Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Appraisal of novel azomethine–thioxoimidazolidinone conjugates as ecto-5′-nucleotidase inhibitors: synthesis and molecular docking studies†
Pervaiz Ali Channar‡
ab,
Sehrish Bano‡c,
Sidra Hassanc,
Fouzia Perveend,
Aamer Saeed*a,
Parvez Ali Mahesare,
Imtiaz Ali Khanf and
Jamshed Iqbal *c
*c
aDepartment of Chemistry, Quaid-I-Azam University, Islamabad, 45320, Pakistan. E-mail: aamersaeed@yahoo.com; asaeed@qau.edu.pk
bDepartment of Basic Sciences, Mathematics and Humanities, Dawood University of Engineering and Technology, Karachi 74800, Pakistan
cCentre for Advanced Drug Research, COMSATS University Islamabad, Abbottabad Campus, Abbottabad-22060, Pakistan. E-mail: drjamshed@cuiatd.edu.pk
dResearch Center for Modeling and Simulations, National University of Sciences and Technology (NUST), Islamabad, Pakistan
eInstitute of Chemistry, Shah Abdul Latif University, Khairpur, 66020, Pakistan
fDepartment of Entomology, Agricultural University, Peshawar 25130, Khyber Pakhtunkhwa, Pakistan
First published on 14th June 2022
Abstract
Purinergic signaling is regulated by a group of extracellular enzymes called ectonucleotidases. One of its members i.e., ecto-5′-nucleotidase (h-e5′NT) is involved in the final step of the enzymatic hydrolysis cascade that is the conversion of adenosine monophosphate (AMP) to adenosine and therefore, involves the regulation of adenosine level in extracellular space. The overexpression of h-e5′NT has been observed in various pathological conditions such as hypoxia, inflammation and cancers, and led to various complications. Hence, the identification of a potent as well as selective inhibitor of h-e5′NT is of greater importance in therapeutic treatment of various diseases. Azomethine-thioxoimidazolidinone derivatives were studied for their inhibition potential against e5′NT enzyme along with cytotoxic potential against cancer cell lines possessing overexpression of e5′NT enzyme. The derivative (E)-3-((4-((3-methoxybenzyl)oxy)benzylidene)amino)-2-thioxoimidazolidin-4-one (4g) displayed selective and significant inhibition towards h-e5′NT with an IC50 value of 0.23 ± 0.08 μM. While two other derivatives i.e., (E)-3-(((5-bromothiophen-2-yl)methylene)amino)-2-thioxoimidazolidin-4-one (4b) and 2-thioxo-3-((3,4,5-trimethoxybenzylidene)amino)imidazolidin-4-one (4e), exhibited non-selective potent inhibitory behavior against both human and rat enzymes. Moreover, these derivatives (4b, 4e and 4g) were further investigated for their effect on the expression of h-e5′NT using quantitative real time polymerase chain reaction. Additionally, molecular docking and DFT studies were also performed to determine the putative binding mode of potent inhibitors within the enzyme active site. HOMO, LUMO, ΔE, and molecular electrostatic potential maps were computed by DFT and the charge transfer regions within the molecules were identified to find out the regions for electrophilic and nucleophilic attack.
1. Introduction
Ecto-5′NT is a metallophosphatase enzyme bearing two zinc atoms in its active site. It is glycophosphatidyl anchored extracellular enzyme that works along with NTPDase in catabolism of nucleotide molecules; NTPDase 1 isozyme catalyzes the dephosphorylation of ATP into AMP whereas CD73 takes part in the hydrolysis of AMP into adenosine.1 In addition, e5′NT is also involved in a non-enzymatic role such as cell type-specific functions including epithelial cell transport and tissue barrier function,2,3 inhibition of endothelial permeability,4 reinforcement of lymphocyte–endothelium interactions,5 inhibition of macrophage and mesenchymal cell-mediated inflammation,6,7 hyperpolarization and relaxation of smooth muscle cells, and antinociception via modulation of neuronal activity.8It has been found that the hypoxic conditions of tumor microenvironment results in increased extracellular expression of both ectonucleotidase enzymes i.e., CD39 and CD73, ultimately increases the production of adenosine.9 In tumor microenvironment, the overexpressed adenosine plays an important role in tumor immunoescape mechanism regardless of other immune modulating factors. Additionally, it also increases the tumor angiogenesis, growth and metastasis. Its role has been studied in several carcinomas, for example, colorectal, breast, bladder, pancreas, ovarian, leukemia and melanoma.10 Among them cervical cancer is highly prevalent among women as the fourth major cause of fatality rate worldwide. The cervical cancer is majorly caused by the infection of human papilloma virus (HPV). The defined role of CD73 in cervical cancer cells has not been fully established.11–13 Therefore, CD73 acting as a regulator of adenosine production, has been treated as valuable drug target in several conditions such as autoimmunity, cancer, ischemic-reperfusion injury and allergy. Previous studies have exhibited the importance of CD73 inhibition by using a substrate analogue APCP or a monoclonal antibody with resultant reduction in tumor growth and metastasis. Hence, CD73 has been regarded as a potential therapeutic target to establish new anticancer therapy. In view of pharmacological role of e5'NT enzyme in tumor development, an effort is carried out to find out the potential therapeutical inhibitors with selectivity as well as greater potency.
Here in present study, we try to synthesize a novel CD73 inhibitor with greater activity against the target enzyme. The basic pharmacophore i.e., 2-thioxoimidazolidinone molecule is used to prepare a hybrid conjugate; azomethine-2-thioxoimidazolidinones. 2-Thioxoimidazolidinones, commonly known as thiohydantoins, are sulfur analogs of hydantoins (2,4-imidazolidinediones) in which one or both carbonyl groups have been substituted with thiocarbonyl groups. By adding groups to provide steric bulk, more hydrophilic or hydrophobic contacts, or stacking, the backbone of thiohydantoin may be readily changed to adopt the preferred structural type over another. Previous literature established the importance of thioxoimidazolidinones in medicinal chemistry as an antitumor, antischistosomiatic, anti-inflammatory, hypoglycemic, antiviral, antitubercular, analgesic, anticonvulsant, antimicrobial, and antifungal agents.14–22 It has not been previously used or tested against CD73 target. Additionally, 3,5-disubstituted-2-thioxoimidazolidinones and their glycoside derivatives displayed higher activity against herpes simplex (HSV) and human immunodeficiency viruses (HIV), leukemia, and prostate cancer.23,24 Combination of two or more bioactive nuclei is considered as a clever approach in drug designing. Azomethines are widely known as Schiff bases; which represent a pharmacologically active class of organic compounds. Herein, we have coupled medicinally active 2-thioxoimidazolidinones with azomethine to seek their biological potential as azomethines were previously reported as ecto-5′-nucleotidase inhibitors. The compounds containing azomethine linkage (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) such as imines, hydrazones, and oximes are extremely valuable compounds in the field of synthetic organic chemistry.
N) such as imines, hydrazones, and oximes are extremely valuable compounds in the field of synthetic organic chemistry.
2. Experimental
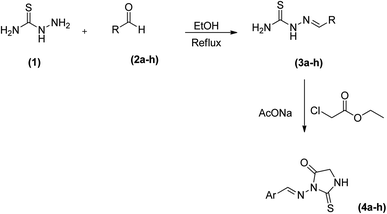
2.1 General procedure for the synthesis of ((aryl) methylene) hydrazine-1-carbothioamides (3a–h)
To a stirred solution of thiosemicarbazide (1) (1.0 mM) in 25 mL absolute ethanol, suitably substituted aldehyde, or ketone (2) (1.0 mM) along with 1–2 drops of conc. Sulfuric acid were added and the mixture was refluxed for 12 h. On completion indicated by TLC, the reaction mixture was cooled to room temperature, and the solids obtained were filtered and recrystallized from ethanol to give compounds (3a–h).2.1.1.1 (E)-3-((4-Chloro-3-nitrobenzylidene)amino)-2-thioxoimidazolidin-4-one (4a). Yield: 72%; mp 285 °C, Rf = 0.51 IR; (KBr, cm−1): 3292 (N–H), 3154 (sp2CH), 1570 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1560 (Ar–C
N), 1560 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1H NMR: (DMSO-d6, 300 MHz): 11.60 (s, 1H, NH), 8.43 (s, 1H, CH
C), 1H NMR: (DMSO-d6, 300 MHz): 11.60 (s, 1H, NH), 8.43 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.54 (d, 2H, J = 8.1 Hz, Ar–H), 7.4 (s, 1H), 3.98 (S, 2H, CH2N) 13C NMR (75 MHz, DMSO-d6): δ (ppm): 178.5, (C
N), 7.54 (d, 2H, J = 8.1 Hz, Ar–H), 7.4 (s, 1H), 3.98 (S, 2H, CH2N) 13C NMR (75 MHz, DMSO-d6): δ (ppm): 178.5, (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 173.1, (C
S) 173.1, (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 154.5, (CH
O), 154.5, (CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 147.7, 143.3, 141.5, 139.5, 137.5, 129.9, 56.9. Anal. calcd for C10H7ClN4O3S C, 40.21; H, 2.36; N, 18.76; S, 10.73 found C, 40.20; H, 2.37; N, 18.74; S, 10.75 found: 392.
N) 147.7, 143.3, 141.5, 139.5, 137.5, 129.9, 56.9. Anal. calcd for C10H7ClN4O3S C, 40.21; H, 2.36; N, 18.76; S, 10.73 found C, 40.20; H, 2.37; N, 18.74; S, 10.75 found: 392.
2.1.1.2 (E)-3-(((5-Bromothiophen-2-yl)methylene)amino)-2-thioxoimidazolidin-4-one (4b). Yield: 75%; mp 222 °C, Rf = 0.66 IR; (KBr, cm−1): 3251 (N–H), 3162 (sp2CH), 1652 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1630 (Ar–C
N), 1630 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1H NMR: (DMSO-d6, 300 MHz): 11.94 (s, 1H, NH), 8.41 (s, 1H, CH
C), 1H NMR: (DMSO-d6, 300 MHz): 11.94 (s, 1H, NH), 8.41 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.32 (dd, 1H), 7.20 (dd, 1H), 3.91 (S, 2H, CH2N) 13C NMR (75 MHz, DMSO-d6): 174.4, 165.7, 150.2, 141.3, 132.7, 131.9, 116.09, 33.5. Anal. calcd for C8H6BrN3OS2C, 31.59; H, 1.99; N, 13.81; S, 21.08 found: C, 31.57; H, 1.98; N, 13.80; S, 21.06 found: 302.
N), 7.32 (dd, 1H), 7.20 (dd, 1H), 3.91 (S, 2H, CH2N) 13C NMR (75 MHz, DMSO-d6): 174.4, 165.7, 150.2, 141.3, 132.7, 131.9, 116.09, 33.5. Anal. calcd for C8H6BrN3OS2C, 31.59; H, 1.99; N, 13.81; S, 21.08 found: C, 31.57; H, 1.98; N, 13.80; S, 21.06 found: 302.
2.1.1.3 (E)-3-(((5-Methylfuran-2-yl)methylene)amino)-2-thioxoimidazolidin-4-one (4c). Yield: (75%): mp 240 °C Rf: 0.42; IR; (KBr, cm−1): 3414 (NH2), 3262 (N–H), 3123 (sp2CH), 2932 (sp3CH), 1601 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1585 (Ar–C
N), 1585 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1H NMR; (300 MHz, DMSO): δ 10.52 (s, 1H, NH), 7.53 (s, 1H, CH
C), 1H NMR; (300 MHz, DMSO): δ 10.52 (s, 1H, NH), 7.53 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 6.18 (dd, 1H), 7.6 (dd, 1H), 3.70 (S, 2H, CH2N) 2.20 (s, 3H). 13C NMR (75 MHz): δ 181.1, (C
N), 6.18 (dd, 1H), 7.6 (dd, 1H), 3.70 (S, 2H, CH2N) 2.20 (s, 3H). 13C NMR (75 MHz): δ 181.1, (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) 170.2, (C
S) 170.2, (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 148.5, (CH
O), 148.5, (CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 155.76, 147.9, 108.7, 110.5, 56.1, 14.4 anal. calcd for C9H9N3O2S C, 48.42; H, 4.06; N, 18.82; S, 14.36 found: C, 48.40; H, 4.08; N, 18.81; S, 14.37 found: 223.
N) 155.76, 147.9, 108.7, 110.5, 56.1, 14.4 anal. calcd for C9H9N3O2S C, 48.42; H, 4.06; N, 18.82; S, 14.36 found: C, 48.40; H, 4.08; N, 18.81; S, 14.37 found: 223.
2.1.1.4 (E)-3-(((9,10-Dihydropyren-4-yl)methylene)amino)-2-thioxoimidazolidin-4-one (4d). Yield: (80%), mp 205 °C; Rf: 0.51; IR; (KBr, cm−1): 3223 (N–H), 1600 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1583 (Ar–C
N), 1583 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1082 (C
C), 1082 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), cm−1; 1H NMR: (DMSO-d6, 300 MHz): δ: 10.42 (s, 1H, NH), 8.2–8.5 (m, 3H, Ar–H), 7.97 (s, 1H, CH
S), cm−1; 1H NMR: (DMSO-d6, 300 MHz): δ: 10.42 (s, 1H, NH), 8.2–8.5 (m, 3H, Ar–H), 7.97 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.94–7.92 (m, 2H, Ar–H), 7.7–7.62 (m, 4H, Ar–H), 3.77 (S, 2H, CH2N) 13C NMR (75 MHz) δ: 184.0 (C
N), 7.94–7.92 (m, 2H, Ar–H), 7.7–7.62 (m, 4H, Ar–H), 3.77 (S, 2H, CH2N) 13C NMR (75 MHz) δ: 184.0 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 171.2, (C
S), 171.2, (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 141 (C
O), 141 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 127, 130, 132, 133, 134, 137, 140, 143, 14558.1, calcd for C20H15N3OS: C, 69.54; H, 4.38; N, 12.17; S, 9.28 found: C, 69.56; H, 4.36; N, 12.19; S, 9.26 found: 345.
