Open Access Article
Open Access ArticleOne-step synthesized amphiphilic carbon dots for the super-resolution imaging of endoplasmic reticulum in live cells†
Jiajia Li‡
a,
Longdi Zhang‡a,
Juan Chena,
Ruilong Zhang a,
Zhengjie Liua,
Jun Zhao
a,
Zhengjie Liua,
Jun Zhao b,
Bianhua Liu
b,
Bianhua Liu b,
Ming-yong Han
b,
Ming-yong Han b,
Guangmei Han
b,
Guangmei Han *a and
Zhongping Zhang
*a and
Zhongping Zhang ab
ab
aInformation Materials and Intelligent Sensing Laboratory of Anhui Province, School of Chemistry and Chemical Engineering, Institutes of Physical Science and Information Technology, Anhui University, Hefei, 230601, P. R. China. E-mail: gmhan@ahu.edu.cn
bKey Lab of Photovoltaic and Energy Conservation Materials, Institute of Solid State Physics, HFIPS, Chinese Academy of Sciences, Hefei, 230031, P. R. China
First published on 5th July 2022
Abstract
Stimulated emission depletion (STED) microscopy provides a powerful tool for visualizing the ultrastructure and dynamics of subcellular organelles, however, the photobleaching of organelle trackers have limited the application of STED imaging in living cells. Here, we report photostable and amphiphilic carbon dots (Phe-CDs) with bright orange fluorescence via a simple one-pot hydrothermal treatment of o-phenylenediamine and phenylalanine. The obtained Phe-CDs not only had high brightness (quantum yield ∼18%) but also showed excellent photostability under ultraviolet irradiation. The CDs can quickly penetrate into cells within 2 min and are specific for intracellular ER. The further investigations by Phe-CDs revealed the reconstitution process of ER from loosely spaced tubes into a continuously dense network of tubules and sheets during cell division. Importantly, compared with the standard microscopy, STED super-resolution imaging allowed the tracking of the ER ultrastructure with a lateral resolution less than 100 nm and the pores within the ER network are clearly visible. Moreover, the three dimensional (3D) structure of ER was also successfully reconstructed from z-stack images due to the excellent photostability of Phe-CDs.
Introduction
Endoplasmic reticulum (ER) is the largest membrane organelle in eukaryotic cells and stretches from the nuclear envelope to cell membrane and plays important roles in various physiological activities,1,2 such as the assembly, post-modification and transport of proteins, the storage of Ca2+ ions and the synthesis of lipids for new cellular membranes. The defects of ER in morphology and imbalance of internal environment may cause ER autophagy and unfolded protein response, resulting in a variety of diseases, including inflammatory diseases, metabolic disorders, neurodegenerative diseases and even cancer.3–5 Therefore, real-time monitoring of ER morphology and dynamics is greatly significant to reveal the physiological and pathological mechanisms.Fluorescence imaging has been widely used in monitoring the structure and dynamics in living cells,6–8 but unfortunately, standard confocal microscopy has failed to show the ultrastructure of ER due to the limitation of optical diffraction (>250 nm). The invention of super-resolution microscopy including structured illumination microscopy (SIM), stochastic optical reconstruction microscopy (STORM) and stimulated emission depletion (STED), overcomes this limitation, realizing the visualization of the intracellular activity at the nanometer level.9–11 Among them, STED has become invincible in cell biology because it can directly integrate with standard confocal microscopy to provide fast scanning, high resolution and real-time imaging in living cells. For example, the tubulin localization and motion at a resolution of 30 nm was visualized by STED microscopy and electron microscopy techniques using a versatile cyclometalated iridium(III) complex probe.12
Fluorescent probe is the premise to obtain a desired imaging picture. Organic dye molecules, the largest family of fluorescent probes, have been widely applied in the imaging of biological events,13 however, only a few of them are photostable enough for STED-based super-resolution imaging due to the depleted laser beams with extremely high power. Fluorescent carbon dot (FCD), as a new member of carbonaceous nanomaterials, has emerged as promising alternative of molecular probe for intracellular imaging and sensing due to its small size (<10 nm), low biotoxicity, bright and tunable fluorescent properties and easy modification. Up to now, a large number of FCDs with subcellular targeting ability have been developed by selecting specific carbon precursors and exploiting surface modification approaches.14 Wang et al. developed CDs functionalized with amine groups and laurylamine for lysosome and endoplasmic reticulum targeting, respectively.15 Wu et al. prepared mitochondria-targeting CD by a one-pot solvothermal treatment of glycerol and a silane molecule for differentiating cancerous cells from normal cells.16 Huang et al. reported CD functionalized with triphenylphosphine and fluorescein 5-isothiocyanate on the surface for mitochondria-targeted ATP ratiometric imaging in living cells.17 Recently, based on the excellent photostability of CD, researches extended the applications of CDs to the super-resolution imaging in living cells. For example, in our previous work,18 a cationic carbon dot was developed by surface modification with 4-carboxybutyl triphenylphosphonium bromide for super-resolution imaging of chromosomes and RNA network in a single nucleolus. Moreover, Wu et al. synthesized red-emitting CDs by the addition of varied metal ions during the hydrothermal treatment of p-phenylenediamine, realizing STED imaging of nucleolus in living cell with an imaging resolution as high as 146 nm.19 Therefore, carbon dots with specific organelle targeting ability will become a powerful tool for super-resolution imaging in living organisms.
