Open Access Article
Open Access ArticleChemical constituents from the seeds of Nigella glandulifera and their hypoglycemic activities†
Qingqing Li‡
a,
Jing Xu‡a,
Yiyu Chena,
Wenli Xiea,
Gui Meia,
Xueni Lia,
Yu Chen*b and
Guangzhong Yang *a
*a
aSchool of Pharmaceutical Sciences, South-Central Minzu University, Wuhan 430074, P. R. China. E-mail: yanggz888@126.com; Fax: +86 27 6784 1196; Tel: +86 27 6784 1196
bCollege of Chemistry and Material Sciences, South-Central Minzu University, Wuhan 430074, P. R. China. E-mail: chenyuwh888@126.com
First published on 5th July 2022
Abstract
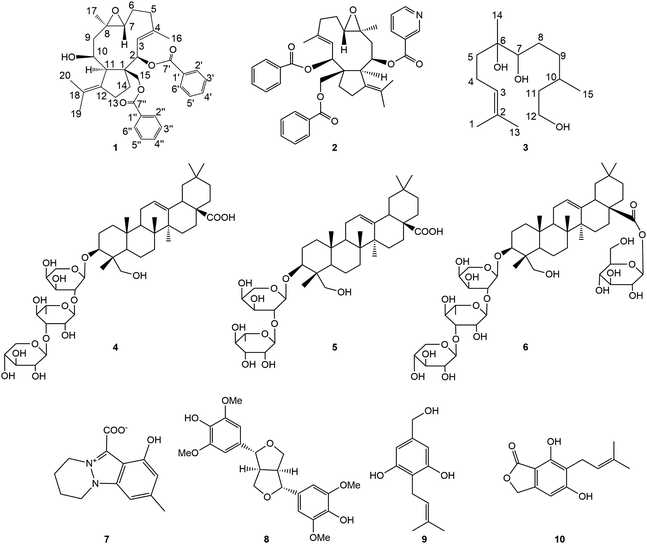
The seeds of Nigella glandulifera Freyn et Sint. are traditional Uygur medicine used for the treatment of diabetes. However, the active anti-diabetic constituents in the seeds of N. glandulifera remain unclear. In the present study, a new delabellane-type diterpene, 8-denicotinoylnigellamine A1 (1), and a new acyclic sesquiterpene, 2,6,10-trimethyl-6,7,12-trihydroxy-dodec-2-ene (3), together with eight known compounds including alkaloids (2 and 7), triterpenoid saponins (4–6), and phenolic compounds (8–10), were isolated from the seeds of N. glandulifera. Their structures were determined by extensive spectroscopic analyses and quantum chemical calculations. We evaluated the potential protective effects of the isolated compounds on an insulin resistant HepG2 (IR-HepG2) cell model. The results showed that compounds 2, 4–8, and 10 could promote the consumption of glucose in IR-HepG2 cells. Those compounds might be responsible for the anti-diabetic effects of the seeds of N. glandulifera.
1. Introduction
Type 2 diabetic mellitus (T2DM) is a chronic metabolic disease associated with the abnormal secretion and action of insulin, characterized by hyperglycemia. It is mainly resulting from insulin resistance (IR) or insufficient insulin secretion, also accompanied by abnormal glucose metabolism.1,2 Improving insulin resistance is an effective method of preventing and treating T2DM. At present, the clinical treatment of T2DM mainly depends on insulin and hypoglycemic drugs by chemical synthesis such as biguanide, sulphonylurea, thiazolidinedione, and meglitinide,3 which have significant effects but are accompanied by some toxic and side effects.4–6 It is important to seek safe, effective, and affordable new drugs for the treatment of T2DM.7 Some natural substances have been considered to be valuable resources for the development of new agents because of their low toxicity and diverse activities.Nigella glandulifera Freyn et Sint. belongs to the genus of Nigella (Ranunculaceae), which is widely distributed in Xinjiang Uygur Autonomous Region of China. The seeds of N. glandulifera are widely used in traditional Uygur medicine for the treatment of diabetes, tinnitus, amnesia, amenorrhea, hypokalaemia, and heat stranguria. Previous phytochemical investigations showed that the active constituents of N. glandulifera are volatile oil, alkaloids,8–10 triterpenoid saponins,11 and flavonoid glycosides.12 Modern pharmacological results showed that the seeds of N. glandulifera had anti-diabetic effects.13,14 Some alkaloids from the seeds of N. glandulifera were reported to have PTP1B inhibitory activity which improved glucose consumption and glycogen synthesis through PI3K/Akt insulin signaling pathway.15 To explore new anti-diabetic compounds from the seeds of N. glandulifera, phytochemical investigations were carried out in the present study. As a result, a new delabellane-type diterpene, 8-denicotinoylnigellamine A1 (1), and a new acyclic sesquiterpene, 2,6,10-trimethyl-6,7,12-trihydroxy-dodec-2-ene (3), together with eight known compounds were isolated from the seeds of N. glandulifera (Fig. 1). Moreover, the hypoglycemic activities of these compounds were investigated using insulin resistance HepG2 cells.