N), 127, 130, 132, 133, 134, 137, 140, 143, 14558.1, calcd for C20H15N3OS: C, 69.54; H, 4.38; N, 12.17; S, 9.28 found: C, 69.56; H, 4.36; N, 12.19; S, 9.26 found: 345.
2.1.1.5 (E)-2-Thioxo-3-((3,4,5-trimethoxybenzylidene)amino)imidazolidin-4-one (4e). Yield: (70%), mp 215 °C; Rf: 0.55; IR; (KBr, cm−1): 3220 (N–H), 1605 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1571 (Ar–C
N), 1571 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1080 (C
C), 1080 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), cm−1; 1H NMR: (DMSO-d6, 300 MHz): δ: 11.96 (s, 1H, NH), 8.32 (s, 1H, CH
S), cm−1; 1H NMR: (DMSO-d6, 300 MHz): δ: 11.96 (s, 1H, NH), 8.32 (s, 1H, CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.09 (s, 2H, Ar–H), 3.83–3.81 (S, 9H, CH3O)3.88 (S, 2H, CH2N) 13C NMR (75 MHz) δ: 174.6 (C
N), 7.09 (s, 2H, Ar–H), 3.83–3.81 (S, 9H, CH3O)3.88 (S, 2H, CH2N) 13C NMR (75 MHz) δ: 174.6 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 165.4, (C
S), 165.4, (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 153.5, 140.1 (C
O), 153.5, 140.1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 130.6, 105.3, 60.6, 56.5, 56.3, 33.4, calcd for C13H15N3O4S: C, 50.47; H, 4.89; N, 13.58; S, 10.37: found C, 50.47; H, 4.89; N, 13.58; S, 10.37 found: 309.
N), 130.6, 105.3, 60.6, 56.5, 56.3, 33.4, calcd for C13H15N3O4S: C, 50.47; H, 4.89; N, 13.58; S, 10.37: found C, 50.47; H, 4.89; N, 13.58; S, 10.37 found: 309.
2.1.1.6 (E)-3-((4-(Benzyloxy)benzylidene)amino)-2-thioxoimidazolidin-4-one (4f). Yield: 72%; solid mp 265 °C, Rf = 0.61 IR; (KBr, cm−1): 3223 (N–H), 3120 (sp2CH), 2964 (sp3CH), 1605 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1593 (Ar–C
N), 1593 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), cm−1 1H NMR: (DMSO-d6, 300 MHz): 11.88 (1H, s, broad, NH), 8.0 (s, 1H, HC
C), cm−1 1H NMR: (DMSO-d6, 300 MHz): 11.88 (1H, s, broad, NH), 8.0 (s, 1H, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.40 (d, 5.4 Hz, 2H), 7.30 (m, 5H), 7.12 (d, 2H, J = 8.1 Hz, Ar–H), 5.11 (s, 2H, OCH2); 3.57 (S, 2H, CH2N) 13C NMR (75 MHz, DMSO-d6): 177.6, (C
N), 7.40 (d, 5.4 Hz, 2H), 7.30 (m, 5H), 7.12 (d, 2H, J = 8.1 Hz, Ar–H), 5.11 (s, 2H, OCH2); 3.57 (S, 2H, CH2N) 13C NMR (75 MHz, DMSO-d6): 177.6, (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 171.2, (C
S), 171.2, (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 161.5, 150.1, (C
O), 161.5, 150.1, (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 143.2, 136.5, 132.2, 131.0, 128.7, 127.5, 114.2, 70.7, 58.3 anal. calcd for C17H15N3O2S: C, 62.75; H, 4.65; N, 12.91; S, 9.85 found: C, 62.73; H, 4.62; N, 12.90; S, 9.83 found: 325.
N), 143.2, 136.5, 132.2, 131.0, 128.7, 127.5, 114.2, 70.7, 58.3 anal. calcd for C17H15N3O2S: C, 62.75; H, 4.65; N, 12.91; S, 9.85 found: C, 62.73; H, 4.62; N, 12.90; S, 9.83 found: 325.
2.1.1.7 (E)-3-((4-((3-Methoxybenzyl)oxy)benzylidene)amino)-2-thioxoimidazolidin-4-one (4g). Yield: 82%; solid mp 280 °C, Rf = 0.61 IR; (KBr, cm−1): 3220 (N–H), 3122 (sp2CH), 2961 (sp3CH), 1600 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1583 (Ar–C
N), 1583 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), cm−1; 1H NMR: (DMSO-d6, 300 MHz): 12.20 (s, 1H, NH), 8.02 (s, 1H, HC
C), cm−1; 1H NMR: (DMSO-d6, 300 MHz): 12.20 (s, 1H, NH), 8.02 (s, 1H, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.80 (d, 6.3 Hz, 2H), 7.60 (6.3 Hz, 2H) 7.4–67.1 (m, 4H), 5.12 (s, 2H, OCH2); 3.90 (s, 3H, OCH3); 3.47 (S, 2H, CH2N) 13C NMR (75 MHz, DMSO-d6): 178.9, (C
N), 7.80 (d, 6.3 Hz, 2H), 7.60 (6.3 Hz, 2H) 7.4–67.1 (m, 4H), 5.12 (s, 2H, OCH2); 3.90 (s, 3H, OCH3); 3.47 (S, 2H, CH2N) 13C NMR (75 MHz, DMSO-d6): 178.9, (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 172.2 (C
S), 172.2 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 161.6, 160.8, 153.5, (C
O), 161.6, 160.8, 153.5, (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 143.8, 132.1, 128.9, 127.3, 122.0, 120.0, 113.9, 109.3, 72.4; 55.9, 55.4. Anal. calcd for C18H17N3O3S: C, 60.83; H, 4.82; N, 11.82; S, 9.02 found C, 60.81; H, 4.84; N, 11.80; S, 9.04 found: 355.
N), 143.8, 132.1, 128.9, 127.3, 122.0, 120.0, 113.9, 109.3, 72.4; 55.9, 55.4. Anal. calcd for C18H17N3O3S: C, 60.83; H, 4.82; N, 11.82; S, 9.02 found C, 60.81; H, 4.84; N, 11.80; S, 9.04 found: 355.
2.1.1.8 (E)-3-((Pyridin-3-ylmethylene)amino)-2-thioxoimidazolidin-4-one (4h). Yield: 61%; solid mp 268 °C, Rf = 0.61 IR; (KBr, cm−1): 3231 (N–H), 3132 (sp2CH), 1605 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1581 (Ar–C
N), 1581 (Ar–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), cm−1 1H NMR: (DMSO-d6, 300 MHz): 12.00 (s, 1H, NH), 8.88 (dd, J = 7.5, 1.3 Hz, 1H), 8.61 (d, J = 1.3 Hz, 1H), 8.46 (dt, J = 7.5, 1.4 Hz, 1H), 8.11 (s, 1H, HC
C), cm−1 1H NMR: (DMSO-d6, 300 MHz): 12.00 (s, 1H, NH), 8.88 (dd, J = 7.5, 1.3 Hz, 1H), 8.61 (d, J = 1.3 Hz, 1H), 8.46 (dt, J = 7.5, 1.4 Hz, 1H), 8.11 (s, 1H, HC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 7.48 (t, J = 7.5 Hz, 1H), 3.90 (S, 2H, CH2N) 13C NMR (75 MHz, DMSO-d6): 174.7 (C
N), 7.48 (t, J = 7.5 Hz, 1H), 3.90 (S, 2H, CH2N) 13C NMR (75 MHz, DMSO-d6): 174.7 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S), 167.0, (C
S), 167.0, (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 154.1, (C
O), 154.1, (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 151.6, 149.6, 134.4, 130.5, 124.4, 33.5 Anal. calcd for C9H8N4OS: C, 49.08; H, 3.66; N, 25.44; S, 14.56 found: C, 49.08; H, 3.66; N, 25.40; S, 14.58 found: 220.
N), 151.6, 149.6, 134.4, 130.5, 124.4, 33.5 Anal. calcd for C9H8N4OS: C, 49.08; H, 3.66; N, 25.44; S, 14.56 found: C, 49.08; H, 3.66; N, 25.40; S, 14.58 found: 220.
2.2 Biochemical assays
2.2.1.1 Preparation of membrane fractions. The confluent transfected cells were collected from the harvesting buffer through scraping and subsequently washed with Tris-saline buffer along with centrifugation by spinning for 5 min at 300 × g 4 °C.25 The obtained cell pellet was allowed to remain suspended in the harvesting buffer with aprotinin (10 μg mL−1) and sonicated. The cells were again centrifuged for 10 min at 800 × g 4 °C to remove the cellular and nuclear debris. Finally, glycerol (7.5%) was incorporated to the resultant supernatant. Bradford microplate assay26 was used for protein quantification employing Bovine serum albumin as standard reference.
2.2.1.2 Ecto-5′-nucleotidase inhibition assay. The ecto-5′-nucleotidase inhibition assay was carried out by using P/ACE MDQ capillary electrophoresis system (Beckman Instruments, Fullerton, CA, USA) according to previously reported method.27 The compound solutions were prepared and analyzed at 0.1 mM concentration in assay buffer (1 mM CaCl2, 10 mM Tris HCl and 2 mM MgCl2, pH 7.4). The 10 μL of each sample was used in assay volume of 100 μL along with 10 μL of h-e5′NT (6.94 μg mL−1) protein extract or r-e5′NT (7.17 μg mL−1) and 70 μL of assay buffer. After 10 min incubation at 37 °C, 10 μL of substrate AMP (adenosine monophosphate) at final concentration of 500 μM was added to start the enzymatic reaction. After 30 min incubation at 37 °C, the reaction was stopped via thermal denaturation by heating the reaction mixture in water bath at 99 °C for 20 min. Finally, 50 μL of this reaction mixture was used for analysis. It was hydrodynamically injected into the capillary under pressure of 0.5 psi for 5 s and 15 kV voltage to allow the separation of substrate and product peaks. The compounds with more than 50% inhibitory activity of either human or rat enzyme were further evaluated for determination of IC50 values. For this purpose, serial dilutions of each active compound were prepared and analyzed to obtain the dose response curve by using the above protocol. All experiments were performed in triplicate. The non-linear regression analysis of program PRISM 5.0 (GraphPad, San Diego, California, USA) was used to calculate the IC50 values.
2.2.2.1 Cell viability assays (MTT assay). The cytotoxic potential of azomethine–thioxoimidazolidine conjugates was carried out according to the previously reported method by employing MTT (Dimethyl-2-thiazolyl-2,5-diphenyl-2H-tetrazolium bromide) reagent.28,29 All the compounds were analyzed against cervical cancer (HeLa). Accordingly, 90 μL of the cell culture (2.5 × 104 cells per mL cells) was seeded/well for overnight in 96 well plates. Then the cells were treated with 10 μL of respective test compounds (at final concentration of 100 μM) and plates were incubated at 37 °C for 24 h in CO2 incubator. Negative control (100 μL culture medium) along with positive (100 μM of carboplatin) control was also examined with the tested compounds. After incubations, 10 μL of MTT reagent (5 mg mL−1) was added to each well and plates were again kept at 37 °C for 4 h. Subsequently 100 μL of stopping reagent (50% isopropanol and 50 mL of 10% sodium dodecyl sulfate) was added to stop the enzyme reaction and solubilized the formazan crystals. The plates were allowed to incubate at room temperature for 30 min under agitation. Finally, absorbance was measured using a 96 well microplate reader (Bio-Tek ELx 800™, Instruments, Inc. Winooski, VT, USA) at 570 nm subtracting the background absorbance at 690 nm. All experiments were carried out in triplicate. Results were reported as mean of three independent experiments (±SEM) and expressed as percent inhibitions.
3. Results and discussion
3.1 Chemistry
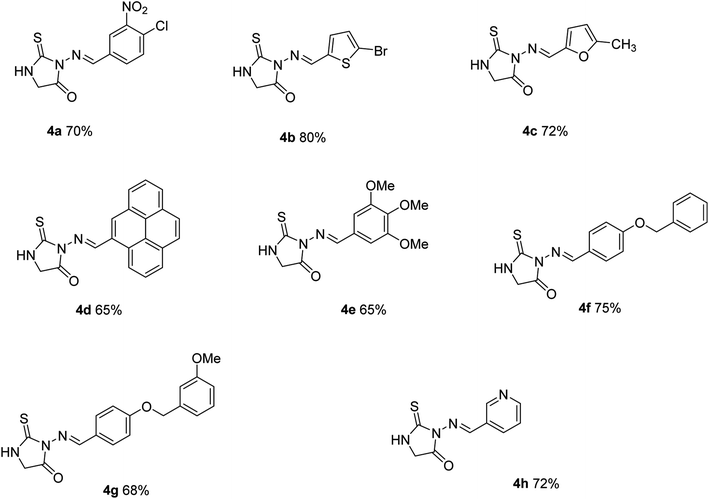
Thiosemicarbazide (1) was reacted with suitably substituted aldehyde in dry ethanol under reflux conditions to afford solid products which were purified by recrystallization using ethanol to give thiosemicarbazones (3a–h). Reaction of the latter (3a–h) with an equimolar amount of ethyl chloroacetate in the ethanol in the presence of fused sodium acetate followed by recrystallization from ethanol afforded (4a–h) in excellent yields (Scheme 1).3.2 Structure activity relationship
The synthesized compounds were tested for their inhibitory potential against both human and rat e-5′NT activity. These derivatives possessed different aromatic substituent with their inhibitory value in the range of 0.18 to 42.8 μM and 0.24 to 57.6 μM against human and rat e5′NT, respectively (Table 1). All derivatives displayed inhibition against h-e5′NT whereas compound 4e showed maximum potential with IC50 values 0.18 ± 0.05 μM. The detailed study of this derivative revealed that it possessed sterically hindered 1,2,3-trimethoxyphenyl ring. The presence of methoxyl groups (–OCH3) increased the electron density of phenyl ring thus, possibly increased its reactivity towards the enzyme and therefore, responsible for the maximum inhibition. Similarly, in case of derivative 4g, the attachment of substituent 3-(phenoxymethyl)phenol resulted in comparable inhibition potential (IC50 ± SEM = 0.23 ± 0.08 μM) as compared to compound 4e. The substitution effect of 1-chloro-2-nitrobenzene group (4a) resulted in significant loss of inhibitory potential (IC50 ± SEM = 4.12 ± 0.12 μM). It might be due to the decreased reactivity of phenyl ring with substitution of electron withdrawing group such as nitro and chloro-group. The thioxoimidazolidine derivatives possessing aromatic ring other than benzene (4b, 4c, 4h) were less potent than derivative 4e and 4g. Among them, the derivative (4b) containing 2-bromo-thiphene ring substituent was more active with IC50 ± SEM = 2.69 ± 0.06 μM value as compared to other derivatives (4c and 4h). The thiophene ring possessed greater aromatic behavior along with greater reactivity against its target than other aromatic substituent. Substituent such as furan (4c) and pyridine ring (4h) exhibited less inhibitory effect with IC50 ± SEM 11.2 ± 0.32 and 42.8 ± 0.64 μM, respectively. Another derivative 4d displayed less inhibitory behavior with IC50 ± SEM = 4.17 ± 0.17 μM. When these derivatives were tested against rat isozymes it displayed different inhibitory behavior. Among all derivatives, two derivatives (4d, 4g) remained inactive while three derivatives (4a, 4b, 4e) showed good inhibition. The derivative 4e displayed maximum inhibition towards rat enzyme with IC50 ± SEM = 0.24 ± 0.03 μM which is nearly 322-fold higher than the control inhibitor sulfamic acid. The derivative 4a showed not as much of inhibition (IC50 ± SEM = 1.03 ± 0.11 μM) as compared to compound 4e that might be due to the substitutional effect of 1-chloro-2-nitrobenzene group. The replacement of phenyl ring substitution with 2-bromo-thiophene ring (4b) resulted in comparable inhibition with IC50 ± SEM = 0.37 ± 0.06 μM as compared to 4e. Other derivatives such as 4c and 4h exhibited less inhibitory activity with IC50 ± SEM values 57.6 and 19.7 μM respectively. Hence, the phenyl ring substitution resulted in greater inhibitory behavior against both h-e5′NT and r-e5′NT enzyme. The previously identified potent compound of our lab i.e., sulfamic acid is used as positive control as there is currently no drug available in market as CD73 inhibitor. Although, some ADP analogue molecules are presently in clinical trial stages (Scheme 2).| Compound | Enzyme inhibition data | Cytotoxic potential | ||
|---|---|---|---|---|
| h-e5′NT | r-e5′NT | HeLa | BHK-21 | |
| μM ± SEM | % inhibition value | |||
| a The IC50 value shows the concentration of tested compound required to inhibit 50% enzyme level (used during assay).b Represents the percent inhibition value. The experiment was performed in triplet (n) and results are given with standard error of mean (S.E.M) which shows the reliability of results and assay sensitivity. | ||||
| (4a) | 4.12 ± 0.12a | 1.03 ± 0.11a | 90.2%b | 4.31%b |
| (4b) | 2.69 ± 0.06a | 0.37 ± 0.06a | 84.3%b | 2.57%b |
| (4c) | 11.2 ± 0.32a | 57.6 ± 1.27a | 87.0%b | 6.72%b |
| (4d) | 4.17 ± 0.17a | 31.4%b | 72.1%b | 3.88%b |
| (4e) | 0.18 ± 0.05a | 0.27 ± 0.03a | 60.0%b | 5.78%b |
| (4g) | 0.28 ± 0.08a | 42.5%b | 71.3%b | 10.2%b |
| (4h) | 42.8 ± 0.64a | 19.7 ± 0.98a | 82.2%b | 12.6%b |
| Sulfamic acid | 42.1 ± 7.8a | 77.3 ± 7.0a | — | — |
| Carboplatin | — | — | 85.2%b | 18.4%b |
3.3 Cytotoxic potential (cell viability assay, MTT)
The cytotoxic potential of synthesized derivatives was tested against the target cell line having higher expression of e5′NT enzyme and also against the normal cell line BHK-21. The obtained results have been mentioned in Table 1. It was found from Table 1 that all these derivatives exhibited cytotoxic potential against the cervical cancer cell line while no activity against the normal one i.e., BHK-21. These derivatives exhibited cytotoxic potential in the range of 60 to 90% against the HeLa cell line at final concentration of 100 μM. In present experiment, the carboplatin was used as reference standard having cytotoxic potential of 85% at 100 μM. All derivatives were tested at 50 μm concentration and their potential was determined after 24 h. Overall, the synthesized derivatives that showed non-selective enzymatic activity against human and rat e5′NT also displayed better cytotoxic potential (80–90%) against cervical cancer cell line. The other two derivatives possessing selective inhibition data against human e5′NT exhibited somewhat lower cytotoxic potential i.e., 70%.3.4 CD73 mRNA expression analysis using qRT-PCR
CD73 represent a vital function in the modulation of the action of adenosine in the progression of tumor. This nucleoside, by sensitizing adenosine receptors, can favor tumor growth and stimulate angiogenesis, immune response suppression and cell proliferation. So, this enzyme is involved in over expression of cancer cells. Cancer cells (HeLa cell line) was treated with the most potent compounds 4b, 4e and 4g for different time interval i.e., 24, 48 and 72 h and measured the expression level of CD73 via relative quantification RT-PCR method. Our results found that relative mRNA expression of CD73 was 83% downregulated after 24 h and 95% down-regulation was observed after 72 h treatment with compound 4b, while compound 4g showed 80 to 85% down-regulation. Compound 4e showed 88% down-regulation after 24 h and 96% down-regulation was observed after 72 h of treatment in comparison with positive control (β-actin). Carboplatin significantly reduced the CD73 expression after 24, 48 and 72 h treatment as shown in Fig. 1. Untreated cervical cancer cells showed increased up-regulation of CD73.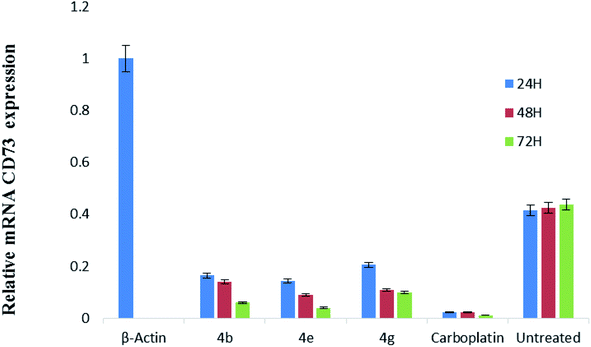 | ||
| Fig. 1 Relative mRNA expression of CD73. The graph showing the reduced expression of CD73 in comparison with control (β-actin) after treating with the compound 4b, 4e and 4g at 24, 48 and 72 h. | ||
3.5 Molecular docking studies
Molecular docking studies for the most potent selective inhibitors were performed to assess the molecular interactions between the ligands and the enzymes. As suggested by the in vitro results, 4g compound exhibited the profound selective inhibition for h-e5′NT while 4b showed the most potent selective inhibition for r-e5′NT enzyme. The amino group of thioxoimidazolidinone ring formed hydrogen bond with Asp85, Asn117, His118, His243, Thr446 and Gly447 residues of the protein, while alkyl groups of hybrid displayed π–π interactions with Leu184, Phe417 and Phe500 residues to collaborate in enzyme inhibition. The two Zn2+ ions in the active pocket of the protein were also seen to chemically interact with the sulfur group of thioxoimidazolidinone of compound 4g (Fig. 2a). For r-e5′NTenzyme, the compound 4b was found the most potent selective inhibitor. Sulfur and amino group of thioxoimidazolidinone ring exhibited hydrogen bonding with His245, Asn247, Gly394 and Asp508 residues of the protein while sulfur group from thiophene ring formed π–sulfur interactions with His40, His120 and Glu182 residues of the protein (Fig. 2b). These interactions were sufficing to correlate the inhibition of the enzymes with these compounds as indicated in the in vitro analysis.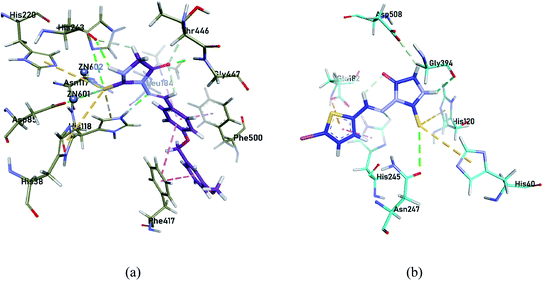 | ||
| Fig. 2 Compound 4g inside the active pocket of h-e5′NT (a) and compound 4b inside the active pocket of r-e5′NT (b). | ||
3.6 Computational structure analysis by DFT studies (4a–h)
Quantum chemical DFT investigations have substantial importance to illustrate the electronic structure, charge distribution and evaluation of related parameters. Herein; Optimized geometries, HOMO-orbital, LUMO-orbital and their respective frontier orbital energies (EHOMO and ELUMO), and surface electrostatic potential map are shown in Fig. 3–5 respectively. Optimized structures of compounds (4a–h) have shown to have non-planner geometries which have also been depicted in molecular docking studies as well. Energies of frontier molecular orbitals (FMOs), i.e., EHOMO and ELUMO, provide an estimate of electron-donating and electron-accepting nature of a molecular system. EHOMO and ELUMO gap (ΔE) determine the kinetic stability of a molecular system, chemical reactivity, and resistance to change in electronic distribution of a molecule.37 Fig. 4 reveals lowest ΔE value for 4e determing its higher reactivity and electron transfer capability as compared to other compounds . A EHOMO–ELUMO energy gap has inverse relationship with the binding tendency of an investigated compound, hence 4e is greater binding affinity.38 Fig. 4 indicated ΔE as 1.8878 eV, 1.8594 eV, 0.7184 eV, 1.9485 eV, 2.7333 eV, 2.2287 eV, 2.2631 eV, 2.3591 eV respectively. Lesser HOMO–LUMO energy gap (ΔE) indicated to have a greater tendency of electronic overlap with enzymes due to presence of 1,2,3-trimethoxyphenyl ring and inferred to have greater binding strength. Compound 4c and 4h was found to have highest ΔE and lowest reactivity due to maximum electronic distribution of LUMO on methyl pyridyl ring and pyridyl ring respectively. Due to higher density distribution at LUMO, tendency of electron transfer from HOMO is restricted thus reducing it reactivity for interactions with enzyme. | ||
| Fig. 3 Quantum chemically optimized geometries of compounds (4a–h) using DFT at GGA-PBE functional and TZP basis set with numerical accuracy of 3.0. | ||
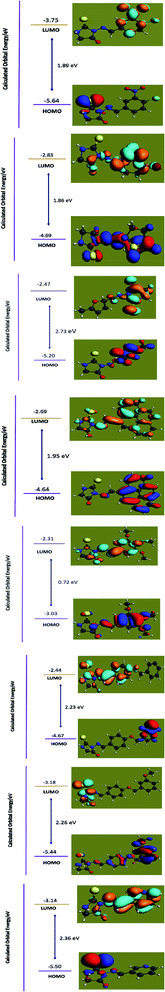 | ||
| Fig. 4 Frontier molecular orbital energy gap b/w HOMO and LUMO of compound (4a–h) from top to bottom at GGA-PBE functional and TZP basis set with numerical accuracy of 3.0. | ||
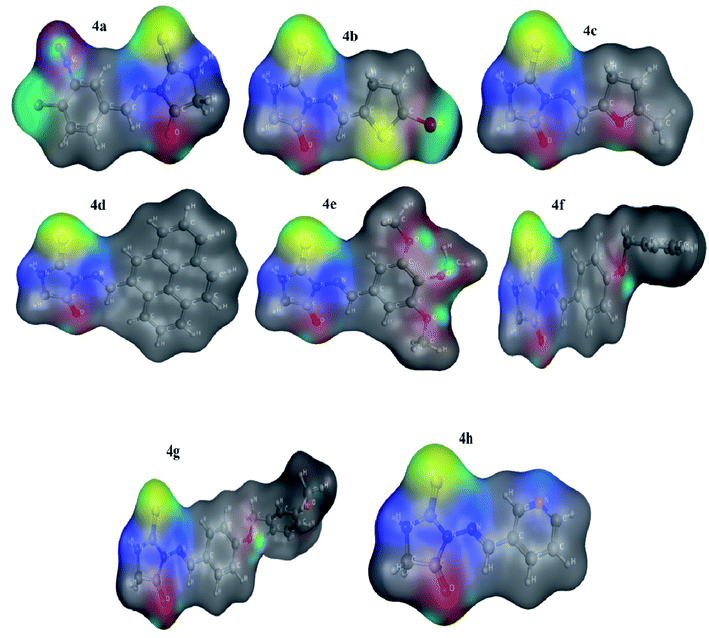 | ||
| Fig. 5 Molecular electrostatic potential surfaces (MESP) of compounds determined at LDA GGA: PBE using TZP basis set. | ||
3.7 Molecular electrostatic potential analysis
The MESP which is measure of the electronic density distribution is a very valuable property to describe electrophilic and nucleophilic centers in a chemical compound. It also responsible to furnish hydrogen-bonding interactions between the molecules.39 Molecular Electrostatic Potential Surfaces (MESP) of optimized geometries of eight compounds at GGA: PBE level of theory was determined to map the electrophilic and nucleophilic regions of compounds as indicated in Fig. 5. It is apparent from Fig. 3 that negative potential is concentrated on oxygen (red), hence representing nucleophilic character to O atoms with molecular structure. A positive potential is strenuous on C as well as H atoms depicting their electrophilic nature while taking part in a chemical process. Negative and positive potential regions give us information about the sites involved in intermolecular interactions denoting –O atom as having electron donating character in compounds. Dispersion of potentials for titled compounds was found to be ranging from −0.0853 to 0.0853 esu.Electrostatic potential maps of compounds (4a–h) have shown the high negative charge density on two nitrogen atoms and less negative charge density on S atom of the optimized compounds and hence S atom has a greater possibility to develop interactions with electron rich amino acids of enzyme, resulting in the stability of compound–enzyme complex.
4. Conclusion
A small set of azomethine–thioxoimidazolidin derivatives was examined for their enzyme inhibition potential against both human and rat isozyme was evaluated. These derivatives displayed significant inhibitory potential against h-e5′NT as compared to r-e5′NT. They also possessed significant anticancer potential against different cancer cell lines such as, MCF-7 and HeLa cells. Compound 4g displayed selective and potential inhibition towards h-e5′NT with IC50 value of 0.23 ± 0.08 μM. The compound 4h displayed nonselective as well as potent enzyme inhibition against both human and rat e5′NT. Compound 4e downregulated the CD73 expression level up to 95%. DFT studies were performed at LDA-GGA level of theory which revealed that EHOMO–ELUMO gap (ΔE) determined higher reactivity of 4e. MESP potential surface analysis indicated electrophilic nature of –S atom in compounds for their interactions with enzyme residues. Moreover, molecular docking studies were also performed to determine their putative binding sites. Hence, these derivatives can be further evaluated for their therapeutic importance in the management of various diseases.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the financial support for this research provided by the Higher Education Commission of Pakistan (HEC) via NRPU project No. 20-15846/NRPU/R&D/HEC/2021, German-Pakistani Research Collaboration Programme and Equipment Grant funded by DAAD, Germany.References
- H. Zimmermann, M. Zebisch and N. Sträter, Cellular function and molecular structure of ectonucleotidases, Purinergic Signalling, 2012, 8, 437–502 CrossRef CAS PubMed.
- J. L. Bowser and R. R. Broaddus, CD73s protection of epithelial integrity: thinking beyond the barrier, Tissue Barriers, 2016, 4(4), 220–238 CrossRef PubMed.
- S. P. Colgan, H. K. Eltzschig, T. Eckle and L. F. Thompson, Physiological roles for ecto-5’-nucleotidase (CD73), Purinergic Signalling, 2006, 2, 351 CrossRef CAS PubMed.
- L. F. Thompson, H. K. Eltzschig, J. C. Ibla, C. J. Van De Wiele, R. Resta, J. C. Morote-Garcia and S. P. Colgan, Crucial role for ecto-5′-nucleotidase (CD73) in vascular leakage during hypoxia, J. Exp. Med., 2004, 200, 1395–1405 CrossRef CAS PubMed.
- L. Airas, J. Hellman, M. Salmi, P. Bono, T. Puurunen, D. J. Smith and S. Jalkanen, CD73 is involved in lymphocyte binding to the endothelium: characterization of lymphocyte-vascular adhesion protein 2 identifies it as CD73, J. Exp. Med., 1995, 182, 1603–1608 CrossRef CAS PubMed.
- P. S. Murphy, J. Wang, S. P. Bhagwat, J. C. Munger, W. J. Janssen, T. W. Wright and M. R. Elliott, CD73 regulates anti-inflammatory signaling between apoptotic cells and endotoxin-conditioned tissue macrophages, Cell Death Differ., 2017, 24(3), 559–570 CrossRef CAS PubMed.
- M. Ohta, K. Toyama, D. D. Gutterman, W. B. Campbell, V. Lemaître, R. Teraoka and H. Miura, Ecto-5′-nucleotidase, CD73, is an endothelium-derived hyperpolarizing factor synthase, Arterioscler. Thromb. Vasc. Biol., 2013, 33(3), 629–636 CrossRef CAS PubMed.
- A. R. Cappellari, L. Rockenbach, F. Dietrich, V. Clarimundo, T. Glaser, E. Braganhol, A. L. Abujamra, R. Roesler, H. Ulrich and A. M. Liveira Battastini, Characterization of ectonucleotidases in human medulloblastoma cell lines: ecto-5′ NT/CD73 in metastasis as potential prognostic factor, PLoS One, 2012, 7, 47468 CrossRef PubMed.
- M. S. Longhi, S. C. Robson, S. H. Bernstein, S. Serra and S. Deaglio, Biological functions of ecto-enzymes in regulating extracellular adenosine levels in neoplastic and inflammatory disease states, J. Mol. Med., 2013, 91, 165–172 CrossRef CAS PubMed.
- R. Sadej and A. C. Skladanowski, Dual, enzymatic and non-enzymatic, function of ecto- 5′-nucleotidase (eN, CD73) in migration and invasion of A375 melanoma cells, Acta Biochim. Pol., 2012, 59, 647–652 CrossRef CAS PubMed.
- K. Synnestvedt, G. T. Furuta, K. M. Comerford, N. Louis, J. Karhausen, H. K. Eltzschig, K. R. Hansen, L. F. Thompson and S. P. Colgan, Ecto-5′-nucleotidase (CD73) regulation by h ypoxia-inducible factor-1 mediates permeability changes in intestinal epithelia, J. Clin. Invest., 2002, 110, 993–1002 CrossRef CAS PubMed.
- L. Antonioli, P. Pacher, E. S. Vizi and G. Haskó, CD39 and CD73 in immunity and inflammation, Trends Mol. Med., 2013, 19, 355–367 CrossRef CAS PubMed.
- E. Coppi, Purines as transmitter molecules, in Electrophysiological studies on purinergic signalling in different cell systems, Firenze University Press, 2009 Search PubMed.
- M. C. Caterina, I. A. Perillo, L. Boiani, H. Pezaroglo, H. Cerecetto, M. Gonzalez and A. Salerno, Imidazolidines as new anti-Trypanosoma cruzi agents: Biological evaluation and structure–activity relationships, Bioorg. Med. Chem., 2008, 16, 2226–2234 CrossRef CAS PubMed.
- Z. Moussa, M. A. M. S. El-Sharief and A. M. S. El-Sharief, Synthesis and characterization of new types of halogenated and alkylated imidazolidineiminothiones and a comparative study of their antitumor, antibacterial, and antifungal activities, Eur. J. Med. Chem., 2011, 46, 2280–2289 CrossRef CAS PubMed.
- J. Pandey, V. K. Tiwari, S. S. Verma, V. Chaturvedi, S. Bhatnagar, S. Sinha, A. N. Gaikwad and R. P. Tripathi, Synthesis and antitubercular screening of imidazole derivatives, Eur. J. Med. Chem., 2009, 44, 3350–3355 CrossRef CAS PubMed.
- T. Y. Reddy, N. P. Reddy, S. Koduru, C. Damodaran and P. A. Crooks, Aplysinopsin analogs: Synthesis and anti-proliferative activity of substituted (Z)-5-(N-benzylindol-3-ylmethylene) imidazolidine-2, 4-diones, Bioorg. Med. Chem., 2010, 18(10), 3570–3574 CrossRef PubMed.
- R. Sharma, M. Chouhan and V. A. Nair, A novel one-pot synthesis of 2-benzoylpyrroles from benzaldehydes, Tetrahedron Lett., 2010, 51, 2039–2043 CrossRef CAS.
- G. L. Khatik, A. Pal, S. M. Mobin and V. A. Nair, Stereochemical studies of 5-methyl-3-(substituted phenyl)-5-[(substituted phenyl) hydroxy methyl]-2-thiooxazolidin-4-ones, Tetrahedron Lett., 2010, 51, 3654–3657 CrossRef CAS.
- G. L. Khatik, A. Pal, T. D. Apsunde and V. A. Nair, A Highly Efficient Methodology for 5-Methyl-3-aryl-2-thiooxazolidin-4-ones Using Lithium Perchlorate in DIPEA Mediated Synthesis, J. Heterocycl. Chem., 2010, 47, 734 CAS.
- G. G. Muccioli, N. Fazio, G. K. E. Scriba, W. Poppitz, F. Cannata, J. H. Poupaert, J. Wouters and D. M. Lamber, Substituted 2-thioxoimidazolidin-4-ones and imidazolidine-2, 4-diones as fatty acid amide hydrolase inhibitors templates, J. Med. Chem., 2006, 49, 417–425 CrossRef CAS PubMed.
- N. Cachet, G. Genta-Jouve, E. L. Regalado, R. Mokrini, P. Amade, G. Culioli and O. P. Thomas, Hydantoin Alkaloids from the Mediterranean Sea Anemone Parazoanthus axinellae. Journal of natural products, J. Nat. Prod., 2009, 72, 1612–1615 CrossRef CAS PubMed.
- C. Tran, S. Ouk, N. J. Clegg, Y. Chen, P. A. Watson, V. Arora, J. Wongvipat, P. M. Smith-Jones, D. Yoo, A. Kwon, T. Wasielewska, D. Welsbie, C. D. Chen, C. S. Higano, T. M. Beer, D. T. Hung, H. I. Scher, M. E. Jung and C. L. Sawyers, Development of a second-generation antiandrogen for treatment of advanced prostate cancer, Science, 2009, 324, 787–790 CrossRef CAS PubMed.
- J. Conradie, Redox behaviour of [Ru(beta-diketonato)(3)] compounds, Electrochim. Acta, 2020, 337, 135801 CrossRef CAS.
- F. Kukulski, S. Lévesque, E. Lavoie, J. Lecka, F. Bigonnesse, A. Knowles, S. Robson, T. Kirley and J. Sévigny, Comparative hydrolysis of P2 receptor agonists by NTPDases 1, 2, 3 and 8, Purinergic Signalling, 2005, 1, 193–204 CrossRef CAS.
- M. M. Bradford, A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS.
- R. Raza, A. Saeed A, J. Lecka, J. Sevigny and J. Iqbal, Identification of small molecule sulfonic acids as ecto-50 -nucleotidase inhibitors, Med. Chem., 2012, 8, 1133–1139 CAS.
- T. Mosmann, Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
- M. Nikš and M. Otto, Towards an optimized MTT assay, J. Immunol. Methods, 1990, 130, 1149–1151 CrossRef.
- E. Braganhol, A. S. Tamajusuku, A. Bernardi, M. R. Wink and A. M. Battastini, Ecto-5′-nucleotidase/CD73 inhibition by quercetin in the human U138MG glioma cell line, Biochim. Biophys. Acta, Gen. Subj., 2007, 1770(9), 1352–1359 CrossRef CAS PubMed.
- K. Knapp, M. Zebisch, J. Pippel, A. El-Tayeb, C. E. Müller and N. Sträter, Crystal structure of the human ecto-50 -nucleotidase (CD73): insights into the regulation of purinergic signal, Structure, 2012, 20, 2161–2173 CrossRef CAS.
- P. A. Channar, S. J. A. Shah, S. Hassan, J. Lecka, J. Sévigny, J. Bajorath, A. Saeed and J. Iqbal, Isonicotinohydrazones as inhibitors of alkaline phosphatase and ecto-50 -nucleotidase, Chem. Biol. Drug Des., 2017, 89, 365–370 CrossRef CAS PubMed.
- ChemAxon. 2017 MarvinSketch, see https://www.chemaxon.com.
- Chemical Computing Group Inc, Molecular Operating Environment (MOE), 2014.08, Chemical Computing Group Inc, Montreal, Canada, 2016, see https://www.chemcomp.com/MOEMolecular_Operating_Environment.htm Search PubMed.
- Accelrys Software Inc. 2013 Discovery studio modeling environment, release 4.0. Accelrys Discovery Studio. San Diego, CA: Accelrys Software Inc Search PubMed.
- J. Conradie J, Redox behaviour of [Ru (β-diketonato) 3] compounds, Electrochim. Acta, 2020, 20(337), 135801 CrossRef.
- B. Nosheen, F. Perveen, Z. Ashraf, A. Bais and T. Noor, Charge transfer and opto-electronic properties of some newly designed polycatenar discotic liquid crystal derivatives: a DFT study, J. Mol. Model., 2020, 26(10), 291 CrossRef CAS PubMed.
- S. Li, Semiconductor physical electronics, Springer, Berlin, 2nd edn, 2006 Search PubMed.
- L. J. Bartolotti and K. Flurchick, An introduction to density functional theory, Rev. Comput., 1996, 7, 187–260 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra02675a |
| ‡ Both these authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |