 Open Access Article
Open Access ArticleRhodamine-based cyclic hydrazide derivatives as fluorescent probes for selective and rapid detection of formaldehyde†
Sung Yeon Kim‡
 ,
Sang-Hyun Park‡,
Chang-Hee Lee,
Jinsung Tae* and
Injae Shin
,
Sang-Hyun Park‡,
Chang-Hee Lee,
Jinsung Tae* and
Injae Shin *
*
Department of Chemistry, Yonsei University, Seoul 03722, Republic of Korea. E-mail: injae@yonsei.ac.kr; jstae@yonsei.ac.kr
First published on 11th August 2022
Abstract
We describe fluorescent probes to detect formaldehyde (FA) in aqueous solutions and cells. The probes rapidly respond to FA in aqueous solutions and have great selectivity toward FA over other biologically relevant analytes. The results of cell studies reveal that probe 1 can be utilized to monitor endogenous and exogenous FA in live cells.
Reactive carbonyl species (RCS) produced through metabolic processes are highly reactive and, thus, their overproduction causes damage to a variety of organisms.1 Formaldehyde (FA), the simplest RCS, is a human toxin and carcinogen as a result of its ability to crosslink DNA and proteins.2 FA is generated in cells by several metabolic events, including methanol oxidation, histone demethylation and N-methylamine deamination.3 During normal metabolic processes, the concentration of FA is maintained at physiological levels in the range from 100 μM in blood to 200–400 μM in brain,4 where it is involved in spatial memory formation and cognition.5 However, upregulation of FA-producing enzymes or exposure to exogenous FA (e.g., industrial pollutants, cigarette smoke, and natural products) can lead to abnormal elevation of FA levels up to as much as 800 μM.4,5 Elevated levels of FA cause memory impairments, cancers, diabetes and neurodegenerative disorders.6 Owing to the physiological and pathological significance of FA, selective and sensitive tools to monitor this RCS in cells are in critical demand.
Fluorescence imaging is a powerful method to detect intracellular analytes (e.g., ions, reactive species and biomolecules) owing to its advantageous features such as operational simplicity, sensitivity and non-invasive properties.7 Several fluorescent probes for detection of FA in cells, which are based on specific chemical reactions including 2-aza-Cope rearrangement, formimine reaction and aminal formation, have been devised thus far.8 However, most of these probes have drawbacks such as low selectivities over other aldehydes and/or slow fluorescence responses to FA.
To develop FA-responsive fluorescent probes that do not suffer from the limitations described above, we designed rhodamine-based cyclic hydrazide derivatives 1 and 2 (Scheme 1), which should be weakly fluorescent owing to the absence of an appropriate fluorophore. We reasoned that the tethered amine groups in 1 and 2 would react with FA to form iminium ions A, which would then undergo intramolecular addition of the hydrazide NH to generate cyclic aminals B. We also envisaged that rapid opening of spirocyclic moiety in B would take place to generate highly fluorescent xanthenes C.9
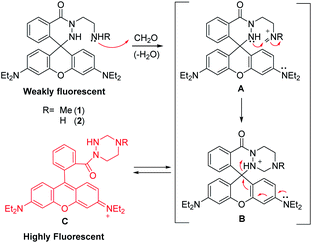 | ||
| Scheme 1 The proposed mechanism of fluorescence sensing of formaldehyde by rhodamine cyclic hydrazide-based probes 1 and 2. | ||
On the basis of this strategy, the new FA-reactive fluorescent probes 1 and 2 were synthesized using reactions of rhodamine B acid chloride with the corresponding amine-appended hydrazines (Schemes S1–S3†). All new compounds were characterized using standard spectroscopic methods. To assess the design strategy displayed in Scheme 1, we measured time-dependent changes in the intensities of fluorescence arising from the probes following treatment with FA at a physiologically relevant concentration (10 equiv., 100 μM) in PBS buffer (1% DMSO, pH 7.4).4 As shown in the spectra and plots (Fig. 1a and S1†), the secondary amine tethered probe 1 underwent an immediate fluorescence response to FA and the emission intensity reached a maximum within 5 min. In the case of probe 2 containing a primary amine appendage, the fluorescence intensity promoted by treatment with FA reached a plateau after 2 min but a lesser extent than that from 1 (Fig. S2†). The pseudo-first-order rate constants for the fluorescence-monitored reactions of 1 and 2 with FA were determined to be k = 1.8 × 10−2 M−1 s−1 and 4.6 × 10−2 M−1 s−1, respectively (Fig. S3†).10 The FA-concentration dependencies of reactions of probes 1 and 2 in aqueous buffer were determined by measuring fluorescence intensities, 10 min after treatment of the probes with 10 equiv. FA. The results showed that 1 and 2 exhibit a respective 15- and 6.5-fold enhancement in fluorescence intensity after addition of 50 equiv. FA (Fig. 1b and S4†).
The selectivity of fluorescence responses of the probes toward FA was then assessed by individually treating 1 and 2 (10 μM) with various biologically relevant analytes, including reactive carbonyl species (FA, acetaldehyde, benzaldehyde, 4-hydroxybenzaldehyde, ethyl pyruvate, ethyl glyoxalate, propionaldehyde, glucose), reactive oxygen species (H2O2, HOCl, NO˙, 1O2, O2˙−, ˙OH) and cations (Cu2+, Fe2+, Fe3+, K+, Zn2+). The results showed that both probes respond to FA but not to the other analytes (Fig. 2 and S5†). Taken together, the above findings indicate that probes 1 and 2 respond rapidly and selectively to FA in aqueous buffer.
To shed light on the mechanistic basis for the responses of probes to FA, the product generated by reaction of 1 and FA was isolated (see ESI† for the detailed procedure) and characterized by using spectroscopic methods. Analysis of the 1H and 13C NMR spectra of the isolated product in CDCl3 suggests that a 1,2,4-triazinane ring system exists, as judged from chemical shifts that correspond to bridging methylene protons (CH2) and carbon (3.24 ppm (s, 2H) and 72.5 ppm, respectively) (Scheme 2). Also, the spectral analysis suggests that the isolated product contains a spirocyclic ring system because of the presence of a signal at 61.3 ppm in the 13C NMR spectrum, which is expected for a quaternary carbon in a spiro ring. Furthermore, analysis of UV-Vis absorption and fluorescence spectra revealed that the product in CH2Cl2 displays very weak absorbance at 560 nm as well as very weak fluorescence at 583 nm (Fig. S6†). These observations led us to conclude that the product in CH2Cl2 has the spirocyclic structure represented by 3 (Scheme 2).
 | ||
| Scheme 2 Solvent (nonpolar organic solvent versus aqueous buffer) dependence of the equilibrium between ring-closed (3) and open (4) forms of the product generated by reaction of 1 with FA. | ||
In contrast, the isolated product in aqueous buffer (1% DMSO, pH 7.4) had a strong absorbance at 560 nm and intense fluorescence at 583 nm (Fig. S7†), spectral properties that are quite similar to those of the substance generated by treatment of 1 with FA in aqueous buffer. Collectively, the results suggest that while the product of the reaction of 1 with FA exists in the ring-closed form 3 in nonpolar organic solvents, in aqueous buffer it exists in the ring-opened xanthene containing form 4 (Scheme 2). As a consequence, it is reasonable to conclude that the reaction responsible for fluorescence generation when 1 is treated with FA in aqueous buffer involves formation of 4. In addition, extinction coefficients (ε514), quantum yields and fluorescence outputs (quantum yield × ε514) of 1, 2 and 3 were determined (Table S1†). Furthermore, the limits of detection of 1 and 2 for FA were calculated to be 1.24 μM and 0.59 μM, respectively (Fig. S8†).
Next, the utility of 1 to image FA in live cells was evaluated. Because 1 displayed a larger increase in fluorescence intensity upon treatment with FA than does 2, 1 was utilized in the cell studies described below. To determine the optimal conditions for cell imaging, HeLa cells (human cervical cancer cells) were incubated with 10 μM 1 for various times and with various concentrations of 1 for 1 h. Analysis of confocal fluorescence microscopy images showed that cells exposed to 10 μM 1 for 0.5–1 h display the intense fluorescence signal (Fig. S9†). In addition, based on the results of an MTT assay as well as the observation of an intact nucleus morphology, 1 had negligible cell death activity under these treatment conditions (Fig. S10†).
We also probed the FA concentration-dependence of the fluorescence response of 1. For this purpose, HeLa cells were first treated with 10 μM 1 and then incubated with concentrations of FA (0–1.0 mM) that are in a physiologically relevant concentration range (ca. 400 μM in normal cells and up to 700–800 μM in cancer tissues).4,11 The results showed that fluorescence signals arising from 1 in cells increase gradually as the FA concentration increases (Fig. 3), indicating the ability of 1 to serve as a probe for FA in live cells.
To evaluate the detection of endogenous FA in cells, several cell lines, including HeLa, MRC-5 (human fibroblast cell line derived from normal lung tissue), HaCaT (human keratinocytes), HEK293T (human embryonic kidney cells) and MCF-7 cells (human breast cancer cells), were incubated with 1 for 1 h. Analysis of cell images revealed that whereas the treated HEK293T cells display the low fluorescence intensity,8i the other four cell lines exhibit similarly strong fluorescence (Fig. 4 and S11†). The findings indicate that while HEK293T cells produce a low level of FA, the other four cells generate high levels of FA.
It is known that FA is generated in cells by the actions of several demethylases and oxidase enzymes.3 The enzyme lysine-specific demethylase 1 (LSD1) catalyzes the removal of one or two methyl groups from modified lysines to produce free lysine and FA.3,12 Also, it is known that GSK-LSD1 serves as a potent inhibitor of LSD1.13 As a result, production of FA by LSD1 in cells was evaluated by incubating MCF-7 cells with 1 in the absence and presence of GSK-LSD1. The results revealed that the intensity of the fluorescence arising from probe 1 in MCF-7 cells is slightly attenuated when GSK-LSD1 is present (Fig. 5 and S12†).8b The findings suggest that LSD1-promoted generation of FA in cells does not occur at high levels in comparison to the amounts formed by several other metabolic events. Taken together, the findings demonstrate that the rhodamine-based probe 1 is capable of detecting endogenous and exogenous FA in live cells.
In conclusion, we have developed novel rhodamine-based cyclic hydrazide derivatives as fluorescent probes for the detection of FA in both aqueous media and live cells. Upon addition of FA to the probes in aqueous buffer, a fluorescence enhancement occurs within a few minutes. In addition, the probes respond to FA but not to other biologically relevant species, indicating that they have a high selectivity toward FA. Furthermore, the results of cell studies demonstrate that probe 1 can be employed to image exogenous and endogenous FA in live cells. As a result, this probe should be useful in efforts aimed at gaining a more detailed understanding of FA-associated biological processes.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported financially by the National Research Foundation of Korea (grant no. 2020R1A2C3003462).Notes and references
- S. W. Hwang, Y.-M. Lee, G. Aldini and K.-J. Yeum, Molecules, 2016, 21, 280 CrossRef PubMed.
- (a) R. C. Grafstrom, A. J. Fornace Jr, H. Autrup, J. F. Lechner and C. C. Harris, Science, 1983, 220, 216 CrossRef CAS PubMed; (b) T. Tayri-Wilk, M. Slavin, J. Zamel, A. Blass, S. Cohen, A. Motzik, D. E. Shalev, O. Ram and N. Kalisman, Nat. Commun., 2020, 11, 3128 CrossRef CAS PubMed.
- (a) H. Kalász, Mini-Rev. Med. Chem., 2003, 3, 175 CrossRef; (b) Y. L. Dorokhov, A. V. Shindyapina, E. V. Sheshukova and T. V. Komarova, Physiol. Rev., 2015, 95, 603 CrossRef CAS; (c) Y. Shi, F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, R. A. Casero and Y. Shi, Cell, 2004, 119, 941 CrossRef CAS PubMed; (d) K. Tulpule, M. C. Hohnholt and R. Dringen, J. Neurochem., 2013, 125, 260 CrossRef CAS PubMed.
- (a) H. D. Heck, M. Casanova-Schmitz, P. B. Dodd, E. N. Schachter, T. J. Witek and T. Tosun, Am. Ind. Hyg. Assoc. J., 1985, 46, 1 CrossRef CAS; (b) Z. Tong, C. Han, W. Luo, X. Wang, H. Li, H. Luo, J. Zhou, J. Qi and R. He, Age, 2013, 35, 583 CrossRef.
- (a) L. Ai, T. Tan, Y. Tang, J. Yang, D. Cui, R. Wang, A. Wang, X. Fei, Y. Di, X. Wang, Y. Yu, S. Zhao, W. Wang, S. Bai, X. Yang, R. He, W. Lin, H. Han, X. Cai and Z. Tong, Commun. Biol., 2019, 2, 446 CrossRef CAS PubMed; (b) Z. Tong, C. Han, W. Luo, H. Li, H. Luo, M. Qiang, T. Su, B. Wu, Y. Liu, X. Yang, Y. Wan, D. Cui and R. He, Sci. Rep., 2013, 3, 1807 CrossRef; (c) K. Tulpule and R. J. Dringen, Neurochem, 2013, 127, 7 CrossRef CAS PubMed; (d) J. Liu, F.-Y. Liu, Z.-Q. Tong, Z.-H. Li, W. Chen, W.-H. Luo, H. Li, H.-J. Luo, Y. Tang, J.-M. Tang, J. Cai, F.-F. Liao and Y. Wan, PLoS One, 2013, 8, e58957 CrossRef CAS; (e) S. Hayami, M. Yoshimatsu, A. Veerakumarasivam, M. Unoki, Y. Iwai, T. Tsunoda, H. I. Field, J. D. Kelly, D. E. Neal, H. Yamaue, B. A. J. Ponder, Y. Nakamura and R. Hamamoto, Mol. Cancer, 2010, 9, 59 CrossRef PubMed; (f) I. Ferrer, J. M. Lizcano, M. Hernandez and M. Unzeta, Neurosci. Lett., 2002, 321, 21 CrossRef CAS; (g) M. Naya and J. Nakanishi, Regul. Toxicol. Pharmacol., 2005, 43, 232 CrossRef CAS; (h) Z. Tong, W. Luo, Y. Wang, F. Yang, Y. Han, H. Li, H. Luo, B. Duan, T. Xu, Q. Maoying, H. Tan, J. Wang, H. Zhao, F. Liu and Y. Wan, PLoS One, 2010, 5, e10234 CrossRef PubMed.
- (a) C. Protano, G. Buomprisco, V. Cammalleri, R. N. Pocino, D. Marotta, S. Simonazzi, F. Cardoni, M. Petyx, S. Lavicoli and M. Vitali, Cancers, 2021, 14, 165 CrossRef; (b) T. Tan, Y. Zhang, W. Luo, J. Lv, C. Han, J. N. R. Hamlin, H. Luo, H. Li, Y. Wan, X. Yang, W. Song and Z. Tong, FASEB J., 2018, 32, 3669 CrossRef CAS; (c) Z. Tong, C. Han, M. Qiang, W. Wang, J. Lv, S. Zhang, W. Luo, H. Li, H. Luo, J. Zhou, B. Wu, T. Su, X. Yang, X. Wang, Y. Liu and R. He, Neurobiol. Aging, 2015, 36, 100 CrossRef CAS; (d) M. Unzeta, M. Sole, M. Boada and M. J. Hernandez, J. Neural Transm., 2007, 114, 857 CrossRef CAS.
- (a) S.-H. Park, N. Kwon, J.-H. Lee, J. Yoon and I. Shin, Chem. Soc. Rev., 2020, 49, 143 RSC; (b) L. Wu, C. Huang, B. P. Emery, A. C. Sedgwick, S. D. Bull, X.-P. He, H. Tian, J. Yoon, J. L. Sessler and T. D. James, Chem. Soc. Rev., 2020, 49, 5110 RSC; (c) J. Li, Y. Zhang, P. Wang, L. Yu, J. An, G. Deng, Y. Sun and J. S. Kim, Coord. Chem. Rev., 2021, 427, 213559 CrossRef CAS; (d) X. Chen, F. Wang, J. Y. Hyun, T. Wei, J. Qiang, X. Ren, I. Shin and J. Yoon, Chem. Soc. Rev., 2016, 45, 2976 RSC; (e) Z. Guo, S. Park, J. Yoon and I. Shin, Chem. Soc. Rev., 2014, 43, 16 RSC.
- (a) A. Roth, H. Li, C. Anorma and J. Chan, J. Am. Chem. Soc., 2015, 137, 10890 CrossRef CAS PubMed; (b) T. F. Brewer and C. J. Chang, J. Am. Chem. Soc., 2015, 137, 10886 CrossRef CAS PubMed; (c) Y. Tang, X. Kong, A. Xu, B. Dong and W. Lin, Angew. Chem., Int. Ed., 2016, 55, 3356 CrossRef CAS PubMed; (d) J. Xu, Y. Zhang, L. Zeng, J. Liu, J. M. Kinsella and R. Sheng, Talanta, 2016, 160, 645 CrossRef CAS PubMed; (e) A. Bi, T. Gao, X. Cao, J. Dong, M. Liu, N. Ding, W. Liao and W. Zeng, Sens. Actuators, B, 2018, 255, 3292 CrossRef CAS; (f) W. Yuan, X. Zhong, Q. Han, Y. Jiang and J. Shen, J. Photochem. Photobiol., A, 2020, 400, 112701 CrossRef CAS; (g) H. Song, S. Rajendiran, N. Kim, S. K. Jeong, E. Koo, G. Park, T. D. Thangadurai and S. Yoon, Tetrahedron Lett., 2012, 53, 4913 CrossRef CAS; (h) T. Cao, D. Gong, S.-C. Han, A. Iqbal, J. Qian, W. Liu, W. Qin and H. Guo, Talanta, 2018, 189, 274 CrossRef CAS PubMed; (i) Y. Du, Y. Zhang, M. Huang, S. Wang, J. Wang, K. Liao, X. Wu, Q. Zhou, X. Zhang, Y.-D. Wu and T. Peng, Chem. Sci., 2021, 12, 13857 RSC.
- (a) M. Kim, S.-K. Ko, H. Kim, I. Shin and J. Tae, Chem. Commun., 2013, 49, 7959 RSC; (b) H. Moon, J. Park and J. Tae, Chem. Rec., 2016, 16, 124 CrossRef CAS.
- K. J. Bruemmer, R. R. Walvoord, T. F. Brewer, G. Burgos-Barragan, N. Wit, L. B. Pontel, K. J. Patel and C. J. Chang, J. Am. Chem. Soc., 2017, 139, 5338 CrossRef CAS.
- M. E. Andersen, H. J. Clewell 3rd, E. Bermudez, D. E. Dodd, G. A. Willson, J. L. Campbell and R. S. Thomas, Toxicol. Sci., 2010, 118, 716 CrossRef CAS PubMed.
- B. Perillo, A. Tramontano, A. Pezone and A. Migliaccio, Exp. Mol. Med., 2020, 52, 1936 CrossRef CAS PubMed.
- A. O. Muñoz, M. C. T. Fyfe, M. M. Pedemonte, M. de Los Angeles Martínez, N. V. Vidal, G. Kurz and J. C. C. P. Laria, (Hetero)aryl Cyclopropylamine Compounds as LSD1 Inhibitors, US Pat., 2015/0025054 A1, 2015 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra02104h |
| ‡ Two authors equally contributed to this study. |
| This journal is © The Royal Society of Chemistry 2022 |