2. Result and discussion
2.1 Structural elucidation of the isolated compounds
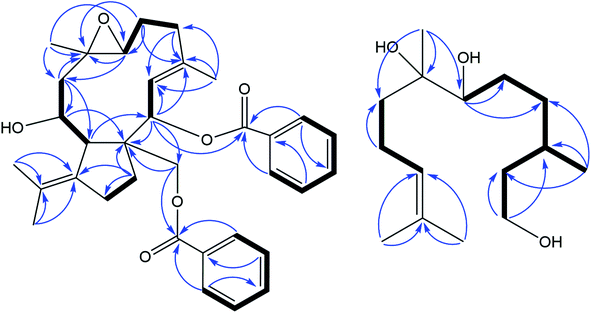
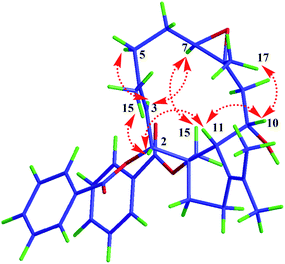
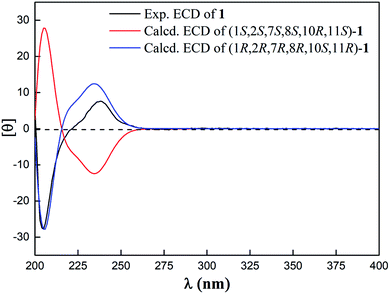
Compound 1 was isolated as a white amorphous powder. Its molecular formula was deduced as C34H40O6 from HR-ESI-MS spectrum with a pseudo molecular ion peak at m/z 567.27148 ([M + Na]+, calcd for C34H40O6Na 567.27171), corresponding to 15 degrees of unsaturation (DOUs). Careful analysis of the NMR data of 1 (Table 1) displayed the presence of two benzoyl groups. In addition to these NMR data described above, the 1H-NMR spectrum of 1 indicated that 1 contained four methyls [δH 1.42 (3H, s); 1.76 (3H, s); 1.80 (3H, s); 1.83 (3H, s)], an oxygenated methylene [δH 5.35 (1H, d, J = 10.8 Hz); 4.75 (1H, d, J = 10.8 Hz)], three oxygenated methines [δH 5.45 (1H, d, J = 10.2 Hz); 3.10 (1H, d, J = 9.0 Hz); 4.23 (1H, dd, J = 12.6, 4.8 Hz)], one olefinic proton [δH 5.75 (1H, d, J = 10.2 Hz)]. The 13C-NMR spectra together with HSQC spectrum indicated the presence of 20 carbon signals, including 5 methines, 4 methyls, 6 methylenes, and 5 quaternary carbons. The above-mentioned evidence indicated that 1 belonged to a dolabellane-type diterpene with two benzoyl groups. Compared with NMR data of nigellamine A1, 1 had one less nicotinoyl signal, suggesting that 1 was denicotinoylated of nigellamine A1.16 The chemical shifts of H-10 being shifted upfield from δH 5.70 in nigellamine A1 to δH 4.23 in 1 confirmed the above inference. HMBC correlations (Fig. 2) from H-2 to C-7′, and H2-15 to C-7′′ confirmed two benzyloxy groups were connected to C-2 and C-15 respectively. Consequently, the planar structure of 1 was determined. Detailed analysis of ROESY spectrum (Fig. 3) suggested that the relative configuration of 1 was the same as that of nigellamine A1. ROESY correlations of H-2 with H-11; H-10 with CH3-17 and H-11; H-7 with H-3 and H2-15 showed that H-2, H-10, H-11, and CH3-17 were cofacial and arbitrarily determined as α-configuration. Instead, H-7 and H2-15 were on the other side and arbitrarily determined as β-configuration. Therefore, the relative configuration of 1 was accurately determined as shown in Fig. 1. Based on the modified allylic benzoate rule and ECD calculations, the absolute configuration of 1 was resolved.16,17 The CD spectrum of 1 displayed a positive cotton effect (CE) at 238 nm, and the optical rotation value of 1 showed dextrorotation, which was contrary to nigellamine A1 (2). These findings suggested that the absolute configuration of 1 was the same as the enantiomer of nigellamine A1. Furthermore, the ECD calculations for (1S, 2S, 7S, 8S, 10R, 11S)-1a and its enantiomer (1R, 2R, 7R, 8R, 10S, 11R)-1a′ were carried out by the TDDFT/ECD method at the B3LYP/6-31+G(d) level. The results showed that the calculated ECD data of (1R, 2R, 7R, 8R, 10S, 11R)-1a′ was consistent with the experimental ECD data. Thus, the absolute configuration of 1 was determined as depicted in Fig. 4.| No | 1H-NMR | 13C-NMR | No | 1H-NMR | 13C-NMR |
|---|---|---|---|---|---|
| 1 | 58.0 | 18 | 126.3 | ||
| 2 | 5.45 (1H, d, 10.2) | 74.7 | 19 | 1.76 (3H, s) | 22.8 |
| 3 | 5.75 (1H, d, 10.2) | 125.3 | 20 | 1.83 (3H, s) | 22.6 |
| 4 | 141.0 | 1′ | 131.5 | ||
| 5 | 2.47 (1H, m) | 38.9 | 2′ | 7.72 (1H, dd, 8.4, 1.2) | 130.7 |
| 2.35 (1H, m) | |||||