Direct synthesis of CDs with specific organelle-targeting ability without further surface modification is of great significance. More recently, amino acids have become ideal precursors to produce desired FCDs without further modification thanks to their heteroatoms such as nitrogen and sulfur atoms to tune the fluorescent properties of CDs.20–23 We herein report ER specific carbon dots (Phe-CDs) via a simple one-pot hydrothermal treatment of o-phenylenediamine and phenylalanine. The as-prepared Phe-CDs show an excitation-independent fluorescence emission at 580 nm and excellent photostability under UV irradiation. Combined with standard and STED super-resolution microscopy, the reconstitution process of ER during cell mitosis and the visualization of ultrastructure of ER network at nanometer scale are realized by the Phe-CDs probe.
Experimental
Materials and instruments
The o-phenylenediamine (o-PD) and phenylalanine were purchased from Sigma. 3-(4,5-Dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) and the commercial dyes (ER-Tracker Blue-White, Hoechst 33342 and Tubulin Tracker Deep red) were purchased from Thermo Fisher. Other chemical reagents were obtained from Sinopharm Chemical Reagent Co., Ltd.UV-vis absorption spectra and fluorescent spectra were measured by HITACHI UH5300 spectrometer and HITACHI F-7000 fluorescence spectrophotometer, respectively. Fluorescent images and stimulated emission depletion (STED) images were acquired on a Leica TCS SP8X microscope equipped with a STED-ONE unit. The morphology of Phe-CDs was examined by a transmission electron microscope (TEM, JEM-2100, JEOL, Tokyo, Japan). Fourier transform infrared (FTIR) spectroscopy was recorded employing a Nexus 870 FTIR spectrometer. The X-ray photoelectron spectroscopy (XPS) spectra were obtained on an ESCALAB250 XPS.
Synthesis of Phe-CDs
The Phe-CDs were prepared via a one-pot hydrothermal method using o-PD and phenylalanine as raw materials. In brief, o-phenylenediamine (0.15 g) and phenylalanine (0.3 g) were dissolved in 60 mL of ultrapure water and sonicated for 15 min to completely dissolve them. Then, the mixture was transferred into a 100 mL Teflon lined reaction kettle and kept at 180 °C for 8 h, and then cooled naturally to room temperature. The turbid reaction liquid was filtered to collect water-insoluble brownish-black solid. The solid was further purified by column chromatography with dichloromethane and ethyl acetate as the eluent (collecting the component when the ratio of dichloromethane and ethyl acetate is 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The orange red part was collected by thin layer chromatography purification and liquid chromatography with methylene chloride and ethyl acetate mixture (methylene chloride
1). The orange red part was collected by thin layer chromatography purification and liquid chromatography with methylene chloride and ethyl acetate mixture (methylene chloride![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate = 3
ethyl acetate = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). The orange-red part was scraped off and dissolved in methanol, ultrasonic for 30 min, and filtered repeatedly for three times to remove the neutral silica. The remaining solution was concentrated by rotary evaporation, and then dried in an oven at 70 °C to obtain the brown-red solid powder (Phe-CDs).
1). The orange-red part was scraped off and dissolved in methanol, ultrasonic for 30 min, and filtered repeatedly for three times to remove the neutral silica. The remaining solution was concentrated by rotary evaporation, and then dried in an oven at 70 °C to obtain the brown-red solid powder (Phe-CDs).
Cell culture and MTT assay
The cells used here were cultured in DMEM (high glucose) supplemented with fetal bovine serum (FBS, 10%), penicillin–streptomycin (1%) at 37 °C in a 5% CO2 incubator. The HeLa cells and HEK cells were chosen For the MTT assay. The live cells were first cultured in a 96-well plate overnight, then the old medium was replaced by fresh culture medium containing various concentrations of Phe-CDs probe for 24 h. Subsequently, 10 μL of 5 mg mL−1 MTT solution was added into each well and incubated for additional 4 h. Finally, 150 μL DMSO was added into each well and shaken for 15 min. The absorbance in each well was measured at 492 nm use of a microplate reader (Biotek, USA).Cell co-localization methods
Different types of cell lines (5637, HEK-293, MCF-10a, SHSY5Y and WI-38) were seeded into the glass-bottomed dishes, and cultured for 24 h. The cells were incubated with commercial dye (e.g. 1 μM ER-Tracker Blue-White) for 15 min and washed with PBS solution for three times. Subsequently, the cells were incubated with medium containing Phe-CDs (2 μg mL−1) for 30 min, also washed with PBS solution for three times. Finally, fluorescence imaging was performed in normal medium.STED super-resolution imaging
HeLa cells were incubated with 2 μg mL−1 Phe-CDs for 30 min and the STED super-resolution imaging were carried out by using the 660 nm depletion laser at 2% power. The imaging pixels were set as 2048 by 2048, and the images were processed by the ‘deconvolution wizard’ function available in the Huygens Professional software (version: 16.05).Results and discussion
Synthesis and characterization of Phe-CDs
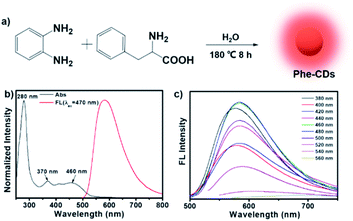
Here, we chose the o-phenylenediamine, a commonly used carbon precursor in the “bottom-up” method, and phenylalanine to prepare the carbon dots (Phe-CDs) via a one-pot hydrothermal treatment at 180 °C for 8 h (Fig. 1a). Here, phenylalanine is introduced into the reaction system to regulate the photoluminescence properties of CDs. On the one hand, it has a benzene ring itself, and on the other hand, the amino and carboxyl groups are easily cross-linked via amide bond, both of which promote the formation of carbonaceous graphitic core to make the red-shift of the fluorescence emission.24 As shown in Fig. 1b, the UV-vis absorption spectrum (black line) of Phe-CDs showed three main absorption peaks at 280, 370 and 460 nm, which can be assigned to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond of carbonaceous graphitic core, C
C bond of carbonaceous graphitic core, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/C
O/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bonds and other chemical functional groups on the edge of aromatic sp2 system, respectively.25,26 The fluorescence spectrum of Phe-CDs (red line) showed the maximum emission at 584 nm of orange fluorescence under the optimal excitation at 470 nm and an excitation-independent emission behavior in the excitation range of 380–560 nm (Fig. 1c). As expected, the emission maxima of Phe-CDs red-shift to 584 nm compared to the CDs obtained by hydrothermal treatment of o-phenylenediamine alone (emission maxima at 535 nm, green fluorescence).27 Moreover, the absolute fluorescence quantum yield of Phe-CDs is ∼18% (Fig. S1†), providing an efficient and bright fluorescence emission for imaging and sensing. And then the Phe-CD was placed on a GL-3120 benchtop UV analyzer (365 nm, 48 W) for continuous irradiation. It is worth mentioning that the fluorescence intensity of Phe-CDs remained about 80% after continuous UV irradiation for 100 min (Fig. S2†), demonstrating that Phe-CDs, as a nanofluorescent probe, had excellent photostability. We also measured the effects of pH on the fluorescence intensity of Phe-CDs. It can be seen that the fluorescence of Phe-CDs is not affected by the pH (Fig. S3†).
N bonds and other chemical functional groups on the edge of aromatic sp2 system, respectively.25,26 The fluorescence spectrum of Phe-CDs (red line) showed the maximum emission at 584 nm of orange fluorescence under the optimal excitation at 470 nm and an excitation-independent emission behavior in the excitation range of 380–560 nm (Fig. 1c). As expected, the emission maxima of Phe-CDs red-shift to 584 nm compared to the CDs obtained by hydrothermal treatment of o-phenylenediamine alone (emission maxima at 535 nm, green fluorescence).27 Moreover, the absolute fluorescence quantum yield of Phe-CDs is ∼18% (Fig. S1†), providing an efficient and bright fluorescence emission for imaging and sensing. And then the Phe-CD was placed on a GL-3120 benchtop UV analyzer (365 nm, 48 W) for continuous irradiation. It is worth mentioning that the fluorescence intensity of Phe-CDs remained about 80% after continuous UV irradiation for 100 min (Fig. S2†), demonstrating that Phe-CDs, as a nanofluorescent probe, had excellent photostability. We also measured the effects of pH on the fluorescence intensity of Phe-CDs. It can be seen that the fluorescence of Phe-CDs is not affected by the pH (Fig. S3†).
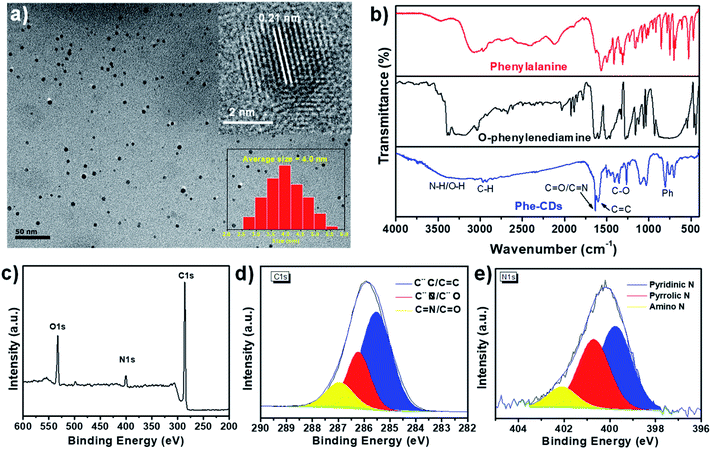
Then, the Phe-CDs were systematically characterized by transmission electron microscopy (TEM), Fourier transform infrared (FT-IR) spectra and X-ray photoelectron spectra (XPS). As shown in Fig. 2a, TEM micrograph showed that Phe-CDs present size distributions from 2.5 to 5.5 nm, with an average diameter of 4.0 nm (inset of Fig. 2a). And the high-resolution TEM (HRTEM) image shows the characteristic lattice spacing of 0.21 nm assigned to the (100) crystal plane of graphene, indicating the formation of carbonaceous graphitic core in Phe-CD. The FT-IR spectra (Fig. 2b) demonstrated the existence of benzene ring (698, 757 and 803 cm−1), C–H (2881 and 2994 cm−1), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/C
O/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N (1635 cm−1), C–O (1349 cm−1), the broad and relative weak peak around 3400 cm−1 belonged to the stretching vibrations of O–H and N–H.26,28 The elemental composition of Phe-CDs was further certified by XPS examination. The full-survey XPS spectrum suggested the existence of C, N and O elements in Phe-CDs (Fig. 2c) and the relative ratios of these elements were listed in Table 1. The high resolution XPS spectrum of C 1s (Fig. 2d) can be deconvoluted into three binding energies (i.e., 285.5 eV (C–C/C
N (1635 cm−1), C–O (1349 cm−1), the broad and relative weak peak around 3400 cm−1 belonged to the stretching vibrations of O–H and N–H.26,28 The elemental composition of Phe-CDs was further certified by XPS examination. The full-survey XPS spectrum suggested the existence of C, N and O elements in Phe-CDs (Fig. 2c) and the relative ratios of these elements were listed in Table 1. The high resolution XPS spectrum of C 1s (Fig. 2d) can be deconvoluted into three binding energies (i.e., 285.5 eV (C–C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 286.2 eV (C–N/C–O) and 286.9 eV (C
C), 286.2 eV (C–N/C–O) and 286.9 eV (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N/C
N/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)).29,30 In addition, the high-resolution XPS spectrum of N 1s (Fig. 2e) shows three peaks at 399.7, 400.7 and 402.1 eV, which can be assigned for pyridinic-N, pyrrolic-N and amino-N, respectively.31 Combining all these results, there may be a large number of benzene ring and nitrogen heterocyclic ring inside the Phe-CDs core and a certain number of hydrophilic groups such as amino and hydroxyl, contributing to its amphiphilic properties. The oil–water separation coefficient (log
O)).29,30 In addition, the high-resolution XPS spectrum of N 1s (Fig. 2e) shows three peaks at 399.7, 400.7 and 402.1 eV, which can be assigned for pyridinic-N, pyrrolic-N and amino-N, respectively.31 Combining all these results, there may be a large number of benzene ring and nitrogen heterocyclic ring inside the Phe-CDs core and a certain number of hydrophilic groups such as amino and hydroxyl, contributing to its amphiphilic properties. The oil–water separation coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) of Phe-CDs was further measured as 1.221 (Fig. S4†), also confirming its amphiphilicity.
P) of Phe-CDs was further measured as 1.221 (Fig. S4†), also confirming its amphiphilicity.
| Sample | C (at%) | N (at%) | O (at%) |
|---|---|---|---|
| Phe-CDs | 80.13 | 5.72 | 14.15 |
Selectivity and sensitivity of Phe-CDs
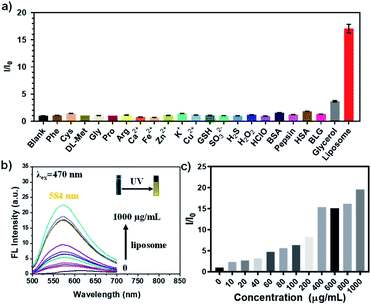
Next, we studied the fluorescent selectivity of the Phe-CDs probe in complicated biological environment. The typical biomolecules in living cells including metal ions, amino acids, proteins, reactive oxygen species (ROS) and liposomes were chosen for selectivity test. As shown in Fig. 3a, the fluorescence intensity of Phe-CDs was significantly increased by the introduction of liposomes, whereas the others except glycerol had negligible influence on the fluorescence intensity of Phe-CDs, confirming the high selectivity of Phe-CDs to the amphiphilic liposome. The glycerol, a proxy for intracellular viscosity, also caused a ∼4-fold fluorescence enhancement. The TEM images of Phe-CDs after mixing with glycerol and liposome showed the presence of Phe-CD aggregates (Fig. S5†), indicating that the fluorescence enhancement was caused by aggregation-induced enhancement (AIE). The titration experiments in Fig. 3b exhibited that the fluorescence intensity of Phe-CDs is gradually enhanced after adding liposomes. When the concentration of liposomes increased to 1 mg mL−1, the fluorescence intensity of Phe-CDs increased by 20-fold (Fig. 3c), a change that is visually detectable as shown in the inset of Fig. 3b.Intracellular distribution of Phe-CDs
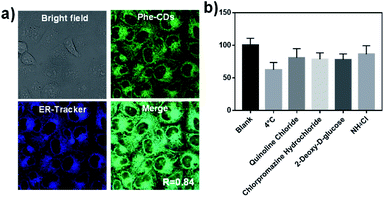
We next explored the applications of the Phe-CDs probe in biological imaging. It was worth mentioning that the Phe-CDs channel in confocal imaging was set with the excitation at 488 nm and the emission collection from 530 to 630 nm, but the pseudo color of Phe-CDs channel here was set to green instead of orange, which is better for display when merged with other commercial dyes later.The low cytotoxicity of Phe-CDs in both normal cells (HEK cell) and cancer cells (HeLa cell) was confirmed by using the classic MTT method at the concentration range of 0.5–20 μg mL−1 (Fig. S6†). Considering the fluorescence intensity and cytotoxicity, the probe culture concentration was optimized to 2 μg mL−1 (Fig. S7†) and the culture time was 30 min (Fig. S8†). It can be seen that the Phe-CDs could enter cells very quickly within 2 min and achieve maximum fluorescence intensity at 30 min. Moreover, the co-localization experiment with commercial dye was performed to demonstrate the intracellular distribution of Phe-CDs. As shown in the Fig. 4a, after incubation with 2 μg mL−1 Phe-CDs for 30 min, the HeLa cells showed strong fluorescence signal (excitation: 488 nm, emission: 530–630 nm, pseudo color: green) and overlapped well with the ER-Tracker channel (excitation: 405 nm, emission: 420–460 nm, pseudo color: blue). Here, the overlap coefficient “R”, a parameter given directly by the confocal software, was used to evaluate the co-localization of Phe-CDs and ER-Tracker. The overlap coefficient is 0.84 in HeLa cells, indicating the Phe-CDs were enriched in ER region. Because the cellular uptake pathway of nanomaterials may play a predominate role in their subcellular localization.32,33 We have further investigated the Phe-CDs cellular uptake mechanism. The low temperature (4 °C) and different biochemical inhibitors (chloroquine, chlorpromazine, 2-deoxy-D-glucose, NH4Cl) were used to estimate the cellular uptake pathway of Phe-CDs. As shown in Fig. 4b, the fluorescence intensity decreases to ∼60% at 4 °C. This maybe because that decreasing temperature reduces the cell membrane fluidity and the transport-related enzyme activity.34 At the same time, the effect of inhibitors on internalization of Phe-CDs is not particularly obvious (Fig. S9†). And taken into consideration that the Phe-CD can quickly penetrate into cells within 2 min. These results suggest that Phe-CDs penetrate into cells and accumulate in ER mainly through a passive diffusion manner, due to their small size and the amphiphilicity property make the more easily interact with the amphiphilic phospholipid bilayer membrane.33
Universality of Phe-CDs as an ER-Tracker
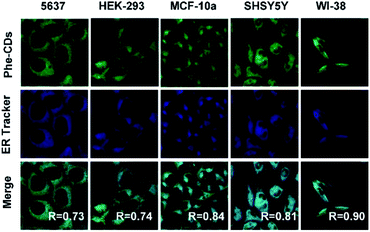
Taken the diversity of cells into consideration, five typical cell lines including normal and cancer cells (5637, HEK-293, MCF-10a, SHSY5Y and WI-38) were cultured to further verify the ER targeting ability of Phe-CDs. As shown in Fig. 5, the morphology of all cells remained normal after incubated with Phe-CDs and bright fluorescence was detected. The merge images showed the fluorescence of Phe-CDs overlapped well with the fluorescence of ER-Tracker Bue-White in 5637, HEK-293, MCF-10a, SHSY5Y and WI-38 cells. The overlap coefficients were 0.73, 0.74, 0.84, 0.81 and 0.90 for 5637, HEK-293, MCF-10a, SHSY5Y and WI-38 cells, respectively, indicating that the Phe-CDs localization in ER disregarding the kind of cell. Taken together, the above results demonstrated that Phe-CDs can serve as a universal ER-targeting probe in living cells.ER dynamics during cell division
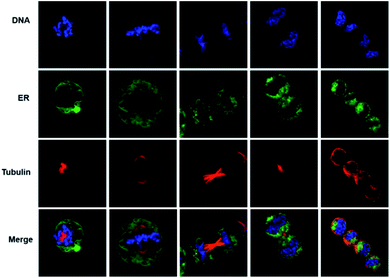
Based on the excellent ER-targeting ability, we applied the Phe-CDs probe in tracking the morphology changes of ER during the cell division. The commercial DNA label dye (Hoechst 33342, DNA channel) was used to indicate the cell division period. It is well known that the morphology of ER will change during the cell division because of the disappearance of nuclear membrane.35,36 As shown in Fig. 6, during the prophase and metaphase, the green fluorescence of Phe-CDs channel showed that the ER become loosely spaced tubes with the disappear of nuclear membrane. As the cell division progressed to the telophase and finally divide into two daughter cells with new nuclear, the ER also recovered into a continuously dense network consisting of sheets and tubules aided by the microtubule (the red fluorescence of tubulin channel labeled with Tubulin Tracker Deep red). These results demonstrate that the Phe-CDs is a powerful tool to monitor the dynamics of ER in living cells.STED super-resolution imaging and 3D reconstruction
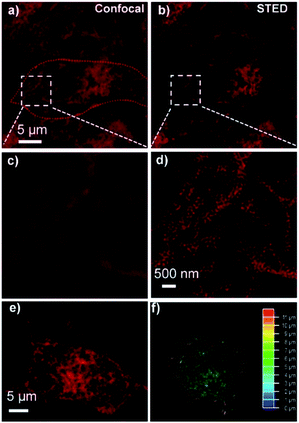
Finally, we explored the application of Phe-CDs probe in STED super-resolution imaging. As shown in Fig. 7a and b, the HeLa cells both showed bright fluorescence in the confocal and STED images after incubated with 2 μg mL−1 Phe-CDs probe for 30 min, indicating that the Phe-CDs was photostable enough for the STED imaging even under the intense laser power density of STED microscopy. Further, as shown in the magnified image of confocal image (Fig. 7c), the ER appeared as thick tubes and large sheets. However, the magnified STED image in Fig. 7d revealed that these tubes and sheets were actually a dense network that consists of a clustering of tubules and the nanoscopic pores within the network were clearly visualized in a resolution of ∼100 nm. Moreover, as shown in Fig. 7e, the STED super-resolution image of ER in an intact single cell displayed that the ER was an expansive, continuous and membrane network and can contact with nearly every other cellular organelle. Therefore, it is of great significance to analyze the interaction between ER and other organelles in three-dimensional (3D) spatial scale. However, 3D reconstruction image often requires prolonged exposure to high intensity laser, but most of small molecule dyes cannot meet this requirement due to their intrinsic photobleaching. Fortunately, as shown in Fig. 7f, our photostable Phe-CDs allowed the 3D reconstruction of ER structures from dozens of z-stack photographs. These results demonstrated that the Phe-CDs can serve as a powerful tool for super-resolution imaging to analyze ER related biology activities at the nanoscale level.Conclusions
In summary, we have developed amphiphilic carbon dots specific for ER in living cells through a simple one-pot hydrothermal method. The CDs also features the small size and good photostability, and thus can quickly enter cells in the free diffuse manner and finally accumulate in ER. Significantly, the Phe-CDs can serve as a universal ER-Tracker in different cell lines and photostable sufficient for STED super-resolution imaging. We therefore envision that the Phe-CDs will become a powerful tool for basic cell biology as well as the diagnosis and treatment of ER related diseases.Author contributions
Conceptualization: JJL, RLZ, GMH and ZPZ. Data curation and methodology: JJL, LDZ and JC. Formal analysis: ZJL, JZ, BHL and HMY. Writing-original draft: JJL, LDZ. Writing – review & editing: RLZ, GMH and ZPZ.Conflicts of interest
The authors declare no competing interests.Acknowledgements
This work was supported by the National Natural Science Foundation of China (22074001, 21974001, 22104001, 21874137 and 21976183) and the open found of Information Materials and Intelligent Sensing Laboratory of Anhui Province (Grant No. IMIS202005).References
- D. Ron and P. Walter, Nat. Rev. Mol. Cell Biol., 2007, 8, 519–529 CrossRef CAS PubMed.
- M. Wang and R. J. Kaufman, Nature, 2016, 529, 326–335 CrossRef CAS PubMed.
- K. Zhang and R. J. Kaufman, Nature, 2008, 454, 455–462 CrossRef CAS PubMed.
- I. Tabas and D. Ron, Nat. Cell Biol., 2011, 13, 184–190 CrossRef CAS PubMed.
- G. S. Hotamisligil, Cell, 2010, 140, 900–917 CrossRef CAS PubMed.
- G. Hong, A. L. Antaris and H. Dai, Nat. Biomed. Eng., 2017, 1, 0010 CrossRef CAS.
- H. Zhu, J. Fan, J. Du and X. Peng, Acc. Chem. Res., 2016, 49, 2115–2126 CrossRef CAS PubMed.
- R. H. Newman, M. D. Fosbrink and J. Zhang, Chem. Rev., 2011, 111, 3614–3666 CrossRef CAS PubMed.
- G. Vicidomini, P. Bianchini and A. Diaspro, Nat. Methods, 2018, 15, 173–182 CrossRef CAS PubMed.
- S. van de Linde, A. Löschberger, T. Klein, M. Heidbreder, S. Wolter, M. Heilemann and M. Sauer, Nat. Protoc., 2011, 6, 991–1009 CrossRef CAS PubMed.
- L. Schermelleh, M. Carlton Peter, S. Haase, L. Shao, L. Winoto, P. Kner, B. Burke, M. C. Cardoso, A. Agard David, G. L. Gustafsson Mats, H. Leonhardt and W. Sedat John, Science, 2008, 320, 1332–1336 CrossRef CAS PubMed.
- X. Tian, C. De Pace, L. Ruiz-Perez, B. Chen, R. Su, M. Zhang, R. Zhang, Q. Zhang, Q. Wang, H. Zhou, J. Wu, Z. Zhang, Y. Tian and G. Battaglia, Adv. Mater., 2020, 32, 2003901 CrossRef CAS PubMed.
- J. Chan, S. C. Dodani and C. J. Chang, Nat. Chem., 2012, 4, 973–984 CrossRef CAS PubMed.
- H. Ali, S. Ghosh and N. R. Jana, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2020, 12, e1617 CAS.
- S. E, Q. X. Mao, X. L. Yuan, X. L. Kong, X. W. Chen and J. H. Wang, Nanoscale, 2018, 10, 12788–12796 RSC.
- G. Gao, Y. W. Jiang, J. Yang and F. G. Wu, Nanoscale, 2017, 9, 18368–18378 RSC.
- R. S. Li, J. Liu, Y. Yan, C. Su, T. Lai, Y. Liao, Y. F. Li, N. Li and C. Z. Huang, Anal. Chem., 2021, 93, 11878–11886 CrossRef CAS PubMed.
- G. Han, J. Zhao, R. Zhang, X. Tian, Z. Liu, A. Wang, R. Liu, B. Liu, M.-Y. Han, X. Gao and Z. Zhang, Angew. Chem., Int. Ed., 2019, 58, 7087–7091 CrossRef CAS PubMed.
- X.-W. Hua, Y.-W. Bao, J. Zeng and F.-G. Wu, ACS Appl. Mater. Interfaces, 2019, 11, 32647–32658 CrossRef CAS PubMed.
- S. Pei, J. Zhang, M. Gao, D. Wu, Y. Yang and R. Liu, J. Colloid Interface Sci., 2015, 439, 129–133 CrossRef CAS PubMed.
- Z. Wang, C. Xu, Y. Lu, X. Chen, H. Yuan, G. Wei, G. Ye and J. Chen, Sens. Actuators, B, 2017, 241, 1324–1330 CrossRef CAS.
- Y.-W. Zeng, D.-K. Ma, W. Wang, J.-J. Chen, L. Zhou, Y.-Z. Zheng, K. Yu and S.-M. Huang, Appl. Surf. Sci., 2015, 342, 136–143 CrossRef CAS.
- J. Jiang, Y. He, S. Li and H. Cui, Chem. Commun., 2012, 48, 9634–9636 RSC.
- Y. Liu, J. H. Lei, G. Wang, Z. Zhang, J. Wu, B. Zhang, H. Zhang, E. Liu, L. Wang, T. M. Liu, G. Xing, D. Ouyang, C. X. Deng, Z. Tang and S. Qu, Adv. Sci., 2022, e2202283, DOI:10.1002/advs.202202283.
- K. Jiang, Y. Wang, C. Cai and H. Lin, Adv. Mater., 2018, 30, 1800783 CrossRef PubMed.
- S. Lu, L. Sui, J. Liu, S. Zhu, A. Chen, M. Jin and B. Yang, Adv. Mater., 2017, 29, 1603443 CrossRef PubMed.
- K. Jiang, S. Sun, L. Zhang, Y. Lu, A. Wu, C. Cai and H. Lin, Angew. Chem., Int. Ed. Engl., 2015, 54, 5360–5363 CrossRef CAS PubMed.
- J. Liu, Y. Geng, D. Li, H. Yao, Z. Huo, Y. Li, K. Zhang, S. Zhu, H. Wei, W. Xu, J. Jiang and B. Yang, Adv. Mater., 2020, 32, 1906641 CrossRef CAS PubMed.
- K.-K. Liu, S.-Y. Song, L.-Z. Sui, S.-X. Wu, P.-T. Jing, R.-Q. Wang, Q.-Y. Li, G.-R. Wu, Z.-Z. Zhang, K.-J. Yuan and C.-X. Shan, Adv. Sci., 2019, 6, 1900766 CrossRef PubMed.
- H. Ding, J. Xu, L. Jiang, C. Dong, Q. Meng, S. u. Rehman, J. Wang, Z. Ge, V. Y. Osipov and H. Bi, Chin. Chem. Lett., 2021, 32, 3646–3651 CrossRef CAS.
- L. Jiang, H. Ding, M. Xu, X. Hu, S. Li, M. Zhang, Q. Zhang, Q. Wang, S. Lu, Y. Tian and H. Bi, Small, 2020, 16, 2000680 CrossRef CAS PubMed.
- M.-S. Wu, Z.-R. Zhou, X.-Y. Wang, B.-B. Chen, M. E. Hafez, J.-F. Shi, D.-W. Li and R.-C. Qian, Anal. Chem., 2022, 94, 2882–2890 CrossRef CAS PubMed.
- S. E, C. He, J.-H. Wang, Q. Mao and X. Chen, ACS Nano, 2021, 15, 14465–14474 CrossRef PubMed.
- W. Tai and X. Gao, Biomaterials, 2018, 178, 720–727 CrossRef CAS PubMed.
- D. J. Anderson and M. W. Hetzer, J. Cell Biol., 2008, 182, 911–924 CrossRef CAS PubMed.
- D. S. Schwarz and M. D. Blower, Cell. Mol. Life Sci., 2016, 73, 79–94 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra02705d |
| ‡ JJL and LDZ contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |