully.19 Among these, protein-directed synthesis is more popular as due to its environmentally benign and biocompatible nature.20 Along with the combination of strong reducing small molecules, proteins can serve as a better surface passivating agent toward the colloidal stability of as-synthesized fluorescent Cu NCs in an aqueous medium. However, the effect of time, temperature, pH, and relative amount on their PL behavior is not yet well understood. Also, their relatively poor stability in aqueous medium due to surface oxidation and irreversible aggregation reduce their PL intensity, and hence the subsequent applications related to luminescent NCs have been curtailed to date.21 Post-synthetic modification with electron-rich ligands was established to achieve a better stability, although this method was time-consuming and complicated.22 Therefore, there is an utmost necessity for a robust Cu NCs synthesis protocol for retaining their strong PL intensity and superior colloidal stability, which could offer a better solution for the drawbacks to be overcome.
The wavelength region ∼630 nm to 1350 nm of the electromagnetic spectrum is commonly known as the “diagnostic window” due to its several advantages over the visible range in a bioimaging context.23 First, photons in the visible range cannot penetrate deep into the tissue due to the scattering and presence of high-level endogenous absorbers, such as lipids, hemoglobin, and water.24 Second, elimination of the background signal, such as tissue autofluorescence, in the imaging can be achieved in this region, where the absorption coefficients of the body fluids are at their minimum.25 Thus, this regime provides benefits toward several biological studies, such as in vivo and in vitro imaging and labeling, deep tissue imaging, and photodynamic therapy.26 Therefore, the development of red-emitting stable, nontoxic Cu NCs as a model fluorescent probe would be of great interest.
Metallic Cu or its ionic form has much biological relevance, such as in transcriptional regulators, storage, cell surface compartment transporter and receptors, oxidoreductase behavior, free radical scavenging, and electron transfer.27 Also, Cu is known to be removed easily from the body. However, the evaluation of such materials in both in vivo and in vitro environments would enhance their potential for translational research. In that respect, the roundworm Caenorhabditis elegans (C. elegans) has emerged as a powerful tool to assess the toxicity and biocompatibility of various chemical compounds, including nanomaterials.28 Low-cost maintenance and ease of performing experiments have seen C. elegans favored over other model systems to evaluate novel nanomaterials for various biomedical applications.29 Also, its short lifespan (average of 3 weeks) enables its use as an ideal model organism to study the effect of nanomaterials in the aging process.30 Hence, we sought to understand the toxicity and fluorescent properties of Cu NCs in C. elegans.
The creation of appreciable PL with superior stability as well as involving a relatively less studied coinage metal suggest that Cu NCs would be an excellent candidate for an alternative diagnostic probe. Herein, a facile synthetic strategy to obtain highly luminescent Cu NCs was demonstrated. A clear understanding of their optical properties was well documented, where the influence of the reaction parameters on the outcome was analyzed. We believe that the reduction of the amount of the reducing agent played a crucial role in not only their enhanced PL behavior but also in their exceptional stability. To the best of our knowledge, the reported colloidal stability and quantum yield of our as-synthesized Cu NCs are the highest reported in the literature to date, where the emission wavelength is in the red region. A theoretical model was established to understand the effect of the reducing agent on the surface of Cu NCs and their effective surface passivation. Subsequently, their usefulness as diagnostics probes in in vitro and in vivo model systems was thoroughly investigated. We further showed that our approach could be extended to other synthetic conditions as well, where the varied concentration of reducing agent might lead to altering the PL intensity of Cu NCs.
2. Results and discussion
2.1 Synthesis and characterization of the Cu nanoclusters
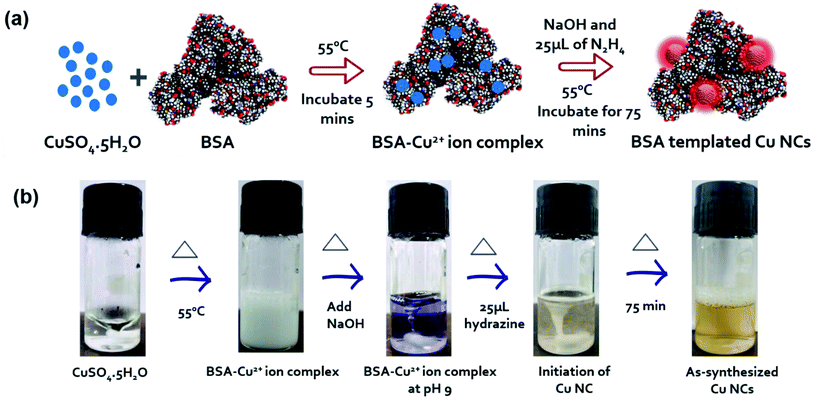
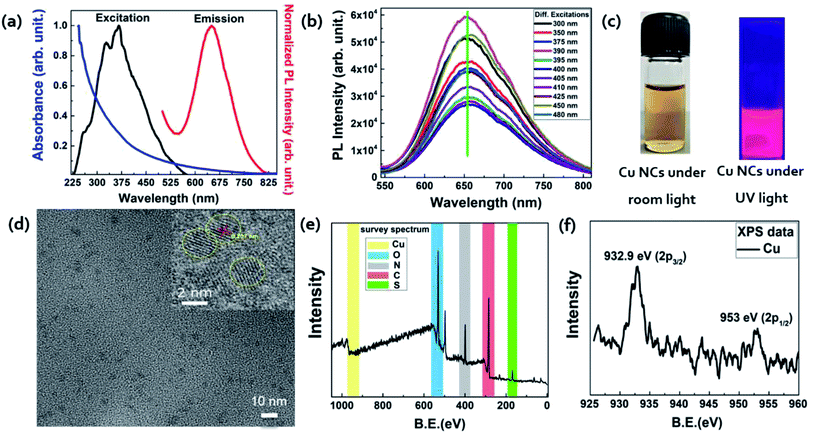
The synthesis of Cu NCs was carried out via an optimized protocol based on the literature.31 A detailed schematic showing each step together with digital images of the structures is in Scheme 1. Initially, 5 mM of copper salt (CuSO4, 5H2O) was mixed with 150 mg of bovine serum albumin (BSA) protein in 5 mL of water. The pH of the solution was adjusted to 9 with the help of NaOH (1 M) solution before introducing a second reducing agent, i.e., 25 μL of N2H4 solution. Finally, the reactants were allowed to react for 75 min at 55 °C under continuous stirring, to obtain colloidally stable, high luminescent Cu NCs. We carefully excluded the addition of other strong reducing agents, such as NaBH4 in this case, as they might lead to the formation of larger nanoparticles in no time (Fig. S1†). The process of stabilizing Cu2+ ions in the aqueous medium and their subsequent Cu NCs formation is depicted in Scheme 1(b). Various functional groups on the protein backbone, –NH2, –COOH, –OH, and –SH, were coordinated32 with the Cu2+ ion to form a white colored gel, and at pH 9, they became a colloidally stable colorless solution. Finally, the initiation of Cu NC formation was started with the help of N2H4 solution, and after 75 min, a yellow color solution confirmed their successful synthesis. This process was relatively simple and robust. As seen in the digital image (Scheme 1(b)), BSA-stabilized Cu NCs showed a nice dispersion in aqueous medium and a light yellow color. The BSA-stabilized Cu NCs were purified through a Vivaspin 20 (MWCO30k) centrifugal concentrator to remove unreacted small molecules used in the synthesis. Here, we assume statistically that all the BSA molecules were attached with Cu NCs. Further, they were analyzed by UV-Vis absorption, photoluminescence, and other advanced characterization techniques.Optical and structural characterizations of the as-synthesized Cu NCs are depicted in Fig. 1. No sharp absorbance peak was observed for Cu NCs, which eliminated the possibility of the formation of larger-sized Cu nanoparticles that generally originate through surface plasmon resonance at ∼540 nm (Fig. S1†). It was observed that these NCs showed the highest excitation peak at ∼390 nm and emission maximum at ∼650 nm (Fig. 1(a)). This larger Stoke's shift between the excitation and emission behavior could be very beneficial for further bioimaging applications, as the incoming excitation source would have minimal interaction with the emission detector. Also, red/near infrared emission is advantageous in bioimaging where deep tissue penetration is necessary. To understand the nature of the emission peak, we excited the Cu NCs samples under various excitation sources, ranging from 300 to 480 nm. A clear excitation independent PL profile was recorded where the emission peak was not shifted (Fig. 1(b)). However, the observed intensities were varied depending on their absorption cross-section. Taken together, we can conclude that better surface passivation was achieved with minimal occurrence of surface defects despite the extremely small regime. A digital image (Fig. 1(c)) showed that the as-synthesized Cu NCs were yellowish in color under room light and emitted red color under UV light irradiation (365 nm). From dynamic light scattering (DLS) experiments, we observed that the hydrodynamic diameter of these Cu NCs was ∼4.7 nm, which is similar to BSA protein itself, and no other aggregates were found. The capping layer of BSA was useful for the prevention of agglomeration in water. These NCs were colloidally stable, and had a zeta potential value of ∼−27.99 mV, which clearly indicated their stability in aqueous medium. This could be beneficial for their use in bioimaging applications. A high-resolution transmission electron microscopy (HRTEM) image of such Cu NCs is presented in the inset of Fig. 1(d), where the crystalline nature of the NCs was clearly evident. The calculated lattice spacing of 0.207 nm confirmed the existence of the Cu (111) plane33 in their structure. Their average size was calculated to be 2.2 ± 0.2 nm, which was obtained after careful consideration of ∼40 representative such small NCs in the TEM images (Fig. 1(d)). Size comparisons of Cu NCs measured from TEM and DLS measurements were carried out to show that both sizes were well correlated and matched. The slight increase in the hydrodynamic size compared to the projected size might be ascribed to the presence of ligand shell/functional groups on the surface of nanoparticles in the aqueous state, which is a well-accepted proposition for colloidal stability. To understand the surface chemical state of the NCs, X-ray photoelectron spectroscopy (XPS) characterization was performed. The initial survey spectrum (Fig. 1(e)) revealed the presence of C, N, S, and O atoms, which generally originated from the BSA protein backbone present with the Cu NCs. A closer analysis of the binding energy peaks near the Cu atoms revealed the existence of 932.9 and 953 eV peaks for Cu-2p3/2 and Cu-2p1/2 respectively. However, the absence of a 942 eV peak confirmed the non-appearance of Cu2+ peaks, proving all the Cu2+ precursors were completely reduced in our reaction condition to produce Cu NCs. Also, C 1s peak analysis of BSA-Cu NCs showed three peaks corresponding to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C or C–C (284.79 eV), C–C/COOH/C–N (285.99 eV), and C
C or C–C (284.79 eV), C–C/COOH/C–N (285.99 eV), and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/C
O/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N (287.84 eV), respectively (Fig. S2(a)†). Similarly, the N 1s peaks confirmed the presence of NH3 (400.33 eV) and CH3–C–N (398.98 eV), whereas O 1s showed three peaks attributed to O**
N (287.84 eV), respectively (Fig. S2(a)†). Similarly, the N 1s peaks confirmed the presence of NH3 (400.33 eV) and CH3–C–N (398.98 eV), whereas O 1s showed three peaks attributed to O**![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–OH (531.37 eV), O
C–OH (531.37 eV), O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–O**H (532.98 eV), and O2/C (536.98 eV) (Fig. S2(b and c)†). Taken together, all three elements confirmed the presence of a protein backbone in the as-prepared Cu NCs. We further compared the Fourier-transform infrared spectroscopy (FTIR) data of Cu-BSA and BSA alone. No observable change was observed, indicating that the Cu NCs were completely embedded inside the protein backbone (Fig. S3†) and the surface structure of BSA was largely unaltered.31
C–O**H (532.98 eV), and O2/C (536.98 eV) (Fig. S2(b and c)†). Taken together, all three elements confirmed the presence of a protein backbone in the as-prepared Cu NCs. We further compared the Fourier-transform infrared spectroscopy (FTIR) data of Cu-BSA and BSA alone. No observable change was observed, indicating that the Cu NCs were completely embedded inside the protein backbone (Fig. S3†) and the surface structure of BSA was largely unaltered.31
2.2 Improvement of the quantum yield of the Cu nanoclusters
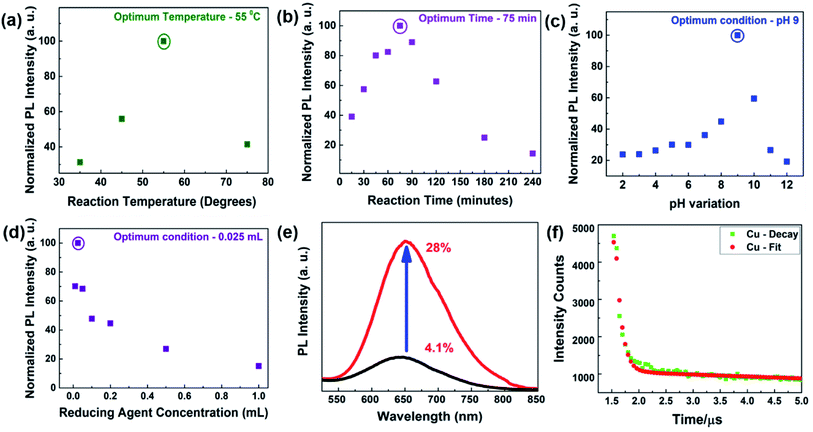
The above-mentioned reaction conditions were thoroughly optimized through careful variation of all the parameters involved in the synthesis. The key parameters, such as temperature, time, pH, reducing agent amount, and surface capping agent, were taken into consideration to yield a better PL intensity and impart better colloidal stability. In each of the cases, there was no apparent shift in the emission peak position, clearly confirming that neither the capping layer was degraded nor was aggregation through irreversible ligand loss observed in these experimental conditions. A remarkable improvement in the quantum yield (QY) of ∼7 times (from 4.1% to 28%) was recorded under our optimum condition. Fig. 2(a–d) clearly demonstrate the change in their PL behaviors in the varied conditions (Fig. S4–S8†), where we plotted the normalized area under each PL spectra. Since, most the literature studies on protein-templated Cu NCs synthesis kept the amount of our secondary reducing agent N2H4 solution as 1 mL in their synthesis,34,35 we also followed the same in our comparison study. Fig. 2(a) shows the highest PL intensity occurred at 55 °C, while comparing it with other synthesis temperatures. This could be attributed to the fact that a relatively higher energy was needed for the acceleration of the reduction dynamics of Cu2+ salt compared with the standard biological temperature (37 °C). However, very high temperatures led to the formation of larger-sized Cu nanoparticles (NPs) with no fluorescence emission. The optimum time to get better Cu NCs formation was recognized to be around ∼75 min in Fig. 2(b). We believe this period would be sufficient to obtain better surface passivation with BSA with an appropriate size regime, while a longer time might lead to the formation of larger-sized Cu NCs with no PL emission. As pH is a vital parameter for synthesizing any nanomaterials to be used in bioapplications,36,37 we synthesized our Cu NCs at 55 °C for 75 min under various pH conditions ranging from pH 2–12 (Fig. S6†). The better PL intensity was confirmed for the pH 9 solution, as displayed in Fig. 2(c). It was worth mentioning that our reaction condition proved to be very robust as we observed appreciable PL in every pH solution. In the literature, PVP-coated Cu NCs38 showed better PL intensity at pH 4, whereas the change in pH to 7 led to aggregation and no fluorescent was observed. Finally, we evaluated the role of reducing agents for the obtained optical behavior of Cu NCs. Interestingly, their amount was of utmost importance, and we observed better quality NCs while using 0.025 mL of N2H4 solution, as depicted in Fig. 2(d). To the best of our knowledge, this is the highest reported QY (∼28%) for protein-coated Cu NCs emitting around the red region (∼650 nm) of the visible spectrum, so far. Fig. 2(e) presents the change in PL intensity of the as-synthesized Cu NCs in BSA, where the amount of reducing agent was used as 1 mL and 0.025 mL in gray and red color spectra, respectively. The slight shift in PL emission peak may be attributed to the fact that amount of –N2H4 influenced the surface passivation. This observation was supported later by the radiative recombination dynamics. Fig. 2(f) shows the lifetime decay curve of the as-synthesized Cu NCs, and it was fitted with three exponential decay profiles. An average lifetime of 2.17 μs was recorded in the standard time-correlated single-photon counting (TCSPC) measurements. An improvement of ∼4 order of magnitude lifetime value was observed while comparing the PL decay of 1 mL reducing agent (Fig. S9†). We compare in Table 1 our synthesis method, reaction time, the temperature of the synthesis, capping ligand, the lifetime of as-synthesized NCs, their QYs, corresponding excitation, and emission wavelength mainly focused on red fluorescent Cu NCs with others reported in the literature. As depicted, our protocol yielded a better QY, which was further substantiated by their longest lifetime. We believe poor passivation, increased surface trap states, or the destruction of the structural properties of the protein backbone in the case of a higher amount of reducing agent may influence their PL behavior. A further hypothesis on the effect of this low concentration of their observed PL behavior is explained through density functional theory (DFT) calculations in the next section.| Serial no. | Synthesis method | Reaction time | Temperature (°C) | Capping agent | Lifetime | QY (%) | λex/λem (nm) | Ref. |
|---|---|---|---|---|---|---|---|---|
| 1 | UV irradiation | 3 h | — | Poly(methacrylic acid) | — | 2.2 | 360/630 | 39 |
| 2 | UV irradiation | 2 h | — | Poly(acrylic acid)graft-mercaptoethylamine | — | 5.7 | 365/630 | 40 |
| 3 | Stirring | 30 min | — | PVP/dihydrolipoic acid | 0.8 μs | 10.8 | 365/435![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 650 650 |
38 |
| 4 | Stirring | 4 h | 55 | BSA | — | 4.1 | 365/620 | 31 |
| 5 | Stirring | 2 h | 40 | Lysozyme | — | 5.6 | 365/600 | 41 |
| 6 | Stirring | 2 h | 37 | Papain | — | 14.3 | 370/620 | 42 |
| 7 | Stirring | 3.5 h | 25 | Transferrin | 15.2 ns | 6.2 | 508/670 | 43 |
| 8 | Shaking | — | — | Glutathione | 15.2 μs | 1.9 | 340/627 | 44 |
| 9 | Stirring | — | RT | Glutathione | — | 3 | 340/620 | 45 |
| 10 | Stirring | 10 min | 80 | Glutathione | — | 5 | 410/610 | 46 |
| 11 | Stirring | 1 h | 37 | Glutathione | — | 3.6 | 360/600 | 47 |
| 12 | Stirring | 5 h | RT | Glutathione | — | — | 345/620 | 48 |
| 13 | Stirring | 90 min | RT | D-Penicillamine | 150 μs | 16.6 | 345/640 | 49 |
| 14 | Stirring | 4 h | RT | Human serum albumin | — | — | 390/646 | 35 |
| 15 | Stirring | 15 min | 45 | Glutathione | — | 2.1 | 394/585 | 50 |
| 16 | Grinding | — | 35 | Phenylethanethiol | — | — | 405/605 | 51 |
| 17 | Stirring | 5 h | RT | BSA | — | 3.7 | 365/630 | 52 |
| 18 | Colloidal | 75 min | 55 | BSA | 2.17 μs | 28 | 375/650 | This work39 |
2.3 Theoretical modeling of the Cu nanoclusters
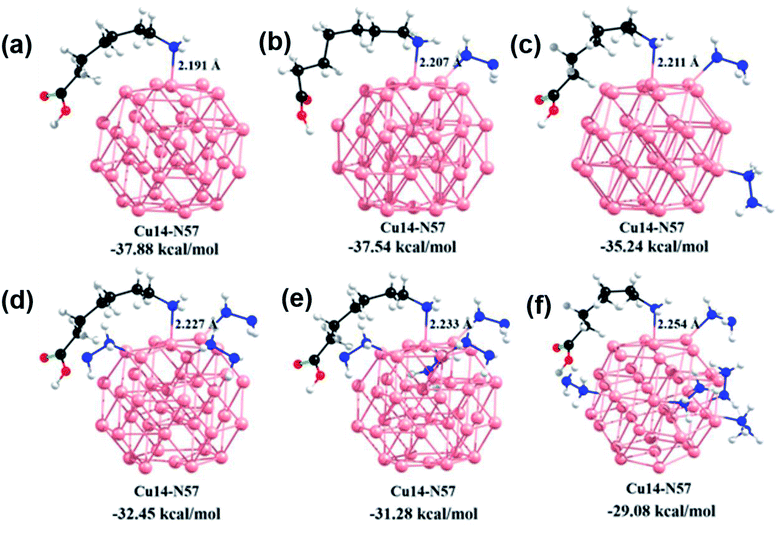
In order to gain insights into the role of hydrazine on the Cu surface and the capping agent's interactions, DFT calculations were carried out by considering six model systems. Each model system consisted of a Cu (38) cluster and a small alkyl chain containing an amine and a carboxylic acid functional group to mimic the capping groups. Furthermore, to understand the impact of hydrazine molecules on the interaction between the Cu cluster and capping group, geometry optimizations of the model systems were carried out in the presence and absence of hydrazine molecules. The model systems geometries were optimized using the B3LYP method using two different basis sets, namely the LANL2DZ basis set for Cu atoms and 6-31g* basis set for C, H, N, and O atoms. The optimized geometries of the model systems considered in the study are depicted in Fig. 3. In the absence of hydrazine (Fig. 3(a)), the intermolecular distance between the nitrogen atom in amine functional group and Cu atom in the Cu cluster was ∼2.19 Å. The same value increased from 2.19 Å to 2.25 Å when more hydrazine molecules were allowed to interact with the Cu cluster.Further, we calculated the binding energy (BE) between the alkyl chain and the Cu cluster with and without hydrazine molecules. As shown in Fig. 3, the calculated BE between the Cu cluster and capping group was 37.8 kcal mol−1 in the absence of hydrazine molecules. The same value came down to 29.08 kcal mol−1 when five hydrazine molecules were adsorbed on the Cu cluster surface. We observed a clear relationship between the number of hydrazine molecules adsorbed and BEs between the alkyl chain and the Cu cluster. Overall, these results clearly indicated that the introduction of more hydrazine molecules weakened the interaction between the capping groups and Cu clusters. We believe that a gradual addition or effective increase of hydrazine molecules near the Cu surface might influence the poor surface passivation and therefore open more surface trap states, which would finally lead to a QY and lifetime decrease.
2.4 Stability and cytotoxicity study of the Cu nanoclusters
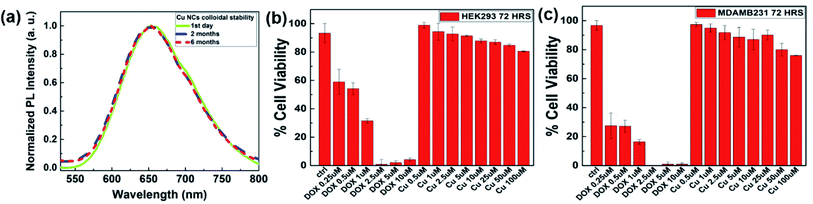
Another promising phenomenon of our protocol was the exceptional stability of the as-synthesized Cu NCs. In general, literature reports stated that Cu NCs or Cu NPs were not stable under ambient conditions in the open air as they are very prone to surface oxidation, which restricts their usefulness in potential bioimaging applications. To improve their stability, various techniques, such as the addition of Zn2+ or S2− (ref. 53) or the incorporation of pyrimidine-based ligands,54 have been employed. Such various capping groups and their stability times are highlighted in Table 2. To the best of our knowledge, our as-synthesized Cu NCs possessed remarkable optical stability of ∼6 months, where the emission intensity was found to be similar (with no peak shift) compared with the first-day sample's results (Fig. 4(a)). In addition, we observed little decrease in the emission intensity in a 10 months-old sample (Fig. S10†). This kind of phenomenon could be further rationalized via their better surface passivation at the higher pH 9 in our synthesis protocol. Also at pH 9, the di-sulfide bonds in the BSA protein backbone might open, which could stabilize/passivate the cluster further through Cu–S interactions.55| S. no. | Capping agent | λexc/λem (nm) | Stability time | Ref. |
|---|---|---|---|---|
| 1 | PVP/dihydrolipoic acid | 365/435![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 650 650 |
3 days | 38 |
| 2 | Egg white | 395/621 | 48 hours | 57 |
| 3 | 5-Methyl-2-thiouracil | 402/670 | 3 months | 54 |
| 4 | Cysteine | 365/660 | 15 days | 58 |
| 5 | BSA | 395/645 | 2 months | 59 |
| 6 | BSA | 365/630 | 24 hours | 44 |
| 7 | BSA | 365/643 | 18 days | 34 |
| 8 | GSH | 358/593 | 2 months | 60 |
| 9 | ssDNA | 340/645 | 30 minutes | 22 |
| 10 | BSA | 365/650 | 6 months | This work |
Before evaluating the potential of the Cu NCs in bioimaging as a diagnostic probe, we assessed their stabilities under various biological media. Respective PL intensity curves were monitored to eliminate the possibilities of PL loss or aggregation under those conditions. We incubated our Cu NCs with fetal bovine serum (FBS), PBS, and RPMI 1640 media for ∼6 h, and recorded the subsequent PL spectra. In all conditions, the existence of similar fluorescent intensities proved the tolerance of our NCs (Fig. S11†). Since the as-prepared Cu NCs were protected by the BSA layer, we expected them to be low or nontoxic toward cells at a suitable concentration. Thus, their toxicity evaluation (MTT assays) was performed through in vitro cell viability experiments, where two representative cell lines, namely HEK 293 (human embryonic kidney) and MDA-MB 231 (human breast adenocarcinoma), were employed. The Cu NCs were added to the two cancerous cell lines at various concentrations from ∼0.5 μM to 100 μM. Largely, our Cu NCs largely displayed nontoxicity toward those cell lines as they maintained their normal morphology after incubation. Even at a higher concentration, such as ∼100 μM Cu concentration, ∼80% cells were viable, where the negative control anticancer chemotherapeutic doxorubicin (DOX) showed proficient killing of cells at ∼2.5 μM concentration (Fig. 4(b and c)). Furthermore, we used confocal microscopy to investigate the intracellular uptake of tailored Cu NCs in the MDA-MB 231 cell line.56
Here, MDA-MB 231 adenocarcinoma cells were treated with Cu NCs at 10 μM concentration for an intracellular morphology and uptake study using a fluorescence confocal microscope as displayed in Fig. S12.† It was visibly observed that there was no effective change in the morphology of the MDA-MB 231 cells after the above treatments compared to the control (untreated-UT) cells. From the images, it could be clearly comprehended that the Cu NCs were localized inside the cell efficiently. The red fluorescence signal was found to be stable but relatively low at the 24 h time-point. These observations noticeably indicate the biocompatible nature and operative use of the Cu NCs compound as a fluorescence cell marker and delivery agent upon future studies (Fig. 4).
2.5 Whole-body disseminations and the effect on the life span of Cu nanoclusters on C. elegans
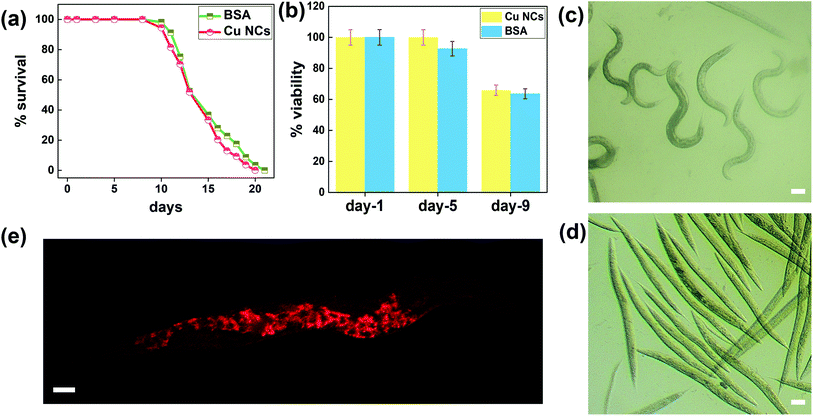
To assess whether Cu NCs possess intact fluorescent properties in biological samples, we performed in vivo experiments to study the biodistribution of these nanoclusters in C. elegans. Worms exposed to a solution of Cu NCs synthesized in a reducing agent of higher concentration died immediately (where 1 mL N2H4 was used for the reduction) compared to animals incubated in Cu NCs synthesized using our modified protocol (Fig. 5(d)), with the corresponding video given in the ESI (Fig. S13†). Hence all the subsequent in vivo assessments in C. elegans were performed with the as-synthesized Cu NCs (when 25 μL N2H4 was added for the reduction) and they exhibited exceptional low acute toxicity (Fig. 5(c)). The corresponding video is attached in the ESI (Fig. S14†). We first incubated worms in Cu NCs solution for various time periods. Fixed worms were imaged using fluorescent microscopy. We found strong fluorescent signals (OPTIKA B1000 fluorescent microscope ITALY 40 × 0.75 lens, excitation channel 405 nm) arising from Cu NCs accumulation in various tissues, such as the pharynx, gut, and tail region, in C. elegans. Interestingly, we could not detect any signal arising from the embryos in the gonad (Fig. 5(e)).Though several studies have reported the acute toxicity of various nanomaterials, the effect of nanoclusters on the organismal lifespan is poorly documented. To assess the effect of Cu NCs exposure on the lifespan, we scored the survivorship of C. elegans exposed to Cu NCs (1.25 mM for 12 h, Fig. 5(a)). We found no significant changes in the mean lifespan of animals exposed to Cu NCs as compared to animals incubated in BSA (Fig. S15†). This finding suggests that Cu NCs synthesized using the chemical reduction method and optimal N2H4 reducing agent concentration did not show a toxic effect on organismal survivorship (Fig. 5(b)). Taken together, these phenomena could open new possibilities of using Cu NCs as a unique diagnostic probe compared to others, such as easily photobleachable organic dyes, expensive noble metal NCs, and toxic Cd-based quantum dots.10
As copper is known for its natural antimicrobial properties, we hypothesized that the as-synthesized Cu NCs would also exhibit the same property.61 To demonstrate this, antimicrobial assays were performed using the disk diffusion technique. The optimized Cu NCs-I exhibited excellent antimicrobial properties at higher concentrations only, due to their nontoxicity at lower concentrations not exhibiting decent antimicrobial properties (Fig. S16†).
2.6 Extension of our approach toward different reductive environments
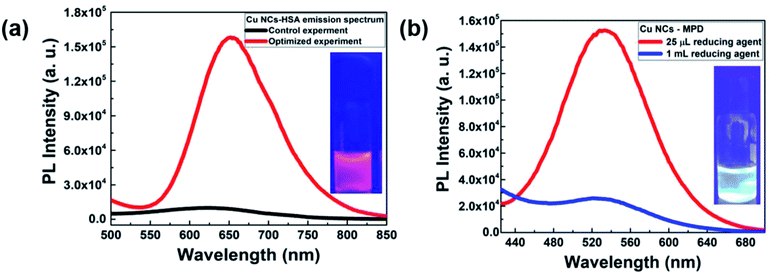
To extend and perceive the wide applicability of our approach, we employed a similar concept to prepare Cu NCs with other ligands. Our representative candidates were other red-emitting and green-emitting Cu NCs where human serum albumin (HSA) and meta-phenylenediamine (MPD) were used as the primary surfactant, respectively. In both, while reducing the effective amount of the second reducing agent, i.e., hydrazine to a low amount, a significant increase in emission intensity was recorded. We believe that “the reduction of the reducing agent amount” concept holds promise in other reductive environments at large for obtaining better QYs in Cu NCs preparation (Fig. 6).3. Conclusions
In summary, a facile synthetic strategy for obtaining Cu NCs was demonstrated, where careful control over various reaction parameters was evaluated. In particular, the amount of the second reducing agent, i.e., N2H4, played a crucial role in obtaining highly fluorescent Cu NCs. Moreover, a theoretical model was established, where more N2H4 could introduce a weak interaction between the capping groups and the Cu NCs, leading to poor stability as well as a low QY. Our optimized samples showed excellent biocompatibility toward solid cancer cell lines and C. elegans in in vitro and in vivo environments, respectively. Taken together, the potential of such Cu NCs as a diagnostic probe was evaluated using C. elegans as a model organism. In particular, the as-synthesized Cu NCs with a low amount of N2H4 imparted almost no toxicity toward the health span of C. elegans. Also, these NCs showed potential in terms of an antibacterial effect, which could be explored further. Finally, successful implementation of these reaction parameters control was applied for improvement of the PL properties while synthesizing Cu NCs using another set of ligand systems. Thus, a transition from a low QY to better QY with nontoxic and colloidal stable metal nanoclusters was observed, providing an innovative way to control the reaction parameters for their use in diagnostically relevant probes in the biological environment. We believe other strategies, such as altering the surface charge, different metal compositions, various intrinsic structures, proper choice of environment, could open up avenues to explore the known methodologies with careful optimization for yielding better attributes.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
K. B. B., K. J., S. B., J. P. G., S. H., M. K. R., M. T. and S. C. would like to thank SRM, AP for their support. K. B. B., S. C., would like to thank S. H., M. K. R., for theoretical modeling. K. B. B., S. C., would like to thank S. B., J. P. G., M. T., for antibacterial studies. K. B. B., S. C., would like to thank K. B., N. P., for In vitro studies. K. B. B., S. C., would like to thank K. J., M. T. for In vivo studies. M. T. thanks DST-INSPIRE grant (DST/INSPIRE/04/2015/002416) for supporting C. elegans experiments conducted in this manuscript. N. P. acknowledges the DST-Inspire faculty scheme for financial support (GAP-0631). N. P. also thanks to the Gujarat State Biotechnology Mission, Govt. of Gujarat, India, grant number (GSBTM/JD(R&D)/618/21-22/00003673).References
- Z. Lin, N. Goswami, T. Xue, O. J. H. Chai, H. Xu, Y. Liu, Y. Su and J. Xie, Adv. Funct. Mater., 2021, 31, 2105662 CrossRef CAS.
- T. Chen, H. Lin, Y. Cao, Q. Yao and J. Xie, Adv. Mater., 2021, 2103918 CrossRef PubMed.
- T. Higaki, Q. Li, M. Zhou, S. Zhao, Y. Li, S. Li and R. Jin, Acc. Chem. Res., 2018, 51, 2764–2773 CrossRef CAS PubMed.
- W. F. Lai, W. T. Wong and A. L. Rogach, Adv. Mater., 2020, 32, 1–21 Search PubMed.
- K. Zheng and J. Xie, Trends Chem., 2020, 2, 665–679 CrossRef CAS.
- A. Mathew and T. Pradeep, Part. Part. Syst. Charact., 2014, 31, 1017–1053 CrossRef CAS.
- X. Kang and M. Zhu, Chem. Soc. Rev., 2019, 48, 2422–2457 RSC.
- K. B. Busi, M. Palanivel, K. K. Ghosh, W. B. Ball, B. Gulyás, P. Padmanabhan and S. Chakrabortty, Nanomaterials, 2022, 12, 301 CrossRef PubMed.
- S. Chakrabortty, J. A. Yang, Y. M. Tan, N. Mishra and Y. Chan, Angew. Chem., Int. Ed., 2010, 49, 2888–2892 CrossRef CAS PubMed.
- Y. Wu, S. Chakrabortty, R. A. Gropeanu, J. Wilhelmi, Y. Xu, K. Shih Er, S. Ling Kuan, K. Koynov, Y. Chan and T. Weil, J. Am. Chem. Soc., 2010, 132, 5012–5014 CrossRef CAS PubMed.
- C. N. Loynachan, A. P. Soleimany, J. S. Dudani, Y. Lin, A. Najer, A. Bekdemir, Q. Chen, S. N. Bhatia and M. M. Stevens, Nat. Nanotechnol., 2019, 14, 883–890 CrossRef CAS PubMed.
- Z. Wang, B. Chen and A. L. Rogach, Nanoscale Horiz., 2017, 2, 135–146 RSC.
- S. Shahsavari, S. Hadian-Ghazvini, F. Hooriabad Saboor, I. Menbari Oskouie, M. Hasany, A. Simchi and A. L. Rogach, Mater. Chem. Front., 2019, 3, 2326–2356 RSC.
- Y. Liu, Z. Li, W. Zhong, L. Zhang, W. Chen and Q. Li, Nanoscale Res. Lett., 2014, 9, 1–8 CrossRef PubMed.
- H. Xu and K. S. Suslick, Adv. Mater., 2010, 22, 1078–1082 CrossRef CAS PubMed.
- I. Chakraborty and T. Pradeep, Chem. Rev., 2017, 117, 8208–8271 CrossRef CAS PubMed.
- J. Yang and R. Jin, ACS Mater. Lett., 2019, 1, 482–489 CrossRef CAS.
- Y. S. Lin, Y. F. Lin, A. Nain, Y. F. Huang and H. T. Chang, Sensors and Actuators Reports, 2021, 3, 100026 CrossRef.
- A. Baghdasaryan and T. Bürgi, Nanoscale, 2021, 13, 6283–6340 RSC.
- J. Xie, Y. Zheng and J. Y. Ying, J. Am. Chem. Soc., 2009, 131, 888–889 CrossRef CAS PubMed.
- Z. Zhao and Y. Li, Colloids Surf., B, 2020, 195, 111244 CrossRef CAS PubMed.
- Z. Qing, X. He, D. He, K. Wang, F. Xu, T. Qing and X. Yang, Angew. Chem., Int. Ed., 2013, 52, 9719–9722 CrossRef CAS PubMed.
- J. Li, J. J. Zhu and K. Xu, TrAC, Trends Anal. Chem., 2014, 58, 90–98 CrossRef CAS.
- R. Datta, T. M. Heaster, J. T. Sharick, A. A. Gillette and M. C. Skala, J. Biomed. Opt., 2020, 25, 1 Search PubMed.
- J. M. An, S. H. Kim and D. Kim, Org. Biomol. Chem., 2020, 18, 4288–4297 RSC.
- G. S. Heo, Y. Zhao, D. Sultan, X. Zhang, L. Detering, H. P. Luehmann, X. Zhang, R. Li, A. Choksi, S. Sharp, S. Levingston, T. Primeau, D. E. Reichert, G. Sun, B. Razani, S. Li, K. N. Weilbaecher, F. Dehdashti, K. L. Wooley and Y. Liu, ACS Appl. Mater. Interfaces, 2019, 11, 19669–19678 CrossRef CAS PubMed.
- A. Dutta, U. Goswami and A. Chattopadhyay, ACS Appl. Mater. Interfaces, 2018, 10, 19459–19472 CrossRef CAS PubMed.
- P. R. Hunt, J. Appl. Toxicol., 2017, 37, 50–59 CrossRef CAS PubMed.
- T. Wu, H. Xu, X. Liang and M. Tang, Chemosphere, 2019, 221, 708–726 CrossRef CAS PubMed.
- M. Markaki and N. Tavernarakis, Biotechnol. J., 2010, 5, 1261–1276 CrossRef CAS PubMed.
- C. Wang, C. Wang, L. Xu, H. Cheng, Q. Lin and C. Zhang, Nanoscale, 2014, 6, 1775–1781 RSC.
- R. Singh, S. Majhi, K. Sharma, M. Ali, S. Sharma, D. Choudhary, C. S. P. Tripathi and D. Guin, Chem. Phys. Lett., 2021, 139226 Search PubMed.
- N. Goswami, A. Giri, M. S. Bootharaju, P. L. Xavier, T. Pradeep and S. K. Pal, Anal. Chem., 2011, 83, 9676–9680 CrossRef CAS PubMed.
- R. Rajamanikandan and M. Ilanchelian, Anal. Methods, 2018, 10, 3666–3674 RSC.
- R. Rajamanikandan and M. Ilanchelian, Mater. Sci. Eng., C, 2019, 98, 1064–1072 CrossRef CAS PubMed.
- S. S. Skeeters, A. C. Rosu, Divyanshi, J. Yang and K. Zhang, ACS Appl. Mater. Interfaces, 2020, 12, 50203–50211 CrossRef CAS PubMed.
- S. Chakrabortty, M. Sison, Y. Wu, A. Ladenburger, G. Pramanik, J. Biskupek, J. Extermann, U. Kaiser, T. Lasser and T. Weil, Biomater. Sci., 2017, 5, 966–971 RSC.
- R. Ghosh, U. Goswami, S. S. Ghosh, A. Paul and A. Chattopadhyay, ACS Appl. Mater. Interfaces, 2015, 7, 209–222 CrossRef CAS PubMed.
- H. Zhang, X. Huang, L. Li, G. Zhang, I. Hussain, Z. Li and B. Tan, Chem. Commun., 2012, 48, 567–569 RSC.
- R. Gui, J. Sun, X. Cao, Y. Wang and H. Jin, RSC Adv., 2014, 4, 29083–29088 RSC.
- C. Wang, S. Shu, Y. Yao and Q. Song, RSC Adv., 2015, 5, 101599–101606 RSC.
- H. Miao, D. Zhong, Z. Zhou and X. Yang, Nanoscale, 2015, 7, 19066–19072 RSC.
- T. Zhao, X. W. He, W. Y. Li and Y. K. Zhang, J. Mater. Chem. B, 2015, 3, 2388–2394 RSC.
- Y. Huang, W. Liu, H. Feng, Y. Ye, C. Tang, H. Ao, M. Zhao, G. Chen, J. Chen and Z. Qian, Anal. Chem., 2016, 88, 7429–7434 CrossRef CAS PubMed.
- J. Liu, Q. M. Zhang, Y. Feng, Z. Zhou and K. Shih, ChemPhysChem, 2016, 17, 225–231 CrossRef CAS PubMed.
- C. Wang, L. Ling, Y. Yao and Q. Song, Nano Res., 2015, 8, 1975–1986 CrossRef CAS.
- Y. Luo, H. Miao and X. Yang, Talanta, 2015, 144, 488–495 CrossRef CAS PubMed.
- X. Jia, J. Li and E. Wang, Small, 2013, 9, 3873–3879 CrossRef CAS PubMed.
- X. Jia, X. Yang, J. Li, D. Li and E. Wang, Chem. Commun., 2014, 50, 237–239 RSC.
- L. Kong, X. Chu, W. Liu, Y. Yao, P. Zhu and X. Ling, New J. Chem., 2016, 40, 4744–4750 RSC.
- A. Ganguly, I. Chakraborty, T. Udayabhaskararao and T. Pradeep, J. Nanopart. Res., 2013, 15, 1522 CrossRef.
- X. G. Li, F. Zhang, Y. Gao, Q. M. Zhou, Y. Zhao, Y. Li, J. Z. Huo and X. J. Zhao, Biosens. Bioelectron., 2016, 86, 270–276 CrossRef CAS PubMed.
- D. Li, Z. Chen, Z. Wan, T. Yang, H. Wang and X. Mei, RSC Adv., 2016, 6, 34090–34095 RSC.
- N. Singh, K. P. Raul, A. Poulose, G. Mugesh and V. Venkatesh, ACS Appl. Bio Mater., 2020, 3, 7454–7461 CrossRef CAS PubMed.
- S. Maity, D. Bain and A. Patra, J. Phys. Chem. C, 2019, 123, 2506–2515 CrossRef CAS.
- P. Mukherjee, A. Kumar, K. Bhamidipati, N. Puvvada and S. K. Sahu, ACS Appl. Bio Mater., 2020, 3, 869–880 CrossRef CAS PubMed.
- D. Li, Y. Zhao, Z. Chen, X. Mei and X. Qiu, Mater. Sci. Eng., C, 2017, 78, 653–657 CrossRef CAS PubMed.
- K. Basu, S. Paul, R. Jana, A. Datta and A. Banerjee, ACS Sustainable Chem. Eng., 2019, 7, 1998–2007 CrossRef CAS.
- J. Xu and B. Han, Nano, 2016, 11, 1650108 CrossRef CAS.
- Q. Du, X. Hu, X. Zhang, H. Cao and Y. Huang, Anal. Methods, 2019, 11, 3446–3451 RSC.
- K. Giannousi, K. Lafazanis, J. Arvanitidis, A. Pantazaki and C. Dendrinou-Samara, J. Inorg. Biochem., 2014, 133, 24–32 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra02819k |
| This journal is © The Royal Society of Chemistry 2022 |