| 6 | 1.92 (1H, m) | 24.1 | 3′ | 7.09 (1H, t, 7.8) | 129.5 |
| 1.76 (1H, m) | |||||
| 7 | 3.10 (1H, d, 9.0) | 67.3 | 4′ | 7.44 (1H, m) | 134.2 |
| 8 | 60.9 | 5′ | 7.09 (1H, t, 7.8) | 129.5 | |
| 9 | 2.31 (1H, m) | 47.6 | 6′ | 7.72 (1H, dd, 8.4, 1.2) | 130.7 |
| 1.45 (1H, t) | |||||
| 10 | 4.23 (1H, dd, 12.6, 4.8) | 73.7 | 7′ | 168.1 | |
| 11 | 2.45 (1H, m) | 50.9 | 1′′ | 132.1 | |
| 12 | 140.3 | 2′′ | 8.09 (1H, dd, 8.4, 1.2) | 131.1 | |
| 13 | 2.57 (1H, m) | 30.0 | 3′′ | 7.44 (1H, m) | 129.9 |
| 2.36 (1H, m) | |||||
| 14 | 2.25 (2H, m) | 32.6 | 4′′ | 7.63 (1H, td, 7.2, 1.2) | 134.5 |
| 15 | 5.35 (1H, d, 10.8) | 68.8 | 5′′ | 7.44 (1H, m) | 129.9 |
| 4.75 (1H, d, 10.8) | |||||
| 16 | 1.80 (3H, s) | 17.0 | 6′′ | 8.09 (1H, dd, 8.4, 1.2) | 131.1 |
| 17 | 1.42 (3H, s) | 19.1 | 7′′ | 168.4 |
Compound (3) was purified as a slight yellow oil and assigned the molecular formula as C15H30O3 by HR-ESI-MS spectrum with a pseudo molecular ion peak at m/z 281.20871 ([M + Na]+, calcd for C15H30O3Na, 281.20872), corresponding to one degree of unsaturation (DOUs). In the UV spectrum, the maximum absorption observed at 205 nm showed that 3 had no obvious conjugate system. The 1H-NMR spectrum of 3 (Table 2) exhibited one olefin proton [δH 5.32 (td, J = 7.2, 1.2 Hz)], three tertiary methyl groups [δH 1.61 (s), 1.67 (s), 1.49 (s)], one secondary methyl [δH 0.99 (d, J = 6.6 Hz)], an oxygenated methylene [δH 3.96 (2H, m)], one oxygenated methines [δH 3.82 (1H, d, J = 10.8 Hz)]. Detail analysis of 13C-NMR, DEPT combined with HSQC spectrum displayed 15 carbon signals assignable to four methyls, three methines (including one sp2 carbon at δC 126.6 and one oxygenated carbon at δC 78.8), six methylenes (including one oxygenated carbon at δC 60.6), one oxygenated tertiary carbon at δC 74.6, one sp2 quaternary carbon at δC 131.0. The above-mentioned data indicated that 3 belonged to an acyclic sesquiterpene with three hydroxy groups and one double bond. Further analysis of the 1H–1H COSY, HSQC, and HMBC spectra led to the elucidation of two structural fragments as a 4-methyl-3-pentenyl and 4-methyl-1,6-hexanediol group. The HMBC correlations from H3-14 to C-5, C-6, and C-7 showed that CH3-14 and 4-methyl-3-pentenyl linked to 4-methyl-1,6-hexanediol group through one oxygen tertiary carbon C-6 [δC 74.6 (s)]. Therefore, the structure of compound 3 was determined as shown.
| No | 1H-NMR | 13C-NMR | No | 1H-NMR | 13C-NMR |
|---|---|---|---|---|---|
| 1 | 1.61 (3H, s) | 18.0 | 8 | 2.01 (1H, m) | 29.8 |
| 1.81 (1H, m) | |||||
| 2 | 131.0 | 9 | 1.87 (1H, m) | 35.7 | |
| 1.70 (1H, m) | |||||
| 3 | 5.32 (1H, td, 7.2, 1.2) | 126.6 | 10 | 1.93 (1H, m) | 30.5 |
| 4 | 2.59 (1H, m) | 23.1 | 11 | 1.90 (1H, m) | 41.6 |
| 2.42 (1H, m) | 1.62 (1H, m) | ||||
| 5 | 2.10 (1H, td, 13.2, 4.8) | 39.1 | 12 | 3.96 (2H, m) | 60.6 |
| 1.83 (1H, m) | |||||
| 6 | 74.6 | 13 | 1.67 (3H, s) | 26.2 | |
| 7 | 3.82 (1H, d, 10.8) | 78.8 | 14 | 1.49 (3H, s) | 23.5 |
| 15 | 0.99 (3H, d, 6.6) | 20.3 |
The remaining eight known compounds were identified as nigellamine A1 (2),16 3-O-[β-D-xylopyranosyl-(1 → 3)-α-L-rhamnopyranosyl-(1 → 2)-α-L-arabinpyranosyl]-hederagenin (4),18 α-hederin (5),18 decaisoside E (6),19 nigellicine (7),20 yangambin (8),21 nigephenol A (9),22 5,7-dihydroxy-6-(3-methybut-2-enyl)isobenzofuran-(13H)-one (10)23 by comparison spectroscopic data with the literature data.
2.2 Glucose consumption of IR-HepG2 cells
The seeds of N. glandulifera exhibit multiple biological activities, such as being anti-inflammatory, anti-diabetic, antioxidative and hepatoprotective.24–27 N. sativa, another commonly used medical plant belongs to the same genus, exerts anti-diabetic and immune-modulatory effects.28,29 Based on the distinct hypoglycemic activities, anti-diabetic effects of the constituents from these plants were further analyzed. The alkaloids, such as nigelladines A-C, exert an anti-diabetic effect by activating PI3K/Akt insulin signaling pathway and inhibiting PTP1B.13 In the present study, phytochemical investigation of the seeds of N. glandulifera led to the isolation of three structural types of natural products, including terpenes (1–6), alkaloids (7), and phenolic compounds (9 and 10). The anti-diabetic effects of these compounds were still unknown. The major contributor to the pathogenesis of T2DM is IR, which refers to the decreased responsiveness of peripheral blood cells (liver, muscle, and adipose tissue) to glucose metabolism, resulting in an abnormal homeostatic response to blood glucose.30 For the establishment of IR model, human hepatoblastoma HepG2 cells were used as reported.31 Moreover, metformin was chosen as the positive control because it could promote hepatic glucose consumption in IR HepG2 cells.32–34 The hypoglycemic activity of these isolated compounds was determined using the IR-HepG2 cell model. Firstly, cell viabilities of all isolated compounds were evaluated by CCK-8 method. As shown in Table 3, all isolated compounds showed no significant effects on the viability of HepG2 cells at concentrations below 20 μM. Therefore, five concentrations of 2.5, 5, 10, 12.5, and 15 μM (cell viability > 90%) were used for further analysis. As shown in Table 4, compared to the normal group, the glucose consumption of the model group was significantly decreased, which demonstrated IR-HepG2 cell model was successfully built up. At a concentration of 5 μM of compounds 2, 4–8, 10, the glucose consumption increased to 5.93–9.15 mM which had no significant difference from the normal group. Among them, the glucose consumption of compound 10 at 12.5 and 15 μM were 10.57 ± 0.84 and 11.01 ± 0.86 mM, respectively, both of which were higher than the positive control of metformin (9.49 ± 0.51 mM). Triterpenoid saponins (4–6) significantly increased glucose consumption in IR-HepG2 cell model at 15 μM. These findings showed that these compounds could improve the IR of HepG2 cells and may have anti-diabetic effects.| Compound | Cell viability | IC50 (μM) | |||||
|---|---|---|---|---|---|---|---|
| 0 μM | 10 μM | 20 μM | 30 μM | 40 μM | 50 μM | ||
| 1 | 100.00% | 97.27 ± 3.45% | 93.91± 3.05% | 90.71 ± 2.77% | 76.58 ± 2.77% | 68.00 ± 2.91% | 68.07 |
| 2 | 100.00% | 97.19 ± 1.04% | 93.08 ± 1.43% | 91.01 ± 0.97% | 87.66 ± 0.84% | 87.70 ± 1.57% | 308.3 |
| 3 | 100.00% | 94.76 ± 1.94% | 83.50 ± 3.68% | 60.27 ± 3.33% | 36.31 ± 1.62% | 33.36 ± 1.90% | 33.95 |
| 4 | 100.00% | 96.11 ± 2.98% | 93.05 ± 3.98% | 85.90 ± 0.77% | 82.95 ± 0.51% | 76.91 ± 2.78% | 120.2 |
| 5 | 100.00% | 98.57 ± 0.57% | 93.39 ± 1.23% | 89.38 ± 1.16% | 76.11 ± 1.73% | 73.33 ± 2.58% | 98.49 |
| 6 | 100.00% | 96.33 ± 1.34% | 92.23 ± 0.55% | 87.40 ± 1.81% | 84.46 ± 3.47% | 69.35 ± 1.71% | 185.4 |
| 7 | 100.00% | 99.30 ± 1.45% | 92.32 ± 1.02% | 78.38 ± 2.34% | 73.81 ± 3.72% | 68.36 ± 2.16% | 72.56 |
| 8 | 100.00% | 94.52 ± 5.42% | 92.59 ± 1.39% | 80.22 ± 3.79% | 76.22 ± 1.66% | 68.57 ± 2.12% | 78.61 |
| 9 | 100.00% | 96.59 ± 2.83% | 99.78 ± 2.92% | 89.44 ± 2.28% | 78.22 ± 3.65% | 62.37 ± 4.10% | 56.08 |
| 10 | 100.00% | 94.50 ± 2.24% | 93.27 ± 1.28% | 91.06 ± 0.79% | 87.28 ± 2.54% | 75.33 ± 1.89% | 96.76 |
| Compound | NG | MG | Met | Concentration | ||||
|---|---|---|---|---|---|---|---|---|
| 2.5 μM | 5.0 μM | 10.0 μM | 12.5 μM | 15 μM | ||||
| a NG: normal group; MG: model group; Met: metformin. | ||||||||
| 1 | 5.90 ± 0.56 | 3.47 ± 0.77 | 8.49 ± 0.79 | 3.92 ± 0.61 | 4.59 ± 0.45 | 4.71 ± 0.17 | 5.60 ± 0.47 | 5.56 ± 0.69 |
| 2 | 6.08 ± 0.30 | 3.74 ± 0.62 | 9.90 ± 0.42 | 4.77 ± 0.79 | 7.01 ± 0.72 | 6.98 ± 0.72 | 7.90 ± 0.52 | 8.56 ± 0.58 |
| 3 | 5.82 ± 0.81 | 1.33 ± 0.43 | 9.42 ± 0.41 | 2.06 ± 1.27 | 2.43 ± 0.67 | 2.66 ± 0.56 | 2.86 ± 0.71 | 2.44 ± 0.60 |
| 4 | 5.26 ± 0.53 | 2.09 ± 0.66 | 8.82 ± 0.53 | 5.75 ± 0.39 | 6.95 ± 0.71 | 7.38 ± 0.62 | 8.32 ± 0.61 | 8.59 ± 0.73 |
| 5 | 5.49 ± 0.40 | 2.90 ± 0.48 | 9.48 ± 0.64 | 6.31 ± 0.96 | 6.95 ± 0.22 | 7.75 ± 0.84 | 8.97 ± 0.37 | 9.99 ± 0.95 |
| 6 | 6.25 ± 0.40 | 2.91 ± 0.84 | 9.52 ± 0.26 | 5.67 ± 0.76 | 5.93 ± 0.94 | 8.59 ± 1.46 | 9.73 ± 0.39 | 11.97 ± 0.72 |
| 7 | 5.53 ± 0.29 | 2.45 ± 0.55 | 9.20 ± 0.59 | 6.75 ± 0.51 | 8.01 ± 0.56 | 8.47 ± 0.46 | 9.01 ± 0.50 | 9.48 ± 0.93 |
| 8 | 5.81 ± 0.17 | 2.45 ± 0.86 | 9.78 ± 0.83 | 6.47 ± 0.17 | 7.97 ± 0.57 | 8.25 ± 0.34 | 8.97 ± 0.47 | 9.76 ± 0.48 |
| 9 | 5.67 ± 0.59 | 3.50 ± 0.40 | 9.87 ± 0.79 | 4.64 ± 0.57 | 4.24 ± 0.92 | 4.77 ± 0.47 | 4.79 ± 0.65 | 4.51 ± 0.76 |
| 10 | 5.98 ± 0.20 | 2.97 ± 0.93 | 9.49 ± 0.51 | 8.96 ± 0.90 | 9.15 ± 0.90 | 9.15 ± 0.38 | 10.57 ± 0.84 | 11.01 ± 0.86 |
3. Experimental section
3.1 General experimental procedures
Optical rotations were measured in MeOH on an Autopol IV polarimeter (Rudolph Research Analytical, Hackettstown, NJ, USA). UV spectra was obtained on a UH5300 UV-VIS Double Beam spectrophotometer (Hitachi Co., Tokyo, Japan). ECD spectra was recorded on a Chirascan Plus spectrometer (Applied Photophysics Ltd, London, England). 1D and 2D NMR spectra were recorded on a Bruker AVANCE IIITM 500 MHz or 600 MHz spectrometers (Bruker, Ettlingen, Germany) using tetramethylsilane (TMS) as an internal reference standard. Chemical shifts (δ) have been expressed in ppm and the coupling constants (J) have been given in Hz. High-resolution electrospray mass spectroscopy was performed on a Thermo Scientific Q Exactive Orbitrap LC-MS/MS System (HR-ESI-MS) (Thermo Scientific, Waltham, MA, USA). High-performance liquid chromatography (HPLC) was conducted on an Ultimate 3000 HPLC system (Dionex Co., Sunnyvale, CA, USA) equipped with an Ultimate 3000 pump and Ultimate 3000 Variable Wavelength detector, as well as a semi-preparative YMC-Pack ODS-A column (250 × 10 mm, 5 μm) and a preparative YMC-Pack ODS-A column (250 × 20 mm, 5 μm) from YMC Co., Ltd (Kyoto, Japan), column chromatography (CC) was conducted on silica gel (200–300 mesh and 300–400 mesh, Qingdao Haiyang Chemical Industry Co., Ltd., Qingdao, China). Chromatographic grade acetonitrile was purchased from Chang Tech Enterprise Co., Ltd (Taiwan, China). Dulbecco's Modified Eagle Medium (DMEM) and 0.25% pancreatin were obtained from Wuhan Procell Life Science Technology Co., Ltd. (Wuhan, China). The glucose kit was obtained from Shanghai Rongsheng Biotech Co., Ltd. (Shanghai, China). Metformin hydrochloride tablets were obtained from Sino-American Shanghai Squibb Pharmaceuticals Ltd. (Shanghai, China). Cell counting kit-8 (CCK8) was obtained from ABclonal Technology Co., Ltd. (Wuhan, China). Glucose solution and palmitate acid (PA) were obtained from Sigma-Aldrich Co., Ltd. (St. Louis, MO, USA). Fetal bovine serum (FBS) was obtained from Zhejiang Tianhang Biotechnology Co., Ltd. (Hangzhou, China).3.2 Plant materials
Dried seeds of N. glandulifera were acquired in October 2018 from Xinjiang Uygur Autonomous Region, P. R. China, and identified by Prof. Xinqiao Liu, School of Pharmaceutical Sciences, South-Central Minzu University. The voucher specimens were deposited in the herbarium of School of Pharmaceutical Sciences, South-Central Minzu University.3.3 Extraction and isolation
Dried seeds of N. glandulifera (9.8 kg) were defatted with petroleum ether three times, and then extracted with 50% EtOH three times (12 h each time) at room temperature. The combined EtOH extracts were evaporated under vacuum to yield crude extract, and then suspended in the distilled water, and extracted successively with ethyl acetate (EtOAc), n-butanol to yield EtOAc extract (120 g), and n-butanol extract (140 g). The EtOAc extract was isolated by silica gel column chromatography, using petroleum ether-CH2Cl2–MeOH gradient as https://fanyi.so.com/?src=onebox to yield 7 fractions (Fr. A.1 ∼ Fr. A.7). Fr. A.3 (2.8 g) was repeatedly fractionated by normal and reversed phase silica gel chromatographic column and further purified by semi-preparative HPLC to obtain compound 1 (1.0 mg, MeOH-0.1% formic acid, 78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22, tR = 40.2 min), 2 (1.0 mg, MeOH-0.1% formic acid, 85
22, tR = 40.2 min), 2 (1.0 mg, MeOH-0.1% formic acid, 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15, tR = 28.0 min), 8 (2.0 mg, MeCN–H2O, 78
15, tR = 28.0 min), 8 (2.0 mg, MeCN–H2O, 78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22, tR = 38.5 min). Fr. A.5 (2.0 g) was subjected to repeated ODS CC with MeOH–H2O (30
22, tR = 38.5 min). Fr. A.5 (2.0 g) was subjected to repeated ODS CC with MeOH–H2O (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 to 90
70 to 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) and semi-preparative HPLC to afford compound 3 (2 mg, MeCN–H2O, 37
10) and semi-preparative HPLC to afford compound 3 (2 mg, MeCN–H2O, 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 63, tR = 19.1 min), 7 (2 mg, MeCN-0.1% formic acid = 23
63, tR = 19.1 min), 7 (2 mg, MeCN-0.1% formic acid = 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 77, tR = 25.8 min) and 9 (2 mg, MeCN–H2O, 25
77, tR = 25.8 min) and 9 (2 mg, MeCN–H2O, 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75, tR = 30.1 min). Fr. A.6 (2.8 g) was purified by ODS CC, using MeOH–H2O gradient (30
75, tR = 30.1 min). Fr. A.6 (2.8 g) was purified by ODS CC, using MeOH–H2O gradient (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 to 90
70 to 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) as eluent, to yield 7 fractions (Fr. A.6.1–Fr. A.6.7). Fr. A.6.2 was separated by semi-preparative HPLC to afford compound 10 (2 mg, MeCN–H2O, 42
10) as eluent, to yield 7 fractions (Fr. A.6.1–Fr. A.6.7). Fr. A.6.2 was separated by semi-preparative HPLC to afford compound 10 (2 mg, MeCN–H2O, 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 58, tR = 22.3 min). The n-butanol extract was applied to D101 macroporous resin using H2O–EtOH as eluent to give 12 fractions. Fr. B.11 and Fr. B.12 were purified by repeated semi-preparative HPLC to afford compound 4 (50.0 mg, MeOH-0.1% formic acid, 77
58, tR = 22.3 min). The n-butanol extract was applied to D101 macroporous resin using H2O–EtOH as eluent to give 12 fractions. Fr. B.11 and Fr. B.12 were purified by repeated semi-preparative HPLC to afford compound 4 (50.0 mg, MeOH-0.1% formic acid, 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 23, tR = 26.1 min), 5 (10.0 mg, MeOH-0.1% formic acid, 77
23, tR = 26.1 min), 5 (10.0 mg, MeOH-0.1% formic acid, 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 23, tR = 28.2 min), 6 (1.0 mg, MeOH-0.1% formic acid, 70
23, tR = 28.2 min), 6 (1.0 mg, MeOH-0.1% formic acid, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, tR = 20.0 min).
30, tR = 20.0 min).
8-denicotinoy.lnigellamine A1 (1): white amorphous powder; [α]24D = +25.0° (c 0.19, MeOH); UV (MeOH) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε): 230 (4.46), 270 (3.46) nm; ECD (3.47 × 10−3 M, MeOH) λ(θ): 204 (−27.40), 238 (+7.62) nm; 1H-NMR (600 MHz, CDCl3) and 13C-NMR (150 MHz, CDCl3): see Table 1; HR-ESI-MS m/z 567.27148 [M + Na]+(calcd for C34H40O6Na, 567.27171).
ε): 230 (4.46), 270 (3.46) nm; ECD (3.47 × 10−3 M, MeOH) λ(θ): 204 (−27.40), 238 (+7.62) nm; 1H-NMR (600 MHz, CDCl3) and 13C-NMR (150 MHz, CDCl3): see Table 1; HR-ESI-MS m/z 567.27148 [M + Na]+(calcd for C34H40O6Na, 567.27171).
2,6,10-trimethyl-6,7,12-trihydroxy-dodec-2-ene (3): light yellow oil; [α]24D = +11.3° (c 0.20, MeOH); UV (MeOH) λmax (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε): 205 (3.22) nm; 1H-NMR (600 MHz, C5D5N) and 13C-NMR (150 MHz, C5D5N) see Table 2; HR-ESI-MS m/z 281.20871 [M + Na]+(calcd for C15H30O3Na, 281.20872).
ε): 205 (3.22) nm; 1H-NMR (600 MHz, C5D5N) and 13C-NMR (150 MHz, C5D5N) see Table 2; HR-ESI-MS m/z 281.20871 [M + Na]+(calcd for C15H30O3Na, 281.20872).
3.4 ECD calculation
The absolute configuration of 1 was determined by quantum chemical TDDFT calculation of its theoretical ECD spectrum according to a previously reported method.353.5 Cell viability
HepG2 cells (1.0 × 105 cells/well) were grown in a 96-well plate at 37 °C in a DMEM medium containing 15% fetal bovine serum. After treatment with different concentrations of compounds 1–10 (10, 20, 30, 40, and 50 μM) for 24 h, the supernatant was removed. 100 μL DMEM and 10% CCK-8 were added to each well, and incubated at 37 °C for 30 min. The absorbance was measured at 450 nm with a microplate analyzer to calculate the cell viability.3.6 Glucose consumption in IR-HepG2 cells
The IR-HepG2 cell model was induced by palmitic acid (PA) as previously reported.36,37 Briefly, after 12 h serum-free starvation, HepG2 cells were cultured in high glucose (30 mM) DMEM plus PA (0.25 mM) as the model group, and high glucose DMEM plus PA in the presence of compounds 1–10 with different concentrations (2.5, 5.0, 10, 12.5 and 15 μM) for 24 h. The glucose concentration in the medium was determined using a glucose assay kit (Nanjing Jiancheng Bioengineering Institute) according to the instruction. The amount of glucose consumption was calculated by the initial glucose level minus the glucose level remaining in the medium. Metformin hydrochloride (2 mM) was used as the positive control.4. Conclusion
In summary, phytochemical studies on the seeds of N. glandulifera resulted in the separation of two previously undescribed compounds (1 and 3) together with eight known compounds (2, 4–10), including two dolabellane diterpenes (1 and 2), one acyclic sesquiterpene (3), triterpene saponins (4–6), one alkaloid (7), and three phenolic compounds (8–10). Moreover, the hypoglycemic activity of the isolated compounds was determined by the IR-HepG2 cell model. The results showed compounds 2, 4–8, 10 could promote the glucose consumption of IR-HepG2 cell. These findings illuminate the active compounds of the seeds of N. glandulifera in the treatment of T2DM and may offer new strategies for alleviating the hepatic IR of T2DM.Author contributions
YC and GY: conceptualization, writing, reviewing and editing; QL: purification and isolation of compounds, writing-original draft; XJ: hypoglycemic activity assay; WX: ECD calculations; YYC, GM and XL: formal analysis, data curation.Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
This work was supported by the Research and Development Program of Hubei Province (2021ACB003), and the Major Scientific, Technological Project of Hubei Province (2020ACA007) and the Special Fund for Basic Scientific Research of Central Colleges, South-Central University for Nationalities (CZY22002).References
- D. D. Huang, G. J. Shi, Y. P. Jiang, C. Chao and C. L. Zhu, Biomed. Pharmacother., 2020, 125, 109767 CrossRef CAS PubMed.
- N. Ning, E. Kim, B. Li, H. B. Pan, T. T. Tong, C. S. Yang and Y. Y. Tu, J. Agric. Food Chem., 2019, 67, 5361–5373 CrossRef PubMed.
- G. E. C. Sun, B. J. Wells, K. Yip, R. Zimmerman, D. Raghavan, M. W. Kattan and S. R. Kashyap, Diabetes, Obes. Metab., 2014, 16, 276–283 CrossRef CAS PubMed.
- S. Okayasu, K. Kitaichi, A. Hori, T. Suwa, Y. Horikawa, M. Yamamoto, J. Takeda and Y. Itoh, Biol. Pharm. Bull., 2012, 35, 933–937 CrossRef CAS PubMed.
- D. Desilets, A. Shorr, K. Moran and K. Holtzmuller, Am. J. Gastroenterol., 2001, 96, 2257–2258 CrossRef CAS PubMed.
- F. W. Germino, Clin. Ther., 2011, 33, 1868–1882 CrossRef CAS PubMed.
- Z. Z. Gao, Q. W. Li, X. M. Wu, X. M. Zhao, L. H. Zhao and X. L. Tong, J. Immunol., 2017, 2017, 1813086 Search PubMed.
- J. Tian, C. Han, W. Guo, Y. Yin, X. Wang, H. Sun, H. Yao, Y. Yang, C. Wang, C. Liu, M. Yang and L. Kong, Org. Lett., 2017, 19, 6348–6351 CrossRef CAS PubMed.
- Y. Liu, L. Sun, Q. Liu, S. Lu and B. Chen, Biochem. Syst. Ecol., 2013, 49, 43–46 CrossRef.
- W. Guo, X. Li, S. Huang, M. Yang and L. Kong, J. Asian Nat. Prod. Res., 2017, 19, 9–14 CrossRef CAS PubMed.
- X. Xin, H. A. Aisa, H. Q. Xue and H. Q. Wang, Chem. Nat. Compd., 2008, 44, 134–136 CrossRef CAS.
- B. Boubertakh, X. G. Liu, X. L. Cheng and P. Li, J. Chem., 2013, 2013, 820183 Search PubMed.
- Y. L. Zheng, Q. Zhang and X. J. Hu, Med. Chem. Res., 2020, 29, 1168–1186 CrossRef CAS.
- Y. Niu, L. Zhou, L. Meng, S. Chen, C. Ma, Z. Liu and W. Kang, J. Evidence-Based Complementary Altern. Med., 2020, 6756835 Search PubMed.
- Y. Ma, J. Li, F. Tong, X. L. Xin and H. A. Aisa, Ind. Crops Prod., 2020, 153, 112592 CrossRef CAS.
- T. Morikawa, F. M. Xu, Y. Kashima, H. Matssuda, K. Ninomiya and M. Yoshikawa, Org. Lett., 2004, 6, 869–872 CrossRef CAS PubMed.
- T. Morikawa, F. M. Xu, K. Ninomiya, H. Matssuda and M. Yoshikawa, Chem. Pharm. Bull., 2004, 52, 494–497 CrossRef CAS PubMed.
- Y. M. Liu, J. S. Yang and Q. H. Liu, China J. Chin. Mater. Med., 2005, 30, 980–983 CAS.
- J. Kong, X. H. Yang, R. H. Lu, Y. P. Wang and H. Q. Wang, Indian J. Chem., 1999, 37b, 882–884 Search PubMed.
- A. Rahman, S. Malik, C. H. He and J. Clardy, Tetrahedron Lett., 1985, 26, 2759–2762 CrossRef.
- N. M. Lima, L. M. Cursino-Hron, A. M. Lima, J. B. V. Souza, A. C. Oliveira, J. V. N. Marinhoa and C. V. Nunez, Rev. Bras. Farmacogn., 2018, 28, 697–702 CrossRef CAS.
- L. Sun, Y. M. Liu, B. Q. Chen and Q. H. Liu, Bioorg. Med. Chem. Lett., 2015, 25, 3864–3866 CrossRef CAS PubMed.
- G. H. Li, L. Li, M. Duan and K. Q. Zhang, Chem. Biodiversity, 2006, 3, 210–216 CrossRef CAS PubMed.
- D. Tang, Q. B. Chen, X. L. Xin and H. A. Aisa, Biomed. Pharmacother., 2017, 87, 145–152 CrossRef CAS PubMed.
- J. B. Gao, X. J. Zhang, R. H. Zhang, L. L. Zhu, D. B. Pu, X. L. Li, H. L. Li, M. Xu and W. L. Xiao, Planta Med., 2018, 84, 1013–1021 CrossRef CAS PubMed.
- D. Maimaitiyiming, M. Kamilijiang, X. Xiaerfuding, S. W. Hui, M. Abudureyimu, Hajinisha and A. Aikemu, Pak. J. Pharm. Sci., 2017, 30, 1567–1571 CAS.
- H. Jiang, F. Xu, L. Zeng, C. Li, Y. Chen, L. Wang, Z. Li and R. Liu, J. Ethnopharmacol., 2022, 283, 114714 CrossRef CAS PubMed.
- J. Dong, Q. Liang, Y. Niu, S. Jiang, L. Zhou, J. Wang, C. Ma and W. Kang, Int. J. Biol. Macromol., 2020, 159, 725–738 CrossRef CAS PubMed.
- Y. Niu, B. Wang, L. Zhou, C. Ma, G. I. N. Waterhouse, Z. Liu, A. F. Ahmed, D. Sun-Waterhouse and W. Kang, Front. Nutr., 2021, 8, 722813 CrossRef PubMed.
- J. C. Chen, L. Li, X. Zhang, L. T. Wan, Q. S. Zheng, D. Xu, Y. T. Li, Y. Liang, M. S. Chen, B. Li and A. Y. Chen, J. Funct. Foods, 2021, 82, 104478 CrossRef CAS.
- Z. C. Yang, W. Huang, J. S. Zhang, M. Xie and X. W. Wang, Eur. J. Pharmacol., 2019, 854, 187–193 CrossRef CAS PubMed.
- Q. Wei, Y. Zhan, B. Chen, B. Xie, T. Fang, S. Ravishankar and Y. Jiang, Food Sci. Nutr., 2020, 8, 332–339 CrossRef CAS PubMed.
- K. A. Akinyede, H. A. Oyewusi, G. D. Hughes, O. E. Ekpo and O. O. Oguntibeju, Molecules, 2021, 27, e155 CrossRef PubMed.
- A. P. A. Nwakiban, S. Cicolari, S. Piazza, F. Gelmini, E. Sangiovanni, G. Martinelli, L. Bossi, E. Carpentier-Maguire, A. D. Tchamgoue, G. Agbor, J. R. Kuiate, G. Beretta, M. Dell'Agli and P. Magni, Metabolites, 2020, 10, e182 CrossRef PubMed.
- X. Tan, X. Y. Han, H. D. Teng, Q. Q. Li, Y. Chen, X. X. Lei and G. Z. Yang, J. Nat. Prod., 2021, 8, 972–978 CrossRef PubMed.
- J. R. Nie, Y. N. Chang, Y. Li, Y. Zhou, J. Qin, Z. Sun and H. B. Li, J. Agric. Food Chem., 2017, 65, 9041–9053 CrossRef CAS PubMed.
- Y. Zhang, S. G. Chen, C. Y. Wei, J. C. Chen and X. Q. Ye, J. Funct. Foods, 2017, 29, 217–225 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra02628g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |